Abstract
BAFF and APRIL are innate immune mediators that trigger immunoglobulin (Ig) G and IgA class switch recombination (CSR) in B cells by engaging the receptor TACI. The mechanism underlying CSR signaling by TACI remains unknown. Here, we found that the cytoplasmic domain of TACI encompasses a conserved motif that bound MyD88, an adaptor protein that activates NF-κB signaling pathways via a Toll-interleukin-1 receptor (TIR) domain. TACI lacks a TIR domain, yet triggered CSR via the DNA-editing enzyme AID by activating NF-κB through a TLR-like MyD88–IRAK-1-IRAK-4–TRAF6–TAK1 pathway. TACI-induced CSR was impaired in mice and humans lacking MyD88 or IRAK-4, indicating that MyD88 controls a B cell-intrinsic, TIR-independent, TACI-dependent pathway for Ig diversification.
INTRODUCTION
Diversification is essential for the generation of immune protection1. Bone marrow B cell precursors generate antigen recognition diversity by recombining VHDJH and VLJL exons encoding antigen-binding immunoglobulin (Ig) heavy (H) and light (L) chain variable regions from individual V (variable), D (diversity) and J (joining) gene segments2. Mature B cells emerging from the bone marrow further diversify their Ig gene repertoire through somatic hypermutation (SHM) and class switch DNA recombination (CSR). SHM introduces point mutations at high rates into recombined VHDJH and VLJL exons, thereby providing a structural correlate for the selection of higher affinity Ig variants by antigen, whereas CSR endows Ig molecules with new effector functions by replacing the heavy chain constant region (CH) of IgM with that of IgG, IgA or IgE without changing antigen specificity3.
CSR involves an exchange of an upstream donor Cμ gene with a downstream acceptor CH gene through a recombinatorial process guided by switch (S) regions. Located 5′ of each CH gene, S regions are preceded by a short intronic (I) exon and a promoter that initiates germline CH gene transcription when the B cell is exposed to appropriate stimuli3. Actively transcribed S regions become substrate of activation-induced cytidine deaminase (AID)4, an enzyme that initiates CSR by introducing double-strand DNA breaks within targeted S regions3. Subsequent deletion of the intervening DNA between recombined S regions juxtaposes the VHDJH exon to a new CH gene3.
In general, CSR requires a primary signal from a tumor necrosis factor (TNF) family member such as CD40 ligand (CD40L; http://www.signaling-gateway.org/molecule/query?afcsid=A000536), B cell-activating factor of the TNF family (BAFF; http://www.signaling-gateway.org/molecule/query?afcsid=A000383) or a proliferation-inducing ligand (APRIL; http://www.signaling-gateway.org/molecule/query?afcsid=A000305), and a co-signal from cytokines5. Most antigens trigger CSR in the germinal center (GC) of lymphoid follicles by promoting interaction of CD40L on CD4+ T cells with CD40 on B cells6. The ensuing oligomerization of CD40 [http://www.signaling-gateway.org/molecule/query?afcsid=A000031] triggers recruitment of TNF receptor associated factor (TRAF) adaptor proteins to its cytoplasmic domain7. These TRAFs activate an IκB kinase (IKK) complex comprising two α and β catalytic subunits and a γ regulatory subunit8. By phosphorylating inhibitor of nuclear factor-κB (IκBα, http://www.signaling-gateway.org/molecule/query?afcsid=A000097), which retains the transcription factor NF-κB in an inactive cytoplasmic state under resting conditions, IKK elicits ubiquitination and proteasome-dependent degradation of IκBα, thereby allowing nuclear translocation of NF-κB8. In the presence of cytokine-induced signal transducer and activator of transcription (STAT) proteins, CD40-induced NF-κB initiates the transcription of targeted CH genes as well as AICDA, which encodes AID3.
CD40 signaling leads to the induction of class-switched and hypermutated memory B cells as well as Ig-secreting plasma cells from follicular B cells, which provide long-term protection9. This T cell-dependent (TD) pathway takes five to seven days — a period too long for the control of rapidly replicating pathogens10. As a means of overcoming this limitation, extrafollicular B cells produce IgM, IgG and IgA through a faster T cell-independent (TI) pathway involving BAFF and APRIL11–15. These factors are released by innate immune cells in response to microbial signals emanating from Toll-like receptors (TLRs) and activate NF-κB by recruiting TRAFs through the receptor transmembrane activator and calcium modulator and cyclophilin ligand interactor (TACI; http://www.signaling-gateway.org/molecule/query?afcsid=A002248), and two receptors related to TACI, B cell maturation antigen (BCMA), and BAFF receptor (BAFF-R, or BR3)16. Of these receptors, TACI mediates CSR, at least in mouse B cells15,17,18.
Deleterious mutations of the TACI gene are common in human populations, particularly in patients with common variable immune deficiency (CVID), a disorder in which the production of IgG, IgA and IgM is impaired19–22. The mechanism by which TACI triggers CSR remains unknown, but previous findings raise the intriguing possibility that BAFF-induced IgG production involves a TI pathway comprising MyD88 [http://www.signaling-gateway.org/molecule/query?afcsid=A003535] (ref. 23). This adaptor protein regulates innate immunity by activating NF-κB and other transcription factors through the Toll-interleukin-1 receptor (TIR) domain of TLRs and IL-1 receptor (IL-1R)24.
We show here that BAFF and APRIL promoted recruitment of MyD88 to a conserved cytoplasmic motif of TACI distinct from the TIR domain of TLRs. TACI–MyD88 interaction induced CSR by triggering NF-κB activation, germline CH gene transcription and AICDA expression through a TIR-independent pathway that was impaired in mice and humans lacking MyD88 or IL-1R-associated kinase 4 (IRAK-4), a signal transducer that binds MyD88 (ref. 24). Thus, we propose that MyD88 enhances Ig diversification and production by linking the innate and adaptive immune systems through TACI.
RESULTS
TACI signals CSR in cooperation with TLRs
Patients carrying mutations in the TNFRSF13B gene encoding TACI show reduced IgG and IgA production19,20,22,25. We investigated the function of human TACI in more detail by visualizing TACI expression in lymphoid organs from healthy subjects through immunohistochemistry. Follicular B cells, which usually mediate TD (CD40-dependent) Ig responses, displayed higher TACI expression in IgD− GCs than in the IgDhi mantle zone (Fig. 1a). Extrafollicular IgDlo B cells, which mediate TI (CD40-independent) Ig responses, showed higher TACI expression than follicular mantle IgDhi B cells. TACI was particularly abundant in IgDlo B cells from the subepithelial area of the tonsils and the marginal zone (MZ) of the spleen.
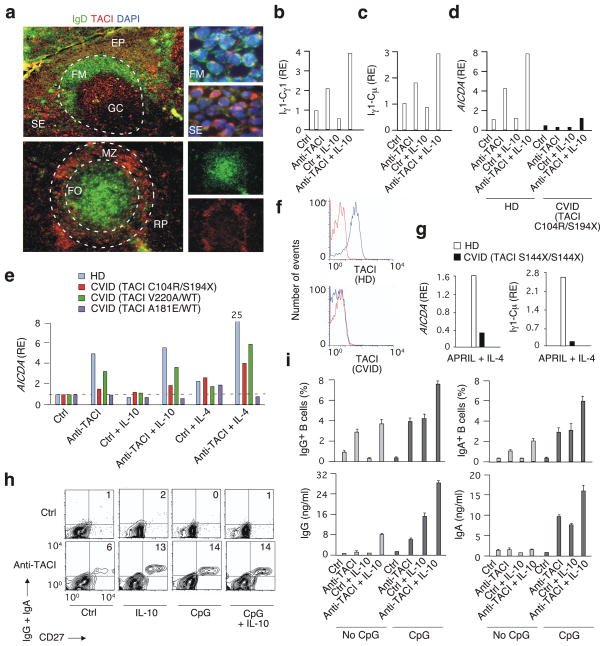
Figure 1. TACI triggers CSR by cooperating with TLR ligands.
(a) Immunofluorescence staining of tonsil (top) and splenic (bottom) tissues for IgD (green), TACI (red), and nuclei (blue). Dashed line, follicle; EP, epithelium; FM, follicular mantle; FO, follicle; GC, germinal center; MZ; marginal zone; SE, sub-epithelium; RP, red pulp. Original magnification, ×10 (left panels) or ×63 (right panels). (b–e) QRT-PCR of Iγ1-Cγ1, Iγ1-Cμ and AICDA in naïve (b–d) or lymphoblastoid (e) B cells from healthy donors (HD) or CVID patients with various heterozygous TACI substitutions cultured for 2 or 4 days with or without anti-TACI, IL-10 and/or IL-4. Results are normalized to ACTB (encoding β-actin) mRNA; RE, relative expression compared to B cells incubated with a control antibody (ctrl). (f) Flow cytometry of TACI on primary CD19+CD27+ B cells from a HD or CVID patient with homozygous S144X/S144X TACI substitution. Red histograms, ctrl; blue histograms, anti-TACI. (g) AICDA and Iγ1-Cμ in primary naive B cells from CVID case shown in f incubated with BAFF or APRIL plus IL-4 for 6 d. (h) Flow cytometry of IgG, IgA and CD27 on primary naive B cells incubated for 7 days with ctr, anti-TACI, IL-10 and/or CpG DNA. Numbers indicate percentages. (i) Flow cytometry of IgG and IgA (upper panels) and ELISA of secreted IgG and IgA (bottom panels) from B cells stimulated as in h. *P < 0.05 (one-tailed unpaired Student’s t-test). Data are from one of three experiments with similar results (a–h) or summarize three experiments (i; error bars, s.d.).
Following our detection of TACI on extrafollicular B cells, we reasoned that TACI might initiate CD40-independent CSR. We tested this hypothesis by culturing primary preswitched IgD+ B cells from the peripheral blood of healthy subjects in the presence or absence of a cross-linking agonistic antibody directed against TACI (anti-TACI) and then analyzing the induction of CSR-related events by quantitative real-time-polymerase chain reaction (qRT-PCR), flow cytometry and enzyme-linked immunosorbent assay (ELISA). IgD+ B cells induced germline Iγ1-Cγ1 transcripts, an early marker of IgG1 CSR, as early as two days after exposure to anti-TACI (Fig. 1b). Switch Iγ1-Cμ transcripts and AICDA transcripts encoding AID, two hallmarks of ongoing IgG1 CSR13, were induced by anti-TACI after four days (Fig. 1c,d). Anti-TACI alone induced some IgG2, but not IgG3 or IgG4 CSR (not shown).
