Abstract
In a previous study, a surprising drying transition was observed to take place inside the nanoscale hydrophobic channel in the tetramer of the protein melittin. The goal of this paper is to determine if there are other protein complexes capable of displaying a dewetting transition during their final stage of folding. We searched the entire protein data bank (PDB) for all possible candidates, including protein tetramers, dimers, and two-domain proteins, and then performed the molecular dynamics (MD) simulations on the top candidates identified by a simple hydrophobic scoring function based on aligned hydrophobic surface areas. Our large scale MD simulations found several more proteins, including three tetramers, six dimers, and two two-domain proteins, which display a nanoscale dewetting transition in their final stage of folding. Even though the scoring function alone is not sufficient (i.e., a high score is necessary but not sufficient) in identifying the dewetting candidates, it does provide useful insights into the features of complex interfaces needed for dewetting. All top candidates have two features in common: (1) large aligned (matched) hydrophobic areas between two corresponding surfaces, and (2) large connected hydrophobic areas on the same surface. We have also studied the effect on dewetting of different water models and different treatments of the long-range electrostatic interactions (cutoff vs PME), and found the dewetting phenomena is fairly robust. This work presents a few proteins other than melittin tetramer for further experimental studies of the role of dewetting in the end stages of protein folding.
1. Introduction
Hydrophobicity plays an important role in molecular self-assembly processes such as the formation of membranes and micelles, protein folding, protein association, ligand binding, and nucleic acid folding.1,2 Since water has the unique property of being tetrahedrally coordinated in bulk, hydrophobicity at small and large length scale is quite different.3 Hydrogen bonding of water persists around small hydrophobic solutes by reorganization but is depleted near large hydrophobic surfaces. Stillinger4 first proposed this water depletion around large hydrophobic solutes. When two strongly hydrophobic nanoscale sized objects are brought together to a critical separation, often large enough to accommodate several layers of water molecules, the water is expelled from the gap between them. This is the so-called dewetting (water drying) transition. The dewetting induced long-range attraction between hydrophobic objects is of broad interest. It was previously proposed that hydrophobic attraction is due to bridging of the thin vapor layers3 or the preexisting nanosized bubbles5 at each hydrophobic surface.
A number of experiments have provided evidence for water depletion in the gap between two hydrophobic surfaces.6–8 Atomic force microscopy (AFM) in the tapping mode (AFM)9 has been used to directly observe nanobubbles on hydrophobic surfaces. Noting that AFM itself may nucleate the bubbles, Steitz et al.,10 using a less invasive technique based on neutron reflectivity together with AFM, detected a water depletion layer with a thickness of 2–5 nm. This roughly agrees with observed jump-in distance between two hydrophobic surfaces in water upon first approach of the two surfaces using AFM.11 Jensen et al.12 observed a similar depletion of water density around a large hydrophobic paraffin surface floating on water using X-ray reflectivity. This depletion layer is less than 15 Å, a result consistent with the predictions of their molecular simulations.13 Recently, Brinker and co-workers14 directly observed cavitation between superhydrophobic surfaces (silicon substrate) which interact attractively over a distance more than 30 times greater than any reported value using interfacial-force microscopy.14 However, no direct observation of dewetting has yet been experimentally observed in any biological system.
Molecular dynamics (MD) and Monte Carlo (MC) simulations are widely used to study dewetting of extended hydrophobic surfaces in water.3,13,15–18 A strong dewetting (water drying) transition has been observed between two nanoscale hydrophobic plates when they are closer than a certain critical separation.3,19,20 The existence of this drying transition is found to be sensitive to the solute–solvent attractions.21,22 Proteins are much more complicated than simple hydrophobic solutes. Simulations have shown that on hydrophobic surfaces of proteins, certain hydrophobic residues do not break neighboring water hydrogen bonds whereas other residues do,23 so that, protein surfaces can be characterized by regions that are heterogeneously “small” or “large”. Is there a similar strong dewetting transition preceding hydrophobic collapse in protein folding, as seen in idealized hydrophobic plates?20 We found in a previous study that there is no strong drying transition in the collapse of the BphC enzyme, a two-domain protein due to solute–solvent attractions.24 We also found that the protein–water attractions, particularly the electrostatic interactions, play a significant role in the protein hydrophobic collapse. Thus, the general conclusion was that the dewetting transition might not play a role in protein folding. Much to our surprise, a dramatic water drying transition was then observed inside the nanoscale channel formed by the protein melittin tetramer, with a channel size of up to 2–3 water diameters.25 This study shows that even in the presence of the polar protein backbone, sufficiently hydrophobic protein surfaces and unique surface topologies can induce a liquid–vapor transition which might then provide an enormous driving force toward further collapse. The question thus arises “Is this melittin tetramer unique in terms of dewetting or are there other protein complexes showing similar dewetting behavior?” Our quest in this paper is to identify other proteins exhibiting this behavior.
It is known that both morphology and structure are important to the existence and kinetics of dewetting between hydrophobic surfaces.7,18 Many studies of protein–protein interfaces1,26–30 show the importance of interface texture to the hydrophobic effect; however, none of them address the question of which protein surfaces will display dewetting transition, if any. To find the correlation between protein surface hydrophobicity and dewetting, we propose a hydrophobic scoring function based on the distribution of hydrophobic areas (residues) on the protein domain–domain or oligomer–oligomer contact surfaces By using this informatics tool, we identify those protein complexes with large aligned hydrophobic surfaces between two domains or oligomers. All top candidates show not only large matched hydrophobic areas between two surfaces but also large connected hydrophobic areas on the same surface. We then performed MD simulations on the top candidates identified by our scoring function to determine whether or not they display a drying transition, and indeed found that several of them show dewetting transitions. By giving detailed pictures of the dewetting phenomena of these proteins, it may help further experimental study of the role of dewetting at the end of stage of folding.
In Section 2 of this paper we present the results of this study, including discussions of the kinetics of hydrophobic aggregation. In Section 3 we conclude with a discussion of the results. In Section 4, we describe the informatics tool that we devised for searching dewetting candidates from the protein PDB database, and we also describe the MD methodology used to perform simulations of these candidates.
