Abstract
Self-renewal and differentiation are cardinal features of stem cells. Asymmetric cell division provides one fundamental mechanism by which stem cell self-renewal and differentiation are balanced1,2. A failure of this balance could lead to diseases such as cancer3–6. During asymmetric division of stem cells, factors controlling their self-renewal and differentiation are unequally segregated between daughter cells. Numb is one such factor that is segregated to the differentiating daughter cell during the stem-cell-like neuroblast divisions in Drosophila melanogaster7, where it inhibits self-renewal8,9. The localization and function of Numb is cell-cycle-dependent7,10–12. Here we show that Polo (ref. 13), a key cell cycle regulator, the mammalian counterparts of which have been implicated as oncogenes as well as tumour suppressors14,15, acts as a tumour suppressor in the larval brain. Supernumerary neuroblasts are produced at the expense of neurons in polo mutants. Polo directly phosphorylates Partner of Numb (Pon, ref. 16), an adaptor protein for Numb, and this phosphorylation event is important for Pon to localize Numb. In polo mutants, the asymmetric localization of Pon, Numb and atypical protein kinase C are disrupted, whereas other polarity markers are largely unaffected. Overexpression of Numb suppresses neuroblast over-proliferation caused by polo mutations, suggesting that Numb has a major role in mediating this effect of Polo. Our results reveal a biochemical link between the cell cycle and the asymmetric protein localization machinery, and indicate that Polo can inhibit progenitor self-renewal by regulating the localization and function of Numb.
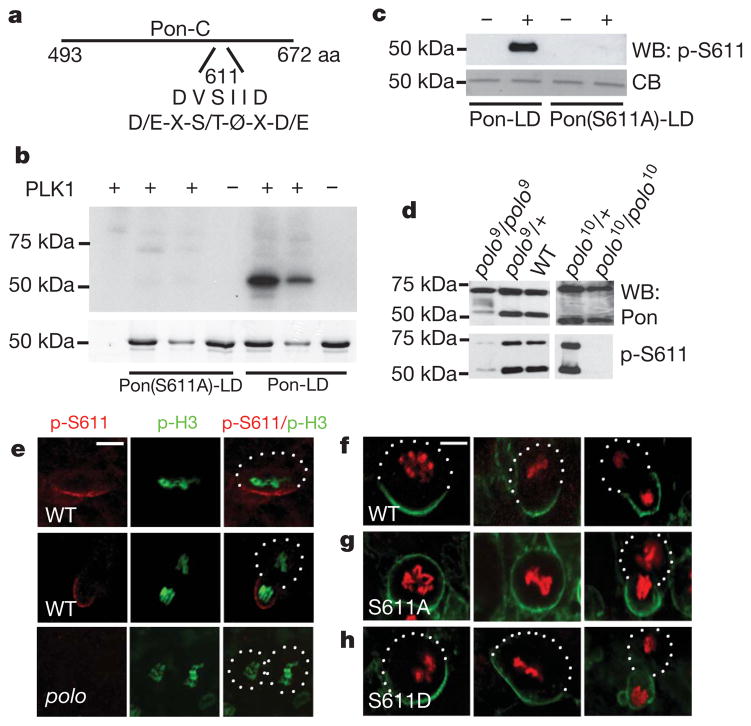
Asymmetric localization of Numb depends on its adaptor protein Pon. The Pon localization domain (Pon-LD) is located at the carboxy terminus of the protein17. The Ser 611 (S611) residue in this domain matches the consensus phosphorylation site for Polo (Fig. 1a), a principal orchestrator of cell cycle events. Because the localization of Pon is cell-cycle-dependent, we tested whether Polo can directly phosphorylate Pon. Pon-LD, but not Pon(S611A)-LD, in which S611 was mutated to Ala, was readily phosphorylated by mammalian Polo-like kinase 1 (Plk1) in vitro (Fig. 1b), demonstrating that Pon S611 is a Polo phosphorylation site.