We further characterized the CSR-inducing function of TACI in CVID patients and healthy individuals carrying TNFRSF13B gene mutations. AICDA induction by anti-TACI was attenuated in primary IgD+ B cells and lymphoblastoid B cells from CVID patients carrying heterozygous C104R/WT and A181E/WT substitutions or compound heterozygous C104R/S194X substitutions affecting the extracellular (C104R), transmembrane (A181E) or cytoplasmic (S194X) domain of TACI (Fig. 1d,e). C104R and A181E may affect TACI oligomerization in response to ligation of BAFF or APRIL, whereas S194X generates a truncated TACI protein lacking most of the cytoplasmic domain26. A heterozygous V220A/WT substitution targeting the cytoplasmic domain of TACI had little or no effect on TACI-induced AICDA expression, which was consistent with previously published studies showing that V220A/WT is usually present in healthy individuals22. The involvement of TACI in CSR was further demonstrated in one individual carrying a homozygous S144X/S144X mutation generating a truncated TACI protein lacking both the transmembrane and cytoplasmic domains. B cells from this patient expressed no TACI (Fig. 1f) and induced less AICDA and Iγ1-Cμ in response to IL-4 and cognate TACI ligands such as APRIL (Fig. 1g) or BAFF (not shown).
When combined with IL-10, IL-4 or IL-21, three cytokines providing CSR-inducing co-signals13,14,27, anti-TACI increased the generation of Iγ1-Cγ1, Iγ1-Cμ and AICDA transcripts more effectively than anti-TACI alone, at least in primary B cells. Lymphoblastoid B cells showed less cooperation between anti-TACI and IL-10, probably because these cells constitutively express large amounts of endogenous IL-10. After five days, anti-TACI and cytokines also induced a higher frequency of B cells expressing the effector-memory antigen CD27 and class-switched IgG and IgA (Fig. 1h,i). Finally, anti-TACI and cytokines augmented the secretion of IgG after seven days (Fig. 1i and Supplementary Fig. 1). This IgG comprised IgG1 and some IgG2, but not IgG3 or IgG4 (not shown).
Given that TLRs induce BAFF and APRIL expression by innate immune cells5, TACI may cooperate with TLRs to induce TI CSR. Flow cytometry showed that extrafollicular B cells expressed TACI as well as TLR5, TLR7 and TLR9 more strongly than follicular B cells (Supplementary Fig. 2). In addition, extrafollicular B cells displayed stronger upregulation of TACI expression in response to two-day engagement of TLR5 (by flagellin), TLR7 (by imiquimod) and TLR9 (by deoxycytidylate phosphate deoxyguanylate-containing DNA, CpG DNA), but not CD40 (by CD40L; not shown). Furthermore, CpG DNA and imiquimod (not shown) enhanced the induction of class-switched effector B cells by anti-TACI (Fig. 1h). Finally, TLR ligands increased the secretion of IgG and induced the secretion of IgA in B cells exposed to anti-TACI and IL-10 (Fig. 1i). Thus, human TACI triggers TI pathways of IgG and IgA CSR and production by cooperating with both cytokines and TLR ligands.
TACI signals CSR in cooperation with CD40
As TACI was expressed on follicular B cells, we also investigated its involvement in CD40-dependent CSR. A combination of anti-TACI and CD40L induced more class-switched CD27+ B cells and IgG secretion than anti-TACI or CD40L alone (Supplementary Fig. S3). The addition of IL-10 further increased the ability of anti-TACI to stimulate CD40-mediated IgG and IgA production. Consistent with these findings, GCs from the spleen of a CVID patient with compound heterozygous C104R/S194X TACI substitutions had less expression of AID, whereas expression of Pax5, a B cell specific transcription factor, and the proliferation nuclear protein Ki-67 were normal. Furthermore, CVID patients with deleterious heterozygous C104R/WT, R72H/WT, A181E/WT or V220A/WT, homozygous S144X/S144X, or compound heterozygous C104R/S194X substitutions affecting the extracellular or intracellular domain of TACI had a lower class-switched IgD−/unswitched IgD+ B cell ratio, as did hyper-IgM (HIGM) patients with deleterious CD40L or AID substitutions (Supplementary Fig. 3 and Supplementary Table 1). Thus, human TACI mediates both TI and TD pathways of CSR.
TACI but not CD40 interacts with MyD88
The functional cooperation of TACI with TLRs and CD40 may stem from the convergence of these receptors on both proximal and distal signaling proteins, such as TRAF6 and NF-κB, respectively16,24. Given the important role of MyD88 in both TI and TD antibody responses28,29, we hypothesized that TACI, TLRs and CD40 may also converge on MyD88. We generated a chimeric GST-TACI protein comprising glutathione S-transferase (GST) fused to the cytoplasmic domain of TACI (residues 187-293) or CD40 (residues 216-277) for initial immunoprecipitation and immunoblotting experiments. These assays were performed with protein extracts from 2E2, a subclone of an IgD+ CL-01 human B cell line that undergoes MyD88-dependent class switching in response to TLR signals30. Like other B cell lines, 2E2 B cells display some constitutive TACI signaling as a result of autocrine BAFF and APRIL production31. We found that GST-TACI coimmunoprecipitated with MyD88, whereas GST-CD40 did not (Fig. 2a). The interaction of GST-TACI with MyD88 was specific, because GST-TACI but not GST-CD40 was also associated with calcium modulator and cyclophilin ligand (CAML), which is a specific TACI-binding protein32. Furthermore, GST-BCMA and GST-BAFF-R fusion proteins encompassing the cytoplasmic domains of BCMA (residues 58-184) and BAFF-R (residues 100-184) associated with TRAF2, TRAF5 and TRAF6 consistent with previously described interaction patterns33,34, but did not immunoprecipitate MyD88 or CAML.
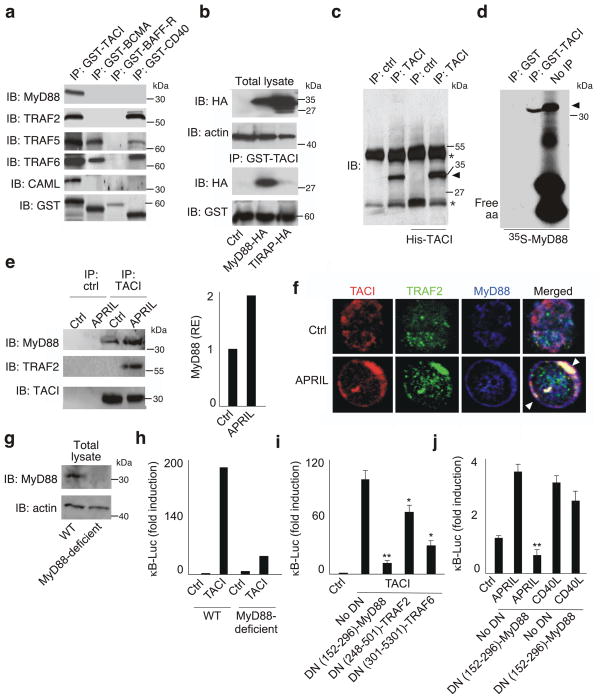
Figure 2. TACI interacts with MyD88.
(a) GST-TACI, GST-BCMA, GST-BAFF-R or GST-CD40 immunoprecipitation (IP) of 2E2 B cell lysates, followed by immunoblotting (IB) of MyD88, TRAF2, TRAF5, TRAF6, CAML, or GST. kDa, kilodaltons. (b) Upper gel: IB of HA or actin in total lysates from 293 cells expressing control plasmid (ctrl), MyD88-HA or TIRAP-HA. Bottom gel: GST-TACI IP of 293 cell lysates, followed by IB of HA or GST. (c) Anti-TACI or anti-His IP of lysates from 2E2 B cells or TACI-His-expressing 293 cells, followed by IB for MyD88 or TACI. Asterisks, heavy (upper) and light (lower) chains of IP antibody; arrowhead, MyD88. (d) GST or GST-TACI IP of 35S-MyD88. Rightmost lane, 35S-MyD88 before IP; arrow, MyD88; free aa, free amino acids. (e) Anti-TACI or control antibody (ctrl) IP of lysates from human primary naïve B cells cultured for 15 min with medium (ctrl) or APRIL, followed by IB of MyD88, TRAF2 and TACI. Bars show intensity of MyD88 band relative to TACI in unstimulated B cells. (f) Confocal microscopy of TACI (red), TRAF2 (green) and MyD88 (blue) in primary naïve B cells exposed to medium (ctrl) or APRIL for 15 min. Arrowheads indicate co-localization. (g) IB of MyD88 and actin (loading control) from WT or MyD88-deficient 293 cells. (h,i) NF-κB reporter assay in WT or MyD88-deficient 293 cells transfected with TACI or a control empty plasmid (ctrl) in the presence or absence of DN-MyD88, DN-TRAF2 or DN-TRAF6. (j) NF-κB reporter assay in 2E2 B cells transfected with DN-MyD88 or a control empty plasmid (no DN) and incubated with medium (ctrl), APRIL or CD40L for 2 d. *P < 0.05 and ** P < 0.005 (one-tailed unpaired Student’s t-test). The data shown are from one of three experiments giving similar results (a–h) or summarize three experiments (i,j; error bars, SEM).
GST-TACI also coimmunoprecipitated MyD88 but not TIR domain-containing adaptor protein (TIRAP, also known as Mal), a TLR-associated molecule related to MyD88 (refs. 24,35), from total lysates of 293 cells transfected with hemagglutinin (HA)-tagged or FLAG-tagged MyD88 and TIRAP expression plasmids (Fig. 2b and Supplementary Fig. 4). In addition, anti-TACI coimmunoprecipitated TACI and MyD88 from total lysates of 2E2 B cells as well as histidine-tagged TACI and MyD88 from total lysates of transfected 293 cells, whereas an isotype-matched control antibody did not (Fig. 2c). Coimmunoprecipitation of TACI with MyD88 probably resulted from direct physical interaction between these two proteins, because GST-TACI was also capable of immunoprecipitating in vitro translated and radiolabeled MyD88 in a cell-free system (Fig. 2d). Thus, human TACI can interact with MyD88.
TACI recruits MyD88 upon ligation
TLRs recruit MyD88 after sensing cognate microbial ligands24. The demonstration of ligand-induced recruitment of MyD88 to TACI in B cells is complicated by the constitutive occupation of TACI by autocrine and paracrine cognate ligands, particularly in B cell lines, and by the spontaneous oligomerization of TACI via its pre-ligand-binding assembly domain31,36,37. To circumvent these limitations, we performed immunoprecipitation experiments using primary preswitched IgD+ B cells from the spleen of healthy subjects. These B cells exhibit less constitutive TACI signaling compared to B cell lines and specifically generate signals from TACI in response to APRIL because they lack BCMA37.