2. Results
2.1. Dewetting in Two-Domain Proteins or Protein Oligomers
In this paper, our aim is to identify other proteins besides the melittin tetramer capable of undergoing dewetting transitions during their final stage of folding. We search the protein data bank (PDB) for the protein complexes with the sequence identity smaller than 30% based on different complex types, such as protein tetramer, protein dimer, and two-domain proteins. A protein dataset is obtained consisting of 40 protein tetramers, 200 protein dimers, and 165 two-domain proteins out of PDB. However it is not feasible to analyze all of these proteins by molecular dynamics simulations, as this would require too many computing resources (even with IBM Blue-Gene/L). So we propose a surface hydrophobicity scoring function (see Section 4) and use it to search our database. The top protein candidates are chosen based on them having large matched hydrophobic areas between two corresponding surfaces and large connected hydrophobic areas on the same surface, features are believed to be necessary for a protein to display a dewetting transition. We subject the top 10 protein tetramers (Table 1), the top 20 protein dimers (Table 2), and the top 20 two-domain proteins (Table 3) to molecular dynamics simulations to determine which can display strong dewetting transitions.
TABLE 1.
The Selected PDB Candidates of Protein Tetramers Based on Surface Hydrophobicity Analysisa
| PDB ID | Am | Amc | dewetting |
|---|---|---|---|
| 1g5y | 552 | 285 | no |
| 1fe6 | 549 | 207 | F |
| 1j2w | 290 | 123 | yes |
| 1ub3 | 279 | 101 | no |
| 2mlt | 244 | 134 | yes |
| 1tvx | 178 | 140 | no |
| 4aah | 144 | 130 | no |
| 1xz4 | 138 | 49 | no |
| 1tlf | 126 | 31 | no |
| 1plf | 81 | 37 | no |
Am and Amc are defined in eqs 2 and 4, respectively. The existence of the dewetting phenomenon in MD simulations when dimer–dimer distance, D = 4 Å, is also shown. “F” denotes fluctuation. In this case, water density has large fluctuations, and large cavities are observed in the region between two dimers, but most of this region remains wet in the MD simulations.
TABLE 2.
The Selected PDB Candidates of Protein Dimmers Based on Surface Hydrophobicity Analysisa
| PDB ID | Am | Amc | dewetting |
|---|---|---|---|
| 1k2e | 259 | 259 | no |
| 1m4i | 220 | 148 | no |
| 1j3q | 214 | 214 | yes |
| 1hsi | 192 | 143 | no |
| 1j30 | 172 | 78 | no |
| 1i4s | 162 | 162 | no |
| 1f4n | 161 | 59 | yes |
| 1jvl | 157 | 157 | no |
| 1eyv | 146 | 96 | F |
| 1cmb | 140 | 140 | no |
| 1jr8 | 138 | 132 | no |
| 1hul | 133 | 93 | no |
| 1bbh | 132 | 132 | no |
| 1g6u | 132 | 132 | yes |
| 1d1g | 130 | 42 | yes |
| 1ipi | 128 | 128 | no |
| 1gfw | 123 | 65 | F |
| 1bja | 122 | 122 | no |
| 1k94 | 120 | 103 | no |
| 1bj3 | 112 | 46 | no |
Am and Amc are defined in eqs 2 and 4, respectively. The existence of the dewetting phenomenon in MD simulations when monomer–monomer distance, D = 4 Å, is also shown. “F” denotes fluctuation. In this case, water density has large fluctuations, and large cavities are observed in the region between two monomers, but most of this region remains wet in the MD simulations.
TABLE 3.
The Selected PDB Candidates of Multidomain Protein Based on Surface Hydrophobicity Analysisa
| PDB ID | Am | Amc | dewetting |
|---|---|---|---|
| 1ldm | 312 | 251 | no |
| 1fsz | 224 | 224 | Yes |
| 2mbr | 222 | 170 | no |
| 1aco | 220 | 124 | no |
| 1han | 220 | 213 | no |
| 1dhy | 216 | 182 | no |
| 1plq | 211 | 158 | no |
| 1a5z | 205 | 172 | no |
| 5ldh | 182 | 89 | F |
| 1mdr | 159 | 95 | no |
| 1pgs | 157 | 82 | no |
| 1pkp | 154 | 154 | no |
| 1cpo | 151 | 134 | no |
| 1boh | 150 | 120 | no |
| 1bg5 | 147 | 83 | no |
| 1akl | 137 | 98 | no |
| 1cne | 128 | 55 | no |
| 1bli | 127 | 81 | no |
| 1hyt | 126 | 55 | no |
| 1clc | 121 | 118 | no |
Am and Amc are defined in eqs 2 and 4, respectively. The existence of the dewetting phenomenon in MD simulations when domain–domain distance, D = 4 Å, is also shown. “F” denotes fluctuation. In this case, water density has large fluctuations, and large cavities are observed in the interdomain region, but most of the interdomain region remains wet in the MD simulations.
We summarize the simulation results for proteins which display dewetting transitions in Table 4 in terms of critical distance, treatment of electrostatics, and water models used. Roughly the same results on dewetting critical distances are found for these proteins in different water models (SPC, TIP3P, and SPC/E) and different long-range electrostatic treatments (Cutoff and PME). So we believe that we have identified several proteins, other than melittin tetramer, which display dewetting transitions at the final stage of folding.
TABLE 4.