Figure 1. Polo phosphorylates Pon and this phosphorylation event regulates the asymmetric localization of Pon.
a, A diagram of the putative Polo phosphorylation site (S611) in Pon-LD, which matches the consensus Polo phosphorylation site (D/E-X-S/T-Ø-X-D/E, where Ø is a hydrophobic amino acid and X is any amino acid). b, In vitro kinase assays. Upper panel, autoradiography; lower panel, Coomassie blue (CB) staining of the same gel. c, Western blot (WB) analysis testing the specificity of the p-S611 antibody. d, Western blot analysis showing loss of p-S611-positive Pon in polo mutants. Note that, in addition to the 72 kDa full-length Pon protein, both the general Pon antibody and the p-S611 antibody recognized a ~50 kDa band—a putative proteolytic product. The level of the 50 kDa band seems to be dependent on Polo activity. e, Staining of endogenous Pon in wild-type (WT, upper and middle panels) and polo mutant (lower panels) cells using the p-S611 antibody. p-H3, p-histone H3. f–h, Localization of GFP–Pon-LD (WT) (f), GFP–Pon(S611A)-LD (g) or GFP–Pon(S611D)-LD (h) in embryos at stages 9–12. Neuroblasts at different cell cycle stages are shown. GFP–Pon-LD proteins were detected by immunostaining with anti-myc antibody (green). Anti-p-histone H3 staining (red) was used to indicate mitotic stages. Scale bars, 5 μm.
To test whether Pon S611 is normally phosphorylated in vivo, we generated an antibody against S611-phosphorylated (p-S611) Pon. The specificity of this antibody was shown by its ability to recognize a glutathione S-transferase–Pon-LD fusion protein (GST-Pon-LD) only after the fusion protein was pre-phosphorylated by Plk1 (Fig. 1c). It did not recognize GST–Pon(S611A)-LD in the same assay. Next, larval brain extracts prepared from wild type as well as heterozygotes (polo9(+/−) and polo10(+/−)), and homozygotes (polo9(−/−) and polo10(−/−)) of two different polo loss-of-function alleles were analysed by western blotting using this p-S611-specific antibody. p-S611-positive Pon was clearly detected in wild-type animals and in heterozygotes, but was barely detectable in homozygous mutant animals (Fig. 1d), demonstrating that Pon is phosphorylated at S611 in vivo in a Polo-dependent fashion.
We also used immunohistochemistry to verify S611 phosphorylation in vivo and to visualize phospho-Pon localization. p-S611-positive endogenous Pon was detected in wild-type larval brains as a crescent in metaphase neuroblasts, and was segregated to the ganglion mother cell (GMC, the daughter committed to the differentiation pathway) after division (Fig. 1e, upper and middle panels; n = 58, 100%). In the polo9 mutant, however, p-S611-positive Pon was undetectable (Fig. 1e, lower panels; n = 43, 100%). The p-S611 antibody also reacted with Pon-LD, but not with Pon(S611A)-LD, in transgenic larval brain (Supplementary Fig. 1).
To test for a functional role of S611 phosphorylation, we mutated S611 to a non-phosphorylatable Ala residue (S611A) or to a phospho-mimetic Asp residue (S611D). Wild-type and phospho-mutant Pon-LD were fused to green fluorescent protein (GFP) and expressed in embryonic neuroblasts. Both GFP–Pon-LD (Fig. 1f; n = 35, 100%) and GFP–Pon(S611D)-LD (Fig. 1h; n = 35, 100%) showed the expected basal localization. In contrast, the localization of GFP–Pon(S611A)-LD was defective. At prometaphase and metaphase, it showed either uniformly cortical (80%) or basally enriched but apically detectable cortical (20%) distribution (Fig. 1g, left and middle panels; n = 56). At anaphase and telophase, however, it formed basal crescents in most neuroblasts (Fig. 1g, right panel; n = 42, 93%). This ‘telophase rescue’ seemed to be partially mediated by endogenous Pon, because less rescue was observed in pon mutant neuroblasts, with 58% (n = 17) neuroblasts mis-segregating GFP–Pon(S611A)-LD at late anaphase/telophase (Supplementary Fig. 2). It is unlikely that the S611A mutation affects Pon localization by altering the charge or global conformation and folding of the protein, because mutation of an adjacent Ser residue (S616) or triple mutations at three potential atypical protein kinase C (aPKC) phosphorylation sites (S644A/S648A/S652A) had no effect on Pon-LD localization (Supplementary Fig. 3), suggesting that S611 represents a unique regulatory point in Pon localization.
