Abstract
Many cellular processes are involved in optimizing protein function for specific neuronal tasks; here we focus on alternative pre-mRNA splicing. Alternative pre-mRNA splicing gives cells the capacity to modify and selectively re-balance their existing pool of transcripts in a coordinated way across multiple mRNAs, thereby effecting relatively rapid and relatively stable changes in protein activity. Here we report on and discuss the coordinated regulation of two sites of alternative splicing, e24a and e31a, in P-type CaV2.1 and N-type CaV2.2 channels. These two exons encode 4 and 2 amino acids, respectively, in the extracellular linker regions between transmembrane spanning segments S3 and S4 in domains III and IV of each CaV2 subunit. Recent genome-wide screens of splicing factor-RNA binding events by Darnell and colleagues show that Nova-2 promotes inclusion of e24a in CaV2.2 mRNAs in brain. We review these studies and show that a homologous e24a is present in the CaV2.1 gene, Cacna1a, and that it is expressed in different regions of the nervous system. Nova-2 enhances inclusion of e24a but represses e31a inclusion in CaV2.1 and CaV2.2 mRNAs in brain. It is likely that coordinated alternative pre-mRNA splicing across related CaV2 genes by common splicing factors allows neurons to orchestrate changes in synaptic protein function while maintaining a balanced and functioning system.
Key words: N-type, P-type, alternative splicing, Nova, neuronal splicing, splicing factors, intron, exon
Introduction
Genes that encode the CaVα1 pore-forming subunits of voltage-gated calcium channels are large. Each CaVα1 gene includes 40–45 constitutive exons that are invariable in processed mRNA, as well as several alternative exons that are optional and cell-specific. Alternatively spliced (AS) exons serve two major functions. First, inclusion or exclusion of certain AS exons can shift the reading frame of mRNAs and result in early termination of protein translation, which may result in mRNA degradation by nonsense mediated decay.1–3 Cells use these exons to regulate protein expression levels downstream of transcription.4,5 Second, AS exons can modify protein sequence and thereby modify protein function. This is achieved by the inclusion, exclusion or substitution of alternatively spliced exons that encode as few as one and up to ∼100 amino acids in key domains of the channel protein.6 In this paper we focus on the regulation of alternatively spliced exons that give rise to protein splice isoforms of CaV2 channels with unique expression patterns and functions.
The decision to include, skip or substitute a given AS exon usually occurs in the cell's nucleus (although see ref. 7) and depends on interactions among the spliceosome, splicing factor proteins and the transcribed pre-mRNA. The spliceosome, a highly specialized macromolecular complex, is composed of five small nuclear RNA proteins (snRNPS) and hundreds of associated proteins and is common to all cell's nucleus (reviewed in ref. 8). Components of the spliceosome bind at intron-exon boundaries and catalyze intron removal and exon joining. Additionally, cis elements in pre-mRNAs and the cell-specific splicing factors that bind to them direct cell-specific AS. Collectively they define the cell's unique mRNA isoform profile.
Cell-specific splicing factors enhance or repress AS exons, and their expression patterns are tissue-, activity- and cell-specific.9 Our discussion here focuses on existing and new evidence that Nova, one of the earliest discovered tissue-specific splicing factors, regulates the pattern of AS exons in CaV2 channels.10 Our emphasis on Nova is driven by two important facts. First, Nova is the first—and to date the only—splicing factor validated as a regulator of AS splicing of CaV2 pre-mRNAs.11 Second, Nova-2 knockout mice exist, providing a way to test directly the consequences of Nova-2 loss on AS exon composition in CaV2 mRNAs.12
Nova Proteins Regulate Alternative Splicing of Synaptic Proteins
Nova proteins, originally discovered as the target antigens in the autoimmune neurological disorder paraneoplastic opsoclonus myoclonus ataxia (POMA), were the first mammalian neuron-specific splicing factors identified.10,13,16 Two genes encode Nova proteins: Nova1 and Nova2, although there is some evidence that other members of the Nova family await discovery.14 Nova-1 and Nova-2 proteins bind similar motifs, but their expression patterns differ. Nova-1 proteins are limited to subcortical CNS structures in developing and adult mice. Nova-2 proteins, by contrast, are expressed throughout the CNS and particularly in cortex, olfactory bulb, thalamus, inferior colliculus, inferior olive, and the internal and external granule cell layers of the cerebellum.10 Like other splicing factors, Nova-1 and Nova-2 bind to consensus sequences close to target AS exons on pre-mRNAs. The three KH domains mediate Nova binding to clusters of YCAY motifs on target pre-mRNAs.10,15
Nova-1 and Nova-2 knockout mice die 2–3 weeks after birth, but tissue from early postnatal animals shows that Nova is especially important for correct alternative splicing of two inhibitory receptor mRNAs in brain: glycine alpha 2 and GABAA.16,17 Nova-1-/- mice and POMA patients share a phenotypic similarity—weakness and tremulousness due to motor neuron atrophy—that led Darnell and colleagues to hypothesize a role for disrupted AS of inhibitory synaptic proteins in POMA disease pathology.16 Evidence is accumulating that misregulation of alternative splicing of synaptic proteins, including voltage-gated calcium channels, can result in disease.18–24
CaV2.2 is a Nova Target
Darnell and colleagues have transformed our understanding of the extent, mechanisms, and potential functional importance of Nova-regulation of AS exons in the mammalian brain. By combining protein-RNA crosslinking and Nova-immunoprecipitation, in a technique called CLIP (cross-linking and immunoprecipitation) they were able to identify new Nova RNA targets.25,26 Their studies generated the most comprehensive view to date of the extent and the mechanism of action of a cell-specific splicing factor. Nova binds YCAY clusters in introns generally within 500 nucleotides of target AS exons. When Nova binds clusters upstream of a target AS exon, it generally represses inclusion by blocking U1 snRNP binding; when Nova binds downstream of the target AS exon, it generally enhances inclusion (Fig. 1).11 Subsequent studies have indicated that this appears to be a general feature of alternative splicing regulators, evident for example with not only Nova but Fox2, MBNL, hnRNP L, hnRNP C, TIA1 and PTB.27 Microarray analyses showed that the majority of Nova-regulated exons are located in genes encoding proteins with important synaptic functions.12 Furthermore, they combined the CLIP technique with high throughput sequencing (HITS-CLIP) and mapped more than 16,000 putative sites of Nova-2 binding onto the mouse genome.28
Figure 1.