The Simulation Results of the Protein Candidates Capable of Displaying Dewetting Transitions in Terms of Protein Complex Type, Critical Distance, Electrostatic Treatment, and Water Modelsa
| PDB ID | complex type |
critical distance |
electrostatic treatment |
water model |
|---|---|---|---|---|
| 1j2w | tetramer | 5–6Å | cutoff, PME | SPC, TIP3P, SPC/E |
| 1j3q | dimer | 4–5Å | cutoff, PME | SPC, TIP3P, SPC/E |
| 1f4n | dimer | 4–5Å | cutoff | SPC |
| 1g6u | dimer | 4–5Å | cutoff | SPC |
| 1d1g | dimer | 4–5Å | cutoff | SPC |
| 1fsz | two-domain | 4–5Å | cutoff | SPC |
2.1.1. Protein Tetramers
Based on our surface hydrophobicity analysis of buried protein surfaces, we list the 10 most hydrophobic tetramer proteins in Table 1. Three out of the first five proteins in the table display either a drying transition or large fluctuations of water density in the region between two dimers. The previously studied melittin tetramer (PDB ID: 2mlt) ranks no. 5 in our list. It has a fairly large Am score, considering its small size (each monomer has only 26 residues). A strong dewetting transition is observed in this protein.25 The protein tetramer with PDB ID 1g5y, ranking no. 1 in our list, does not show dewetting, however. Even though this protein complex has a high score of Am, its dimer–dimer interface has a strange shape and the effective hydrophobic interfaces between two dimers are not large when viewed with VMD. The number 2 candidate in our list is the RHCC tetramer (PDB ID 1fe6),31 which does not show a strong drying transition either; however, it does exhibit large fluctuations in water density inside the confined region. This is found to be largely related to the fact that the RHCC tetramer (PDB ID 1fe6) contains a large hydrophobic cavity which is filled with water molecules.
2.1.1.1. 1j2w
The MD simulation results for the 2-Deoxyribose-5-phosphate aldolase from Thermus thermophilus HB8 (TtDERA)32 (PDB ID 1j2w) are shown in Figure 2. It is the third most hydrophobic protein in our list, with the matched hydrophobic area Am = 290Å2, and the matched and connected hydrophobic area Amc = 123 Å2. As mentioned above, the vector connecting the two dimer center-of-masses (COM) serves as the z-direction, and the midpoint between the COMs of the two dimers serves as the origin (see Figure 2a). The water density in the interdimer region (−6 Å < x < 6 Å, −8 Å < y < 8 Å) versus simulation time for the different dimer–dimer separations D = 4 Å, D = 5 Å and D = 6 Å are plotted in Figure 2b. The water density is defined as the number of water molecules divided by Nmax, where Nmax is the maximum number of water molecules which can fill the interdimer region in our simulation. For an initial separation of 4 Å (−5 Å < z < 5 Å) with about 30 water molecules between the two dimers, we have found that the region dries completely in less than 100 ps. Although water molecules refill and empty the confined region a few times due to large fluctuations, the system stays dry most of the time during the simulation. When D = 5 Å (−5.5 Å < z < 5.5 Å), the system prefers to remain in the dry state as shown in red in Figure 2b. Even when D is increased to 6 Å (−6 Å < z < 6 Å), the system still dewets, although it takes as long as 2000 ps for the drying transition to occur (Figure 2b (blue)). However, when D = 7 Å, the system stays in “wet” state during entire simulation (data not shown here).
Figure 2.
(a). The coordinate system for protein tetramer with PDB ID 1j2w is shown and the interdimer region is defined and filled with water. (b). The water density inside interdimer region (see text for definition) versus time is shown. The maximum number of water filling the interdimer region is Nmax = 30 when dimer dimer distance D = 4 Å (shown in black). Nmax = 38 when D = 5 Å (shown in red). Nmax = 46 when D = 6 Å (shown in blue).
2.1.2. Protein Dimers
The top 20 protein dimers with highest hydrophobic scores are listed in Table 2. There are four protein dimers (PDB ID: 1j3q, 1f4n, 1g6u, and 1d1g) in this list that display the drying transition when the monomer–monomer gap has been enlarged by at least 4 Å. For two other targets (PDB ID: 1eyv and 1gfw), the water density exhibits large fluctuations and vapor cavities are observed in the intermonomer region.
2.1.2.1. 1j3q
The results for a phosphoglucose isomerase (PDB ID: 1j3q) are shown in Figure 3. One monomer has 185 residues, and the other one has 187 residues. As shown in Table 2, this protein has very large Am and Amc scores, indicating that it has very hydrophobic surfaces buried between the two monomers. Surface hydrophobicity distributions defined in eq 1 for this protein are displayed in Figure 1b. It is clear that the cells located in the central region on both of the two buried monomer surfaces are mostly hydrophobic. Furthermore, these hydrophobic cells on the two different surfaces are matched well. The results of MD simulations for this protein are shown in Figure 3. In Figure 3b, the black curve shows the time evolution of the water density in the intermonomer region (−6 Å < x < 10 Å, −8 Å < y < 8 Å) when D = 4 Å (−5 Å < z < 5 Å). It is obvious that the water density in the intermonomer region decreases from 1 to about 0.23 g/cm3 in about 2500 ps. After that, the water density in the gap region fluctuates, but the average density stays around 0.23 g/cm3. The system remains dry for the rest of the simulation. A strong drying transition is observed at this monomer–monomer distance. If the protein is allowed to move, there will be a large hydrophobic force causing the two monomers to collapse. When the monomer–monomer distance is increased to D = 5 Å (−5.5 Å < z < 5.5 Å), the system first dries with the water density decreasing from ~0.90 to ~0.20 g/cm3 in about 1200 ps, and then wets (water refills the region between two monomers) at about 6000 ps. During the whole 11 ns simulation, we find that the system oscillates between the “dry” and “wet” states as shown in Figure 3b (red). This indicates that the critical distance for this system is approximately D = 5 Å. When the distance between the two monomers is even larger (D = 6 Å) (−6 Å < z < 6 Å), the system remains in the “wet” state during the entire simulation (see Figure 3d (blue)).
Figure 3.
(a). Protein dimer with PDB ID 1j3q, the intermonomer region is filled with water. (b). The water density inside the region between the two monomers (see text for definition) versus time is shown. The maximum number of water filling in the intermonomer region is Nmax = 34 when monomer–monomer distance D = 4 Å (shown in black). Nmax = 41 when D = 5 Å (shown in red). Nmax = 48 when D = 6 Å (shown in blue).
Figure 1.
(a). The coordinates system is shown. C1 and C2 are the centers of mass of two domains (or two protein oligomers) respectively. R is the geometry center of a residue on the protein surface. (b). Surface hydrophobicity distributions of (A). Monomer 1 and (B). Monomer 2 of a protein dimer (PDB ID: 1j3q).
