Substance use disorders are maladaptive patterns of substance use leading to inability to control use despite significant consequences.1 When the impairment is in multiple areas of life or the individual has significant signs of tolerance or withdrawal, the diagnosis changes to substance dependence. These general definitions apply to all drugs of abuse, including alcohol, nicotine, and cocaine. Substance use disorders are common: alcohol dependence/abuse has a prevalence of 13.5%, nicotine dependence has a prevalence of 13%, and all other drug dependence/abuse has a prevalence of 6.1%.2,3
Substance use disorders are highly comorbid with many other mental illnesses. In one study, the odds of having an addictive disorder not related to nicotine increased by a factor of 2.7 in the presence of a mental disorder.3 In addition, the presence of an addictive disorder increased the risk for other mental illness. Specifically, 37% of subjects with alcohol addiction had a comorbid mental disorder, and 53% of subjects with other drug addiction had a comorbid mental disorder, compared with a baseline prevalence of non–substance-related mental disorders of 22.5%.3 This comorbidity leads to more severe illness. In a five-state study of Medicaid recipients, the subjects with both a severe mental illness and a substance use disorder had a higher odds of using inpatient and emergency psychiatric facilities compared with subjects with the respective mental illness alone.4
Although many studies distinguish nicotine dependence from other substance-related disorders, nicotine dependence is also highly comorbid with mental disorders and negatively affects their severity and prognosis. There are increased rates of life-time smoking in subjects with mental illness compared with controls (59% vs 39%, respectively, P<.001).5 The prevalence of nicotine dependence is 13% in the general population and 30% to 70% in the presence of other psychiatric disorders.2 Although these data indicate the strong need for smoking cessation in the psychiatrically ill population, quit rates are substantially lower for subjects with active mental illness compared with smokers without mental illness (31% quit rate vs 43% quit rate).5
Not only is a significant proportion of nicotine dependence comorbid with psychiatric illness but a significant portion of morbidity in psychiatric illness may be attributed to nicotine dependence. The greatest mortality risk of mental illness is premature death caused by heart disease and cancer. In an 8-state comparison of deaths of clients at public mental health clinics, public mental health clients lived 13 to 30 years less than their general public counterparts.6,7 Although rates of suicide and accidental death are higher in this population compared with controls, the primary causes of the deaths are medical illnesses such as heart disease and cancer. Therefore, the largest modifiable risk factor for mortality in the mental health population is cigarette smoking. In addition, nicotine dependence is associated with disease-specific poor outcomes. For example, lifetime smoking in bipolar disorder is associated with earlier age of onset of symptoms, greater severity of symptoms, poorer functioning, history of suicide attempts, comorbid anxiety, and substance use disorders.8
The more severe course of illness and increased mortality highlights the importance of integrating the study of addiction with the study of other psychiatric and medical illnesses. This importance is emphasized by recent genetic overlap found between alcohol dependence and aerodigestive cancers, and between nicotine dependence and lung disease. This article chronicles the genetic associations found in alcohol dependence, including related findings in aerodigestive cancers, and the genetic associations found in nicotine dependence, including related findings in lung disease. The article then briefly discusses the genetic findings in cocaine dependence and psychiatric comorbidities with substance dependence.
GENETICS OF ALCOHOL DEPENDENCE
Alcohol dependence is a complex disease, with genetic and environmental risk factors. Alcohol dependence (commonly known as alcoholism) necessarily has a strong environmental component because exposure to and consumption of alcohol is required for the disorder. But, there is a substantial heritable component to the risk for alcoholism. Among first-degree relatives of alcohol-dependent individuals, the risk of alcohol dependence is 3 to 8 times the baseline population risk.9
Alcohol dependence was the first behavioral disorder to have validated genetic findings. In 1972, subjects of Asian descent were noted to have facial flushing and decreased alcohol tolerance compared with subjects of European origin.10 This was hypothesized to be genetic rather than cultural, based on observations that infants of Asian descent who were exposed to a small amount of alcohol were more likely to flush than infants of European descent.10 The flushing reaction to ingestion of alcohol was found to be secondary to a deficiency of aldehyde dehydrogenase (specifically ALDH2), an enzyme involved in the metabolism of ethanol.11 The prevalence of ALDH2 deficiency was then examined in the Japanese population and found to be 41% in the general population and 2% in alcoholics, suggesting a protective role for the deficiency of ALDH2 in alcoholism.12
Since these initial findings in the early 1980s, much has been learned about the genetics of alcohol metabolism. Ethanol metabolism occurs predominantly in the liver in 2 steps: (1) oxidation to acetaldehyde catalyzed by alcohol dehydrogenases (ADHs), and (2) oxidation of acetaldehyde to acetate by ALDH. Multiple genetic variants of ADH and ALDH influence rates of alcohol metabolism and alcohol dependence.13 The mechanism through which variants of these enzymes influence risk of alcohol dependence is believed to be related to elevation of acetaldehyde levels, leading to facial flushing, nausea, and tachycardia. It is hypothesized that people with the genetic variants of ADH and ALDH leading to increased acetaldehyde levels would be less likely to drink excessively because of the discomfort of ingesting a small amount of ethanol. This behavioral aversion to alcohol with increased acetaldehyde levels is exploited by administering disulfiram, a medication that interferes with ALDH, to alcoholics. Disulfiram blocks ALDH, leading to a build-up of acetaldehyde, causing symptoms including nausea, vomiting, flushing, shortness of breath, and headache when alcohol is ingested. It is hoped that the administration of disulfiram to alcoholics decreases cravings and discourages them from using alcohol.