In unstimulated primary IgD+ B cells, some constitutive coimmunoprecipitation occurred between TACI and MyD88 but not TRAF2 (Fig. 2e), perhaps due to ligand-independent aggregation of TACI through its pre-ligand-binding assembly domain26. Stimulation with APRIL for 15 min increased the association of TACI with MyD88 by two-fold and also induced coimmunoprecipitation of TACI with TRAF2. Consistent with these results, confocal microscopy showed minute plasma membrane foci of TACI and MyD88 protein colocalization in the absence of APRIL. These scattered foci developed into large and polarized membrane patches of TACI, MyD88, TRAF2, TRAF6 and CAML colocalization upon exposure to APRIL (Fig. 2f and Supplementary Fig. 5). Such patches also contained flotillin-1, a protein associated with plasma membrane lipid-rich microdomains specialized in the compartmentalization of signaling molecules38.
Since TACI engagement by APRIL (or BAFF) is followed by activation of the CSR-inducing transcription factor NF-κB, we also verified whether MyD88 was required for the activation of NF-κB by TACI. Immunoblotting and luciferase reporter assays showed impaired MyD88 expression in I3A cells (Fig. 2g), a subclone of 293 cells unable to respond to IL-1 (ref. 39). I3A cells also showed decreased NF-κB-dependent gene transcription upon overexpression of TACI for 48 h (Fig. 2h). Similarly, TACI induced little or no NF-κB-dependent gene transcription in 293 cells cotransfected with dominant-negative (DN)-MyD88 (Fig. 2i). Of note, DN-TRAF2 and DN-TRAF6 reduced TACI-mediated NF-κB-dependent gene transcription less than DN-MyD88 did. DN-MyD88 also hampered NF-κB-dependent gene transcription in 2E2 B cells stimulated with APRIL, but not CD40L (Fig. 2j). Thus, human TACI recruits MyD88 in a ligand-dependent fashion to initiate downstream signaling events, including NF-κB activation.
TACI requires MyD88 to activate NF-κB
To confirm ligand-induced recruitment of MyD88 to TACI, we stimulated resting B cells from the spleen of wild-type or MyD88-deficient C57BL/6J (B6) mice with APRIL. This ligand increased TACI stable interaction with MyD88, TRAF2, TRAF5, TRAF6, CAML and IRAK4 as early as 5 min after B cell stimulation in wild-type mice (Fig. 3a). After 15 min, confocal microscopy showed colocalization of TACI, MyD88 and TRAF2 within polarized cellular compartments (Fig. 3b). Consistent with previous reports showing intracellular expression of TACI36,37, APRIL triggered cytoplasmic-to-membrane translocation of not only MyD88 and TRAF2, but also some TACI. We then assessed the role of MyD88 in early NF-κB signals emanating from TACI by immunoblotting. APRIL induced phosphorylation of IKKα, IKKβ and IκBα (triggering its degradation, not shown) as early as 5 min in wild-type cells, whereas no phosphorylation occurred in MyD88-deficient B cells (Fig. 3c). Phosphorylation of p38, a mitogen-activated kinase signal transducer associated with many receptors, including TLRs24, was not induced by APRIL.
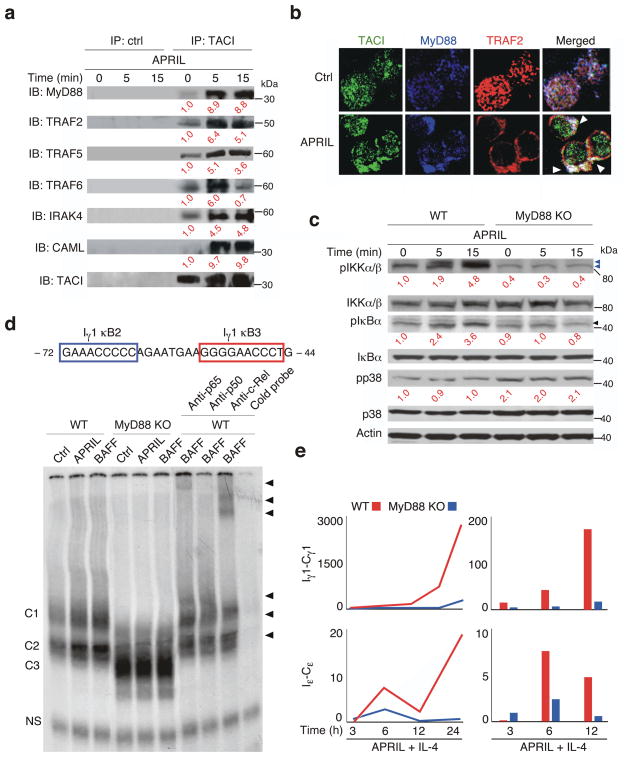
Figure 3. TACI requires MyD88 to activate NF-κB.
(a) Immunoblotting (IB) of MyD88, TRAF2, TRAF5, TRAF6, IRAK-4, CAML and TACI upon control (ctrl) antibody or anti-TACI immunoprecipitation (IP) of lysates from primary mouse B cells cultured for 0, 5 or 15 min with APRIL. Red numbers indicate band intensity relative to TACI in unstimulated B cells. kDa, kilodaltons. (b) Confocal microscopy of TACI (green), TRAF2 (red) and MyD88 (blue) in primary mouse B cells exposed to medium (ctrl) or APRIL for 15 min. Arrowheads point to co-localization. (c) IB of phosphorylated (p) IKKα/β, IKKα/β, pIκBα, pIκBα, pp38, p38 and actin in primary WT or MyD88 KO B cells incubated with APRIL for 0, 5 or 15 min. Blue arrowheads point to pIKKβ (upper) and pIKKα (lower); black arrowhead points to pIκBα. Red numbers indicate band intensity relative to IKKα/β, IκBα and p38 in unstimulated B cells. (d) EMSA of NF-κB binding to the Iγ1 promoter in mouse primary B cells cultured with medium (ctrl), BAFF or APRIL for 3 h. Numbers indicate nucleotide positions compared to transcription initiation site. C1-C3, specific protein-DNA complexes; black arrowheads, supershifts; NS, non-specific band. (e) QRT-PCR of Iγ1-Cγ1 and Iε-Cε transcripts in primary WT (red) or MyD88 KO (blue) naïve B cells incubated with APRIL and IL-4 for various time points. Results are normalized to ACTB (encoding β-actin) mRNA; RE, relative expression compared to unstimulated B cells. Curves, time course; bars, earliest time points. Data are from one of three experiments giving similar results.
Nuclear extracts obtained from wild-type or MyD88-deficient B cells stimulated or not with APRIL or BAFF for 3 h were used in electrophoretic mobility shift assays with a probe derived from κB2, a key NF-κB-bindingκB site from the Cγ1 gene (also called Iγ1) promoter3. Wild-type B cells induced nuclear translocation of C1 and C2 complexes containing p50, p65 and c-Rel upon exposure to BAFF or APRIL, whereas MyD88-deficient B cells did not (Fig. 3d). In MyD88-deficient mice, κB2 was associated with a distinct C3 complex that lacked p50, p65 and c-Rel in both unstimulated and stimulated B cells (Supplementary Fig. 6). In mice, optimal transcription of Cγ1 and Cε a gene functionally related to Cγ1, requires a second signal provided by IL-4 (ref. 3). Compared to APRIL or IL-4 stimulation alone, a combination of APRIL and IL-4 induced a robust expression of germline Iγ1-Cγ1 and Iγ-Cγ transcripts, detected by qRT-PCR, as early as 6 h after stimulation (Fig. 3e and Supplementary Fig. 7). This early induction of transcription did not occur in the absence of MyD88. Thus, TACI requires MyD88 for the early initiation of NF-κB-dependent germline CH gene transcription.
TACI signals through a TIR-less MyD88-binding site
We then investigated the molecular requirements for the interaction of human TACI with MyD88. Comparison of the cytoplasmic domain of TACI from different species led to the identification of five highly conserved domains with a high hydrophobicity index (Supplementary Fig. 8), which has been linked to protein-protein interactions. These conserved regions included PTQES, which is a canonical TRAF2-binding site. Given that TRAF2 is critical for the induction of CSR by CD40 (ref. 34), we used PTQES as a reference to generate progressive TACI deletion mutants, which were subsequently used in immunoprecipitation assays to map the MyD88-binding site of TACI (Fig. 4a). GST-TACI interacted with MyD88 through a 217–239 region upstream from the TRAF2-binding site that lacked any similarity with TIR (Fig. 4b). Additional assays further narrowed the MyD88-binding site to a conserved 227–239 segment that was named TACI highly conserved (THC) domain.
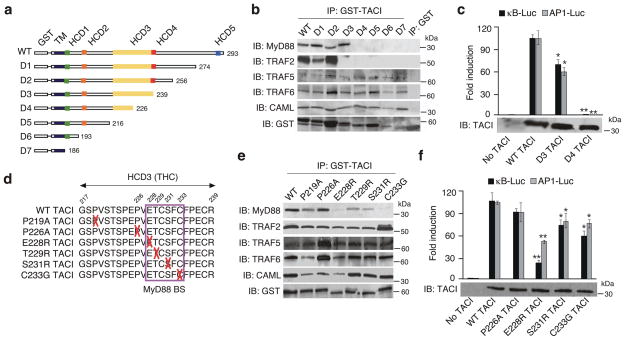
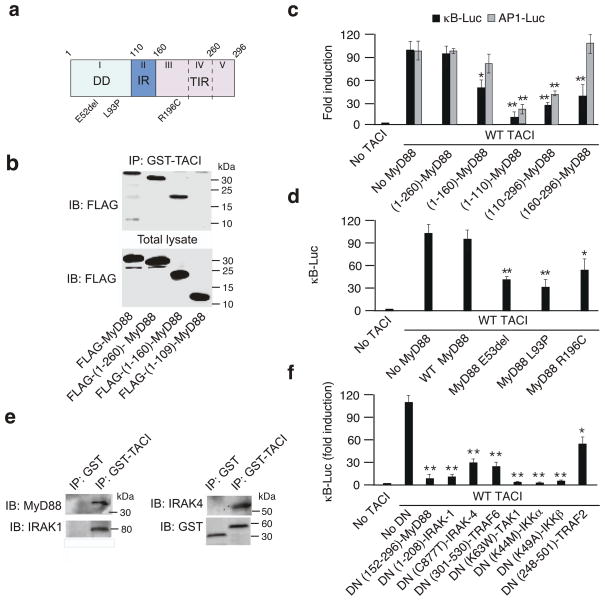
Figure 4. TACI binds MyD88 through a THC domain.
