Abstract
The rodent-borne Arenaviruses are divided into two major antigenic groups: the Old World and New World complexes. Of the 15 known New World arenaviruses, four (Junin, Machupo, Sabia, and Guanarito) have been associated with hemorrhagic fever in humans. It has been difficult to assess the pathogenic or epidemic potential of the remaining viruses and the threat of emerging disease. We obtained full-length small (S) segment sequence data, encoding the nucleoprotein (NP) and glycoprotein precursor (GPC), from all American arenaviruses to predict their evolutionary and functional relationships. Phylogenetic analysis of NP or GPC amino acid sequences from all New World arenaviruses revealed three lineages and that Tamiami and Whitewater Arroyo viruses were probably derived from a single recombinant progenitor. The results imply that arenaviruses have been evolving independently for a very long time, leading to very diverse groupings that do not correlate with geography, rodent host, or human epidemic potential.
INTRODUCTION
All recognized members of the Arenaviridae family were first divided into two complexes based primarily on serologic cross-reactivity and geographic distribution (Coimbra et al., 1994). The Old World complex contains Lymphocytic choriomeningitis virus (LCMV), the only known arenavirus with worldwide distribution, as well as the four African viruses, Lassa (LASV), Mobala (MOBV), Mopeia (MOPV), and Ippy (IPPV) in the LCMV, or Old World, complex. The New World, or Tacaribe, complex is composed of 15 viruses from North and South America: Allpahuayo (ALLV), Amapari (AMAV), Flexal (FLEV), Guanarito (GUAV), Junin (JUNV), Latino (LATV), Machupo (MACV), Oliveros (OLIV), Parana (PARV), Pichinde (PICV), Pirital (PIRV), Sabia (SABV), Tacaribe (TACV), Tamiami (TAMV), and Whitewater Arroyo (WWAV). Some of these viruses have the potential to cause hemorrhagic fevers with high mortalities in humans; all are transmitted by distinct species of rodents.
Arenaviruses have a host-derived, lipid envelope with a single-stranded, bisegmented, ambisense-coding RNA genome. The two segments, named large (L) and small (S), are an average 7100 and 3400 nucleotides in length, respectively. The L RNA segment encodes two proteins, the viral polymerase and zinc-finger-like protein, from two nonoverlapping open reading frames (ORF) of opposite polarity. The S RNA segment encodes the nucleoprotein (NP) from the 3′ end, in a genome complementary sense, and the glycoprotein precursor (GPC) from the 5′ end, in the genomic sense from nonoverlapping ORFs. The two ORFs are separated by a noncoding intergenic region that is predicted to fold into very stable secondary structures, in the form of stem loops (Southern, 1996). The GPC later undergoes posttranslational cleavage to generate the two mature glycoproteins, GP1 and GP2, which are inserted in the viral envelope.
As new arenaviruses have been discovered, partial sequencing has been used in addition to standard serologic methods for typing new strains (Anonymous, 2000; Coimbra et al., 1994; Fulhorst et al., 1996, 1997; Mills et al., 1996; Moncayo et al., 2001; Salas et al., 1991). Previous work with partial NP gene sequences (613–649 nucleotides) described three phylogenetic lineages within the New World complex: A, B, and C (Bowen et al., 1996b). Understanding the genetics and comparing the structures of these viruses could lead to important information regarding their pathogenesis and epidemiology in addition to allowing us to design effective vaccines. In this study, the full-length S RNA sequence was determined for 10 of the 15 New World arenaviruses (ALLV, AMAV, FLEV, GUAV, LATV, MACV, PARV, PIRV, TAMV, WWAV) and used to generate more robust phylogenies for the GPC and NP genes of all the known New World arenaviruses. Analysis of the intergenic region secondary structure was also performed to determine if grouping according to geography or pathogenic potential was apparent.
RESULTS
Arenavirus nucleotide sequences for the S segment
The complete sequence data obtained in this study include two ambisense open reading frames, encoding GPC and NP, and an intergenic region which are all characteristic of Arenaviruses (Southern, 1996): in total 3382 nucleotides for ALLV, 3341 nucleotides for AMAV, 3376 nucleotides for FLEV, 3408 nucleotides for GUAV, 3475 nucleotides for LATV, 3440 nucleotides for MACV, 3406 nucleotides for PARV, 3393 nucleotides for PIRV, 3533 nucleotides for TAMV, and 3316 nucleotides for WWAV. The 19 nucleotides at the 3′ and 5′ termini were not directly determined in this study as they were obscured by the primers used to generate the sequence template, but are expected to be complementary as is characteristic of Arenaviruses (Southern, 1996).
For each of the 10 American arenaviruses sequenced, the GPC was encoded in a single long ORF, 1440 to 1557 nucleotides in length, at the 5′ end of the viral S RNA, initiated by an AUG codon and terminating at an in-frame stop codon (see Table 1 for details). The intergenic region (i.e., untranslated span of nucleotides between the stop codons of GPC and NP) varied from 71 to 182 nucleotides in length for the 10 arenaviruses sequenced. The NP was encoded also in a single long nonoverlapping ORF, 1656 to 1713 nucleotides in length at the 3′ end of the viral S RNA, with initiation at the AUG codon and termination at an in-frame stop codon.