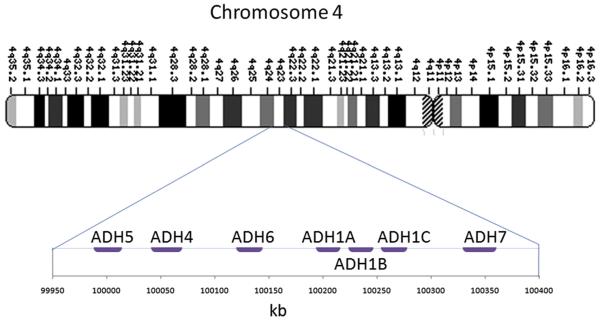
There are multiple ADH and ALDH genes, and many variants of these genes have been examined for their association with alcohol dependence. Most of the studied variants involve changes of single nucleotide polymorphisms (SNPs), in coding and noncoding regions of the DNA. The ADH genes are located in a small region on chromosome 4 (Fig. 1). The ALDH gene associated with alcohol dependence, ALDH2, is on chromosome 12q24.2. This article discusses the major association findings between alcohol metabolism genes and alcoholism. For a more comprehensive review on the genetics of alcohol metabolism, see Edenberg.13
Fig. 1.
Chromosome 4 region q23, including the ADH enzymes ADH1A, ADH1B, ADH1C, ADH4, ADH5, ADH6, and ADH7.
The ALDH2 variant leading to decreased risk of alcohol dependence, ALDH2*2, has been further characterized since its initial discovery in the 1980s. ALDH2*2 is a coding variant resulting from the substitution of lysine for glutamate at position 504, resulting in a nearly inactive ALDH2 enzyme and leading to markedly elevated acetaldehyde levels in the blood with the consumption of small amounts of alcohol. The allele is common in people of East Asian descent, but it is essentially nonexistent in people of European or African descent.14 The protective effect of this allele is strong and has been replicated in multiple studies.15-17 The odds ratio of alcohol dependence for subjects with 1 ALDH2*2 allele is 0.33, and there are almost no documented cases of people with alcohol dependence who are homozygous for ALDH2*2.15-17 This allele interacts with a nonsynonymous gene variant for the ADH1 enzyme, ADH1B*1, by further decreasing the odds ratio of alcohol dependence to 0.05 in the presence of both alleles.15 The protective effect of the ALDH2*2 allele is susceptible to environmental pressures. A study by Higuchi and colleagues18 found that the fraction of Japanese alcoholics carrying the ALDH2*2 allele increased from 3% in 1979 to 13% in 1992. This increase is believed to be due to an overall increase in alcohol consumption in Japanese society during that period.
The most robust association between a variant of an ADH gene and alcohol dependence is with the ADH1B*2 allele (previously known as ADH2*2). This variant is in the ADH1B gene that encodes the β2 subunit of ADH, and results in histidine instead of arginine at position 48.13 It is associated with a more rapid ethanol oxidation to acetaldehyde and is protective against alcohol dependence, with an odds ratio of 0.12 in a Chinese population.13,15 Again, this variant is common in people of East Asian descent, and rare in people of European and African descent. The protective effect of this allele seems to be weaker in European than in Asian populations.19 It is unclear how much of this variability is due to different social and environmental factors rather than unidentified coinherited genes that modify susceptibility.