2.1.2.2. 1f4n
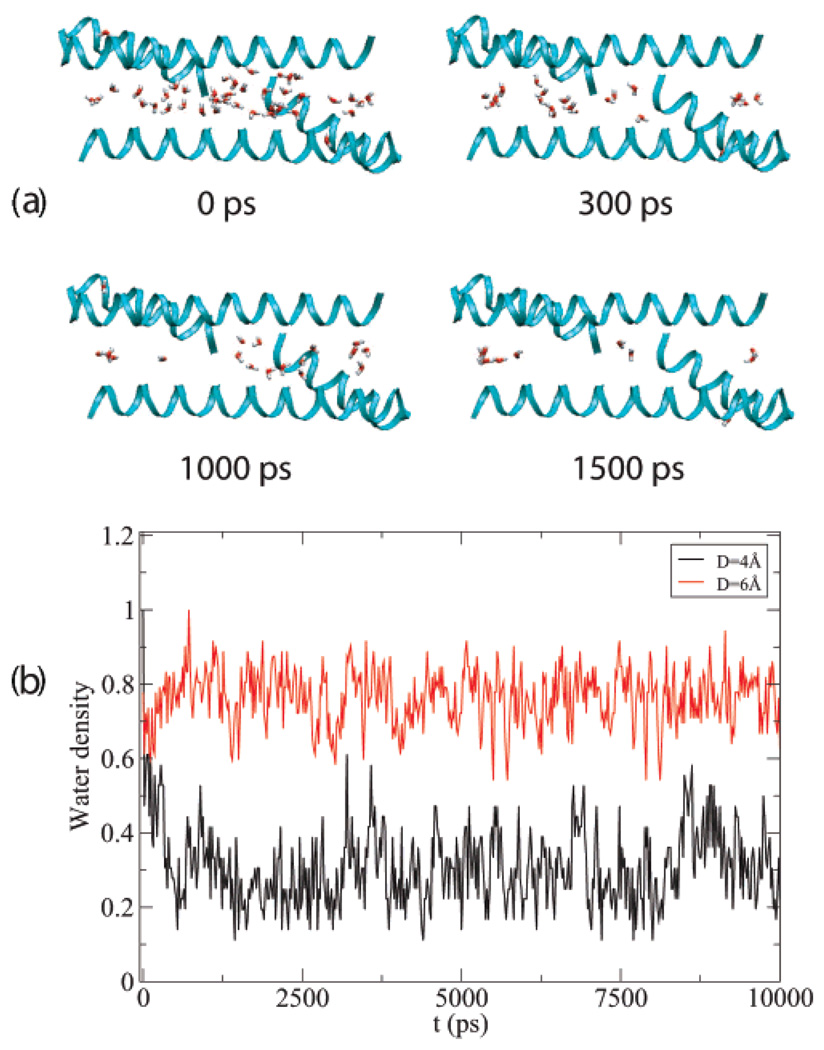
Rop or ROM is an RNA binding protein which is involved in regulation of the copy number of ColE1 plasmids in Escherichia coli. Ala2Ile2 − 6 (PDB ID: 1f4n), a variant of Rop, is a dimer of two helix-turn-helix protomers that form an antiparallel four-helix bundle. The relative reorientation of the two protomers is rotated by 180° which destroys the RNA binding activity.33 The isoleucine knobs on one protomer pack nicely over the ridges connecting the isoleucine knobs on the other protomer and into the holes formed by alanines (see ref 33, Figure 6), which is consistent with its large Am as shown in Table 2. The simulation result is shown in Figure 4. The region between two protomers is defined slightly different from the previous set up, namely as a cylinder with radius r = 4 Å and r = 5 Å for D = 4 Å and D = 5 Å, respectively. The water density (same definition as in aldolase enzyme (PDB ID 1j2w)) in the cylindrical channel decreases dramatically from 1 to 0.3 g/cm3 in the first 200ps when D = 4 Å as shown in Figure 4b (black). At t = 400 ps the channel dries almost completely leaving only a few water molecules at the edge of the channel (see Figure 4a). Although waters eventually return, drying is observed again at t = 1500 ps with remaining waters found only at the edge or two ends of the channel. The system stays dry during most of the simulation. This behavior is likely due to the very hydrophobic isoleucine knobs on the monomer–monomer interfaces. When D is increased to 5 Å, the water density inside the channel exhibits large fluctuations (see Figure 4b (red)). Although the channel stays “wet” for most of the simulation, very large cavities occupying most of the channel region are observed. Wild type protein Rop (PDB ID: 1rpr), which has smaller Am and Amc scores than those of Ala2Ile2 − 6 (data not shown here), is also studied. The region between two monomers is defined as for Ala2Ile2 − 6. Inside the channel between two protomers, the water density undergoes large fluctuations (weak dewetting) for D = 4 Å in contrast to the strong dewetting for Ala2Ile2 − 6 as shown in Figure 4b (blue). This is not surprising since Ala2Ile2 − 6 achieves a more densely packed hydrophobic core than Rop by using an offset packing arrangement with the creation of a low ridge between two isoleucine knobs on both helices of the protomer.33
Figure 6.
(a). Protein dimer with PDB ID 1d1g, the intermonomer region is filled with water. (b). The water density inside the region between the two monomers (see text for definition) versus time is shown. The maximum number of water filling in the intermonomer region is Nmax = 24 when monomer monomer distance D = 4 Å (shown in black). Nmax = 35 when D = 6 Å (shown in red). (c). Snapshots of water molecules inside the region between the two monomers (Water are shown as sticks, while the protein is not shown because of viewing the evolution process of cavities) when D = 4 Å. The green rectangle box represents the xy plane of the region between two monomers.
Figure 4.
(a). Snapshots of water molecules inside the channel between two protomers with PDB ID 1f4n (The protein is shown as ribbons and water as sticks) when monomer monomer distance D = 4 Å. (b). The water density inside the channel (see text for definition) versus time is shown. The maximum number of water filling the channel is Nmax = 17 when D = 4 Å for the protein with PDB ID 1f4n (shown in black). Nmax = 47 when D = 5 Å (PDB ID 1f4n, shown in red). Nmax = 27 when D = 4 Å for the protein with PDB ID 1rpr (shown in blue).