TABLE 1.
Characteristics of the New World Arenaviruses Sequenced in this Study
| Virus | Glycoprotein precursor |
Intergenic region |
Nucleoprotein |
S RNA |
||||
|---|---|---|---|---|---|---|---|---|
| Start codon (nt)a | Stop codon | Length (aa)b | Length (nt) | Start codon (nt) | Stop codon | Length (aa) | Length (nt) | |
| Allpahuayo | 51–53 | UAA | 508 | 70 | 3329–3327 | UAA | 562 | 3382 |
| Amapari | 62–64 | UAG | 482 | 99 | 3288–3286 | UAG | 561 | 3341 |
| Flexal | 59–61 | UAA | 508 | 96 | 3325–3323 | UGA | 560 | 3376 |
| Guanarito | 74–76 | UGA | 480 | 94 | 3289–3287 | UAG | 561 | 3342 |
| Latino | 90–92 | UGA | 516 | 61 | 3400–3398 | UAA | 558 | 3475 |
| Machupo | 89–91 | UAA | 497 | 64 | 3363–3361 | UAA | 565 | 3440 |
| Parana | 53–55 | UAA | 508 | 97 | 3326–3324 | UGA | 560 | 3406 |
| Pirital | 47–49 | UAG | 511 | 87 | 3341–3339 | UAA | 561 | 3393 |
| Tamiami | 81–83 | UAA | 486 | 182 | 3417–3415 | UAG | 566 | 3533 |
| Whitewater Arroyo | 63–65 | UAG | 481 | 71 | 3364–3362 | UGA | 563 | 3416 |
nt = nucleotide.
aa = amino acid.
Phylogenetic analysis
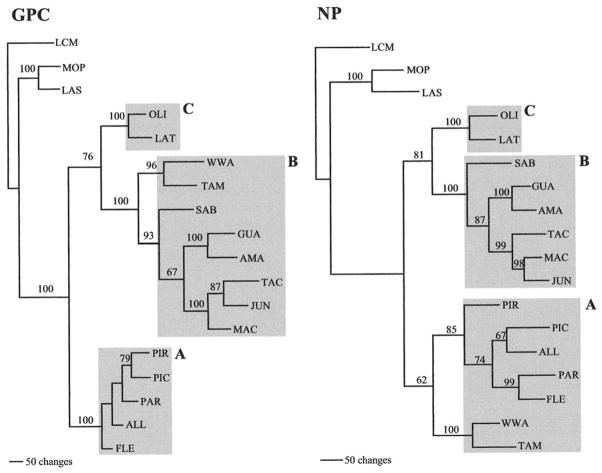
The full-length GPC and NP sequences determined for ALLV, AMAV, FLEV, GUAV, LATV, MACV, PARV, PIRV, TAMV, and WWAV were translated and compared pairwise to amino acids for JUNV, OLIV, PICV, SABV, TACV, LASV, and MOPV; LCMV was chosen as the outgroup taxon because it is the most divergent when based on amino acid sequence comparisons. Phylogenies were not done with nucleotides because divergence was so high among the Arenaviruses that alignments became arbitrary; this is probably due to long-term independent evolution of these viruses (see Discussion). Unweighted maximum parsimony (MP) optimality analysis of the GPC amino acid alignments was carried out using a step matrix found in the PROTPARS example NEXUS file of PAUP* (Swofford, 2000). This step matrix takes into account the number of nucleotide substitutions required to convert one amino acid to another. The data set used 561 characters, of which 398 were parsimony informative. A heuristic search was performed using stepwise addition, with tree-bisection-reconnection branch swapping, and a single most parsimonious tree was generated, shown in Fig. 1. The statistical significance of the branching patterns to the right of each theoretical node ranged from 67 to 100%. Following previous classifications using partial NP sequences (Bowen et al., 1996b), the phylogenetic trees derived here categorized all New World arenaviruses into three lineages, each supported by 100% bootstrap values. FLEV, ALLV, PARV, PICV, and PIRV fell into lineage A. Lineage B contained MACV, JUNV, TACV, AMAV, GUAV, SABV, TAMV, and WWAV. Lineage C contained only OLIV and LATV. Lineage diversity was 61% between A and B, 39% between B and C, and 55% between A and C.
FIG. 1.
Phylogenetic relationships among New World arenaviruses based on maximum parsimony analysis of the amino acid sequences of the complete glycoprotein precursor (GPC) gene (561 characters) and the complete nucleoprotein (NP) gene (581 characters). The numbers above the nodes indicate bootstrap support for branching patterns to the right. Scale indicates 50 changes. Lineages are indicated by A, B, or C.