(a) Scheme of WT TACI and D1-D7 GST-TACI deletion mutants. Numbers, carboxy-terminal residues; TD, transmembrane domain; HCD, highly conserved domain. Complete TACI sequence shown in Supplementary Fig. S8. (b) Immunoprecipitation (IP) of 2E2 B cell lysates with GST, WT GST-TACI, or GST-TACI deletion mutants followed by immunoblotting (IB) of MyD88, TRAF2, TRAF5, TRAF6, CAML, or GST. kD, kilodaltons. (c) NF-κB and AP1 reporter assays in 293 cells expressing no TACI, WT TACI, D3 TACI or D4 TACI. Bottom gel, IB of TACI from total lysates. (d) Site-directed mutations in the THC domain. Upper numbers, residue positions; red crosses, substitutions; purple box, MyD88-binding site (BS). (e) IP of 2E2 B cell lysates with WT GST-TACI or GST-TACI site-directed mutants followed by IB of MyD88, TRAF2, TRAF5, TRAF6, CAML, or GST. (f) NF-κB and AP1 reporter assays in 293 cells expressing no TACI, WT TACI, or TACI site-directed mutants. Bottom gel, IB of TACI from total cell lysates. *P < 0.05 and ** P < 0.005 (one-tailed unpaired Student’s t-test). The data shown are from one of three experiments giving similar results (c,f) or summarize three experiments (d,g; error bars, SEM).
Of note, TRAF5, TRAF6 and CAML but not MyD88 and TRAF2 showed conserved binding to constructs encompassing the transmembrane domain and eventually a fragment of the cytoplasmic region of TACI (D6, D7). Protein sequence analysis led to the identification of two transmembrane domains and a TRAF-binding site within CAML (Supplementary Fig. 9). In addition, CAML co-immunoprecipitated with TRAF5 and TRAF6 from 2E2 B cells with constitutive TACI signaling activity, suggesting that TACI amplifies the recruitment of TRAF5 and TRAF6 by interacting with CAML through one or both its transmembrane domains.
Luciferase reporter assays showed that, compared to wild-type TACI, a TACI deletion mutant (D3) lacking only the TRAF2-binding site induced less effectively NF-κB and activator protein 1 (AP-1) (Fig. 4c), a transcription factor involved in B cell activation but not CSR. Another TACI deletion mutant (D4) lacking both the TRAF2-binding site and the THC domain showed virtually no induction of NF-κB and AP-1 despite retaining TRAF5, TRAF6 and CAML binding activity. We investigated the contributions of MyD88 and TRAF2 to TACI signaling further, by using site-directed mutagenesis to generate E228R, T229R, S231R and C233G substitutions affecting the THC domain but not the TRAF2-binding motif of TACI (Fig. 4d). These substitutions decreased the binding of MyD88 but not TRAF2, TRAF5, TRAF6 and CAML to TACI (Fig. 4e) and attenuated TACI signaling via both NF-κB and AP-1 (Fig. 4f).
A control A181E substitution affecting the transmembrane domain of TACI or R202H, P219A, V220A and P226A substitutions affecting the cytoplasmic domain of TACI outside its MyD88-, TRAF- and CAML-binding sites had little or no inhibitory effect on TACI signaling via NF-κB and AP1 and did not impair TACI interaction with MyD88, TRAF2, TRAF5, TRAF6 or CAML (Fig. 4f and Supplementary Fig. 10). An additional S194X substitution truncating the cytoplasmic tail of TACI upstream from the MyD88- and TRAF2-binding sites had no effect on TACI interaction with TRAF5, TRAF6 and CAML, but abolished TACI binding to MyD88 and TRAF2 and TACI signaling via NF-κB and AP1. Thus, TACI recruits MyD88 and activates NF-κB via a TIR-less THC domain.
TACI interacts with MyD88 in a TIR-independent fashion
MyD88 dimerization occurs upon TLR stimulation, which facilitates interaction of the death domain (DD) of MyD88 with a homologous DD of IRAK-1 and IRAK-4 (Fig. 5a). By recruiting TRAF6, IRAK-4 mediates activation of the kinase TAK1, which in turn activates IKK24. Consistent with the lack of TIR in the cytoplasmic tail of TACI, FLAG-(1-260)-MyD88 and FLAG-(1-160)-MyD88 proteins lacking all or part of the TIR domain retained the ability to interact with GST-TACI and had little or no DN effect on TACI signaling via NF-κB and AP-1 in 293 cells (Fig. 5b,c). By contrast, a FLAG-(1-109)-MyD88 protein lacking both the TIR domain and the intermediary region (IR) showed no binding to GST-TACI, rather this construct displayed potent inhibitory activity against TACI signaling via both NF-κB and AP1. A similar dominant-negative effect was observed with a FLAG-(110-296)-MyD88 protein lacking only the DD, whereas a FLAG-(160-296)-MyD88 protein lacking both the DD and IR inhibited TACI signaling via NF-κB, but spared TACI signaling via AP-1. Of note, DN-MyD88 constructs did not alter NF-κB signaling in 293 cells lacking TACI (Supplementary Fig. 11). TACI signaling was also impaired by MyD88 substitutions E52del, L93P and R196C (Fig. 5d), which mimic those causing MyD88 deficiency and found in patients with an immunodeficiency causing recurrent infections in childhood40.
Figure 5. TACI signals through a TLR-like pathway.
(a) Structure of MyD88; top Arabic numerals, residues delimiting DD, IR and TIR; Roman numerals (I to V), exons; bottom codes, mutations causing MyD88 deficiency. (b) Immunoblotting (IB) of FLAG from 293 cells expressing wt FLAG-MyD88 or FLAG-MyD88 deletion mutants before (total lysate) or after immunoprecipitation (IP) with GST-TACI. (c,d) NF-κB and AP1 reporter assays in 293 cells expressing wt MyD88 or various deletion and site-directed MyD88 mutant proteins in the presence or absence of TACI. *P < 0.05 or **P < 0.005 versus no MyD88 (one-tailed unpaired Student’s t-test). (e) IP of 2E2 B cell lysates with GST or GST-TACI, followed by IB of MyD88, IRAK-1, IRAK-4 or GST (loading control). (f) NF-κB reporter assays in 293 cells with and without wt TACI expression in the presence or absence of DN-MyD88, DN-IRAK-1, DN-IRAK-4, DN-TRAF6, DN-TAK1, DN-IKKα, DN-IKKβ, or DN-TRAF2. *P < 0.05 and ** P < 0.005, versus no DN (one-tailed unpaired Student’s t-test). Data represent one of three experiments giving similar results (b,e) or summarize three experiments (c,d,f; error bars, SEM).
Since TACI recruits IRAK4 and TRAF6 in addition to MyD88, we determined the role of these and other TLR-associated signal transducers in the activation of NF-κB by TACI. Immunoprecipitation assays showed that human GST-TACI bound IRAK1 and IRAK4 from 2E2 B cells (Fig. 5e), whereas luciferase reporter assays demonstrated that DN versions of IRAK-1, IRAK-4, TRAF6, TAK1, IKKα and IKKβ inhibited NF-κB induction by TACI in 293 cells (Fig. 5f). Also DN-TRAF2 had an inhibitory effect on TACI-induced NF-κB activation, but less than DN-MyD88. A similar DN approach confirmed the involvement of IRAK-1, IRAK-4, TRAF6, TAK1 and IKKβ but not TRAF3 in the induction of NF-κB by BAFF in 2E2 B cells (Supplementary Fig. 12). Thus, MyD88 does not make use of TIR-TIR interaction to bind to TACI, yet signals through a TLR-like pathway emanating from TACI and cooperating with TRAF2.
TACI requires MyD88 to trigger CSR
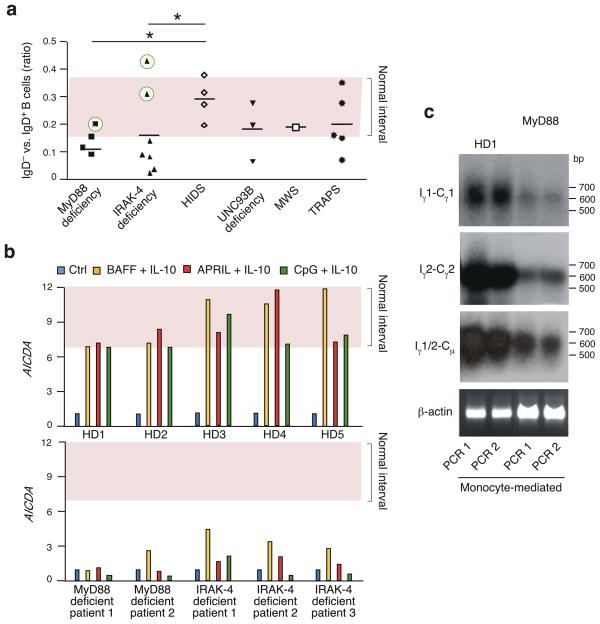
We next wondered whether the lack of MyD88 or IRAK-4 attenuates IgG CSR in humans. In spite of having conserved serum IgG, some MyD88- and IRAK-4-deficient patients had impaired IgG responses to microbial polysaccharides (Supplementary Table 1; ref. 41), a canonical TI antigen. Similar to individuals with TACI defects, these patients also had a lower IgD−/IgD+ B cell ratio than age-matched healthy controls and patients with unrelated inflammatory disorders (Fig. 6a). B cells from two MyD88-deficient patients and three IRAK-4-deficient patients induced AICDA transcripts less strongly upon four-day exposure to BAFF, APRIL or CpG DNA than did B cells from healthy subjects (Fig. 6b). AICDA induction by BAFF and/or APRIL was blunted in two IRAK-4-deficient patients with a normal IgD−/IgD+ B cell ratio, whereas one MyD88-deficient patient showed a paradoxically stronger response to BAFF, APRIL and CpG DNA (Supplementary Fig. 13). Finally, B cells from a MyD88-deficient patient expressed fewer Iγ1-Cγ1, Iγ2-Cγ2 and Iγ1-Cμ transcripts upon four-day exposure to monocytes expressing BAFF and APRIL than did B cells from a healthy control (Fig. 6c and Supplementary Fig. 14).
Figure 6. TACI requires MyD88 to induce CSR in humans.
(a) Flow cytometric analysis of IgD−/IgD+ B-cell ratio in the peripheral blood of patients with MyD88 or IRAK-4 deficiency and control age-matched patients with hyper-IgD syndrome (HIDS), UNC93B deficiency, Mukle-Wells syndrome (MWS) or TNF receptor-associated periodic fever syndrome (TRAPS). Pink area delimits range of IgD−/IgD+ B-cell ratio in healthy donors (normal interval). Circled cases are further studied in Supplementary Fig. S12. *P <0.05 (two-tailed paired Student’s t-test). (b) QRT-PCR of AICDA transcripts in B cells from healthy donors (HD) and patients with MyD88 or IRAK-4 deficiency, after culture for 4 d in the presence or absence of BAFF plus IL-10, APRIL plus IL-10 or CpG DNA plus IL-10. Results are normalized to ACTB (encoding β-actin) mRNA; RE, relative expression compared to control (ctrl) unstimulated B cells. (c) Southern blot analysis of RT-PCR-amplified Iγ1-Cγ1, Iγ2-Cγ2, and Iγ1/2-Cμ transcripts in B cells from healthy or MyD88-deficient subjects exposed to BAFF- and APRIL-expressing monocytes. Additional controls shown in Supplementary Fig. S13. PCR1 and PCR2 indicate independent PCR amplifications; bp, base pairs.