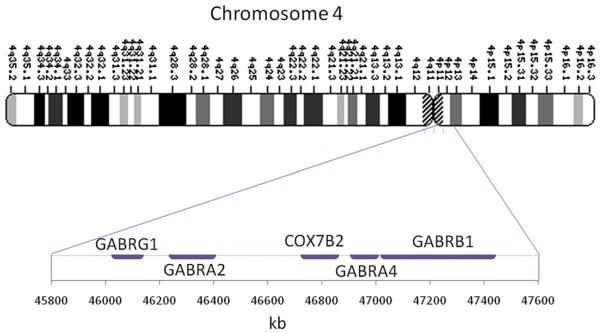
In addition to the association between alcohol dependence and alcohol-metabolizing enzymes, γ-aminobutyric acid (GABA) receptor genes have also been associated with alcohol dependence. This association is of particular relevance because alcohol is an agonist of the GABA receptor. This region includes the subunit genes GABRG1, GABRA2, GABRA4, and GABRB1 (Fig. 2). Edenberg and colleagues20 evaluated this region in detail and found a strong association between GABRA2, encoding the α2 subunit of GABAA, and alcohol dependence. This association was replicated in multiple populations, and was found to be most strong in alcohol-dependent individuals with comorbid dependence on illicit drugs.21-24
Fig. 2.
Chromosome 4 region p12, including the GABA receptor gene cluster.
GABA receptor genes have a clear relationship to alcohol dependence. GABA transmission mediates effects of alcohol, including disruption of motor coordination, anxiolysis, sedation, ethanol preference, and symptoms related to withdrawal.25 Furthermore, rat models have shown GABAA agonists increase ethanol intake and GABAA antagonists decrease ethanol intake.26 Although GABA subunit genes and alcohol-metabolizing genes have been genetically associated with alcohol dependence, the biologic mechanism is different in each case.
The advent of the genome-wide association studies (GWAS) brought increased genomic density and large data sets. GWAS relied on the completion of the International HapMap Project in 2005, in which common SNPs were mapped tightly throughout the genome. Using a case-control design and hundreds of thousands of SNPs spanning the entire genome, GWAS looked for association between SNPs and disease. Although there are more statistical tests of association (1 test per SNP) than there are subjects (typically thousands), the combination of setting a sufficiently low threshold for P value significance and placing a high emphasis on replication of significant results in independent data sets has resulted in discovery of robust associations.
Although GWAS are a technological breakthrough in the density of markers that are tested across the genome, the genome is not completely represented by the SNP arrays used in the GWAS studies. For example, the Affymetrix Genome-Wide Human SNP Array 6.0 genotypes 906,600 SNPs out of an estimated 10,000,000 SNPs in the human genome (approximately 2,000,000 of which are known). The dense coverage in GWAS studies, although an improvement compared with previous methodology, is still not dense enough to ensure that associations are found. Nonetheless, a SNP reaching genome-wide association significance indicates that functional variation is likely to occur through this SNP or 1 of the correlated, untested SNPs.
The first GWAS for alcohol dependence was published by Treutlein and colleagues27 in 2009. Two SNPs reached genome-wide significance, and these investigators nominated another group of candidate SNPs to evaluate in a secondary dataset. Further confirmation of their findings is necessary. The ADH and ALDH genes previously found to be associated with alcohol dependence were not significant. This finding is likely due to low-risk allele frequencies in the European population that was used for the study.
Because alcohol is related to medical illnesses such as aerodigestive cancer and variations in alcohol metabolism are related to the development of alcohol dependence, a study was designed to evaluate whether genetic variation in ADH may contribute to the development of cancer of the upper aerodigestive tract, including several types of head and neck cancer that develop in the context of heavy alcohol and tobacco exposure.28 Based on 3800 subjects with aerodigestive cancer and 5200 controls, variants in ADH1B and ADH7 were found to be protective against aerodigestive cancer.28 Even after stratifying for site of cancer, alcohol consumption and other covariates, the protective effects were still strong. These effects suggest that ADH variants not only decrease the risk for alcohol dependence but also decrease the susceptibility to cancer associated with alcohol dependence, perhaps by modifying the carcinogenic effects of alcohol.
The investigation of genetic associations in alcohol dependence started with the observation of facial flushing in an Asian subpopulation and has developed into genome-wide studies of large populations with alcohol dependence and alcohol-related cancers. Although many years of study have led to improved understanding, fundamental questions of the genetics of alcohol dependence remain unanswered.