Nova acts as a splicing repressor or enhancer depending on its binding site relative to the target exon. Nova binds to YCAY motifs in introns upstream of e31a in CaV2 pre-mRNAs to repress its inclusion, whereas Nova binds to YCAY motifs in introns downstream of e24a in CaV2 pre-mRNAs to enhance its inclusion, during pre-mRNA splicing.
While the importance of these landmark studies rests in the genome-wide scale of the analyses, our interest was piqued by one target: e24a in Cacna1b, the gene that encodes the CaV2.2 channel. With analyses that compared e24a expression in wild-type and Nova-2-/- mice, Darnell and colleagues confirmed that Nova-2 enhances e24a expression in mouse brain, likely by binding YCAY clusters in the 3′ intron downstream of e24a (Figs. 1 and 2 for YCAY motifs located downstream of e24a in mouse Cacna1b).11 Splicing regulation of e24a by Nova is conserved in the homologous Cacna1b genes of zebrafish and chicken, strongly suggesting that this AS exon plays a critical role in optimizing neuronal function in specific regions of the nervous system.28 Additional Nova-2 binding sites have been identified in Cacna1b, motivating us to explore whether Nova-2 coordinates inclusion of other AS exons in CaV2.2 and homologous exons in the CaV2.1 paralog.
Figure 2.
Nova binding motifs are located in the intron downstream of e24a of CaV2.2. The genomic sequence in this region of Cacna1b is shown, and the amino acids encoded by e24a indicated (SFMG). The Nova binding motifs (black squares) are shown below the genomic sequence and were mapped to the UCSC genome browser (July 2007 mouse assembly). Nova binding motifs are shown within 70 nucleotides and downstream of e24a in Cacna1b. The Nova binding site track was provided by SFmap.62 SFmap is a computational tool that uses an algorithm to predict splicing factor binding sites by identifying binding site motif clusters and evaluating the conservation of these clusters across species.
AS Exons in CaV2 Genes
Figure 3 highlights some of the known AS exons in three closely related CaV2 genes—Cacna1a, Cacna1b and Cacna1e—that encode CaV2.1, CaV2.2 and CaV2.3 proteins, respectively. These proteins (in association with accessory subunit proteins) in turn underlie P-type, N-type and R-type currents in neurons. We and others have mapped the tissue distribution and functional consequences of AS exons on channel activity and drug action in mice in vivo.20,21,30–49 Several AS exons are conserved among the three CaV2 genes, and in some cases, for example the homologous exons e18a of Cacna1b and Cacna1e, their inclusion appears to be coregulated.31 Here we review published data from the Darnell lab and show new evidence that the same splicing factor, Nova-2, regulates the splicing of homologous exons, e24a and e31a in CaV2.1 and CaV2.2 channels.
Figure 3.
Alternatively spliced exons in the CaV2 genes. The locations of constitutive exons (black) and conserved alternative exons (colored) in mouse Cacna1a, Cacna1b and Cacna1e genes. Gene diagrams are adapted from the UCSC genome browser (Feb. 2006 mouse assembly). Alternative exons 37a and 37b are found in all three CaV2 channel genes. An alternative exon e18a is present in Cacna1b and Cacna1e, but a homologous exon has not been identified in Cacna1a. Alternative exons e24a and e31a are conserved in Cacna1a and Cacna1b, but equivalent exons have not been identified in Cacna1e.
Nova-2 Regulates e24a and e31a in CaV2 Channels
E24a and e31a encode short sequences in the S3–S4 extracellular linkers of domains III and IV, respectively, of CaV2.1 and CaV2.2. In separate studies, the Snutch and Williams labs showed that inclusion of e31a in CaV2.1 decreases the affinity of ω-agatoxin IVA for the channel ∼10-fold, while inclusion of e31a in either channel slows channel activation and deactivation kinetics.33,49,50 We analyzed gating currents from CaV2.2 channels containing e31a and showed that it affects the putative S4 voltage sensor.41 Most relevant to this discussion, we found e31a is absent (CaV2.2) or at lower levels (CaV2.1) in rat brain compared to peripheral ganglia.36 E24a of Cacna1b encodes four amino acids, SFMG, in domain IIIS3–IIIS4 of mouse and rat CaV2.2 and SFVG in human CaV2.2 (S. Schorge and D. Lipscombe).45 E24a is found in the majority of CaV2.2 mRNAs in brain but is found at lower levels in ganglia. Although it is not currently present in EST or cDNA databases, we also recently discovered a corresponding e24a in Cacna1a (see below).