To assess whether Polo has a role in neuroblast self-renewal and/or asymmetric division, we quantified central brain neuroblast numbers in two strong hypomorphic alleles, polo9 and polo10, using Deadpan (Dpn) and Miranda (Mira) as neuroblast markers. Wild-type larval central brains contained 37.3 ± 5.5 neuroblasts 24 h after larval hatching (ALH) and 90.7 ± 7.0 neuroblasts 96 h ALH (Fig. 2a). polo9 larval central brains had 36.5 ± 4.5 neuroblasts 24 h ALH. However, the number increased dramatically to 253.6 ± 44.5 neuroblasts 96 h ALH (Fig. 2a). Consistent with this increase in neuroblast number (Fig. 2a, and lower panels of b–d), the numbers of BrdU-labelled (lower panel of Fig. 2g), CycE-positive (lower panel of Fig. 2h) or phospho-histone-H3-positive proliferating cells (lower panel of Fig. 2i) were also increased in polo9 mutant brains compared to wild type (upper panels of Fig. 2g–i). Concomitantly, a dramatic reduction of differentiated cells expressing neuronal markers, Embryonic Lethal Abnormal Vision (Elav) (lower panel of Fig. 2e) or nuclear Prospero (Pros) (lower panel of Fig. 2f), was observed in polo9 mutant brains. A similar neuroblast overproliferation phenotype was observed in polo10 and in trans-heterozygotes between polo9 and a deficiency that deletes polo (data not shown). A Polo–GFP genomic construct fully rescued this polo mutant phenotype (Supplementary Fig. 5c), verifying that these defects are caused by polo loss-of-function. Excess self-renewal and proliferation at the expense of neuronal differentiation was also observed in MARCM (mosaic analysis with a repressible cell marker) clones derived from single polo9 mutant neuroblasts (lower panels of Fig. 2j, k, and data not shown). These results indicate that Polo behaves like a tumour suppressor to inhibit neuroblast self-renewal and to promote differentiation. polo mutant GMCs may revert to neuroblast-like cells, as has been shown for brat (brain tumor) mutants5.
Figure 2. Polo functions as a tumour suppressor to inhibit neuroblast self-renewal and to promote neuronal differentiation.
a, Quantification of central brain neuroblast numbers in wild-type and polo9 mutants from 24- to 96-h ALH. n = 20 per time point per genotype. b–i, Confocal single-scanning images of wild-type (b–i, upper panels) and the polo9 mutant (b–i, lower panels) larval brains at 96 h ALH that are stained with neuroblast markers Dpn (b), Insc (c) or Mira (d), neuronal markers Elav (e) or Pros (f), or cell proliferation markers BrdU (g), CycE (h) or phospho-histone H3 (i). j, k, polo9 MARCM clones showed the neuroblast overgrowth phenotype. Single neuroblast clones marked by CD8–GFP (and demarcated by stippled line) were generated for the wild-type (j, k, upper panels) and the polo9 mutant (j, k, lower panels) in larval brains. Wild-type clones contained one neuroblast that was marked by Dpn (j, upper panel; 1.0 ± 0 neuroblast per clone, n = 9), whereas polo9 mutant clones contained multiple neuroblast-like cells (j, lower panel; 10.6 ± 3.4 (s.d.) neuroblasts per clone, n = 7).
We then analysed the physiological role of Polo in regulating Pon localization and function. Most larval neuroblasts were found at metaphase in polo9 mutant brains, and both Pon (Fig. 3a; n = 37, 79%) and Numb (Fig. 3c; n = 32, 87%) were uniformly distributed. In late anaphase/telophase neuroblasts, Pon (Fig. 3b; n = 12, 58.3%) and Numb (Fig. 3d; n = 14, 57.1%) were mis-segregated to both daughter cells. Defective Pon and Numb localization in the polo mutant is unlikely to be a secondary consequence of cell cycle arrest, because arrest of wild-type neuroblasts at metaphase with microtubule-depolymerizing drugs actually increased the number of cells possessing a Numb crescent11.