When MP analysis of the NP amino acid alignment was performed, using the same assumptions, with 581 total characters, 375 of which are parsimony informative, a single most parsimonious tree was generated, shown in Fig. 1. The statistical significance of the branching patterns to the right of each theoretical node ranged from 62 to 100%. Complete NP gene amino acid phylogenies generally followed those reported previously, using partial NP sequences (Bowen et al., 1996b). Lineage C comprised OLIV and LATV. Lineage B grouped SABV, GUAV, AMAV, TACV, JUNV, and MACV. Unlike GPC, in which lineage B contained TAMV and WWAV, the NP gene of these viruses were grouped in lineage A along with FLEV, ALLV, PARV, PICV, and PIRV. Lineage diversity ranged as follows: A to B is 36%, B to C is 28%, and A to C is 37%. The different grouping patterns of TAMV and WWAV genes indicated that the evolutionary history of WWAV and TAMV are linked (with NP genes from lineage A and GPC genes from lineage B), and that probably a single recombination event between two New World progenitors has led to two distinct North American arenaviruses.
RNA folding patterns
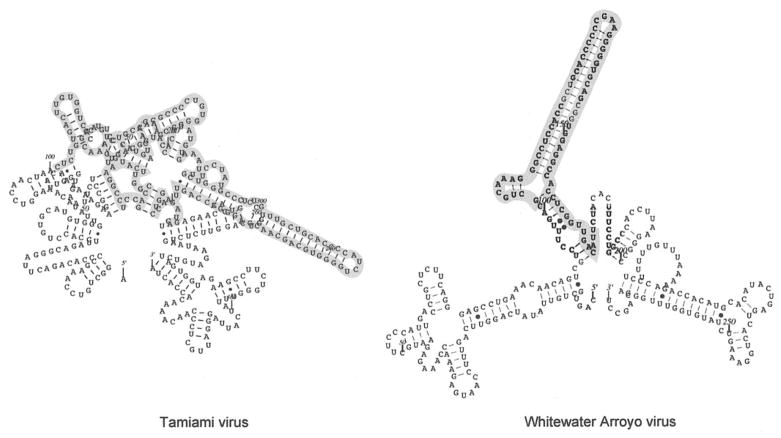
The predicted RNA secondary structures for the intergenic regions of the S segment did not show distinct groupings, but did confirm some previous predictions concerning the number of hairpins for each virus. One intergenic loop was predicted for the arenaviruses ALLV, LASV, LCMV, PICV, PIRV, and WWAV (Auperin et al., 1984, 1986; Charrel et al., 2001; Moncayo et al., 2001; Romanowski and Bishop, 1985). Our study also indicated that PARV has only one predicted intergenic hairpin. Two intergenic loops were previously predicted for JUNV, OLIV, and TACV (Bowen et al., 1996a; Ghiringhelli et al., 1991; Iapalucci et al., 1991) and this group now included the viruses AMAV, FLEV, GUAV, LATV, MACV, and MOPV. RNASTAR analysis confirmed the previous three intergenic loop prediction for SABV using Mfold (Gonzalez et al., 1996). The lengths of the intergenic regions were variable, and arenaviruses with one or two predicted hairpin loops fell into the same lineage. Figure 2 provides two examples of the predicted secondary structures. Some arenaviruses had complex folding patterns (i.e., TAMV, 182 nucleotides in length) while others, such as WWAV, were simpler (71 nucleotides in length), although these structures were not a function of length. The complexity of the predicted folding of the intergenic region in TAMV limits the ability to assess the number of intergenic loops, which may be greater than three. Sequencing the intergenic region of WWAV was made more difficult by what can now be visualized as strong C–G pairs forming at the top of the stem loop; this virus required copying and cloning to resolve 11 nucleotides of the intergenic region, probably because the polymerase would fall off of the template or skip over the stem loop structure during PCR and/or sequencing reactions.
FIG. 2.
RNA secondary structure predicted using RNASTAR for the intergenic nucleotides of Tamiami virus (TAMV), 182 nucleotides, and Whitewater Arroyo virus (WWAV), 71 nucleotides. Sequences were analyzed with the 100 flanking nucleotides on either side of the intergenic region for context.
DISCUSSION
During the last decade, searches for novel human pathogens (hantaviruses and specific arenaviruses) have led to a rise in the discovery of new arenaviruses. Four of these viruses have produced human disease after natural infection, but it is unknown whether all arenaviruses have the potential to cause human disease in natural settings. Several laboratory accidents have led to infection and disease in personnel working with high quantities and concentrations of these viruses, presumably through the inhalation of virus aerosols, and research has been limited by the availability of facilities for safely working with these agents. The understanding of the genetics of these unique viruses should help us develop new therapeutic agents, such as vaccines or antiviral drugs.