NICOTINE DEPENDENCE
The conceptualization of the different stages of smoking is helpful in understanding the varying environmental and genetic contributions to each step in the development of smoking behaviors. To better understand smoking behavior, it is compartmentalized into (1) initiation (the period during which subjects smoke for the first time and experiment with smoking), (2) regular smoking (defined as subjects having smoked at least 100 cigarettes during their lifetime), and (3) nicotine dependence (a psychiatric disorder defined by symptoms of tolerance, withdrawal, and loss of control).1 The heritability of smoking initiation was estimated to be 44%.29 Conversely, the heritability of nicotine dependence was markedly higher at 75%.29
Although smoking initiation and nicotine dependence are related, the genetics of nicotine dependence are of primary interest because of its public health ramifications: nicotine dependence predicts difficulty with cessation and carries most of the morbidity associated with smoking. Specifically, the quantity of cigarettes smoked in a lifetime is associated with lung disease and heart disease, and smoking cessation is a positive prognostic factor for both. Individuals with nicotine dependence account for 13% of the general population but consume 58% of the cigarettes smoked in the United States.2 Thus, understanding the genetics of nicotine dependence can lead to targeted treatments and may ultimately decrease tobacco-associated morbidity and mortality.
In the study of nicotine dependence, it is important to understand the behavioral progression to nicotine dependence when choosing a control group. Subjects who have not had adequate nicotine exposure (ie, who have not smoked enough cigarettes) have not had the opportunity to become nicotine dependent. For this reason, genetic studies of nicotine dependence carefully choose for their control group regular smokers who did not become nicotine dependent. This choice minimizes the possibility that the controls would become dependent if they had adequate nicotine exposure.
The strong heritability of nicotine dependence has led to genetic studies. The initial efforts to identify susceptibility loci primarily used linkage designs. In addition to linkage studies, isolated candidate gene case-control studies targeted various neurotransmitters and their receptors, but these generally had low power and insufficient density of genotypes.
The first GWAS on nicotine dependence was conducted by Bierut and colleagues.30 The study compared 1050 nicotine-dependent subjects and 879 controls (who smoked but did not become dependent). Although no individual SNPs reached genome-wide significance, the data were reanalyzed for a targeted association study with 3713 SNPs in 348 candidate genes.31 Several cholinergic nicotinic receptor genes dominated the top signals after controlling for multiple testing.
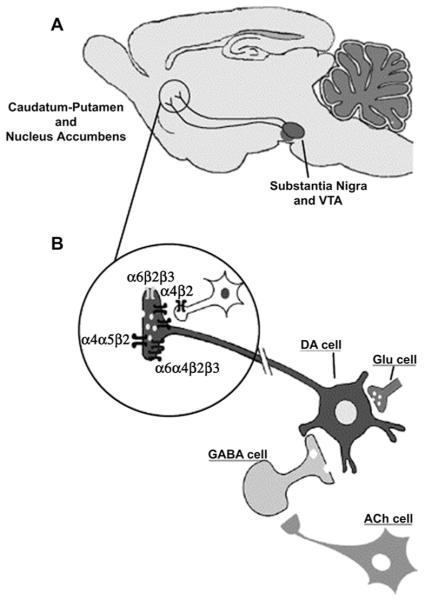
It is biologically relevant that the top genetic associations for nicotine dependence were in genes encoding for nicotinic receptor subunits. Nicotine produces its central and peripheral actions by binding to neuronal nicotinic acetylcholine receptors (nAChRs), a class of neuronal ligand-gated ion channels expressed in the nervous system. nAChRs are made of 5 combinations of α and β subunits constructed around a central pore. The subunits are encoded by 9 α (α2–α10) and 3 β (β2–β4) subunit genes, named CHRNA2–CHRNA10 and CHRNB2–CHRNB4, respectively. The expression of the different subunits in specific anatomic areas leads to hypotheses regarding their functional relevance. The addiction of nicotine is believed to arise from the interaction between dopaminergic and nicotinic neurons in the striatum (Fig. 3). Multiple nicotinic subunits are involved in this interaction, including α4, α5, α6, β2, and β3. This region has been implicated in the reward pathway and is important for the development of substance dependence.
Fig. 3.
nAChRs in the striatum of the rodent central nervous system. (A) The mesostriatal dopaminergic pathway. (B) Subunit composition of the functional nAChRs expressed by dopaminergic nerve terminals (α6β2β3, α6α4β2β3, α4β2, α4α5β2). VTA, ventral tegmental area. (Reproduced from Gotti C, Clementi F. Neuronal nicotinic receptors: from structure to pathology. Prog Neurobiol 2004;74(6):363–96; with permission.)