We recently used RT-PCR to compare the expression pattern of CaV2 channel mRNAs including and lacking e24a and e31a in brains and ganglia of wild-type and Nova-2-/- mice to determine whether Nova-2 regulates tissue-specific expression of these exons.
CaV2.1 and CaV2.2 e31a
Exons 31a in CaV2.1 and CaV2.2 map to the extracellular linker between transmembrane spanning segments 3 and 4 in the IV domain of both channel proteins. An AS exon is located in this region of several CaVα1 subunits and in many species, including human and Drosophila.31 E31a in both CaV2.1 and CaV2.2 encode two amino acids, NP in CaV2.1 and ET in CaV2.2.50 We have shown that e31a is found in CaV2.1 mRNAs in dorsal root ganglia (DRG) and superior cervical ganglia, similar to the distribution of e31a of CaV2.2.50 However, e31a-containing CaV2.1 mRNAs are also expressed in hippocampus and cerebellum albeit at levels lower than in ganglia, an expression pattern that is different from e31a of CaV2.2 which is at very low levels in all regions of the CNS.50
Putative Nova-2 binding sites have been mapped very close to the 31a exons of the Cacna1b and Cacna1a genes. As discussed above, exons 31a of both genes are found at very low levels in brain, suggesting that Nova-2 might repress e31a inclusion. Consistent with this, we found an increase in the percentage of CaV2.1[e31a] transcripts (from 30% to 80%) and CaV2.2[e31a] transcripts (from not detectable to ∼45%) in RT-PCR amplified products from Nova-2 knockout mouse brain cDNA compared to wild-type brain cDNA (Fig. 4). In cerebellum (CB), the increase in e31a-containing CaV2 sequences in Nova-2-/- was smaller (CaV2.1: from 20% to 50% and CaV2.2: ∼10% to 30%) compared to the rest of the brain. But in DRG, the proportion of e31a-containing sequences was not different in wild-type compared to Nova-2-/- mice (∼80–85%); this is consistent with the lack of Nova-2 in this tissue.
Figure 4.
Alternative exons e31a of CaV2.1 and CaV2.2 are repressed in brain by Nova-2. The approximate locations of e31a of CaV2.1 (NP) and CaV2.2 (ET) are mapped on a schematic of CaV2 channels that shows transmembrane and intracellular domains (A and B). RT-PCR products amplified from mouse brain using primers that flank e31a are shown separated in 8% denaturing polyacrylamide gels. Predicted sizes of PCR products are 100 (+31a) and 94 (Δ31a) nts for CaV2.1 (A) and 106 (+31a) and 100 (Δ31a) nts for CaV2.2 (B). RT-PCR amplification was carried out using brain lacking cerebellum (Brain), cerebellum (CB) and dorsal root ganglia (DRG) from wild-type (WT) and Nova-2 knockout mice (KO). Percentages of cDNAs containing e31a of the total amplified cDNA for each reaction are shown below each lane. (A) 30% of cDNAs amplified from wild-type brain contain e31a compared to 80% in Nova-2-/- brain. In cerebellum, 17% of cDNAs amplified from wild-type tissue contain e31a compared to 46% in Nova-2-/- tissue. cDNA amplified from DRG contains a high percentage of e31a-containing sequence similarly in wild-type and Nova-2 knockout tissue (82–87%). (B) cDNAs amplified from wild-type brain lacked e31a compared to 44% representation in cDNAs from Nova-2-/- brain. In cerebellum, 13% of cDNAs amplified from wild-type tissue contain e31a compared to 32% in Nova-2-/- tissue. cDNA amplified from DRG contains a high percentage of e31a-containing sequence (83–84%) similarly in wild-type and Nova-2 knockout tissue. A schematic shown below the gels illustrates how Nova acts to repress e31a inclusion via a binding site in the upstream intron in brain and cerebellum, but not in DRG.
Our data show that Nova-2 is a repressor of e31a in both CaV2.1 and CaV2.2 pre-mRNAs in the mammalian brain (Fig. 1). Based on the very low to undetectable levels of e31a-containing CaV2.2 mRNAs in brain, Nova appears to exert stronger repression of e31a in CaV2.2 than in CaV2.1. Additional studies are needed to explore the precise mechanism of Nova action. For example, Nova repression of e31a in CaV2.2 might be stronger compared to CaV2.1, or the inclusion of e31a in CaV2.1 may also be influenced by other as yet unidentified splicing factors, such as a splicing enhancer that promotes e31a inclusion in specific brain regions.
CaV2.1 and CaV2.2 e24a
Several observations led us to hypothesize the presence of an AS exon in Cacna1a equivalent to e24a of Cacna1b. These include the overall high degree of conservation between Cacna1a and Cacna1b genes, their close functional roles, the conservation of AS exons 31a that are located in the homologous region of the channel in domain IV (IVS3–IVS4), and the location of a putative Nova binding site in the intron between exons 24 and 25 in Cacna1a. We located a region in the intron that is strongly conserved among species (Fig. 5A). We used RT-PCR with primers located in constitutive exons e24 and e25 to amplify CaV2.1 mRNA isolated from different regions of the brains of wild-type mice (Fig. 5B). We observed two differently sized bands when we analyzed PCR cDNA products by gel separation, consistent with the presence of two alternative sequences that differ by only a few nucleotides. We confirmed a 12-nucleotide insert encoding the tetrapeptide sequence SSTR in CaV2.1-derived products by sequencing (Fig. 5A).