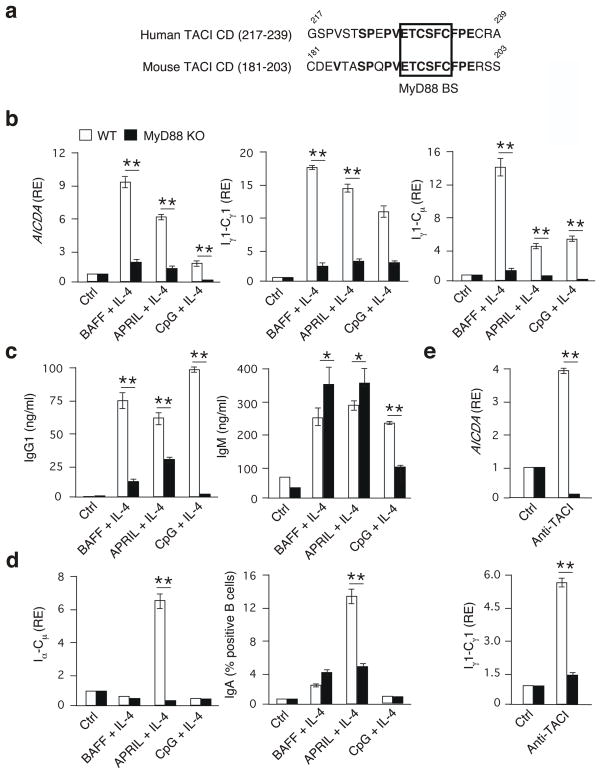
As the MyD88-binding site was identical in human and mouse TACI proteins (Fig. 7a), we sought to confirm in mouse B cells whether MyD88 was required for the induction of CSR by TACI. Naive B cells from the spleen of MyD88-deficient mice induced fewer Iγ1-Cγ1, AICDA and Iγ1-Cμ transcripts and secreted less IgG1 upon two-, four- or seven-day exposure to BAFF, APRIL or CpG DNA and IL-4 than did control B cells (Fig. 7b,c). The lack of MyD88 also impaired the induction of Iα-Cμ and membrane IgA by APRIL and IL-4 (Fig. 7d) or the induction of AICDA and Iγ1-Cγ1 by anti-TACI (Fig. 7e). These effects were specific, because the lack of MyD88 did not hamper the IgM secretion and minimally affected the survival of B cells exposed to BAFF or APRIL (Supplementary Fig. 15). Thus, MyD88 regulates CSR via a TIR-independent pathway emanating from TACI (Supplementary Fig. 16).
Figure 7. TACI requires MyD88 to induce CSR in mice.
(a) MyD88-binding site (BS) in human or mouse TACI. Numbers, amino-acid positions; bold letters, identical amino acids; box, MyD88-binding site in the THC domain. (b) QRT-PCR of AICDA, Iγ1-Cγ1 and Iγ1-Cμ transcripts from WT (open bars) or MyD88 KO (solid bars) mouse B cells cultured for 4 d in the presence or absence of BAFF, APRIL or CpG DNA plus IL-4. Results are normalized to ACTB (encoding β-actin) mRNA; RE, relative expression compared to control (ctrl) unstimulated B cells. (c) ELISA of IgG1 and IgM from WT or MyD88 KO B cells cultured as in c for 8 days. (d,e) QRT-PCR of Iα1-Cμ and flow cytometric analysis of surface IgA from WT or MyD88 KO B cells cultured as in b for 48 h (Iα1-Cμ) or 5 days (IgA). (e) QRT-PCR of AICDA and Iγ1-Cγ1 from WT or MyD88 KO B cells cultured with a ctrl antibody or anti-TACI for 2 d. *P < 0.05, versus wild type (one-tailed unpaired Student’s t-test). The data presented summarize three independent experiments performed by pooling splenic naive B cells from three mice (error bars, SEM).
DISCUSSION
We found that BAFF and APRIL elicited CSR by inducing recruitment of MyD88 to a highly conserved THC cytoplasmic domain of TACI different from the TIR domain of TLRs. Interaction of TACI with MyD88 initiated germline CH gene transcription, AID expression and CSR by activating NF-κB through a TLR-like signaling cascade that was impaired in mice and humans lacking MyD88 or IRAK-4. These findings indicate that MyD88 controls a novel B cell-intrinsic, TACI-dependent, TIR-independent pathway for Ig gene diversification and production.
TLR signaling is commonly visualized as a linear cascade initiated by recruitment of MyD88 to the TIR domain24. Yet, TLRs generate convoluted signaling routes characterized by extensive branching and crosstalk with non-TLR pathways42. By showing ligation-induced recruitment of MyD88 to TACI, our findings point to the existence of an intimate B cell-intrinsic crosstalk between TACI and TLRs and provide a novel molecular framework for recent observations indicating that BAFF and APRIL need MyD88 to generate IgG and IgA23,29,43. Unlike TLRs, TACI did not make use of the TIR domain to interact with MyD88, but rather relied on a highly conserved THC domain. This motif recognized the IR domain of MyD88, which is also required by MyD88 to interact with the interferon-γ receptor44.
Similar to TLRs, TACI utilized a MyD88-IRAK-1-IRAK-4-TRAF6-TAK1-IKK signaling pathway to activate NF-κB in B cells. TACI-induced NF-κB initiated germline Cγ1 gene transcription by trans-activating the Iγ1 promoter upon binding to key cis-regulatory sites such as κB2. In MyD88-deficient B cells, this κB2 site was constitutive occupied by unknown proteins possibly corresponding to transcriptional repressors, suggesting that MyD88 exerts a complex control on CSR. A similar MyD88-dependent pathway likely supported TACI-induced AICDA gene transcription, because lack of MyD88 or IRAK4 impaired AICDA induction in response to TACI ligation. In general, TACI required co-signals from IL-4, IL-10 or IL-21 to elicit optimal CH and AICDA gene transcription and subsequent CSR. Additional co-signals from TLRs were required to initiate IgG and IgA production.
A prominent feature of the MyD88-binding site of TACI was its proximity to a canonical TRAF2-binding site. Deletion of both MyD88- and TRAF2-binding sites abrogated TACI signaling, whereas disruption of either site did not, which suggests that MyD88 and TRAF2 function in a cooperative manner. Such cooperation may be fostered by the ability of TACI of eliciting ligand-dependent co-localization of MyD88 and TRAF2 in flotillin-1-containing plasma membrane microdomains specialized in the organization of signaling complexes38. Flotillin-1-containing areas also comprised TRAF6 and CAML. The contribution of TRAF6 to TACI signaling is unclear, but it may function downstream of MyD88 to facilitate the activation of IKK. TRAF6 also interacted with CAML, an NF-AT (nuclear factor-activating T cells)-inducing protein that binds to the membrane-proximal cytoplasmic domain of TACI32,45. Although capable of interacting also with TRAF5, CAML eventually played a marginal role in the induction of NF-κB and AP-1 activation by TACI, which mostly required MyD88 and TRAF2. Yet, TACI may utilize CAML to enhance B cell proliferation, B cell survival and plasmacytoid differentiation independently of NF-κB and AP-1, perhaps through a pathway involving NF-AT46,47.
In agreement with previously published data15,17,40,41,43,48, TACI signaling via MyD88 and MyD88-interacting proteins such as IRAK-4 may help extrafollicular B cells to mount sustained IgG and IgA responses against TI antigens such as polysaccharides from encapsulated bacteria. Consistent with this interpretation, some patients with MyD88 or IRAK-4 deficiency showed decreased IgG responses to pneumococcal polysaccharides in addition to impaired induction of AID and IgG CSR in B cells exposed to BAFF or APRIL. As suggested by recent findings47, TACI-MyD88 interaction may further enhance TI antibody responses by delivering survival signals to activated extrafollicular B cells, including class-switched plasmablasts.
TACI signaling via MyD88 may also enhance IgG and IgA responses against TD antigens such as common microbial proteins. Accordingly, follicular B cells expressed TACI and augmented IgG production upon dual engagement of TACI and CD40. Moreover, patients carrying deleterious TACI substitutions had decreased AID expression in the germinal center, a B cell area highly dependent on CD40. The involvement of TACI signaling via MyD88 in TD humoral immunity would be consistent with the impairment of TD IgG responses in mice with a B cell-intrinsic efficiency of MyD88, at least in some models28,49,50. In spite of their involvement in TACI-induced CSR, MyD88 and its downstream partner IRAK-4 were not critical for the control of circulating IgG and IgA under steady-state conditions. This circumstance could be explained by the fact that TACI utilizes TRAF2 to compensate for the lack of MyD88 or IRAK-4. Further compensatory signals may be generated by BAFF-R, BCMA and CD40, which indeed did not make use of MyD88.
Recently published data show that lupus-prone T cell-deficient mice require MyD88 to develop IgG autoantibodies in response to a BAFF transgene23, which suggests that TACI signaling via MyD88 may exacerbate autoimmunity by promoting IgG CSR in autoreactive B cells. Should this be the case, small inhibitors of TACI-MyD88 interaction could be used to treat autoimmune disorders associated with pathogenic CSR and dysregulated BAFF and APRIL expression. A more general implication of our findings is that MyD88 has important adaptive functions, given its utilization by B cells downstream from TACI, and therefore cannot be regarded as an adaptor molecule operating exclusively within the innate immune system. By conveying CSR-inducing signals emanating from both TACI and TLRs, MyD88 probably serves as a B cell-intrinsic molecular bridge between innate and adaptive responses. Consequently, the B cell-related phenotypes previously observed in MyD88-deficient mice28,29,50 may not result solely from defective TLR and IL-1R signaling, but are instead likely to involve additional defects in TACI signaling.
ONLINE METHODS
Patients
The patients affected with CVID, MyD88 deficiency, IRAK-4 deficiency, CD40L deficiency (HIGM1), CD40 deficiency (HIGM3), AID deficiency (HIGM2), Muckle-Wells syndrome (MWS), TNF receptor-associated periodic syndrome (TRAPS) and hyper-IgD syndrome (HIDS) studied here have been described elsewhere27,40,48. The Institutional Review Board of Mount Sinai School of Medicine and Institut Municipal d’Investigació Mèdica-Hospital del Mar approved the use of blood and tissue specimens from these patients.