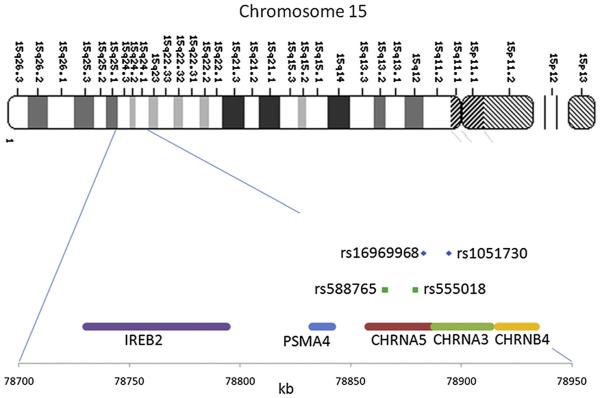
CHRNA5 and CHRNA3 are nicotinic receptor subunit genes on chromosome 15q25 coding for the α5 nicotinic receptor subunit and the α3 nicotinic receptor subunit, respectively (Fig. 4). The coding sequences are adjacent to one another, and SNPs in the 2 genes are in high linkage disequilibrium (LD). The most biologically compelling association with nicotine dependence was found in rs16969968, a nonsynonymous SNP in the α5 nicotinic receptor subunit gene CHRNA5.30,31 This association has been replicated with either rs16969968 or correlated SNPs in many other independent studies.32-36 rs16969968 is seen in Fig. 4 in the coding region of CHRNA5. rs16969968 is a nonsynonymous SNP for which the minor variant results in an amino acid change of aspartic acid to asparagine. The frequency of the minor allele varies between populations. It ranges from 0% in African populations to 37% in European populations. Although rs16969968 is in the coding sequence of CHRNA5, and a biologically plausible relationship exists between CHRNA5 and nicotine dependence, SNPs in high LD with rs16969968 span a large area encompassing the genes IREB2, PSMA4, CHRNA5, CHRNA3, and CHRNB4. Although rs16969968 is a compelling candidate for a causal association with nicotine dependence, other unidentified but related markers cannot be excluded as the cause for the observed association with nicotine dependence.
Fig. 4.
Chromosome 15q25, including the nicotinic receptor gene cluster. The SNPs associated with nicotine dependence are highlighted.
The biologic importance of rs16969968 has been shown in several settings. First, the α5 nicotinic receptor is expressed in the brain. Its expression in the striatum and direct interaction with the dopaminergic pathway is particularly relevant to addiction. Second, the protein sequences of CHRNA5 homologs were examined in multiple species (human, chimpanzee, Bolivian squirrel monkey, domestic cow, mouse, chicken, and African clawed frog) and the aspartic acid residue was present in all species.37 This conservation across species suggests that it has functional importance. Third, an in vitro functional study found that α4α5β2 receptors containing the asparagine amino acid substitution in α5 exhibited decreased response to a nicotine agonist compared with the receptors with the aspartic acid variant in a5.37 This finding suggests that decreased nicotinic receptor function is associated with increased risk for nicotine dependence.
In-depth analyses of the region led to the discovery of additional SNPs that form a haplotype with rs16969968 to modify the expression of CHRNA5.32,36,38 The SNP rs588765 was found to be associated with nicotine dependence and to modify expression of CHRNA5. In addition, together with rs16969968, it forms a haplotype to alter the risk of nicotine dependence. Although rs588765 can lead to high expression and low expression of CHRNA5, and rs16969968 can lead to high risk and low risk for nicotine dependence, only 3 haplotypes exist in the population (Table 1): high risk (rs16969968) and low expression (rs588765), low risk (rs16969968) and low expression (rs588765), and low risk (rs16969968) and high expression (rs588765). The lowest risk of nicotine dependence occurs with the low-expression allele of rs588765 and the low-risk allele of rs16969968, and the highest risk for nicotine dependence occurs with the low-expression allele of rs588765 and the high-risk allele of rs16969968. A similar association was seen in a population of African Americans with the SNP rs555018, an SNP in high LD with rs588765 in European Americans and African Americans, suggesting that rs555018 is associated with variable expression of CHRNA5.32
Table 1.
Risk of nicotine dependence based on haplotypes constructed from rs16969968 and rs588765
| Haplotype Risk of Nicotine Dependence | |||
|---|---|---|---|
| High | Intermediate | Low | |
| rs16969968 | High-risk allele | Low-risk allele | Low-risk allele |
| rs588765 | Low-expression allele | High-expression allele | Low-expression allele |
Alleles of rs16969968 are represented as high-risk allele for nicotine dependence and low-risk allele for nicotine dependence. Alleles for rs588765 are represented as high expression of CHRNA5 and low expression of CHRNA5. The haplotype of high-risk allele (rs16969968) and high-expression allele (rs588765) is not listed because it is rare in the population.