Previous Arenavirus evolution studies used limited data from small regions of the NP gene or did not include all arenaviruses known to date. In 1996, Bowen et al. published the first in-depth analysis of New World arenavirus phylogenies (Bowen et al., 1996b) and first discussed the three lineages now commonly referred to as A, B, and C. But by using only a small region of the NP gene (613–649 nucleotides) for analysis, the recombinant nature of TAMV was not evident, nor were some of the closer relationships that arenaviruses show within the same GPC gene lineage. At that time, sequence information was not available for ALLV, PIRV, or WWAV. A more recent analysis included more viruses, but still used only a 626–631 nucleotide region at the 3′ end of the NP gene (Bowen et al., 1997). This was the first evidence of TAMV pairing with WWAV, but without GPC data in the analysis, the predicted recombination event was undetected. Evidence for virus–host cospeciation was discussed, but some inconsistencies were still apparent. Other work used data representing the entire S RNA nucleotide sequence as well as the amino acid sequence for GPC and NP proteins of five New World arenaviruses to study the possible relationships between New World and Old World arenaviruses (Albarino et al., 1998). With only a few of the New World arenaviruses analyzed, OLIV and PICV grouped with the Old World viruses when the GPC and GP1 (a GPC cleavage product) were used, but the significance values for these monophyletic groupings were low (67 and 61 for GPC and GP1, respectively).
One hypothesis that explained the close relationship of OLIV and PICV with the Old World viruses involves a recombination event within GPC (Albarino et al., 1998), although evidence for Arenavirus recombination was only presented in WWAV phylogenies (Charrel et al., 2001). Comparing the amino acid sequences for both GPC and NP of WWAV with other New World arenaviruses led to the grouping of WWAV GPC with lineage B and NP with lineage A arenaviruses. Further analysis using a computing technique called bootscanning, using bootstrap values initially from a subset of sequence data then incrementally shifting along the alignment, localized the recombination event to the 3′ end of the GPC gene (Charrel et al., 2001). TAMV, aligned and visually analyzed here, seems to have a similar recombination junction in the GPC gene, between amino acids 461 and 486; the precise junction cannot be determined because of variations within this 25 amino acid window. We cannot identify the parents of the recombination event, nor the progenitor of these North American viruses; these viruses may not exist in our collection or may have existed only in the distant past.
The finding that TAMV is also a product of recombination, suggested by its GPC and NP genes segregating into two different lineages when analyzed independently, combined with the recombination also seen in WWAV, illustrates the capacity these bisegmented viruses have for exchanging genetic material with one another. While the natural occurrence of internal recombination is indeed rare within an RNA segment, only one such event would be necessary to generate the two North American viruses. There is similar phylogenetic evidence for recombination events in such virus families as Bunyaviridae, Coronaviridae, Flaviviridae, Picornaviridae, Reoviridae, Retroviridae, and Togaviridae (for review see Worobey and Holmes, 1999). For TAMV and WWAV, recombination occurred between two New World arenaviruses, but does not rule out the possibility that a recombination event involving New World and Old World arenavirus progenitors could have occurred, resulting in the data published for OLIV and PICV (Albarino et al., 1998). Until the viral factors responsible for pathogenesis are better understood, it is still possible to envision these recombination events leading to the acquisition of pathogenicity by a previously nonpathogenic arenavirus.
Reassortment of independent segments has already been described for Old World arenaviruses, albeit in vitro (Lukashevich, 1992), and has been shown to affect pathogenicity of LCMV in guinea pigs, where the L segment from the WE strain (fatal) combined with the S segment of the ARM strain (nonfatal) to produce death (Riviere, 1987; Riviere et al., 1985). To date there is no evidence regarding reassortment within New World arenaviruses, because the L segments have not been analyzed for the majority of the strains.
In this study, phylogenies using full-length amino acid sequence data from either the GPC or the NP gene of all 15 New World arenaviruses showed that known human pathogenic viruses (SABV, GUAV, JUNV, and MACV) fell into only one previously defined lineage, but viruses with weaker associations to disease (e.g., WWAV and FLEV) cloud the correlation between genetic classification and pathogenic or epidemic potential. Previous work (Bowen et al., 1996b) suggested that the highly pathogenic New World arenaviruses (GUAV, JUNV, MACV, SABV) all occupied lineage B and may descend from a common pathogenic progenitor. FLEV (lineage A) was noted as an exception, but it has caused only one nonfatal laboratory infection. The association between WWAV and fatal disease in patients from California (2000), although not confirmed virologically, and its recombinant background does not clarify the evolution of pathogenicity. Work done with PICV variants, passaged through guinea pigs (Zhang et al., 1999), suggests that three missense mutations in the GP1 protein sequence correlate with virulence. If this is consistent throughout the New World arenaviruses, it is possible that by acquiring genome sequences from lineage B, WWAV has acquired pathogenic potential through recombination. Further study is needed to evaluate the connection between viruses of lineage B and pathogenicity. There was no obvious grouping of viruses based on geographic distribution, with viruses of different monophyletic groups being isolated in the same country (i.e., OLIV and JUNV, FLEV and AMAV, PIRV and GUAV, LATV and MACV) (see Table 1).