Figure 5.
Alternative exons e24a of CaV2.1 and CaV2.2 are enhanced in brain by Nova-2. (A) Location and sequence, SSTR, of newly identified e24a (red) in the mouse Cacna1a gene. Constitutive exons e24 and e25 are shown in gray flanking the alternatively spliced e24a (red). Below, the degree of sequence conservation among vertebrate Cacna1a genes is represented. Image is adapted from UCSC genome browser, Feb 2006 mouse assembly. (B) RT-PCR products amplified using primers located in e24 and e25 from RNA isolated from the following regions of adult mouse brain: cortex (CTX), hippocampus (HC), hypothalamus (HP), midbrain (MB), olfactory bulb (OB) and cerebellum (CB). Predicted sizes of products are 152 nts (+24a) and 140 nts (Δ24a). PCR products were separated on 8% denaturing polyacrylamide gels. (C) RT-PCR products amplified from mouse brain using primers that flank e24a are shown separated on a 3% agarose (DRG) or 8% denaturing polyacrylamide gels. Predicted sizes of PCR products are 152 (+24a) and 140 (Δ24a) nts. RT-PCR amplification was carried out using brain lacking cerebellum (Brain), cerebellum (CB) and dorsal root ganglia (DRG) from wild-type (WT) and Nova-2 knockout mice (KO). Percentages of cDNAs containing e24a of the total amplified cDNA pool are shown below each lane. (A) 13% of cDNAs amplified from wild-type brain contain e24a but this isoform was not detectable in Nova-2-/- brain samples. In cerebellum, 23% of cDNAs amplified from wild-type tissue contain e24a compared to 12% in Nova-2-/- tissue. cDNA amplified from DRG contains a very low percentage of e24a-containing sequence similarly in wild-type and Nova-2 knockout tissue (4–5%). A schematic shown below the gels illustrates how Nova acts to enhance e24a inclusion via a binding site in the downstream intron in brain and cerebellum, but not in DRG.
We tested whether e24a of Canca1a was regulated by Nova-2 by comparing its expression levels in wild-type and Nova-2-/- mice. In wild-type brain e24a-containing sequences represented about ∼10% of CaV2.1 sequences, but we were unable to detect e24a containing sequences in PCR amplified products derived from RNA from Nova-2-/- brain (Fig. 5C). Levels of e24a-containing CaV2.1 sequences were very low (∼5%) similarly in DRG of wild-type and Nova-2-/- mice. Our data suggest that Nova has an enhancing action on e24a in brain but not in DRG, where Nova-2 is absent. The analogous e24a in CaV2.2 is found at higher levels in brain compared to peripheral ganglia. Furthermore, Darnell and colleagues showed that e24a in CaV2.2 is enhanced by Nova.11
We would like to know the function of e24a in CaV2.1 and CaV2.2 channels. In CaV2.2, e24a has a relatively minor influence on channel properties. As described above, we only recently identified e24a in CaV2.1 and have not assessed its effects on channel properties.50 However, we speculate that the extracellular location of e24a might point to a role in mediating interactions with extracellular proteins. For example, neurexins have been shown to regulate CaV2.1 and CaV2.2 channel functioning through direct or indirect extracellular coupling.51–53 It is possible that other extracellular proteins might interact with CaV2.1 and CaV2.2 channels in a splice isoform-dependent manner, a possibility that needs to be tested.
Nova Influences a Subset of Alternatively Spliced Exons in CaV2
Other sites of alternative splicing have been studied in detail by our lab (e18a and e37a/e37b in CaV2.2) and by other labs (e37a/e37b in CaV2.1).30,33–35,38,48,54–58 None of these AS exons appears to be regulated by Nova-2. For example, we find no difference in expression patterns of exon 37a of CaV2.2 in Nova-2-/- and wild-type mice (data not shown). Exon 37a of CaV2.2 is enriched in nociceptors of DRG but is expressed at very low levels in other parts of the nervous system. Likewise, the expression pattern of e18a in CaV2.2 is similar in wild-type and Nova-2-/- mice (data not shown). Although we focus on Nova-2 in this report, splicing factors other than Nova must regulate cell-specific inclusion of e18a and e37a/e37b. Furthermore, exon selection in vivo is likely to depend on the concerted actions of splicing enhancers and repressors (Fig. 6).4,58,60 The Darnell group recently concluded that ∼15% of Nova-regulated exons might also be regulated by Fox proteins, another family of neuronal splicing factors.62
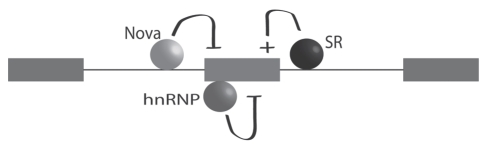
Figure 6.