The GWAS results and corresponding biologic experiments suggest that CHRNA5 is involved in the development of nicotine dependence, but the relationship between changes in structure and expression of CHRNA5 and the development of smoking behavior remains unknown. Various research groups are evaluating the relationship between these genes and smoking initiation, the pleasurable responses associated with smoking, and the age of onset of smoking.39-41
The purpose of understanding the genetics of nicotine dependence is, ultimately, to change behavior and effectively treat the disorder. Therefore, research has been conducted on the genetics of smoking cessation. Because the greatest predictor of smoking cessation is nicotine dependence, a negative correlation would be expected between smoking cessation and the major SNP associated with nicotine dependence, rs16969968. The initial population-based study to assess genetic associations with smoking cessation did not find an association with rs16969968.42 However, 2 recent studies selected participants specifically targeted for smoking cessation, and both found the minor allele of rs16969968 to be significantly associated with inability to quit smoking or high likelihood of relapse.43,44 This finding suggests that rs16969968 increases the risk of nicotine dependence, and decreases the ability to quit.
The CHRNA5/CHRNA3/CHRNB4 region has also been associated with lung cancer and chronic obstructive pulmonary disease (COPD). Because lung cancer and COPD are nearly completely attributable to smoking it is difficult to determine whether the observed association is independent of nicotine dependence. The difficulty with interpretation has been emphasized in multiple studies.
Associations have been found between lung cancer and an SNP in LD with rs16969968, rs1051730.33,34,45,46 Various methodologies have been used to try to separate the environmental contribution of nicotine dependence from a direct genetic contribution to lung cancer of rs1051730 or a related SNP. Several researchers found increased risk for lung cancer after adjusting for smoking, supporting the hypothesis that rs1051730 contributes to lung cancer independent of nicotine dependence.33,45,46 Amos and colleagues46 found an association between lung cancer and rs1051730 in 3 independent samples, robust to adjustment for pack-years of cigarette exposure. However, the association between rs1051730 and lung cancer was absent in nonsmokers. If rs1051730 were related to lung cancer independent of smoking exposure the effect would be present in nonsmokers. Despite this, the repeated evidence of association of this variant with lung cancer independent of degree of smoking exposure among those who have smoked argues that there is an association between rs1051730 (and related SNPs) and lung cancer beyond the impact on risk of nicotine dependence itself.
The relationship between rs1051730, nicotine dependence, and lung cancer was more specifically explored by Spitz and colleagues.43 This was accomplished by using 3 samples: (1) patients with lung cancer and controls with a history of smoking, (2) patients with lung cancer and controls who were lifetime nonsmokers, and (3) patients with bladder cancer and controls with a history of smoking. The investigators found a statistically significant association of rs1051730 with lung cancer in subjects with a history of smoking. When stratified for number of cigarettes smoked per day, the highest risk was seen in the lightest smokers, subjects smoking less than 20 cigarettes per day. There was no association between lung cancer and rs1051730 in subjects who had never smoked and no association between rs1051730 and bladder and renal cancers. The investigators concluded that because the highest risk group was in the subjects who smoked the least, rs1051730 mediated risk to lung cancer beyond the cigarette exposure from nicotine dependence.
A recent GWAS for COPD also found association of rs1051730 with COPD.47 This association remained significant after adjusting for pack-years (P = 5.7 × 10−10). Although this finding suggests that this is a true association with COPD, the investigators acknowledged that the lack of further smoking variables makes it difficult to identify the full contribution of nicotine exposure on COPD.
One explanation for the association of rs16969968 with lung disease independent of cigarette quantity is that the quantity of toxin inhaled when smoking is not well explained by the number of cigarettes smoked per day, suggesting that people with the risk allele of rs16969968 may inhale more deeply. To investigate this possibility, Le Marchand and colleagues48 conducted a study that evaluated the amount of nicotine and carcinogens consumed in a sample of 819 smokers. A positive association was found between the amount of nicotine metabolites seen in urine, a measure of the amount of nicotine consumed, and the minor variant of rs16969968 after adjusting for number of cigarettes smoked per day. This finding indicates that more nicotine and associated carcinogens are absorbed by individuals with the risk allele than individuals without the risk allele. Moreover, the differential absorption is not accounted for by number of cigarettes smoked per day. Because these data argue that number of cigarettes smoked per day is not a complete proxy for nicotine and toxin absorption, the association between lung cancer and rs16969968 may still be confounded by the increased nicotine and toxin intake in nicotine dependence even after adjusting for number of cigarettes smoked per day.