Also, segregation of the New World arenaviruses was not consistent with their known rodent host species. While TAMV and WWAV have been shown to be evolutionarily related, their respective rodent hosts, Sigmodon hispidus and Neotoma species, are evolutionarily distant in some phylogenetic studies (Engel et al., 1998) but closely related according to others (Sullivan et al., 1995). In addition, some viruses from different lineages share similar rodent hosts (i.e., JUNV and MACV from Lineage B and LATV from Lineage C are found in Calomys species, while FLEV, PARV, and PICV from Lineage A and AMAV and GUAV from Lineage B are found in the related Oryzomyini tribe). The evidence for coevolution of arenaviruses with their rodent hosts has been reviewed recently and further studies are required to clarify their relationships (Clegg, 2002).
All New World complex arenaviruses are divergent, but still fall into the same serocomplex when antisera prepared in guinea pigs, hamsters, and mice are used for classification (Howard, 1993; Wulff et al., 1978). This means that although the amino acids encoding GPC were up to 61% divergent across lineages, and those encoding NP were up to 39% divergent across lineages, there are some common antigenic sites that can be detected by in vitro immunologic assays. While there is presently only one effective and licensed vaccine against a New World arenavirus, a live-attenuated Junin virus vaccine (Enria et al., 1999), it seems feasible that we might identify a common antigenic structure with which to stimulate a cross-reactive immune response. We are presently working on the development of consensus vaccine candidates that would provide protection against those known arenaviruses and those that might be discovered in the future (Rico-Hesse, 1999).
MATERIALS AND METHODS
Virus strains
Table 2 lists the viruses and/or sequences used in this study. Some isolates were obtained from our collections at the Southwest Foundation for Biomedical Research, San Antonio, TX: LATV (MARU 10924), PARV (12056), TAMV (CDC W-10777), GUAV (INH-95551), and AMAV (BeAn70563). FLEV (BeAn293022), WWAV (9310141), PIRV (VAV-488), MACV (Carvallo), and ALLV (CLHP-2098) were obtained from Dr. Thomas G. Ksiazek, Special Pathogens Branch, Centers for Disease Control and Prevention, Atlanta, GA. Each virus was grown in monolayers of Vero E6 cells (ATCC CRL-1587) cultured in minimal essential medium supplemented with 2% fetal bovine serum, L-glutamine, and antibiotics. All work with infectious virus was carried out in the biosafety level 3 or biosafety level 4 laboratories of the Southwest Foundation for Biomedical Research, San Antonio, TX.
TABLE 2.
Arenaviruses Used in this Study
| Virus name | Strain | Country of origin | Passage levela |
|---|---|---|---|
| Allpahuayo | CLHP-2098 | Peru | Vero#3 |
| Amapari | BeAn70563 | Brazil | SM#13 Vero#2 |
| Flexal | BeAn293022 | Brazil | SM#2 SH#1 Vero#2 |
| Guanarito | INH-95551 | Venezuela | Vero#1 |
| Junin | MC2 | Argentina | NR |
| Lassa | Josiah | Nigeria | NR |
| Latino | MARU 10924 | Bolivia | SM#8 SH#2 Vero#4 |
| LCMVb | Armstrong 53b | USA | NR |
| Machupo | Carvallo | Bolivia | SH#2, 33H#1 Vero#2 |
| Mopeia | 800150 | Mozambique | Vero#3 |
| Oliveros | RIID 3229 | Argentina | NR |
| Parana | 12056 | Paraguay | SM#8 SH#3 Vero#4 |
| Pichinde | 3739 | Colombia | NR |
| Pirital | VAV-488 | Venezuela | Vero#2 |
| Sabia | SPH114202 | Brazil | NR |
| Tacaribe | clone p2b-2 | Trinidad | NR |
| Tamiami | CDC W-10777 | USA | SM#7 Vero#2 |
| Whitewater Arroyo | 9310141 | USA | Vero#5 |
Vero = African Green monkey kidney cell line; SM = suckling mouse; SH = suckling hamster; NR = not reported.
LCMV = Lymphocytic choriomenigitis virus.
RNA extraction
Total RNA was extracted from 200 μl aliquots of infected cell-culture medium with 1 ml Trizol reagent (Gibco-BRL) and 200 μl chloroform, according to the manufacturer’s instructions. Ethanol-precipitated RNA was recovered by centrifugation and air-dried. The RNA pellet was resuspended in diethyl pyrocarbonate (DEPC)-treated water for the reverse transcriptase-polymerase chain reaction (RT-PCR).