Cellular control of alternative splicing likely involves the net contribution of more than one splicing factor. Schematic shows hypothetical, combinatorial control of an alternatively spliced exon by Nova binding downstream and acting as a repressor, by another serine rich (SR) splicing factor binding downstream and acting as an enhancer, and by a third splicing factor of the hnRNP family binding to the target exon and acting as a repressor of exon inclusion. The relative importance and contribution of each splicing factor depends on several factors, including their expression levels. Thus, tissue specific splicing can arise from the tissue-specific expression profiles of splicing factors.
Summary
The nervous system operates with small error margins and adaptive responses are essential to normal neuronal function. These impact neuronal networks, individual neurons, and individual synapses and involve changes in synapse number, morphology and efficacy. Coordinated and temporal changes in protein activity and function underlie all these adaptive responses. Alternative pre-mRNA splicing has an impressive ability to generate a vast array of functionally distinct proteins. Alternative pre-mRNA splicing gives cells the capacity to modify and selectively re-balance their existing pool of transcripts in a coordinated way across multiple mRNAs, thereby effecting relatively rapid and relatively stable changes in protein activity.
Alternative pre-mRNA splicing events are predicted to occur in ∼95% of multi-exon human genes.63,64 Genome-wide screens of splicing factor-RNA binding events such as HITS-CLIP Nova screens and those looking at binding sites for other splicing factors such as Fox and PTB are providing us the information we need to define cell-specific mechanisms that orchestrate alternative splicing across multiple genes, including CaV2.27,65,66 In this short report we review limited published data and in addition show evidence that four alternative exons in two closely related calcium channel CaV2 subunit genes are regulated by Nova-2. Thus, the ability of Nova to function both as repressor (e31a) and enhancer (e24a), depending on binding location imprinted in each gene, provides an elegant yet simple mechanism to coordinate splicing of homologous exons in related CaV2 genes (Fig. 1). This duality of function, based on RNA binding location relative to the target exon, is a property shared by several splicing factors.27,59 Cells control and coordinate patterns of alternative splicing by regulating the expression levels and activity of individual splicing factors. Coordinated alternative pre-mRNA splicing across related CaV2 genes allows neurons to orchestrate changes in synaptic protein function while maintaining a balanced and functioning system.
Acknowledgements
We are grateful to Gerald Zamponi and Terry Snutch for organizing another fantastic Calcium Ion Channel Meeting where we presented some of these data and ideas. This work was supported by Grants RO1NS29967 (Diane Lipscombe), F31NS066691 (Summer E. Allen), and RO1NS34389 (Robert B. Darnell)
References
- 1.Le Hir H, Seraphin B. EJCs at the heart of translational control. Cell. 2008;133:213–216. doi: 10.1016/j.cell.2008.04.002. [DOI] [PubMed] [Google Scholar]
- 2.Isken O, Maquat LE. Quality control of eukaryotic mRNA: safeguarding cells from abnormal mRNA function. Genes Dev. 2007;21:1833–1856. doi: 10.1101/gad.1566807. [DOI] [PubMed] [Google Scholar]
- 3.McGlincy NJ, Smith CW. Alternative splicing resulting in nonsense-mediated mRNA decay: what is the meaning of nonsense? Trends Biochem Sci. 2008;33:385–393. doi: 10.1016/j.tibs.2008.06.001. [DOI] [PubMed] [Google Scholar]
- 4.Boutz PL, Stoilov P, Li Q, Lin CH, Chawla G, Ostrow K, et al. A post-transcriptional regulatory switch in polypyrimidine tract-binding proteins reprograms alternative splicing in developing neurons. Genes Dev. 2007;21:1636–1652. doi: 10.1101/gad.1558107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Makeyev EV, Zhang J, Carrasco Ma, Maniatis T. The MicroRNA miR-124 promotes neuronal differentiation by triggering brain-specific alternative pre-mRNA splicing. Mol Cell. 2007;27:435–448. doi: 10.1016/j.molcel.2007.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lipscombe D. Neuronal proteins custom designed by alternative splicing. Curr Op Neurobiol. 2005;15:358–363. doi: 10.1016/j.conb.2005.04.002. [DOI] [PubMed] [Google Scholar]
- 7.Glanzer J, Miyashiro KY, Sul JY, Barrett L, Belt B, Haydon P, et al. RNA splicing capability of live neuronal dendrites. Proc Natl Acad Sci USA. 2005;102:16859–16864. doi: 10.1073/pnas.0503783102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jurica MS, Moore MJ. Pre-mRNA splicing: awash in a sea of proteins. Mol Cell. 2003;12:5–14. doi: 10.1016/s1097-2765(03)00270-3. [DOI] [PubMed] [Google Scholar]