Human B cell isolation and cultures
Total pre-switched IgD+ B cells and CD14+ monocytes were obtained from the peripheral blood of healthy donors as previously reported27. Lymphoblastoid B cell lines were obtained by incubating peripheral blood mononuclear cells from healthy donors or CVID patients carrying TACI substitutions as described elsewhere31. 2E2 is a BAFF/APRIL/CpG DNA-responsive subclone of the CL-01 clone30,31, which was derived from a BL16 line with constitutive BAFF and APRIL signaling activity51. Cultures were performed in complete RPMI medium supplemented with 10% (volume/volume) bovine serum. B cells were incubated with CD40L (PeproTech), 500 ng/ml; BAFF (Alexis), 500 ng/ml; APRIL MegaLigand (Alexis), 500 ng/ml; IL-4 (Schering-Plough), 200 U/ml; IL-10 (Peprotech), 50 ng/ml; and IL-21 (PeproTech), 100 ng/ml. TACI was cross-linked with 1 μg/ml mouse biotin-conjugated 11H3 mAb (eBioscience) and anti-biotin-microbeads (Miltenyi). Alternatively, 1 μg/ml mouse 165604 mAb to TACI (R&D Systems) was immobilized on irradiated (7000 rad) CD32 (Fc© receptor II)-expressing mouse lung fibroblasts lacking thymidine kinase (Ltk− cells or L-cells). After irradiation, L-cells were washed, adhered to plastic wells, and incubated with B cells at a 1:5 ratio. A similar strategy was followed to cross-link CD40 with 1 μg/ml mouse 89 mAb (Schering-Plough). Mouse IgG1 mAb with irrelevant binding activity (Santa Cruz Biotechnology) were used as control. TLR5 was activated with 0.5 μg/ml flagellin from Bacillus subtilis (Sigma), TLR7 with 1 μg/ml imiquimod (Invivogen), and TLR9 with 5μg/ml phosphorothioate-modified 5′-tcgtcgttttgtcgttttgtcgtt-3′ oligodeoxynucleotide-2006 (Operon Technologies). In some cultures, B cells were incubated with autologous monocytes at a 1:2 ratio.
Mouse B cell isolation and cultures
Splenic naive IgM+IgD+ B cells were negatively selected from the spleen of 8–12 wk-old WT or Myd88−/− C57BL/6J mice using a modified version of a commercially available mouse B cell isolation kit (Miltenyi Biotec). Briefly, total splenocytes were first depleted of red blood cells with ammonium chloride potassium lysis buffer and then incubated with a biotin-antibody cocktail containing a mix of antibodies to T cells, NK cells and monocytes (Miltenyi Biotec). This mix was supplemented with biotin-conjugated rat A85-1 mAb to IgG1 and rat C10-1 mAb to IgA (BD Biosciences) to separate class-switched B cells. Anti-biotin microbeads and an appropriate magnetic device (Miltenyi Biotec) were used for the final sorting. Cultures were performed in complete RPMI medium in the presence or absence of 10 ng/ml IL-4, 500 ng/ml MegaAPRIL (Alexis), 500 ng/ml BAFF (Alexis) and/or 5 μg/ml CpG ODN (Operon Technologies). In some experiments, cells were treated with 5 μg/ml goat biotin-conjugated pAb to TACI (R&D Systems).
Flow cytometry
B cells were incubated with an Fc blocking reagent (Miltenyi Biotec) or saturating concentrations of purified human IgG and stained, at 4 °C, with appropriate polyclonal antibodies (pAbs) or monoclonal antibodies (mAbs) against IgD, IgM, IgG, IgA and CD27, as reported elsewhere27. Additional stainings were performed with the following unconjugated, fluorochrome-conjugated or biotin-conjugated primary antibodies: mouse mAb 165604 to TACI (R&D Systems), goat 2032-02 pAb to IgD, goat 2050-08 pAb to IgA, goat 2042-08 pAb to IgG, mouse SA-DA4 mAb to IgM (Southern Biotech), mouse M-T271 mAb to CD27 (Ancell), rabbit pAb 36-3900 to TLR5, rabbit pAb 36-6500 to TLR7, or rabbit pAb 52-5197 to TLR9 (Invitrogen). Primary antibodies were detected with appropriate secondary reagents as described14,27,52. In experiments involving TLR measurements, B cells were permeabilized with saponin. 7-AAD was used to exclude dead cells from the analysis. Events were acquired on a FACS Calibur or BD LSR II machine (BD Biosciences) and analyzed with FlowJo (Tree Star).
Immunohistofluorescence
Tonsil samples were from individuals undergoing tonsillectomy due to tonsillar hypertrophy. Lymph node samples were from individuals undergoing biopsies due to reactive lymphoadenopathy. Spleen samples were from individuals undergoing post-traumatic splenectomy. All these samples were obtained as part of routine diagnostic procedures and subsequently archived in local tissue repositories. The Institutional Review Board of Mount Sinai School of Medicine and IMIM-Hospital del Mar approved the use of these specimens. Tissues were stained with the following unconjugated or conjugated primary antibodies: goat 2032-02 pAb to IgD (Southern Biotech), goat C-20 pAb to TACI, goat C-20 pAb to AID, mouse A11 mAb to Pax5 (Santa Cruz Biotechnologies), or mouse MIB-1 mAb to Ki-67 (Dako). Primary antibodies were detected with appropriate secondary reagents as described previously14,27,52. Nuclei were visualized with DAPI, 4′,6-diamidine-2′-phenylindole dihydrochloride (Boehringer Mannheim). Slides were applied with Slow Fade reagent (Molecular Probes) and analyzed with a Zeiss Axioplan 2 microscope (Atto Instruments).
Laser-scanning confocal microscopy
Cells were resuspended in Cell Adhesive Solution as instructed by the manufacturer (Crystalgen), applied onto slides (Gold Seal Products), fixed with 1.6% paraformaldehyde, and permeabilized with 0.2% Triton-100 in phosphate buffer solution. Stainings were performed with the following unconjugated or biotin-conjugated primary reagents: goat pAb N-19 to TACI, goat pAb N-19 to MyD88, rabbit pAb C-20 to TRAF2, goat pAb H-274 to TRAF6 (Santa Cruz Biotechnologies), rabbit pAb AB16529 to MyD88 (Milipore), goat pAb AF174 to TACI (R&D Systems), mouse pAb ab67714 to CAML, goat pAb ab13493 to flotillin-1 (Abcam). Negative controls were performed with primary reagents with irrelevant binding activity and included biotin-conjugated F(ab′)2 pAb and unconjugated goat, mouse or rabbit pAbs (Santa Cruz Biotechnologies). Secondary reagents included streptavidin, Alexa Fluor 488-conjugated anti-mouse pAb, Alexa Fluor 546-conjugated anti-rabbit pAb, or Alexa Fluor 647-conjugated anti-goat pAb (Molecular Probes). Nuclei were visualized with 4′,6-diamidine-2′-phenylindole dihydrochloride (DAPI) (Boehringer Mannheim). Slides were coverslipped with FluorSave reagent (Calbiochem). Fluorescence images were generated with a Leica SP5 DMI upright confocal microscope (Wetzlar) by acquiring at least 3 different xy planes utilizing 63×/1.4 NA objective lenses (Carl Zeiss) with optimal z spacing (~0.016 μm). Views were constructed with maximum projection and exported as 30–40 TIFF images. AutoQuant X2 AutoDeblur software (Media Cybernetics) was used to deconvolve all images and restore detail to datasets. Further processing was performed using Adobe Photoshop software CS3 for Macintosh, version 10 (Adobe Systems).
RT-PCR, qT-PCR, and Southern blotting
To detect human germline Iγ1-Cγ1 and Iγ2-Cγ2 transcripts, switch Iγ1/2-Cμ circle transcripts and β-actin, total RNA was extracted using the RNeasy Mini Kit (Qiagen) and cDNA was synthesized as previously described14,30,52. Transcripts were amplified by standard RT-PCR and hybridized with appropriate radiolabeled probes through Southern blotting as described previously14,30,52. To quantify Iγ1-Cγ1 and AICDA transcripts, total RNA was extracted using TRIzol (Invitrogen). cDNA synthesis and qRT-PCR were performed as previously reported14,30,52. Mouse AICDA, Iγ1-Cγ1, Iγ1-Cμ, Iμ-Cμ and β-actin transcripts were quantified by QRT-PCR using AICDA sense primer 5′-GCCACCTTCGCAACAAGTCT-3′ and AICDA antisense primer 5′-CCGGGCACAGTCATAGCAC-3′; Iγ1-Cγ1 sense primer 5′-GGCCCTTCCAGATCTTTGAG-3′ and Iγ1-Cγ1 antisense primer 5′-GGATCCAGAGTTCCAGGTCACT-3′; Iγ1-Cμ sense primer 5′-GG CCCTTCCAGATCTTTGAG-3′ and Iγ1-Cμ antisense primer 5′-GAAGACATTTGGGAAGGACTGACT-3′; Iα-Cμ sense primer 5′-CCTGGCTGTTCCCCTATGAA-3′ and Iα-Cμ antisense primer 5′-GAAGACATTTGGGAAGGACTGACT-3′; Iε-Cε sense primer 5′-CTGGCCAGCCACTCACTTAT-3′ and Iε-Cε antisense primer 5′-CAGTGCCTTTACAGGGCTTC-3′; and β-actin sense primer 5′-CATG TACGTAGCCATCCAGGC-3′ and β-actin antisense primer 5′-CTCTTTGATGTCACGCACGAT-3′.
Plasmids
Human full-length TACI was generated by cloning a TNFRSF13B cDNA into a modified pcDNA3.0 expression-plasmid encoding a carboxy-terminal 6X histidine (His) tag (Invitrogen) using appropriate primers (Supplementary Table 2). GST-TACI fusion proteins were obtained by cloning a TNFRSF13b cDNA encoding the transmembrane and cytoplasmic domains of human (residues 149-293) or mouse (residues 121-249) TACI into the EcoRI and NotI cloning sites of a pGEX-6P-1 expression-plasmid using appropriate primers (Supplementary Table 2). Human GST-TACI D1, D2, D3, D4, D5, D6 and D7 deletion mutants were PCR-amplified from a GST-TACI-encoding expression plasmid using appropriate primers (Supplementary Table 2). A mouse GST-(121-180)-TACI deletion mutant lacking the cytoplasmic MyD88 binding site was obtained by cloning a TNFRSF13B cDNA encoding the 121-180 fragment of mouse TACI into the EcoRI and NotI cloning sites of a pGEX-6P-1 expression-plasmid using appropriate primers (Supplementary Table 2). Human A181E, S194X, P219A, R202H, V220A, P226A, E228R, T229R, S231R and C233G TACI mutants were PCR-generated from expression plasmids encoding full-length TACI or GST-TACI using the QuickChange II Site-Directed Mutagenesis kit (Stratagene) (Supplementary Table 2). Human BCMA-GST, BAFF-R-GST and CD40-GST fusion proteins were generated by cloning TNFRSF17, TNFRSF13C and TNFRSF5 cDNAs encoding the cytoplasmic domain of BCMA (residues 58-184), BAFF-R (residues 100-184), and CD40 (residues 216-277) into the EcoRI and NotI cloning sites of a pGEX-6P-1 plasmid (Supplementary Table 2). Human full-length MyD88 was obtained by cloning a MYD88 cDNA into either a modified pcDNA3.0 expression vector encoding a carboxy-terminal HA tag or into a pCMV-FLAG2 (Sigma) expression vector encoding an amino-terminal FLAG tag (Supplementary Table 2). Human FLAG-MyD88 1-260, 1-160, 1-110, 110-296 and 160-296 deletion mutants were PCR-amplified from a plasmid encoding full-length MyD88 (Supplementary Table 2). Human E53del, L93P and R196C MyD88 site-directed mutants were generated as described previously40. Human full-length TIRAP was obtained by cloning PCR-amplified TIRAP cDNA into a modified pcDNA3.0 expression vector encoding a carboxy-terminal HA tag (Supplementary Table 2). DN-(152-296)-MyD88, DN-(1-208)-IRAK-1 (Tularik), DN-(C877T)-IRAK-4 (from S. Vogel and A. Medvedev, University of Maryland), DN-(248-501)-TRAF2, DN-(301-530)-TRAF6 (Science Reagents), DN-(K44M)-IKKα, DN-(K49A)-IKKβ, DN-(K429/430A)-NIK (from J.D. Li, University of South California), and DN-(K63W)-TAK1 are shown elsewhere30,53,54.