RT-PCR
Reverse transcription was performed on the virus RNAs using a synthetic oligonucleotide, ARE 3′ END, complementary to the 19 nucleotide sequence conserved at the 3′ terminus of the Arenavirus S genomic segment (Auperin et al., 1982). In the cases of ALLV, FLEV, and GUAV, an oligonucleotide, GUA 3′ END, containing the 19 conserved 3′ terminus and three additional bases was used, with higher yields. RT-PCR was carried out using the TaqPlus Long kit (Stratagene) to ensure amplification of the entire S segment RNA. Each 100 μl RT-PCR reaction mixture contained 1 μM ARE 3′ END primer (5′-CGCACAGTGGATCCTAGGC-3′) or GUA 3′ END primer (5′-CGCACAGTGGATCCTAGGACATTTT-3′), 1× TaqPlus Long low salt buffer, 10 mM deoxynucleoside triphosphates, 40 U RNasin, 1 mM DTT, 200 U Superscript II (Gibco-BRL), and 10 U TaqPlus Long polymerase. After the mixture was incubated at 45°C for 90 min, it was amplified on a Perkin–Elmer 9700 Thermocycler at 94°C for 2 min, 52°C for 30 s, 70°C for 4 min, followed by nine cycles of 94°C for 1 min, 52°C for 30 s, 70°C for 4 min, then 20 cycles of 94°C for 1 min, 52°C for 30 s, 70°C for 3 min, and finished with a 72°C hold for 15 min. Electrophoresis was performed on 10% of each reaction on a 1% agarose gel in Tris–borate–EDTA buffer. The gel was then stained with ethidium bromide and PCR products were visualized by UV translumination to assess product yield.
Sequence determination
The remaining PCR amplification product was purified using Qiaquick PCR purification columns (Qiagen) according to the manufacturer’s instructions. The purified PCR products eluted in 30 μl EB buffer (Qiagen) were sequenced directly using the dye termination cycle sequencing technique (Applied Biosystems, Inc.), using ARE 3′ END or GUA 3′ END primers. Sequencing reactions contained 10 to 33% of the purified product, 3.3 pM primer, and 8 μl premix (Applied Biosystems) for a final volume of 20 μl. Specific primers were then designed based on first-round sequencing data and used in additional rounds of sequence “walking” to complete both strands of the PCR products. A list of the primers used can be found at http://www.sfbr.org/sfbr/departments/virology/arenavirus.html. For samples with high secondary structure, primers, premix, and reaction volumes were doubled to a total of 40 μl with improved results. A 530-nucleotide region of WWAV required cloning into pCR2.1 plasmid (Invitrogen) for resolution of a 52 nucleotide intergenic region. Cloning was performed using manufacturer’s instructions.
Phylogenetic analysis
Sequences were translated using the EditSeq program within the LaserGene package, version 5 (DNA-STAR, Inc.), and aligned with the corresponding NP and GPC gene sequences of LCMV (GenBank Accession No. M20869), LASV (J04324), MOPC (M33879), JUNV (D10072), SABV (U41071), TACV (M20304, M65834), OLIV (U34248), and PICV (K02734), obtained from the GenBank sequence database using the multiple sequence alignment method Clustal W, within the MegAlign program of the LaserGene package (Thompson et al., 1994). Phylogenetic analyses were done independently for each gene, using 581 amino acids encoding NP and 561 amino acids encoding GPC, with the PAUP* program, version 4.0b8, with PROTPARS stepmatrix of amino acid substitution, TBR branch-swapping, and a heuristic search for the most parsimonious trees (Swofford, 2000). The reliability of the two inferred phylogenetic trees were estimated using the bootstrap method, with 1000 replications (Felsenstein, 1985).
Secondary structure analysis
RNA secondary structure predictions, or folding patterns of the intergenic region of the S segment, were performed with the RNASTAR program (Gultyaev et al., 1995). The genetic algorithm files were saved in the bracket format to determine if pseudoknots occurred. The resulting predicted folding patterns were graphically represented with the loopDloop program (Gilbert, 1996) using the polygon multi-loop, straight inner loop format.
Data deposition
The nucleotide sequences reported in this study have been deposited in the GenBank database with the following accession numbers: ALLV (AY081210), AMAV (AF485256), FLEV (AF485257), GUAV (AF485258), LATV (AF485259), MACV (AF485260), PARV (AF485261), PIRV (AF485262), TAMV (AF485263), and WWAV (AF485264).
Acknowledgments
We thank April Hopstetter for editing assistance. This research was funded by NIH Grants AI42795 and AI07522.