- 9.Chen M, Manley JL. Mechanisms of alternative splicing regulation: insights from molecular and genomics approaches. Nature Rev Mol Cell Biol. 2009;10:741–754. doi: 10.1038/nrm2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang YY, Yin GL, Darnell RB. The neuronal RNAbinding protein Nova-2 is implicated as the autoantigen targeted in POMA patients with dementia. Proc Natl Acad Sci USA. 1998;95:13254–13259. doi: 10.1073/pnas.95.22.13254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ule J, Stefani G, Mele A, Ruggiu M, Wang X, Taneri B, et al. An RNA map predicting Nova-dependent splicing regulation. Nature. 2006;444:580–586. doi: 10.1038/nature05304. [DOI] [PubMed] [Google Scholar]
- 12.Ule J, Ule A, Spencer J, Williams A, Hu JS, Cline M, et al. Nova regulates brain-specific splicing to shape the synapse. Nature genetics. 2005;37:844–852. doi: 10.1038/ng1610. [DOI] [PubMed] [Google Scholar]
- 13.Buckanovich RJ, Posner JB, Darnell RB. Nova, the paraneoplastic Ri antigen, is homologous to an RNAbinding protein and is specifically expressed in the developing motor system. Neuron. 1993;11:657–672. doi: 10.1016/0896-6273(93)90077-5. [DOI] [PubMed] [Google Scholar]
- 14.Fletcher CF, Okano HJ, Gilbert DJ, Yang Y, Yang C, Copeland NG, et al. Mouse chromosomal locations of nine genes encoding homologs of human paraneoplastic neurologic disorder antigens. Genomics. 1997;45:313–319. doi: 10.1006/geno.1997.4925. [DOI] [PubMed] [Google Scholar]
- 15.Jensen KB, Musunuru K, Lewis HA, Burley SK, Darnell RB. The tetranucleotide UCAY directs the specific recognition of RNA by the Nova K-homology 3 domain. Proc Natl Acad Sci USA. 2000;97:5740–5745. doi: 10.1073/pnas.090553997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jensen KB, Dredge BK, Stefani G, Zhong R, Buckanovich RJ, Okano HJ, et al. Nova-1 regulates neuron-specific alternative splicing and is essential for neuronal viability. Neuron. 2000;25:359–371. doi: 10.1016/s0896-6273(00)80900-9. [DOI] [PubMed] [Google Scholar]
- 17.Dredge BK, Darnell RB. Nova regulates GABA(A) receptor γ2 alternative splicing via a distal downstream UCAU-rich intronic splicing enhancer. Mol Cell Biol. 2003;23:4687–4700. doi: 10.1128/MCB.23.13.4687-4700.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Faustino NA, Cooper TA. Pre-mRNA splicing and human disease. Genes Dev. 2003;17:419–437. doi: 10.1101/gad.1048803. [DOI] [PubMed] [Google Scholar]
- 19.Cáceres JF, Kornblihtt AR. Alternative splicing: multiple control mechanisms and involvement in human disease. Trends Genet. 2002;18:186–193. doi: 10.1016/s0168-9525(01)02626-9. [DOI] [PubMed] [Google Scholar]
- 20.Eunson LH, Graves TD, Hanna MG. New calcium channel mutations predict aberrant RNA splicing in episodic ataxia. Neurology. 2005;65:308–310. doi: 10.1212/01.wnl.0000169020.82223.dd. [DOI] [PubMed] [Google Scholar]
- 21.Altier C, Dale CS, Kisilevsky AE, Chapman K, Castiglioni AJ, Matthews EA. Differential role of the N-type calcium channel splice isoforms in pain. J Neurosci. 2007;27:10. doi: 10.1523/JNEUROSCI.0307-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baralle D, Baralle M. Splicing in action: assessing disease causing sequence changes. J Med Genet. 2005;42:737–748. doi: 10.1136/jmg.2004.029538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grabowski PJ, Black DL. Alternative RNA splicing in the nervous system. Prog Neurobiol. 2001;65:289–308. doi: 10.1016/s0301-0082(01)00007-7. [DOI] [PubMed] [Google Scholar]
- 24.Liao P, Zhang HY, Soong TW. Alternative splicing of voltage-gated calcium channels: from molecular biology to disease. Pflügers Archiv. 2009;458:481–487. doi: 10.1007/s00424-009-0635-5. [DOI] [PubMed] [Google Scholar]
- 25.Ule J, Jensen K, Mele A, Darnell RB. CLIP: a method for identifying protein-RNA interaction sites in living cells. Methods. 2005;37:376–386. doi: 10.1016/j.ymeth.2005.07.018. [DOI] [PubMed] [Google Scholar]
- 26.Ule J, Jensen KB, Ruggiu M, Mele A, Ule A, Darnell RB. CLIP identifies Nova-regulated RNA networks in the brain. Science. 2003;302:1212–1215. doi: 10.1126/science.1090095. [DOI] [PubMed] [Google Scholar]
- 27.Witten JT, Ule J. Genome-wide studies of splicing regulation by RNA-binding proteins. Trends in Genetics. In press. [Google Scholar]
- 28.Licatalosi DD, Mele A, Fak JJ, Ule J, Kayikci M, Chi SW, et al. HITS-CLIP yields genome-wide insights into brain alternative RNA processing. Nature. 2008;456:464–469. doi: 10.1038/nature07488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jelen N, Ule J, Zivin M, Darnell RB. Evolution of Nova-dependent splicing regulation in the brain. PLoS genetics. 2007;3:1838–1847. doi: 10.1371/journal.pgen.0030173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Adams PJ, Garcia E, David LS, Mulatz KJ, Spacey SD, Snutch TP. CaV2.1 P/Q-type calcium channel alternative splicing affects the functional impact of familial hemiplegic migraine mutations: implications for calcium channelopathies. Channels. 2009;3:110–121. doi: 10.4161/chan.3.2.7932. [DOI] [PubMed] [Google Scholar]