Luciferase reporter assays
293T and I3A cells were transfected using Superfect transfection reagent (Qiagen) and 10 μg κB(2x)-Luc or AP1-Luc reporter plasmid expressing firefly luciferase and 500 ng pRL-TK reporter plasmid expressing renilla luciferase under the control of the thymidine kinase promoter (Promega) in the presence or absence of 1 μg pcDNA 3.0 expression plasmids encoding wild type or mutant TACI or MyD88. 293 were also co-transfected with 10 μg DN-TRAF2, DN-TRAF6, DN-TAK1, DN-IKKα or DN-IKKβ expression plasmids. An empty pcDNA3.0 expression plasmid was used as negative control. Empty pcDNA3.0 was also used to equal the overall amount of plasmid in transfected 293 cells. Luciferase activities were measured after 48 h with Dual Luciferase Assay System (Promega). Firefly luciferase activity was normalized to that of the cotransfected pRL-TK control plasmid.
In vitro translation assay
Transcription-translation reactions were performed at 30 °C for 60 min in 25 μl of rabbit reticulocyte lysate using a TnT T7 Quick Coupled Transcription/Translation System (Promega). The reaction mix contained 10 μCi/μl 35S-methionine, 10 μCi/μl 35S-cysteine, and MyD88-expressing pcDNA3.0. Total lysates were used for pull-down assays with GST-TACI or control GST.
Pull down and co-immunoprecipitation assays
For pull down assays, GST-TACI, GST-BCMA, GST-BAFF-R or GST-CD40 fusion proteins were purified using glutathion-agarose beads according to the manufacturer’s protocol (Sigma) and analyzed for homogeneity by sodium dodecyl sulfate (SDS) -polyacrylamide gel and silver staining. Total human B cell lysates were pulled-down with GST or GST fusion proteins, fractionated onto sodium dodecyl sulphate-polyacrylamide gel, and then transferred into nitrocellulose membranes. After blocking, membranes were immunoblotted for TRAF2, TRAF5, TRAF6, MyD88, CAML or GST. For co-immunoprecipitation assays, total B cell lysates were first pre-cleared with 100 μl Streptavidin Sepharose beads (GE healthcare) for 30 min at 4°C, and then incubated with 2.5 μg control IgG2a mAb or human-reactive biotin-conjugated anti-TACI 11H3 mAb (eBioscience) or mouse-reactive biotin-conjugated BAF104 mAb (R&D Systems) overnight. The next day, lysates were incubated with 100 μl Streptavidin Sepharose beads for 1 h at 4°C to pull down immune complexes. Immunoprecipitated proteins were fractionated onto sodium dodecyl sulphate-polyacrylamide gel and immunoblotted for MyD88, IRAK-4, TRAF2, TRAF5, TRAF6, CAML or TACI.
Immunoblotting
Equal amounts of total or immunoprecipitated proteins were fractionated onto a 10% sodium dodecyl sulfate-polyacrylamide gel and transferred into nylon membranes (BioRad). After blocking, membranes were probed with primary mouse, rabbit or goat pAbs or mAbs to the following human proteins: TACI (11H3), GST tag (B-14), TRAF2 (sc-876), TRAF5 (sc-7220), TRAF6 (sc-7221), p50 (C-19), p65 (C-20), c-Rel (B-6), IκBα (C-21), Oct1 (12F11), actin (I-19) (Santa Cruz Biotechnologies), Ser32p-Ser36p-IκBα (5A5) (Cell Signaling Technology), CAML (67714), 6X His tag (4D11) (AbCam), MyD88 (16527), IRAK-4 (4E9) (Chemicon), FLAG tag (M2) (Sigma), and HA tag (5B1D10) (Invitrogen). Primary mouse, rabbit or goat pAbs or mAbs to the following mouse proteins were also used: MyD88 (AB16527) (Chemicon/Millipore), IRAK4 (sc-7221), TRAF2 (sc-875), TRAF5 (sc-6195), TRAF6 (sc-7221), CAML (sc-7335), IKKα/β (sc-7607), IκBα (sc-371), p38 (sc-535), actin (sc-61) (Santa Cruz Biotechnology), TACI (R&D Systems) (AF1041), phosphoserine 176/phosphoserine 18-IKKα/β, or phosphothreonin 180/phosphotyrosine 182-p38 (28B10) (Cell Signaling). Membranes were then washed and incubated with an appropriate secondary antibody as described previously14,27,52. Proteins were detected with an enhanced chemiluminescence detection system (Amersham). Signal intensity was quantified by using a Quant1 software (Bio-Rad).
EMSA and supershift assays
An oligonucleotide encompassing the κB2 site of the evolutionarily conserved sequence of the mouse Iγ1 promoter was labeled with [α-32P] ATP and used at approximately 30,000 c.p.m. in each electrophoretic mobility-shift assay. Reaction samples were prepared as previously published13 and separated in a 5% non-denaturing polyacrylamide gel. The composition of DNA-bound protein complexes was determined by incubating the reaction mixture with 1 μg pAb C-20 to p65, C-19 to p50 or B-6 to c-Rel (Santa Cruz Biotechnologies) before adding the radiolabeled probe.
ELISA
Human IgG, IgA, BAFF and APRIL were determined by ELISA, as previously described14,27. The concentration of mouse IgM and IgG1 was determined with an ELISA Quantitation Set as instructed by the manufacturer (Bethy Laboratories).
Statistical analysis
Statistical significance was assessed by a one-tailed unpaired or two-tailed paired Student’s t-test.
Supplementary Material
Acknowledgments
We thank S.Y. Zhang for peripheral blood mononuclear cells from UNC93B-deficient patients; S. Vogel and A. Medvedev (University of Maryland) for dominant negative IRAK-4-C877T; J.D. Li (University of South California) for dominant negative IKKα-K44M and IKKβ-K49A; J. Jun Ninomiya-Tsuji for dominant negative TAK1-K63W; Z. Cao (Tularik) for dominant negative IRAK-1-1-208, and the Mount Sinai School of Medicine Microscopy Shared Resource Facility. Supported by US National Institutes of Health R01 AI-05753 and R01 AI-074378 (A.Ce,), Catalan Institute for Research and Advanced Studies (A.Ce.), Fundacio’ IMIM (A.Ce.), Ministerio de Ciencia e Innovación SAF 2008-02725 (A. Ce.), The Irma T. Hirschl Charitable Trust Research Scholar Award (A Ce.), Comissionat per a Universitats i Recerca del Departament d’Innovació, Universitats i Empresa de la Generalitat de Catalunya (R.S.), Programa Juán de la Cierva (I.P.), Fondazione C. Golgi and Centro Immunodeficienze Mario di Martino (A.P.), TÁMOP 4.2.2-08/1-2008-0015 (L.M.), National Institutes of Health-National Cancer Institute shared resources grant 5R24 CA095823, National Institutes of Health shared instrumentation grant S10RR09145, and National Science Foundation Major Research Instrumentation grant DBI-9724504.
Footnotes
AUTHORS’ CONTRIBUTIONS
B.H., R.S., W.X., M.C., K.C., I.P., M.S., and E.M. designed and performed research; H.X. provided spleen from wt and MyD88-deficient mice; J.B.B., A.Ch., A.Pu., J.R., L.M., R.D., J.V., A.I., J.K., G.D., B.G., and C.P. provided blood and tissue samples; X.L. provided reagents, including MyD88-deficient 293 cells; A.P., C.C.R., and J.L.C. provided blood samples, tissue samples and clinical information and discussed data; and A. Ce. designed research, discussed data and wrote the paper.
COMPETING INTEREST STATEMENT
The authors declare that they have no competing financial interests.