References
- Albarino CG, Posik DM, Ghiringhelli PD, Lozano ME, Romanowski V. Arenavirus phylogeny: A new insight. Virus Genes. 1998;16:39–46. doi: 10.1023/a:1007993525052. [DOI] [PubMed] [Google Scholar]
- Anonymous. Fatal illness associated with a New World arenavirus—California, 1999–2000. MMWR Morb Mortal Wkly Rep. 2000;49:709–711. [PubMed] [Google Scholar]
- Auperin DD, Compans RW, Bishop DHL. Nucleotide sequence conservation at the 3′ termini of the virion RNA species of New World and Old World arenaviruses. Virology. 1982;121:200–203. doi: 10.1016/0042-6822(82)90130-1. [DOI] [PubMed] [Google Scholar]
- Auperin DD, Romanowski V, Galinski M, Bishop DHL. Sequencing studies of Pichinde arenavirus S RNA indicate a novel coding strategy, and ambisense viral S RNA. J Virol. 1984;52:897–904. doi: 10.1128/jvi.52.3.897-904.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auperin DD, Sasso DR, McCormick JB. Nucleotide sequence of the glycoprotein gene and intergenic region of the Lassa virus S genome RNA. Virology. 1986;154:155–167. doi: 10.1016/0042-6822(86)90438-1. [DOI] [PubMed] [Google Scholar]
- Bowen MD, Peters CJ, Mills JN, Nichol ST. Oliveros virus: A novel arenavirus from Argentina. Virology. 1996a;217:362–366. doi: 10.1006/viro.1996.0124. [DOI] [PubMed] [Google Scholar]
- Bowen MD, Peters CJ, Nichol ST. Phylogenetic analysis of the Arenaviridae: Patterns of virus evolution and the evidence of cospeciation between arenaviruses and their rodent hosts. Mol Phylogenet Evol. 1997;8:301–316. doi: 10.1006/mpev.1997.0436. [DOI] [PubMed] [Google Scholar]
- Bowen MD, Peters CJ, Nichol ST. The phylogeny of New World (Tacaribe complex) arenaviruses. Virology. 1996b;219:285–290. doi: 10.1006/viro.1996.0248. [DOI] [PubMed] [Google Scholar]
- Charrel RN, de Lamballerie X, Fulhorst CF. The Whitewater Arroyo virus: Natural evidence for genetic recombination among Tacaribe serocomplex viruses (family Arenaviridae) Virology. 2001;283:161–166. doi: 10.1006/viro.2001.0874. [DOI] [PubMed] [Google Scholar]
- Clegg JCS. Molecular phylogeny of the arenaviruses. Curr Top Microbiol Immunol. 2002;262:1–24. doi: 10.1007/978-3-642-56029-3_1. [DOI] [PubMed] [Google Scholar]
- Coimbra TLM, Nassar ES, Burattini MN, Souza LTMd, Ferreira IB, Rocco IM, Travassos da Rosa APA, Vasconcelos PFC, Pinheiro FP, LeDuc JW, Rico-Hesse R, Gonzalez JP, Jarhling PB, Tesh RB. New arenavirus isolated in Brazil. Lancet. 1994;343:391–392. doi: 10.1016/s0140-6736(94)91226-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel SR, Hogan KM, Taylor JF, Davis SK. Molecular systematics and paleobiogeography of the South American Sigmodontine rodents. Mol Biol Evol. 1998;15:35–49. doi: 10.1093/oxfordjournals.molbev.a025845. [DOI] [PubMed] [Google Scholar]
- Enria D, Feuillade M, Levis S, Briggiler A, Ambrosio A, Saavedra M, Becker J, Riera L, Calderon G, Pini N, Sottosanti J, Aviles G, Garcia J, Sabattini M. Impact of vaccination of a high-risk population for Argentine hemorrhagic fever with a live-attenuated Junin virus vaccine. In: Saluzzo J, Dodet B, editors. Factors in the Emergence and Control of Rodent-borne Viral Diseases. Elsevier; Paris: 1999. pp. 273–280. [Google Scholar]
- Felsenstein J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- Fulhorst CF, Bowen MD, Ksiazek TG, Rollin PE, Nichol ST, Kosoy MY, Peters CJ. Isolation and characterization of Whitewater Arroyo virus, a novel North American arenavirus. Virology. 1996;224:114–120. doi: 10.1006/viro.1996.0512. [DOI] [PubMed] [Google Scholar]
- Fulhorst CF, Bowen MD, Salas RA, deManzione NMC, Duno G, Utrera A, Ksiazek TG, Peters CJ, Nichol ST, deMiller E, Tovar D, Ramos B, Vasquez C, Tesh RB. Isolation and characterization of Pirital virus, a newly discovered South American arenavirus. Am J Trop Med Hyg. 1997;56:548–553. doi: 10.4269/ajtmh.1997.56.548. [DOI] [PubMed] [Google Scholar]
- Ghiringhelli PD, Rivera-Pomar RV, Lozano ME, Grau O. Molecular organization of Junin virus S RNA: Complete nucleotide sequence, relationship with other members of the Arenaviridae and unusual secondary structures. J Gen Virol. 1991;72:2129–2149. doi: 10.1099/0022-1317-72-9-2129. [DOI] [PubMed] [Google Scholar]
- Gilbert DG. loopDloop in Java, a Java application for visualizing RNA secondary structure. 1996 http://iubio.bio.indiana.edu/soft/molbio/loopdloop/java.