2.1.2.3. 1g6u
As shown in Table 2, the protein dimer with PDB ID 1g6u has a fairly large Am score, considering its small size (each monomer has only 48 residues). This protein is a domain-swapped dimer (DSD) formed by the monomers with up-down-up topology.34 The hydrophobic core of DSD is exclusively composed of hydrophobic leucine side chains. There are 24 leucine residues out of total 96 residues. The results of MD simulations for this protein are shown in Figure 5. As shown in Figure 5b (black), there are approximately 40 water molecules in the cylindrical channel at time t = 0 ps when D = 4 Å (r = 5.5 Å). The water density in the channel decreases dramatically in the first 100 ps, and fluctuates around 0.55 g/cm3 after that. Large cavities form in the nanoscale channel (see Figure 5a at t = 300 ps). After t = 300 ps, the water density decreases to 0.38 g/cm3. Actually, at this time, a few water molecules reside inside the channel, and most of the remaining water molecules are found near the two ends of the channel. By t = 1500 ps, the channel becomes totally empty. Thus a drying transition is observed inside the nanoscale channel formed by DSD dimer. After that the water density in the cylindrical channel fluctuates with low frequency, but the average density stays around 0.2 g/cm3. This water density fluctuation arises from water molecules near the two ends of the channel. So on average the system remains dry in the rest of simulation. When D is increased to 6 Å (r = 7.5 Å), the channel remains “wet”, although large cavities are observed in the region away from the connection of two monomers (see Figure 5b (red)). The results of simulation for the different monomer–monomer distances confirm that DSD has a very hydrophobic core between the two monomers.
Figure 5.
(a). Snapshots of water molecules inside the channel of the DSD dimer with PDB ID 1g6u (The protein is shown as ribbons and water as sticks) when monomer monomer distance D = 4 Å. (b). The water density inside the channel between the two monomers (see text for definition) versus time is shown. The maximum number of water filling in the channel is Nmax = 36 when D = 4 Å (shown in black). Nmax = 72 when D = 6 Å (shown in red).
2.1.2.4. 1d1g
The protein dimer (PDB ID 1d1g) is Dihydrofolate reductase from the hyperthermophilic bacterium Thermotoga maritima (TmDHFR). It is important in the pharmaceutical industry as a drug target against bacterial, fungal, and protozoan infection, etc.35 Although TmDHFR has large Am score as shown in Table 2 because its monomer–monomer interfaces have many matched small hydrophobic areas, such hydrophobic areas on each dimer interface are not well connected, and thus its Amc value is not very large. The simulation results for TmDHFR are shown in Figure 6. Since NADPH and MTX binding affects neither the overall structure nor the interaction between subunits of TmDHTR,35 they are not included in the MD simulation. As shown in Figure 6b (black), the water density in the intermonomer region (−9 Å < x < 3 Å, −7 Å < y < 8 Å) decreases quickly from 1 to 0.33 g/cm3 in about 400ps for D = 4 Å (−8 Å < z < 7 Å). At t = 0 ps, there is a small cavity formed in the gap because of the protrusions on both of dimer interfaces which cannot accommodate water molecules there. By t = 400 ps, large cavities form as shown in Figure 6c, and the water density in the gap region undergoes large fluctuations around an average density of approximately 0.33 g/cm3. The gap region stays in the “wet” state only for a very short time, but for the remaining part of the simulation it stays dry, with some water molecules distributed at the edges of the gap region. When the monomer distance is increased to 6 Å (−11 Å < z < 6 Å), the gap region remains in the “wet” state during almost the entire simulation as shown in Figure 6b (red).
2.1.3. Two-Domain Proteins
Twenty two-domain proteins with highest hydrophobic scores are listed in Table 3. We note that the BphC enzyme (1dhy)24 is no. 6 in this list, indicating that it is a very hydrophobic protein on the two-domain interface. This is also consistent with the results of a hydrophobicity profiling analysis based on hydrophobic moments.36 MD simulations are performed for each of the proteins listed in Table 3. The results show that the protein Ftsz (PDB ID:1fsz) is the only listed two-domain protein that displays a strong drying transition in the interdomain region.
2.1.3.1. 1fsz
Protein Ftsz is important in the last step of bacterial cell division, in which the constriction of the cell membrane leads to the formation of two daughter cells.37 Protein Ftsz consists of two domains with a long, 23 residue, helix H5 connecting them: Domain 1 (residues 23–231) and Domain 2 (residues 232–356) as shown in Figure 7a. Figure 7b shows snapshots along one trajectory with an interdomain gap distance of 4 Å (−5 Å < z < 5 Å). A large cavity forms in the interdomain region (−7 Å < x < 8 Å, −10 Å < y < 10 Å). The remaining water molecules are found mostly at the edge of the interdomain gap region, leaving the center area empty. It is interesting to note that a bump on one of the domain surfaces might help drying (see Figure 7c).
Figure 7.
(a). The two-domain protein with PDB ID 1fsz, the interdomain region is filled with water. (b). Time evolution of water configurations in the interdomain region of this two-domain protein (Water are shown as sticks, while the protein is not shown because of viewing the evolution process of cavities). Domain–domain distance D = 4 Å. (c). A bump formed by ILE 204 and LEU 203 on one of the domain surfaces is shown in red.
2.2. Folding Kinetics
To investigate the time scale and kinetics of drying in the hydrophobic collapse of two domain proteins or oligomers, we investigate phosphoglucose isomerase (PDB ID: 1j3q), by performing a 5 ns “folding simulation” for three different initial monomer–monomer separations (D = 5, 6, 7 Å) with up to 10 different water configurations for each separation. The kinetics of the collapse of this protein dimer starting from its extended configuration with intermonomer separation D = 6 Å is shown in Figure 8. The number of water molecules inside the intermonomer region decreases rapidly within about 300 ps after which the intermonomer region almost dries completely (see Figure 8a). D decreases very rapidly in the first 250 ps and the collapse of two monomers happens in less than 500 ps (see Figure 8b). This collapse will be even faster, within 200 ps, if the initial monomer–monomer separation is chosen to be very close to the critical distance, D = 5 Å. Even starting at the larger initial separation of D = 7 Å, the time scale of the hydrophobic collapse does not increase much and is found to be approximately 500 ps. There is a large hydrophobic force pushing the two monomers together, as recently observed in the simulation of collapse of melittin tetramer.25 A drying induced hydrophobic collapse is found in some trajectories (one of them is shown in Figure 8c (black circle)) although most of the trajectories show that drying and collapse happen at roughly the same time as was observed in the melittin tetramer case.25 During the entire 5 ns, the two individual monomers remain folded, with root-mean-square displacements of backbone from the starting native structures of less than 2.0–3.0 Å and the fluctuations in the radius of gyration of less than 0.7 Å for each monomer.