Figure 3. Polo kinase is required for the asymmetric localization of Pon and Numb and for correct spindle orientation.
Neuroblasts from wild type (upper panels) and the polo9 mutant (lower panels) were immunostained for Pon (a, b), Numb (c, d), Brat (e), Insc (f) and aPKC (g, h). The neuroblasts shown in b, d and h were at telophase. i–l, abnormal spindle orientation phenotype in the polo mutant. Wild-type (i) and polo mutant (k) neuroblasts were triple-labelled for Insc, α-tubulin (α-Tub) and DNA. Spindle orientation in wild-type (n = 22) and polo9 mutant (n = 53) neuroblasts was quantified in j and l, respectively.
To test whether Polo is specifically required for Pon/Numb localization, we analysed other apical and basal proteins. In polo9 mutant neuroblasts, the basal localization of Brat (Fig. 3e; n = 54, 79.6%; Supplementary Fig. 4d) and Pros (Supplementary Fig. 4a) was relatively normal. Moreover, double-labelling of the same mutant neuroblast showed that the localization of Mira, the adaptor protein for Pros and Brat, was normal, whereas Pon localization was abnormal (Supplementary Fig. 4b). Introduction of a Polo–GFP transgene into polo9 effectively rescued the Pon localization and cell-cycle defects (Supplementary Fig. 5a). Apical proteins such as Insc, Baz, Pins and Discs Large 1 (Dlg1) were localized normally in polo9 mutant neuroblasts (Fig. 3f and Supplementary Fig. 4a, c). The only apical protein we found showing abnormal localization was aPKC (Fig. 3g, h, and Supplementary Fig. 4e), which became distributed uniformly on the cortex and showed cytoplasmic localization (Fig. 3g; n = 38, 60.5%). During telophase, aPKC could be mis-segregated into both daughter cells (Fig. 3h; n = 18, 44.4%).
Polo is localized to centrosomes and is required for centrosome organization and separation. We tested whether Polo has a role in orienting neuroblast mitotic spindles. The tight coupling of spindle orientation with crescent formation seen in wild-type neuroblasts (Fig. 3i, j; n = 24, 100%) was disrupted in polo9 metaphase neuroblasts with two centrosomes (Fig. 3k, l; n = 55, 56%). Therefore, Polo kinase is also required for coupling mitotic spindle orientation with cortical protein asymmetry. This spindle phenotype was fully rescued by the Polo–GFP transgene (Supplementary Fig. 5b).
We next probed the role of Pon phosphorylation in mediating Numb localization. Full-length Pon containing the S611A or S611D mutation was used to assess the effects of S611 phosphorylation. In pon mutant neuroblasts, Numb localization was defective (Fig. 4a), as reported previously16. Introducing wild-type Pon restored Numb asymmetric localization at metaphase and later stages (Fig. 4b; n = 27, 100%). Most pon mutant neuroblasts expressing Pon(S611D) also showed rescue (Fig. 4c; n = 12, 73%). In contrast, pon mutant neuroblasts expressing Pon(S611A) showed largely abnormal Numb localization (Fig. 4d; n = 19, 86%). Polo-mediated phosphorylation is therefore important for Pon to localize Numb. We also analysed the function of Pon in neuroblast self-renewal by generating pon MARCM clones. Interestingly, compared to wild-type clones (Fig. 4e, f), ectopic neuroblast self-renewal was observed in pon mutant clones (Fig. 4g, h).