- Gonzalez JPJ, Bowen MD, Nichol ST, Rico-Hesse R. Genetic characterization and phylogeny of Sabia virus, an emergent pathogen in Brazil. Virology. 1996;221:318–324. doi: 10.1006/viro.1996.0381. [DOI] [PubMed] [Google Scholar]
- Gultyaev AP, van Batenburg FHD, Pleij CWA. The computer simulation of RNA folding pathways using a genetic algorithm. J Mol Biol. 1995;250:37–51. doi: 10.1006/jmbi.1995.0356. [DOI] [PubMed] [Google Scholar]
- Howard C. Antigenic diversity among arenaviruses. In: Salvato M, editor. The Arenaviridae. Plenum Press; New York: 1993. pp. 37–49. [Google Scholar]
- Iapalucci S, Lopez N, Franze-Fernandez MT. The 3′ end termini of the Tacaribe arenavirus subgenomic RNAs. Virology. 1991;182(1):269–278. doi: 10.1016/0042-6822(91)90670-7. [DOI] [PubMed] [Google Scholar]
- Lukashevich IS. Generation of reassortants between African arenaviruses. Virology. 1992;188:600–605. doi: 10.1016/0042-6822(92)90514-p. [DOI] [PubMed] [Google Scholar]
- Mills JN, Oro JGB, Bressler DS, Childs JE, Tesh RB, Smith JF, Enria DA, Geisbert TW, McKee KT, Bowen MD, Peters CJ, Jahrling PB. Characterization of Oliveros virus, a new member of the Tacaribe complex (Arenaviridae: arenavirus) Am J Trop Med Hyg. 1996;54:399–404. doi: 10.4269/ajtmh.1996.54.399. [DOI] [PubMed] [Google Scholar]
- Moncayo AC, Hice CL, Watts DM, Rosa APATd, Guzman H, Russell KL, Calampa C, Gozalo A, Popov VL, Weaver SC, Tesh RB. Allpahuayo virus: A newly recognized arenavirus (Arenaviridae) from arboreal rice rats (Oecomys bicolor and Oecomys paricola) in northeastern Peru. Virology. 2001;284:277–286. doi: 10.1006/viro.2000.0803. [DOI] [PubMed] [Google Scholar]
- Rico-Hesse R. Vaccines for emergent American arenaviruses. In: Saluzzo JF, Dodet B, editors. Factors in the Emergence and Control of Rodent-borne Diseases. Elsevier; Paris: 1999. [Google Scholar]
- Riviere Y. Mapping arenavirus genes causing virulence. Curr Top Microbiol Immunol. 1987;133:59–65. doi: 10.1007/978-3-642-71683-6_5. [DOI] [PubMed] [Google Scholar]
- Riviere Y, Ahmed R, Southern PJ, Buchmeier MJ, Oldstone MBA. Genetic mapping of Lymphocytic choriomeningitis virus pathogenicity: Virulence in guinea pigs is associated with the L RNA segment. J Virol. 1985;55:704–708. doi: 10.1128/jvi.55.3.704-709.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanowski V, Bishop DHL. Conserved sequences and coding of two strains of Lymphocytic Choriomeningitis virus (WE and ARM) and Pichinde arenavirus. Virus Res. 1985;2:35–51. doi: 10.1016/0168-1702(85)90058-9. [DOI] [PubMed] [Google Scholar]
- Salas R, Manzione ND, Tesh RB, Rico-Hesse R, Shope RE, Betancourt A, Godoy O, Bruzual R, Pacheco ME, Ramos B, Taibo ME, Tamayo JG, Jaimes E, Vasquez C, Araoz F, Querales J. Venezuelan haemorrhagic fever. Lancet. 1991;338:1033–1036. doi: 10.1016/0140-6736(91)91899-6. [DOI] [PubMed] [Google Scholar]
- Southern PJ. Arenaviridae: The viruses and their replication. In: Fields BN, Knipe DM, Howley PM, editors. Fields Virology. Lippincott-Raven Publishers; Philadelphia: 1996. pp. 1505–1519. [Google Scholar]
- Sullivan J, Holsinger KE, Simon C. Among-site rate variation and phylogenetic analysis of 12S rRNA in Sigmodontine rodents. Mol Biol Evol. 1995;12:988–1001. doi: 10.1093/oxfordjournals.molbev.a040292. [DOI] [PubMed] [Google Scholar]
- Swofford DL. PAUP*, Phylogenetic Analysis Using Parsimony (*and Other Methods) 4.0b8. Sinauer Associates; Sunderland, MA: 2000. [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worobey M, Holmes EC. Evolutionary aspects of recombination in RNA viruses. J Gen Virol. 1999;80:2535–2543. doi: 10.1099/0022-1317-80-10-2535. [DOI] [PubMed] [Google Scholar]
- Wulff H, Lange JV, Webb PA. Interrelationships among arenaviruses measured by indirect immunofluorescence. Intervirology. 1978;9:344–350. doi: 10.1159/000148956. [DOI] [PubMed] [Google Scholar]
- Zhang L, Marriott K, Aronson JF. Sequence analysis of the small RNA segment of guinea pig-passaged Pichinde virus variants. Am J Trop Med Hyg. 1999;61:220–225. doi: 10.4269/ajtmh.1999.61.220. [DOI] [PubMed] [Google Scholar]