In summary, multiple associations have been found between nicotine dependence and nicotinic receptor subunit genes. The strongest and most robust association is seen with a nonsynonymous SNP in the nicotinic receptor gene CHRNA5, rs16969968. This SNP is also associated with lung cancer and COPD, both smoking-related disorders. The current scientific question is whether there is an association between these smoking-related disorders and rs16969968, independent of nicotine dependence.
COCAINE DEPENDENCE
It is estimated that approximately 2.1% of US residents have used cocaine in the past month, the second most prevalent nonmedical drug to marijuana.49 Similar to nicotine and alcohol dependence, cocaine dependence has a strong component of heritability: siblings of cocaine-dependent probands had an estimated relative risk of 1.8 of developing cocaine dependence compared with probands without cocaine-dependent siblings.50 Because of the hypothesis that common genetic factors may lead to dependence on various classes of drugs, a study evaluated the relationship between cocaine dependence and rs16969968, the nonsynonymous CHRNA5 SNP associated with increased risk for nicotine dependence.51 The minor allele of rs16969968 that is associated with increased risk for nicotine dependence is associated with decreased risk for cocaine dependence. The bidirectional association is hypothesized to be due to the involvement of nAChRs with excitatory and inhibitory modulation of dopaminemediated reward pathways. A negative association was also seen between the CHRNA5 SNP rs588765 and alcohol dependence in a sample that was heavily comorbid with cocaine dependence.52 This association suggests that the CHRNA5 gene may play a dual role in modulating susceptibility to addiction via the different mechanisms of action of cocaine and nicotine.
GENETIC ASSOCIATION OF SUBSTANCE DEPENDENCE WITH OTHER MENTAL ILLNESS
The tendency of subjects with mental illness to use substances has been extensively discussed. A traditional hypothesis was that substance use is a means of self-medicating and dependence is therefore ‘caused’ by mental illness. On the contrary, data have been published that smoking precedes the diagnosis of mental illness and may contribute to the disorder. For example, there is an increased risk of first onset major depression in smokers53-56 and an increase in depressive symptoms in persons who smoked in adolescence.57-59 Ex-smokers do not differ from current smokers in their risk for mood disorders, although daily smoking may be a causal factor in panic disorder and agoraphobia.60 One interpretation of these data is that smoking confers additional risk in the development of psychiatric disorders.
An alternative explanation for the comorbidity of substance dependence and other psychiatric illness is a shared cause. The heritability of major psychiatric illness is high, with estimates of 64% in schizophrenia,61 59% in bipolar disorder,61 and 42% in major depressive disorder.62 The combination of the heritability of substance dependence, the heritability of psychiatric illnesses, and comorbidity of substance dependence with psychiatric illness indicates that specific genes may be pleiotropic (ie, contribute to substance dependence and other psychiatric illness).
Because of the phenotypic complexity of psychiatric illnesses, the search for pleiotropic genes may help clarify psychiatric diagnoses and help understand biologic causes of mental illness. For example, suspicion of shared genetic susceptibilities with bipolar disorder and illegal substance dependence has been the subject of recent research. A bipolar GWAS data set and an illegal substance dependence GWAS data set were compared for overlap of top signals, and evidence was found of shared associations.63 These studies suggest that advances may be made by understanding genetic pleiotropy and interconnections between addiction and other major psychiatric illnesses.
SUMMARY
Addictions are common heritable disorders with multiple psychiatric and medical comorbidities. Persons with substance abuse are approximately twice as likely to suffer psychiatric morbidities as those without substance abuse, and have an increased risk of using inpatient and emergency facilities. In addition, public mental health clients had a decreased life span of 13 to 30 years; most deaths are caused by heart disease and cancer. Cigarette smoking is therefore the largest modifiable risk factor for morbidity and mortality in the general population and in the psychiatric population.
Alcohol dependence was the first behavioral disorder to have validated genetic findings. The alcohol metabolism gene, ALDH2, was initially found in the early 1980s to decrease the risk of alcohol dependence by increasing the levels of acetaldehyde in the blood, leading to uncomfortable physiologic effects including facial flushing, nausea, and tachycardia. A second alcohol-metabolizing gene, ADH1B, also decreases the risk for alcohol dependence by increasing ethanol oxidation to acetaldehyde. In addition to associations between alcoholism and alcohol-metabolizing genes, a GABA receptor gene, GABRA2, has been associated with alcohol dependence. The mechanism for this association is believed to be due to GABA mediation of behavioral affects of alcohol.