Figure 4. Polo controls Numb asymmetry through phosphorylation of Pon, and neuroblast self-renewal through regulation of Numb.
a–d, Phosphorylation of Pon S611 mediates the asymmetric localization of Numb. Numb exhibits uniform cortical localization in pon mutant metaphase neuroblasts (a). The Numb localization defect in pon mutant neuroblasts was rescued by wild-type Pon (b) and Pon(S611D) (c), but not by Pon(S611A) (d). e–h, In wild-type (e, f) and pon (g, h) MARCM clones marked by CD8–GFP and demarcated by stippled lines, multiple neuroblasts (marked by Dpn) were observed in clones derived from single pon mutant neuroblasts (g), whereas only one neuroblast was observed in wild-type clones (e). i–k, Overexpression of Numb, using wor-Gal4 to drive expression of UAS-Numb–GFP (wor-Numb–GFP), suppressed the supernumerary neuroblast phenotype in the polo9 mutant. Neuroblasts were marked by Dpn (green) in wor-Gal4; polo9 (i) and wor-Numb–GFP; polo9 (j) brains. Results from overexpressing wor-Numb are shown in Supplementary Fig. 7. k, Quantitative analysis of central brain neuroblast numbers for wor-Gal4; polo9, wor-Numb–GFP; polo9 and wor-Numb–GFP 96 h ALH at 25 °C (n = 20). l, Loss of function of Notch suppresses neuroblast overgrowth phenotype of polo mutant. Quantification of central brain neuroblast numbers for polo9, Notchts1; polo9 (Nts1; polo9) and Notchts1 (Nts1) was performed 72 h ALH at 29 °C (n = 20). k, l, Error bars are s.d.
We have shown that polo mutations affect Numb and aPKC localization as well as spindle orientation—processes known to affect neuroblast self-renewal to various degrees8,9,18–20. Strikingly, overexpression of either Numb–GFP or Numb effectively suppressed the ectopic neuroblast self-renewal phenotype seen in the polo9 mutant (Fig. 4i–k and data not shown). This effect was not caused by increased neuroblast apoptosis (Supplementary Fig. 6), and overexpression of Numb–GFP or Numb in a wild-type background did not affect the neuroblast number (Fig. 4k, Supplementary Fig. 6 and data not shown). These results indicate that Numb is a principal player downstream of Polo in regulating neuroblast self-renewal. Numb overexpression did not rescue the aPKC mislocalization and spindle misorientation phenotypes of polo mutants (Supplementary Fig. 7). These defects could also contribute to the neuroblast overgrowth phenotype of polo mutants, but their effects might have been masked by Numb overexpression. Consistent with this, introduction of Pon(S611D) into a polo mutant neuroblast did not significantly rescue the neuroblast overgrowth phenotype, despite the partial restoration of Numb localization (Supplementary Fig. 8). Because aPKC localization and spindle orientation defects were not rescued by Pon(S611D) (Supplementary Fig. 8), these defects may account for the inability of Pon(S611D) to rescue the overgrowth phenotype of polo. aPKC has been shown to phosphorylate Numb (ref. 21). Delocalized aPKC at the basal side may be sufficient to inactivate endogenous Numb, but not overexpressed Numb, owing to titration by the overexpressed protein.
Numb was previously shown to inhibit neuroblast self-renewal by antagonizing Notch signalling9. Reducing Notch significantly suppressed the neuroblast overgrowth phenotype of the polo9 mutant (Fig. 4l and Supplementary Fig. 9). Reducing Notch in a wild-type background also led to a partial loss of neuroblasts (Fig. 4l and Supplementary Fig. 9), consistent with Notch being required for progenitor self-renewal. We envision that in polo or pon mutants, owing to the symmetric distribution of Numb, the GMCs receive insufficient Numb to inhibit Notch, thereby causing them to adopt a neuroblast-like fate. To test further the importance of Numb asymmetric localization in neuroblast self-renewal versus differentiation, we analysed the numbS52F mutation, which apparently affects Numb asymmetric localization but not its stability or activity22. In numbS52F neuroblast clones, ectopic neuroblast self-renewal similar to that seen in polo or pon clones was observed (Supplementary Fig. 10). Thus, loss of Numb asymmetric localization is sufficient to cause neuroblast overgrowth.