- 31.Gray AC, Raingo J, Lipscombe D. Neuronal calcium channels: splicing for optimal performance. Cell Calcium. 2007;42:409–417. doi: 10.1016/j.ceca.2007.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lipscombe D, Pan JQ, Gray AC. Functional diversity in neuronal voltage-gated calcium channels by alternative splicing of CaVα1. Molecular neurobiology. 2002;26:21–44. doi: 10.1385/MN:26:1:021. [DOI] [PubMed] [Google Scholar]
- 33.Bourinet E, Soong TW, Sutton K, Slaymaker S, Mathews E, Monteil A, et al. Splicing of α1A subunit gene generates phenotypic variants of P- and Q-type calcium channels. Nature Neurosci. 1999;2:407–415. doi: 10.1038/8070. [DOI] [PubMed] [Google Scholar]
- 34.Thaler C, Gray AC, Lipscombe D. Cumulative inactivation of N-type CaV2.2 calcium channels modified by alternative splicing. Proc Natl Acad Sci USA. 2004;101:5675–5679. doi: 10.1073/pnas.0303402101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Castiglioni AJ, Raingo J, Lipscombe D. Alternative splicing in the C-terminus of CaV2.2 controls expression and gating of N-type calcium channels. J Physiol. 2006;576:119–34. doi: 10.1113/jphysiol.2006.115030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lin Z, Lin Y, Schorge S, Pan JQ, Beierlein M, Lipscombe D. Alternative splicing of a short cassette exon in α1B generates functionally distinct N-type calcium channels in central and peripheral neurons. J Neurosci. 1999;19:5322–5331. doi: 10.1523/JNEUROSCI.19-13-05322.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bell TJ, Thaler C, Castiglioni AJ, Helton TD, Lipscombe D. Cell-specific alternative splicing increases calcium channel current density in the pain pathway. Neuron. 2004;41:127–138. doi: 10.1016/s0896-6273(03)00801-8. [DOI] [PubMed] [Google Scholar]
- 38.Raingo J, Castiglioni AJ, Lipscombe D. Alternative splicing controls G protein-dependent inhibition of N-type calcium channels in nociceptors. Nature Neurosci. 2007;10:285–292. doi: 10.1038/nn1848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Asadi S, Javan M, Ahmadiani A, Sanati MH. Alternative splicing in the synaptic protein interaction site of rat CaV2.2 (α1B) calcium channels changes induced by chronic inflammatory pain. J Mol Neurosci. 2009;39:40–48. doi: 10.1007/s12031-008-9159-2. [DOI] [PubMed] [Google Scholar]
- 40.Kanumilli S, Tringham EW, Payne CE, Dupere JRB, Venkateswarlu K, Usowicz MM. Alternative splicing generates a smaller assortment of CaV2.1 transcripts in cerebellar Purkinje cells than in the cerebellum. Physiological genomics. 2006;24:86–96. doi: 10.1152/physiolgenomics.00149.2005. [DOI] [PubMed] [Google Scholar]
- 41.Lin Y, McDonough SI, Lipscombe D. Alternative splicing in the voltage-sensing region of N-Type CaV2.2 channels modulates channel kinetics. J Neurophysiol. 2004;92:2820–2830. doi: 10.1152/jn.00048.2004. [DOI] [PubMed] [Google Scholar]
- 42.Jiménez C, Bourinet E, Leuranguer V, Richard S, Snutch TP, Nargeot J. Determinants of voltage-dependent inactivation affect Mibefradil block of calcium channels. Neuropharmacology. 2000;39:1–10. doi: 10.1016/s0028-3908(99)00153-7. [DOI] [PubMed] [Google Scholar]
- 43.Ghasemzadeh MB, Pierce RC, Kalivas PW. The monoamine neurons of the rat brain preferentially express a splice variant of α1B subunit of the N-type calcium channel. Journal of neurochemistry. 1999;73:1718–1723. doi: 10.1046/j.1471-4159.1999.731718.x. [DOI] [PubMed] [Google Scholar]
- 44.Kaneko S, Cooper CB, Nishioka N, Yamasaki H, Suzuki A, Jarvis SE, et al. Identification and characterization of novel human CaV2.2 (α1B) calcium channel variants lacking the synaptic protein interaction site. J Neurosci. 2002;22:82–92. doi: 10.1523/JNEUROSCI.22-01-00082.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lin Z, Haus S, Edgerton J, Lipscombe D. Identification of functionally distinct isoforms of the N-type Ca2+ channel in rat sympathetic ganglia and brain. Neuron. 1997;18:153–166. doi: 10.1016/s0896-6273(01)80054-4. [DOI] [PubMed] [Google Scholar]
- 46.Pereverzev A, Leroy J, Krieger A, Malecot CO, Hescheler J, Pfitzer G, et al. Alternate splicing in the cytosolic II–III loop and the carboxy terminus of human E-type voltage-gated Ca2+ channels: electrophysiological characterization of isoforms. Mol Cell Neurosci. 2002;21:352–365. doi: 10.1006/mcne.2002.1179. [DOI] [PubMed] [Google Scholar]