Figure 8.
(a). The number of water molecules inside the intermonomer region versus the simulation time for protein dimer (PDB ID 1j3q). only water molecules within a spherical radius of 10 Å from the center of the enlarged dimer are analyzed. (b). The monomer–monomer distance versus simulation time for the “folding” simulation starting from the initial separation, D = 6 Å. (c). The number of water molecules inside the intermonomer region versus monomer–monomer distance for three folding trajectories starting from the initial separation of 6 Å. One trajectory (black circle) indicates a drying-induced collapse, while the other two (blue square and red triangle up) show drying and collapse happening at roughly the same time.
3. Discussion
The existence of a dewetting transition is sensitive to the strength of the solute–solvent attractions.24 Since even the hydrophobic core of the proteins contains a significant fraction of polar residues, realistic proteins are rarely found to display a drying or dewetting transition. Surprisingly melittin tetramer can undergo a drying transition inside its nanoscale channel. To find out other proteins displaying a drying transition in the end stage of folding, a hydrophobic score proposed by us was tested to search for possible dewetting candidates in all two-domain proteins, protein dimers, and protein tetramers in the PDB. The score is based on the assumption that the top candidates should have (1) large aligned (matched) hydrophobic areas between two corresponding surfaces, and (2) large connected hydrophobic areas on the same surface. Based on our analysis, we subjected the top 20 two-domain proteins (Table 3), the top 20 protein dimers (Table 2), and the top 10 protein tetramers (Table 1) to molecular dynamics simulations to determine which evince strong drying transitions. These large scale molecular dynamics simulations show that indeed more protein complexes display either a strong drying transition or the large water density fluctuations typical of weak drying transitions inside the confined region. We found two two-domain proteins, six protein dimers, and three protein tetramers. A drying transition might play an important role in the last stage of protein folding when the protein complex collapses into its final shape, after each individual domain or oligomer have been formed. Although a high value for our hydrophobicity score is necessary but not sufficient in predicting the dewetting transition, we did successfully identify several other proteins complexes showing dewetting transition which may help the experimental study of the role of dewetting in the last stage of folding.
All the MD simulations discussed above are based on the SPC38 water model. From the macroscopic thermodynamic theory based on Young’s Equation, we know that the critical distance for drying is related to the liquid vapor surface tension γlv, vapor pressure Pv, and the contact angle θ by the equation Dc = 2Δγ/((P − Pv) + bγlv/Rm) where Δγ = −γlv cos θc.22 Since different water models have different γlv, Pv, and θ, the critical distance for the drying transition might be different too. Therefore, we reran all the simulations for two proteins 1j2w (tetramer) and 1j3q (dimer) using TIP3P39 and SPC/E water models.40 In TIP3P water, the protein with PDB ID 1j2w dewets for D ≤ 6 Å, which is consistent with the results in SPC water. For the protein with PDB ID 1j3q, a strong trying transition is found for D = 4 Å in TIP3P water, consistent with that in SPC water. However, when D = 5 Å there’s water fluctuation in the intermonomer region in TIP3P water while in SPC water there’s a drying transition at this monomer separation. For other three proteins (PDB ID: 1g6u, 1f4n, and 1fsz), their systems also fluctuate between “dry” and “wet” states in TIP3P water instead of drying in SPC water at the same domain or oligomer separation. It might due to the fact it is slightly more difficult for the confined region to dry in TIP3P water compared to SPC water. The SPC water model has slightly higher bulk water–water interaction energy than the TIP3P water model.39 In SPC/E water,40 the results on dewetting in the confined regions are consistent with that in SPC water for both protein tetramer 1j2w and protein dimer 1j3q. Overall, these three different water models give roughly the same thermodynamical results on dewetting, even though the time scale for the dewetting transition can be slightly different. Further validations have been done with both the PME and cutoff methods for the long-range electrostatic interactions. Two proteins 1j2w (tetramer) and 1j3q (dimer) have been used for this validation. Both protein exhibit a drying transition (data not shown) in the three water models for D = 4 Å when either PME or cutoff is used. So in general, for these protein candidates, the drying transition is quite robust since it is observed for different water models and for different treatments of electrostatic interactions.
Drying transitions might also be important in ligand-binding. In a recent study, Young et al. observed dewetting in the Cox-2 active site.41 They found that this binding cavity is entirely devoid of water with the size large enough to hold seven water molecules sterically. We have used our surface hydrophobicity analysis tools to search through the protein–ligand database. The preliminary results show that several proteins from different classes (i.e., binding proteins with PDB ID: 1WUB, 1RBP, 1Y9L, 1WBE) exhibit a drying transition, indicating that dewetting might be an important factor to consider in the ligand binding free energy calculation.
4. Method
4.1. Surface Hydrophobicity Distribution Function
To identify other protein candidates capable of displaying a drying transition, we propose a hydrophobic score based on a protein surface hydrophobicity analysis throughout the protein database. Since the hydrophobic residues are normally buried to avoid contact with water, the solvent accessible regions of proteins are mostly hydrophilic. We hypothesize that for a protein to display a drying transitions it should have large matched and connected hydrophobic surface regions on the buried contact protein surfaces. To explore the surface hydrophobicity of different proteins, we have performed a surface hydrophobicity analysis of the protein contact surfaces between two domains or oligomers. In our analysis, we have defined the z axis such that it connects the centers of mass of the two domains or protein oligomers (see Figure 1a). The first step is to project the geometric centers of the surface residues into a plane perpendicular to the z axis (the x – y plane), which is then divided into 5 × 5 Å cells (we have tried other cell lengths and found 5 Å to give slightly better results). The surface hydrophobicity distribution is defined as,
| (1) |
where the sum is over all residues i within cell {x, y}. Here cell {x, y} is that cell which contains the center point (x, y), hi is the Eisenberg hydrophobicity value,36 and ai is the solvent accessible surface area of residue i which is computed by the software of molecular surface package.42 Typical surface hydrophobicity distribution functions f(x, y) for a dimer protein are shown in Figure 1b. f(x, y) is not normalized since we believe the larger hydrophobic surface the protein has, the easier it can display dewetting.