Our results indicate that Polo controls the self-renewal versus differentiation decision of neural progenitors by regulating the localization and activity of Numb and the orientation of mitotic spindles. Polo regulates the localization of Numb by means of Pon. Although immunofluorescence shows that Polo is primarily localized to the centrosomes, a cytosolic pool of Polo probably exists because Polo exhibits dynamic in vivo association with the mitotic apparatus and many non-centrosomal Polo substrates have been identified15,23. Cytosolic localization of the centrosomal kinase Aurora-A has been demonstrated24. How Polo regulates the localization of aPKC, the activity of Numb and the orientation of mitotic spindles awaits further investigation. In addition to the Numb/Notch pathway, other factors such as Pros and Brat are necessary for preventing GMCs from undergoing self-renewing divisions4–6,25. Because these factors are segregated normally in polo neuroblasts, it seems that they are not sufficient to prevent progenitor self-renewal or that activation of Notch is able to override their effects. Intriguingly, some Plks behave as tumour suppressors in mammals, and loss of Numb has also been implicated in the hyperactivation of Notch signalling in breast cancer cells15,26,27. Our results and future studies in models like Drosophila will provide mechanistic insights into these observed tumour-suppressor roles of Polo and Numb.
METHODS SUMMARY
Fly genetics
Fly culture and crosses were performed according to standard procedures and were raised at indicated temperatures. To generate Pon(S611A) and Pon(S611D) transgenic flies, the corresponding complementary DNA constructs generated in pUAST vector were injected into w− embryos to obtain transgenic lines according to established procedures. All other fly stocks and Gal4 lines were obtained from the Bloomington Drosophila stock centre or as described17,23,28,29.
Antibody production and immunohistochemistry
A synthetic peptide with the following sequence CLADVpSIIDTSE was conjugated to the keyhole limpet haemocyanin carrier protein and used to elicit anti-p-S611 antibody production at Open Biosystems. Immunohistochemistry on embryos or larval brain tissues was performed as described16. The primary antibodies used were: anti-myc (1:200), anti-GFP (1:2,000), anti-p-S611 (1:20), anti-Pon (1:1,000), anti-Elav (1:20), anti-CycE (1:10), anti-Dpn (1:1,000), anti-Mira (1:1,000), anti-Numb (1:1,000), anti-Baz (1:500), anti-Insc (1:500), anti-Pins (1:500), anti-Dlg (1:200), anti-aPKC (1:500), anti-Brat (1:100), anti-Pros (1:20) and anti-p-histone (1:500).
In vitro phosphorylation assays
The conditions of the in vitro kinase assays were essentially as described30.
Clonal analysis
To generate single neuroblast clones in MARCM analysis, 24 h ALH larvae were heat-shocked at 37 °C for 90 min and further aged for 3 days at 25 °C.
BrdU labelling
Dissected larval tissue was given a 40-min pulse of 37.5 μg ml−1 BrdU in Shields and Sang 3 M insect medium, and fixed for 15 min in 3.7% formaldehyde. DNA was denatured with 2 M HCl for 40 min, and tissue was washed with PBS and incubated with anti-BrdU.
Supplementary Material
Acknowledgments
We thank D. Glover, F. Matsuzaki, Y. N. Jan, A. Wodarz, C. Doe, J. Knoblich, L. Luo, T. Orr-Weaver, C. Sunkel, B. Edgar, H. Richardson, F. Schweisguth, J. B. Skeath, A. Gould, E. D. Schejter, the Developmental Studies Hybridoma Bank and the Bloomington Stock Center for fly stocks and antibodies. We thank S. Guo for reading the manuscript, and F. Yu and members of the Lu and Chia laboratories for discussion and help. H.W. thanks U. Heberlein for support and S. G. S. Ling for technical help. This was supported by a NIH grant (B.L.) and Temasek Lifesciences Laboratory funding (W.C.)
Footnotes
Full Methods and any associated references are available in the online version of the paper at www.nature.com/nature.
Author Information Reprints and permissions information is available at www.nature.com/reprints. The authors declare no competing financial interests.