- 47.Adams PJ, Rungta RL, Garcia E, van den Maagdenberg aMJM, MacVicar Ba, Snutch TP. Contribution of calcium-dependent facilitation to synaptic plasticity revealed by migraine mutations in the P/Q-type calcium channel. Proc Natl Acad Sci USA. 2010:1–6. doi: 10.1073/pnas.1009500107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Andrade A, Denome S, Jiang YQ, Marangoudakis S, Lipscombe D. Opioid inhibition of N-type Ca2+ channels and spinal analgesia couple to alternative splicing. Nat Neurosci. 2010;13:1249–1256. doi: 10.1038/nn.2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hans M, Urrutia A, Deal C, Brust PF, Stauderman K, Ellis SB, et al. Structural elements in domain IV that influence biophysical and pharmacological properties of human α1A-containing high-voltage-activated calcium channels. Biophys J. 1999;76:1384–1400. doi: 10.1016/S0006-3495(99)77300-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lin Z, Lin Y, Schorge S, Pan JQ, Beierlein M, Lipscombe D. Alternative splicing of a short cassette exon in α1B generates functionally distinct N-type calcium channels in central and peripheral neurons. J Neurosci. 1999;19:5322–5331. doi: 10.1523/JNEUROSCI.19-13-05322.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dudanova I, Sedej S, Ahmad M, Masius H, Sargsyan V, Zhang W, et al. Important contribution of a-neurexins to Ca2+-triggered exocytosis of secretory granules. J Neurosci. 2006;26:10599–10613. doi: 10.1523/JNEUROSCI.1913-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Missler M, Zhang W, Rohlmann A, Kattenstroth G, Hammer RE, Gottmann K, et al. A-neurexins couple Ca2+ channels to synaptic vesicle exocytosis. Nature. 2003;423:939–948. doi: 10.1038/nature01755. [DOI] [PubMed] [Google Scholar]
- 53.Zhang W, Rohlmann A, Sargsyan V, Aramuni G, Hammer RE, Südhof TC, et al. Extracellular domains of α-neurexins participate in regulating synaptic transmission by selectively affecting N- and P/Q-type Ca2+ channels. J Neurosci. 2005;25:4330–4342. doi: 10.1523/JNEUROSCI.0497-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Altier C, Dale CS, Kisilevsky AE, Chapman K, Castiglioni AJ, Matthews EA, et al. Differential role of N-type calcium channel splice isoforms in pain. J Neurosci. 2007;27:6363–6373. doi: 10.1523/JNEUROSCI.0307-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bell TJ, Thaler C, Castiglioni AJ, Helton TD, Lipscombe D. Cell-specific alternative splicing increases calcium channel current density in the pain pathway. Neuron. 2004;41:127–138. doi: 10.1016/s0896-6273(03)00801-8. [DOI] [PubMed] [Google Scholar]
- 56.Chang SY, Yong TF, Yu CY, Liang MC, Pletnikova O, Troncoso J, et al. Age and gender-dependent alternative splicing of P/Q-type calcium channel EF-hand. Neuroscience. 2007;145:1026–1036. doi: 10.1016/j.neuroscience.2006.12.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lipscombe D, Raingo J. Alternative splicing matters: N-type calcium channels in nociceptors. Channels. 2007;1:225–227. doi: 10.4161/chan.4809. [DOI] [PubMed] [Google Scholar]
- 58.Graves TD, Imbrici P, Kors EE, Terwindt GM, Eunson LH, Frants RR, et al. Premature stop codons in a facilitating EF-hand splice variant of CaV2.1 cause episodic ataxia type 2. Neurobiol Disease. 2008;32:10–15. doi: 10.1016/j.nbd.2008.06.002. [DOI] [PubMed] [Google Scholar]
- 59.Li Q, Lee JA, Black DL. Neuronal regulation of alternative pre-mRNA splicing. Nat Rev Neurosci. 2007;8:819–831. doi: 10.1038/nrn2237. [DOI] [PubMed] [Google Scholar]
- 60.Licatalosi DD, Darnell RB. RNA processing and its regulation: global insights into biological networks. Nat Rev Genet. 2010;11:75–87. doi: 10.1038/nrg2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Xiao X, Lee JH. Systems analysis of alternative splicing and its regulation. Wiley Interdiscip Rev Syst Biol Med. 2010;2:550–565. doi: 10.1002/wsbm.84. [DOI] [PubMed] [Google Scholar]
- 62.Zhang C, Frias MA, Mele A, Ruggiu M, Eom T, Marney CB, et al. Integrative modeling defines the Nova splicing-regulatory network and its combinatorial controls. Science. 2010:439. doi: 10.1126/science.1191150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, et al. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Castle JC, Zhang C, Shah JK, Kulkarni AV, Kalsotra A, Cooper TA, et al. Expression of 24,426 human alternative splicing events and predicted cis regulation in 48 tissues and cell lines. Nat Genet. 2008;40:1416–1425. doi: 10.1038/ng.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yeo GW, Coufal NG, Liang TY, Peng GE, Fu XD, Gage FH. An RNA code for the FOX2 splicing regulator revealed by mapping RNA-protein interactions in stem cells. Nat Struct Mol Biol. 2009;16:130–137. doi: 10.1038/nsmb.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Xue Y, Zhou Y, Wu T, Zhu T, Ji X, Kwon Ys, et al. Genome-wide analysis of PTB-RNA interactions reveals a strategy used by the general splicing repressor to modulate exon inclusion or skipping. Mol Cell. 2009;36:996–1006. doi: 10.1016/j.molcel.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Paz I, Akerman M, Dror I, Kosti I, Mandel-Gutfreund Y. SFmap: a web server for motif analysis and prediction of splicing factor binding sites. Nucleic Acids Res. 2010;38:W281–W285. doi: 10.1093/nar/gkq444. [DOI] [PMC free article] [PubMed] [Google Scholar]