When f(x, y) > 0, the surface of the cell located at (x, y) is hydrophobic. We want to find candidates that maximize the corresponding cells on the two opposing domain or oligomer surfaces that are both hydrophobic. Thus, we define a hydrophobic score, Am, which measures the total matched hydrophobic cells,
| (2) |
where fA(x, y) and fB(x, y) are the surface hydrophobicity distributions for domains A and B, the prime indicates that we are summing over matched hydrophobic cells, and Nmatch is the number of matched hydrophobic cell pairs. Again, Am is not averaged by Nmatch because the larger the number of matched hydrophobic cells is, the more probable it is to display dewetting.
Hydrophobic patches defined as clusters of neighboring nonpolar atoms on a protein surface are essential for the protein folding and aggregation.27,43 Here, we define another quantity, the connected hydrophobic surface area Ac, which measures the connected hydrophobic cells. This can be computed from the sum of contiguous hydrophobic cells’ surface hydrophobicity distribution functions (f(x, y)) using a connected component analysis based on 8-connectivity.44
| (3) |
where star indicates the summation is over all the connected hydrophobic cells (only hydrophobic cells but not hydrophilic ones). Nconnect is the number of connected hydrophobic cells on each domain or oligomer surface.
If we consider both the matched and connected hydrophobic surfaces, another similar score, Amc, matched and connected hydrophobic area, can be defined by eq 2:
| (4) |
where double prime indicates that we are summing over not only matched, but also connected hydrophobic cells. Nmatch–connect is the number of matched and connected hydrophobic cell pairs.
Among three parameters defined above (Am, Ac, and Amc), matched hydrophobic areas (Am) and matched connected hydrophobic areas (Amc) are most crucial to find proteins with most hydrophobic buried surfaces. Ac is only of subsidiary importance. In our lists shown in Tables 1–3, the proteins are ranked based on Am.
4.2. MD Simulations
The starting structure of each selected protein is taken from the crystal structure deposited in PDB bank. To test if these proteins can display a drying transition, two domains or protein oligomers of each candidate are extended by a distance D, ranging from 4 to 7 Å, along the direction of the vector connecting the centers of mass of the two domains or protein oligomers to create gaps. The resulting configurations are solvated in water boxes with water molecules at least 8 Å from the protein surface. Counterions are added to make the system electrically neutral. GROMACS45 simulation package is used for its fast speed. The OPLSAA force field is used for the protein.46 In all of the simulations, SPC38 water model for the explicit solvent is used unless explicitly stated (in a few cases, TIP3P39 and SPCE40 water models are also used for validation). A cutoff of 12 Å is adopted for the nonbonded interactions. And in several cases, the particle mesh Ewald (PME) method is used for the long-range electrostatic interactions as for comparison (the dewetting results do not differ much). For each protein system, up to 12 ns NPT MD simulations (1 atm and 298 K) with protein atoms constrained are performed after a conjugated gradient minimization. We use Berendsen methods for both pressure and temperature coupling. 47 For each protein showing drying transition, up to 5 ns NPT MD simulations with no position constraint for protein atoms are performed starting at different initial separations, with up to 10 different initial water configurations for each separation.
Acknowledgment
This work was supported in part by an NIH grant (GM43340) to B.J.B. Part of the simulations were run on IBM BlueGene/L development machines. We thank Jose Castanos and his team for providing the running environment, and Maria Eleftheriou, Bob Walkup, and Ajay Royyuru for the help and support with the BG/L machine.
References and Notes
- 1.Nicholls A, Sharp K, Honig B. Proteins. 1991;11:281–296. doi: 10.1002/prot.340110407. [DOI] [PubMed] [Google Scholar]
- 2.Sorin E, Rhee Y, Pande V. Biophys. J. 2005;88:2516–2524. doi: 10.1529/biophysj.104.055087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lum K, Chandler D, Weeks JD. J. Phys. Chem. B. 1999;103:4570–4577. [Google Scholar]
- 4.Stillinger FH. J. Solution Chem. 1973;2:141. [Google Scholar]
- 5.Attard P. Langmuir. 2000;16:4455. [Google Scholar]
- 6.Israelachvili J. Surf. Sci. Rep. 1992;14:109–159. [Google Scholar]
- 7.Christenson H, Claesson P. Adv. Colloid Interface Sci. 2001;91:391–436. [Google Scholar]
- 8.Ball P. Nature. 2003;423:25–26. doi: 10.1038/423025a. [DOI] [PubMed] [Google Scholar]
- 9.Tyrrell J, Attard P. Phys. Rev. Lett. 2001;87(1–4):176104. doi: 10.1103/PhysRevLett.87.176104. [DOI] [PubMed] [Google Scholar]
- 10.Steitz R, Gutberlet T, Hauss T, Klösgen B, Krastev R, Schemmel S, Simonsen A, Findenegg GH. Langmuir. 2003;19:2409–2418. [Google Scholar]