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
References
- 1.Morrison SJ, Kimble J. Asymmetric and symmetric stem-cell divisions in development and cancer. Nature. 2006;441:1068–1074. doi: 10.1038/nature04956. [DOI] [PubMed] [Google Scholar]
- 2.Jan YN, Jan LY. Asymmetric cell division in the Drosophila nervous system. Nature Rev Neurosci. 2001;2:772–779. doi: 10.1038/35097516. [DOI] [PubMed] [Google Scholar]
- 3.Caussinus E, Gonzalez C. Induction of tumor growth by altered stem-cell asymmetric division in Drosophila melanogaster. Nature Genet. 2005;37:1125–1129. doi: 10.1038/ng1632. [DOI] [PubMed] [Google Scholar]
- 4.Betschinger J, Mechtler K, Knoblich JA. Asymmetric segregation of the tumor suppressor Brat regulates self-renewal in Drosophila neural stem cells. Cell. 2006;124:1241–1253. doi: 10.1016/j.cell.2006.01.038. [DOI] [PubMed] [Google Scholar]
- 5.Lee CY, Wilkinson BD, Siegrist SE, Wharton RP, Doe CQ. Brat is a Miranda cargo protein that promotes neuronal differentiation and inhibits neuroblast self-renewal. Dev Cell. 2006;10:441–449. doi: 10.1016/j.devcel.2006.01.017. [DOI] [PubMed] [Google Scholar]
- 6.Bello B, Reichert H, Hirth F. The brain tumor gene negatively regulates neural progenitor cell proliferation in the larval central brain of Drosophila. Development. 2006;133:2639–2648. doi: 10.1242/dev.02429. [DOI] [PubMed] [Google Scholar]
- 7.Rhyu MS, Jan LY, Jan YN. Asymmetric distribution of numb protein during division of the sensory organ precursor cell confers distinct fates to daughter cells. Cell. 1994;76:477–491. doi: 10.1016/0092-8674(94)90112-0. [DOI] [PubMed] [Google Scholar]
- 8.Lee CY, et al. Drosophila Aurora-A kinase inhibits neuroblast self-renewal by regulating aPKC/Numb cortical polarity and spindle orientation. Genes Dev. 2006;20:3464–3474. doi: 10.1101/gad.1489406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang H, et al. Aurora-A acts as a tumor suppressor and regulates self-renewal of Drosophila neuroblasts. Genes Dev. 2006;20:3453–3463. doi: 10.1101/gad.1487506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tio M, Udolph G, Yang X, Chia W. cdc2 links the Drosophila cell cycle and asymmetric division machineries. Nature. 2001;409:1063–1067. doi: 10.1038/35059124. [DOI] [PubMed] [Google Scholar]
- 11.Knoblich JA, Jan LY, Jan YN. Asymmetric segregation of Numb and Prospero during cell division. Nature. 1995;377:624–627. doi: 10.1038/377624a0. [DOI] [PubMed] [Google Scholar]
- 12.Zhou Y, et al. The mammalian Golgi regulates numb signaling in asymmetric cell division by releasing ACBD3 during mitosis. Cell. 2007;129:163–178. doi: 10.1016/j.cell.2007.02.037. [DOI] [PubMed] [Google Scholar]
- 13.Sunkel CE, Glover D. M polo, a mitotic mutant of Drosophila displaying abnormal spindle poles. J Cell Sci. 1988;89:25–38. doi: 10.1242/jcs.89.1.25. [DOI] [PubMed] [Google Scholar]
- 14.Barr FA, Sillje HH, Nigg EA. Polo-like kinases and the orchestration of cell division. Nature Rev Mol Cell Biol. 2004;5:429–440. doi: 10.1038/nrm1401. [DOI] [PubMed] [Google Scholar]
- 15.van de Weerdt BC, Medema RH. Polo-like kinases: a team in control of the division. Cell Cycle. 2006;5:853–864. doi: 10.4161/cc.5.8.2692. [DOI] [PubMed] [Google Scholar]
- 16.Lu B, Rothenberg M, Jan LY, Jan YN. Partner of Numb colocalizes with Numb during mitosis and directs Numb asymmetric localization in Drosophila neural and muscle progenitors. Cell. 1998;95:225–235. doi: 10.1016/s0092-8674(00)81753-5. [DOI] [PubMed] [Google Scholar]