References
- 1.Cooper MD, Alder MN. The evolution of adaptive immune systems. Cell. 2006;124:815–822. doi: 10.1016/j.cell.2006.02.001. [DOI] [PubMed] [Google Scholar]
- 2.Schlissel MS. Regulating antigen-receptor gene assembly. Nat Rev Immunol. 2003;3:890–899. doi: 10.1038/nri1225. [DOI] [PubMed] [Google Scholar]
- 3.Stavnezer J, Guikema JE, Schrader CE. Mechanism and regulation of class switch recombination. Annu Rev Immunol. 2008;26:261–292. doi: 10.1146/annurev.immunol.26.021607.090248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Muramatsu M, et al. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 2000;102:553–563. doi: 10.1016/s0092-8674(00)00078-7. [DOI] [PubMed] [Google Scholar]
- 5.Cerutti A. The regulation of IgA class switching. Nat Rev Immunol. 2008;8:421–434. doi: 10.1038/nri2322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.MacLennan IC. Germinal centers. Annu Rev Immunol. 1994;12:117–139. doi: 10.1146/annurev.iy.12.040194.001001. [DOI] [PubMed] [Google Scholar]
- 7.Banchereau JF, et al. The CD40 antigen and its ligand. Annu Rev Immunol. 1994;12:881–922. doi: 10.1146/annurev.iy.12.040194.004313. [DOI] [PubMed] [Google Scholar]
- 8.Siebenlist U, Brown K, Claudio E. Control of lymphocyte development by nuclear factor-κB. Nat Rev Immunol. 2005;5:435–445. doi: 10.1038/nri1629. [DOI] [PubMed] [Google Scholar]
- 9.McHeyzer-Williams MG, Ahmed R. B cell memory and the long-lived plasma cell. Curr Opin Immunol. 1999;11:172–179. doi: 10.1016/s0952-7915(99)80029-6. [DOI] [PubMed] [Google Scholar]
- 10.Fagarasan S, Honjo T. T-Independent immune response: new aspects of B cell biology. Science. 2000;290:89–92. doi: 10.1126/science.290.5489.89. [DOI] [PubMed] [Google Scholar]
- 11.Litinskiy MB, et al. DCs induce CD40-independent immunoglobulin class switching through BLyS and APRIL. Nat Immunol. 2002;3:822–829. doi: 10.1038/ni829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Balazs M, Martin F, Zhou T, Kearney J. Blood dendritic cells interact with splenic marginal zone B cells to initiate T-independent immune responses. Immunity. 2002;17:341–352. doi: 10.1016/s1074-7613(02)00389-8. [DOI] [PubMed] [Google Scholar]
- 13.Xu W, et al. Epithelial cells trigger frontline immunoglobulin class switching through a pathway regulated by the inhibitor SLPI. Nat Immunol. 2007;8:294–303. doi: 10.1038/ni1434. [DOI] [PubMed] [Google Scholar]
- 14.He B, et al. Intestinal bacteria trigger T cell-independent immunoglobulin A2 class switching by inducing epithelial-cell secretion of the cytokine APRIL. Immunity. 2007;26:812–826. doi: 10.1016/j.immuni.2007.04.014. [DOI] [PubMed] [Google Scholar]
- 15.Katsenelson N, et al. Synthetic CpG oligodeoxynucleotides augment BAFF- and APRIL-mediated immunoglobulin secretion. Eur J Immunol. 2007;37:1785–1795. doi: 10.1002/eji.200636800. [DOI] [PubMed] [Google Scholar]
- 16.Schneider P. The role of APRIL and BAFF in lymphocyte activation. Curr Opin Immunol. 2005;17:282–289. doi: 10.1016/j.coi.2005.04.005. [DOI] [PubMed] [Google Scholar]
- 17.von Bulow GU, van Deursen JM, Bram RJ. Regulation of the T-independent humoral response by TACI. Immunity. 2001;14:573–582. doi: 10.1016/s1074-7613(01)00130-3. [DOI] [PubMed] [Google Scholar]
- 18.Castigli E, et al. TACI and BAFF-R mediate isotype switching in B cells. J Exp Med. 2005;201:35–39. doi: 10.1084/jem.20032000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Castigli E, et al. TACI is mutant in common variable immunodeficiency and IgA deficiency. Nat Genet. 2005;37:829–834. doi: 10.1038/ng1601. [DOI] [PubMed] [Google Scholar]
- 20.Salzer U, et al. Mutations in TNFRSF13B encoding TACI are associated with common variable immunodeficiency in humans. Nat Genet. 2005;37:820–828. doi: 10.1038/ng1600. [DOI] [PubMed] [Google Scholar]
- 21.Zhang L, et al. Transmembrane activator and calcium-modulating cyclophilin ligand interactor mutations in common variable immunodeficiency: clinical and immunologic outcomes in heterozygotes. J Allergy Clin Immunol. 2007;120:1178–1185. doi: 10.1016/j.jaci.2007.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pan-Hammarstrom Q, et al. Reexamining the role of TACI coding variants in common variable immunodeficiency and selective IgA deficiency. Nat Genet. 2007;39:429–430. doi: 10.1038/ng0407-429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Groom JR, et al. BAFF and MyD88 signals promote a lupuslike disease independent of T cells. J Exp Med. 2007;204:1959–1971. doi: 10.1084/jem.20062567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Takeda K, Kaisho T, Akira S. Toll-like receptors. Annu Rev Immunol. 2003;21:335–376. doi: 10.1146/annurev.immunol.21.120601.141126. [DOI] [PubMed] [Google Scholar]
- 25.Cunningham-Rundles C. Autoimmune manifestations in common variable immunodeficiency. J Clin Immunol. 2008;28 (Suppl 1):S42–S45. doi: 10.1007/s10875-008-9182-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Garibyan L, et al. Dominant-negative effect of the heterozygous C104R TACI mutation in common variable immunodeficiency (CVID) J Clin Invest. 2007;117:1550–1557. doi: 10.1172/JCI31023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen K, et al. Immunoglobulin D enhances immune surveillance by activating antimicrobial, proinflammatory and B cell-stimulating programs in basophils. Nat Immunol. 2009;10:889–898. doi: 10.1038/ni.1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pasare C, Medzhitov R. Control of B-cell responses by Toll-like receptors. Nature. 2005;438:364–368. doi: 10.1038/nature04267. [DOI] [PubMed] [Google Scholar]
- 29.Tezuka H, et al. Regulation of IgA production by naturally occurring TNF/iNOS-producing dendritic cells. Nature. 2007;448:929–933. doi: 10.1038/nature06033. [DOI] [PubMed] [Google Scholar]
- 30.He B, Qiao X, Cerutti A. CpG DNA induces IgG class switch DNA recombination by activating human B cells through an innate pathway that requires TLR9 and cooperates with IL-10. J Immunol. 2004;173:4479–4491. doi: 10.4049/jimmunol.173.7.4479. [DOI] [PubMed] [Google Scholar]
- 31.He B, Raab-Traub N, Casali P, Cerutti A. EBV-encoded latent membrane protein 1 cooperates with BAFF/BLyS and APRIL to induce T cell-independent Ig heavy chain class switching. J Immunol. 2003;171:5215–5224. doi: 10.4049/jimmunol.171.10.5215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.von Bulow GU, Bram RJ. NF-AT activation induced by a CAML-interacting member of the tumor necrosis factor receptor superfamily. Science. 1997;278:138–141. doi: 10.1126/science.278.5335.138. [DOI] [PubMed] [Google Scholar]
- 33.Xu LG, Shu HB. TNFR-associated factor-3 is associated with BAFF-R and negatively regulates BAFF-R-mediated NF-kappa B activation and IL-10 production. J Immunol. 2002;169:6883–6889. doi: 10.4049/jimmunol.169.12.6883. [DOI] [PubMed] [Google Scholar]
- 34.Jabara H, et al. The binding site for TRAF2 and TRAF3 but not for TRAF6 Is essential for CD40-mediated immunoglobulin class switching. Immunity. 2002;17:265–276. doi: 10.1016/s1074-7613(02)00394-1. [DOI] [PubMed] [Google Scholar]
- 35.Horng T, Barton GM, Medzhitov R. TIRAP: an adapter molecule in the Toll signaling pathway. Nat Immunol. 2001;2:835–841. doi: 10.1038/ni0901-835. [DOI] [PubMed] [Google Scholar]
- 36.He B, et al. Lymphoma B cells evade apoptosis through the TNF family members BAFF/BLyS and APRIL. J Immunol. 2004;172:3268–3279. doi: 10.4049/jimmunol.172.5.3268. [DOI] [PubMed] [Google Scholar]
- 37.Chiu A, et al. Hodgkin lymphoma cells express TACI and BCMA receptors and generate survival and proliferation signals in response to BAFF and APRIL. Blood. 2007;109:729–739. doi: 10.1182/blood-2006-04-015958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Browman DT, Hoegg MB, Robbins SM. The SPFH domain-containing proteins: more than lipid raft markers. Trends Cell Biol. 2007;17:394–402. doi: 10.1016/j.tcb.2007.06.005. [DOI] [PubMed] [Google Scholar]
- 39.Li X, Commane M, Jiang Z, Stark GR. IL-1-induced NFkappa B and c-Jun N-terminal kinase (JNK) activation diverge at IL-1 receptor-associated kinase (IRAK) Proc Natl Acad Sci U S A. 2001;98:4461–4465. doi: 10.1073/pnas.071054198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.von Bernuth H, et al. Pyogenic bacterial infections in humans with MyD88 deficiency. Science. 2008;321:691–696. doi: 10.1126/science.1158298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ku CL, et al. Selective predisposition to bacterial infections in IRAK-4-deficient children: IRAK-4-dependent TLRs are otherwise redundant in protective immunity. J Exp Med. 2007;204:2407–2422. doi: 10.1084/jem.20070628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Colonna M. All roads lead to CARD9. Nat Immunol. 2007;8:554–555. doi: 10.1038/ni0607-554. [DOI] [PubMed] [Google Scholar]
- 43.Casola S, et al. B cell receptor signal strength determines B cell fate. Nat Immunol. 2004;5:317–327. doi: 10.1038/ni1036. [DOI] [PubMed] [Google Scholar]
- 44.Sun D, Ding A. MyD88-mediated stabilization of interferon-gamma-induced cytokine and chemokine mRNA. Nat Immunol. 2006;7:375–381. doi: 10.1038/ni1308. [DOI] [PubMed] [Google Scholar]
- 45.Xia XZ, et al. TACI is a TRAF-interacting receptor for TALL-1, a tumor necrosis factor family member involved in B cell regulation. J Exp Med. 2000;192:137–143. doi: 10.1084/jem.192.1.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Castigli E, et al. Transmembrane activator and calcium modulator and cyclophilin ligand interactor enhances CD40-driven plasma cell differentiation. J Allergy Clin Immunol. 2007;120:885–891. doi: 10.1016/j.jaci.2007.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bossen C, et al. TACI, unlike BAFF-R, is solely activated by oligomeric BAFF and APRIL to support survival of activated B cells and plasmablasts. Blood. 2008;111:1004–1012. doi: 10.1182/blood-2007-09-110874. [DOI] [PubMed] [Google Scholar]
- 48.Picard C, et al. Pyogenic bacterial infections in humans with IRAK-4 deficiency. Science. 2003;299:2076–2079. doi: 10.1126/science.1081902. [DOI] [PubMed] [Google Scholar]
- 49.Gavin AL, et al. Adjuvant-enhanced antibody responses in the absence of toll-like receptor signaling. Science. 2006;314:1936–1938. doi: 10.1126/science.1135299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Delgado MF, et al. Lack of antibody affinity maturation due to poor Toll-like receptor stimulation leads to enhanced respiratory syncytial virus disease. Nat Med. 2009;15:34–41. doi: 10.1038/nm.1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cerutti A, et al. CD40 ligand and appropriate cytokines induce switching to IgG, IgA, and IgE and coordinated germinal center-like phenotype differentiation in a human monoclonal IgM+IgD+ B cell line. J Immunol. 1998;160:2145–2157. [PMC free article] [PubMed] [Google Scholar]
- 52.Xu W, et al. Viral double-stranded RNA triggers Ig class switching by activating upper respiratory mucosa B cells through an innate TLR3 pathway involving BAFF. J Immunol. 2008;181:276–287. doi: 10.4049/jimmunol.181.1.276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shuto T, et al. Activation of NF-kappa B by nontypeable Hemophilus influenzae is mediated by toll-like receptor 2-TAK1-dependent NIK-IKK alpha /beta-I kappa B alpha and MKK3/6-p38 MAP kinase signaling pathways in epithelial cells. Proc Natl Acad Sci USA. 2001;98:8774–8779. doi: 10.1073/pnas.151236098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Medvedev AE, et al. Distinct mutations in IRAK-4 confer hyporesponsiveness to lipopolysaccharide and interleukin-1 in a patient with recurrent bacterial infections. J Exp Med. 2003;198:521–531. doi: 10.1084/jem.20030701. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.