- 11.Yakobov G, Butt H-J, Vinogradova O. J. Phys. Chem. B. 2000;104:3407–3410. [Google Scholar]
- 12.Jensen T, Jensen M, Reitzel N, Balashev K, Peters GH, Kjaer K, BjOrnholm T. Phys. Rev. Lett. 2003;90(1–4):086101. doi: 10.1103/PhysRevLett.90.086101. [DOI] [PubMed] [Google Scholar]
- 13.Jensen M, Mouritsen O, Peters GH. J. Chem. Phys. 2004;120:9729–9744. doi: 10.1063/1.1697379. [DOI] [PubMed] [Google Scholar]
- 14.Singh S, Houston J, Swol FV, Brinker CJ. Nature. 2006;442:526. doi: 10.1038/442526a. [DOI] [PubMed] [Google Scholar]
- 15.Hummer G, Garde S. Phys. Rev. Lett. 1998;80:4193–4196. [Google Scholar]
- 16.Wallqvist A, Berne BJ. J. Phys. Chem. 1995;99:2893–2899. [Google Scholar]
- 17.Leung K, Luzar A, Bratko D. Phys. Rev. Lett. 2003;90(1–4):65502. doi: 10.1103/PhysRevLett.90.065502. [DOI] [PubMed] [Google Scholar]
- 18.Luzar A, Leung K. J. Chem. Phys. 2000;113(14):5836–5844. [Google Scholar]
- 19.Wolde PRT, Chandler D. Proc. Natl. Acad. Sci. U.S.A. 2002;99:6539–6543. doi: 10.1073/pnas.052153299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang X, Margulis CJ, Berne BJ. Proc. Nat. Acad. Sci. U.S.A. 2003;100:11953–11958. doi: 10.1073/pnas.1934837100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hummer G, Rasaiah JR, Noworyta JP. Nature. 2001;414:188–190. doi: 10.1038/35102535. [DOI] [PubMed] [Google Scholar]
- 22.Huang X, Margulis C, Berne B. Proc. Natl. Acad. Sci. U.S.A. 2003;100:11953. doi: 10.1073/pnas.1934837100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cheng Y, Rossky PJ. Nature. 1998;392:696–699. doi: 10.1038/33653. [DOI] [PubMed] [Google Scholar]
- 24.Zhou R, Huang X, Margulius CJ, Berne BJ. Science. 2004;305:1605–1609. doi: 10.1126/science.1101176. [DOI] [PubMed] [Google Scholar]
- 25.Liu P, Huang X, Zhou R, Berne BJ. Nature. 2005:437. [Google Scholar]
- 26.Tsai C-J, Nussinov R. Protein Sci. 1997;6:1426–1437. doi: 10.1002/pro.5560060707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lijnzaad P, Argos P. Proteins. 1997;28:333–343. [PubMed] [Google Scholar]
- 28.Jones S, Thornton J. J. Mol. Biol. 1997;272:121–132. doi: 10.1006/jmbi.1997.1234. [DOI] [PubMed] [Google Scholar]
- 29.Tsai C-J, Lin S, Wolfson H, Nussinov R. Protein Sci. 1997;6:53–64. doi: 10.1002/pro.5560060106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Larsen T, Olson A, Goodsell D. Structure. 1998;6:421–427. doi: 10.1016/s0969-2126(98)00044-6. [DOI] [PubMed] [Google Scholar]
- 31.Stetefeld J, Jenny M, Schulthess T, Landwehr R, Engel J, Kammerer RA. Nat. Struct. Biol. 2000;7:772–776. doi: 10.1038/79006. [DOI] [PubMed] [Google Scholar]
- 32.Lokanath N, Shiromizu IN, Ohshima YN, Sugahara M, Yokoyama S, Kuramitsu S, Miyano M, Kunishima N. Acta Cryst. 2004;D60:1816–1823. doi: 10.1107/S0907444904020190. [DOI] [PubMed] [Google Scholar]
- 33.Willis M, Bishop B, Regan L, Brunger A. Structure. 2000;8:1319–1328. doi: 10.1016/s0969-2126(00)00544-x. [DOI] [PubMed] [Google Scholar]
- 34.Ogihara N, Ghirlanda G, Bryson J, Gingery M, Degrado W, Eisenberg D. Proc. Natl. Acad. Sci. U.S.A. 2001;98:1404–1409. doi: 10.1073/pnas.98.4.1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dams T, Auerbach G, Bader G, Jacob U, Ploom T, Huber R, Jaenicke R. J. Mol. Biol. 2000;297:659–672. doi: 10.1006/jmbi.2000.3570. [DOI] [PubMed] [Google Scholar]
- 36.Zhou R, Silverman BD, Royyuru A, Athma P. Proteins. 2003;52:561–572. doi: 10.1002/prot.10419. [DOI] [PubMed] [Google Scholar]
- 37.Lowe J, Amos LA. Nature. 1998;391(8):203. doi: 10.1038/34472. [DOI] [PubMed] [Google Scholar]
- 38.Berendsen HJC, Postma JPM, van Gunsteren WF, Hermans J. In: Pullman B, editor. Intermolecular Forces; Proceedings of the 14th Jerusalem Symposium on Quantum Chemistry and Biochemistry; April 13–16, 1981; Jerusalem, Israel. Dordrecht, Holland: Reidel; 1981. pp. 331–342. [Google Scholar]
- 39.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. J. Chem. Phys. 1983;79:926. [Google Scholar]
- 40.Werder T, Walther J, Jaffe R, Halicioglu T, Koumoutsakos P. J. Phys. Chem. B. 2003;107:1345–1352. [Google Scholar]
- 41.Young T, Abel R, Kim B, Berne B, Friesner RA. Proc. Nat. Acad. Sci. U.S.A. 2007;104(3):808–813. doi: 10.1073/pnas.0610202104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Connolly ML. Molecular Surface Package, Version 3.92. 2002 [Google Scholar]
- 43.Lijnzaad P, Berendsen H, Argos P. Proteins. 1996;26:192–203. doi: 10.1002/(SICI)1097-0134(199610)26:2<192::AID-PROT9>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 44.Fisher R, Perkins S, Walker A, Wolfart E. Hypermedia Image Processing Reference, Version 1. 2000 [Google Scholar]
- 45.Lindahl E, Hess B, van der Spoel D. J. Mol. Model. 2001;7:306–317. [Google Scholar]
- 46.Jorgensen WL, Maxwell D, Tirado-Rives J. J. Am. Chem. Soc. 1996;118:11225–11236. [Google Scholar]
- 47.Berendsen HJC, Postma JPM, van Gunsteren WF, Haak J. J. Chem. Phys. 1984;81:3684. [Google Scholar]