- 17.Lu B, Ackerman L, Jan LY, Jan YN. Modes of protein movement that lead to the asymmetric localization of partner of Numb during Drosophila neuroblast division. Mol Cell. 1999;4:883–891. doi: 10.1016/s1097-2765(00)80218-x. [DOI] [PubMed] [Google Scholar]
- 18.Lee CY, Robinson KJ, Doe CQ. Lgl, Pins and aPKC regulate neuroblast self-renewal versus differentiation. Nature. 2006;439:594–598. doi: 10.1038/nature04299. [DOI] [PubMed] [Google Scholar]
- 19.Izumi Y, Ohta N, Hisata K, Raabe T, Matsuzaki F. Drosophila Pins-binding protein Mud regulates spindle-polarity coupling and centrosome organization. Nature Cell Biol. 2006;8:586–593. doi: 10.1038/ncb1409. [DOI] [PubMed] [Google Scholar]
- 20.Siller KH, Cabernard C, Doe CQ. The NuMA-related Mud protein binds Pins and regulates spindle orientation in Drosophila neuroblasts. Nature Cell Biol. 2006;8:594–600. doi: 10.1038/ncb1412. [DOI] [PubMed] [Google Scholar]
- 21.Smith CA, et al. aPKC-mediated phosphorylation regulates asymmetric membrane localization of the cell fate determinant Numb. EMBO J. 2007;26:468–480. doi: 10.1038/sj.emboj.7601495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bhalerao S, Berdnik D, Torok T, Knoblich JA. Localization-dependent and -independent roles of numb contribute to cell-fate specification in Drosophila. Curr Biol. 2005;15:1583–1590. doi: 10.1016/j.cub.2005.07.061. [DOI] [PubMed] [Google Scholar]
- 23.Moutinho-Santos T, Sampaio P, Amorim I, Costa M, Sunkel CE. In vivo localisation of the mitotic POLO kinase shows a highly dynamic association with the mitotic apparatus during early embryogenesis in Drosophila. Biol Cell. 1999;91:585–596. [PubMed] [Google Scholar]
- 24.Berdnik D, Knoblich J. A Drosophila Aurora-A is required for centrosome maturation and actin-dependent asymmetric protein localization during mitosis. Curr Biol. 2002;12:640–647. doi: 10.1016/s0960-9822(02)00766-2. [DOI] [PubMed] [Google Scholar]
- 25.Choksi SP, et al. Prospero acts as a binary switch between self-renewal and differentiation in Drosophila neural stem cells. Dev Cell. 2006;11:775–789. doi: 10.1016/j.devcel.2006.09.015. [DOI] [PubMed] [Google Scholar]
- 26.Pece S, et al. Loss of negative regulation by Numb over Notch is relevant to human breast carcinogenesis. J Cell Biol. 2004;167:215–221. doi: 10.1083/jcb.200406140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stylianou S, Clarke RB, Brennan K. Aberrant activation of Notch signaling in human breast cancer. Cancer Res. 2006;66:1517–1525. doi: 10.1158/0008-5472.CAN-05-3054. [DOI] [PubMed] [Google Scholar]
- 28.Clarke AS, Tang TT, Ooi DL, Orr-Weaver TL. POLO kinase regulates the Drosophila centromere cohesion protein MEI-S332. Dev Cell. 2005;8:53–64. doi: 10.1016/j.devcel.2004.12.003. [DOI] [PubMed] [Google Scholar]
- 29.Ohshiro T, Yagami T, Zhang C, Matsuzaki F. Role of cortical tumour-suppressor proteins in asymmetric division of Drosophila neuroblast. Nature. 2000;408:593–596. doi: 10.1038/35046087. [DOI] [PubMed] [Google Scholar]
- 30.Nishimura I, Yang Y, Lu B. PAR-1 kinase plays an initiator role in a temporally ordered phosphorylation process that confers tau toxicity in Drosophila. Cell. 2004;116:671–682. doi: 10.1016/s0092-8674(04)00170-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.