Recently, strong genetic associations with nicotine dependence have been found in the nicotinic receptor subunit CHRNA5. It seems that there are at least 2 distinct biologic mechanisms that alter the risk of nicotine dependence. The first biologic mechanism is caused by an amino acid change in CHRNA5, in the nonsynonymous SNP rs16969968. Functional studies have found that expression of the minor variant leads to reduced response to a nicotinic agonist. The second mechanism altering risk of nicotine dependence is through altered expression of the α5 mRNA. A second group of SNPs in the region, including rs588765 and rs555018, is associated with variable expression of CHRNA5. The combination of altered protein and variable mRNA expression leads to different levels of addiction risk.
Although there have been major advances in understanding the genetics of addictions, the current evidence accounts for only a small proportion of the genetic variance. As new statistical and genetic tools allow a better understanding of the mechanism by which multiple genes lead to psychiatric and medical illness, new discoveries will inform and challenge understanding of these illnesses.
Medical disorders that occur in the context of substance dependence seem to have genetic associations that are not entirely explained by the contribution of the genetic variant to substance use. Specifically, gene variants in ADH were protective for cancers of the upper aerodigestive tract, even after adjusting for alcohol consumption. Similarly, SNPs correlated with rs16969968 are associated with an increased risk of COPD and lung cancer, after adjusting for cigarette use. Although the interpretation of these studies is difficult because of the confounding between the medical illness and substance exposure, there is evidence of a genetic interaction in both cases.
Substance dependence is strongly heritable and heavily comorbid with other medical illnesses. Therefore, the importance of understanding the genetics of substance dependence in the context of its many related medical and psychiatric disorders cannot be overstated.
Acknowledgments
Funding support: Collaborative Genetic Study of Nicotine Dependence P01 CA89392 (National Cancer Institute), Study of Addiction: Genetics and Environment U01 HG004422 (National Human Genome Research Institute), Case Control Candidate Gene Study of Addiction R01 DA19963 (National Institute on Drug Abuse), Human Genetics of Addiction: A Study of Common and Specific Factors K02 DA021237 (National Institute on Drug Abuse), Collaborative Study on the Genetics of Alcoholism U10 AA008401 (National Institute on Alcohol Abuse and Alcoholism).
Footnotes
Disclosures: Dr L Bierut is listed as an inventor on a patent “Markers of Addiction” (US 20070258898), covering the use of certain single nucleotide polymorphisms in determining the diagnosis, prognosis, and treatment of addiction. Dr Bierut acted as a consultant for Pfizer, Inc in 2008.
REFERENCES
- 1.American Psychiatric Association. Task Force on DSM-IV . Diagnostic and statistical manual of mental disorders: DSM-IV-TR. 4th edition American Psychiatric Association; Washington, DC: 2000. [Google Scholar]
- 2.Grant BF, Hasin DS, Chou SP, et al. Nicotine dependence and psychiatric disorders in the United States: results from the national epidemiologic survey on alcohol and related conditions. Arch Gen Psychiatry. 2004;61(11):1107–15. doi: 10.1001/archpsyc.61.11.1107. [DOI] [PubMed] [Google Scholar]
- 3.Regier DA, Farmer ME, Rae DS, et al. Comorbidity of mental disorders with alcohol and other drug abuse. Results from the Epidemiologic Catchment Area (ECA) Study. JAMA. 1990;264(19):2511–8. [PubMed] [Google Scholar]
- 4.Clark RE, Samnaliev M, McGovern MP. Treatment for co-occurring mental and substance use disorders in five state Medicaid programs. Psychiatr Serv. 2007;58(7):942–8. doi: 10.1176/ps.2007.58.7.942. [DOI] [PubMed] [Google Scholar]
- 5.Lasser K, Boyd JW, Woolhandler S, et al. Smoking and mental illness: a population-based prevalence study. JAMA. 2000;284(20):2606–10. doi: 10.1001/jama.284.20.2606. [DOI] [PubMed] [Google Scholar]
- 6.Colton CW, Manderscheid RW. Congruencies in increased mortality rates, years of potential life lost, and causes of death among public mental health clients in eight states. Prev Chronic Dis. 2006;3(2):A42. [PMC free article] [PubMed] [Google Scholar]
- 7.Parks J, Svendsen D, Singer P, et al. Morbidity and mortality in people with serious mental illness. National Association of State Mental Health Program Directors; Alexandria (VA): 2006. [Google Scholar]
- 8.Ostacher MJ, Nierenberg AA, Perlis RH, et al. The relationship between smoking and suicidal behavior, comorbidity, and course of illness in bipolar disorder. J Clin Psychiatry. 2006;67(12):1907–11. doi: 10.4088/jcp.v67n1210. [DOI] [PubMed] [Google Scholar]
- 9.Reich T, Edenberg HJ, Goate A, et al. Genome-wide search for genes affecting the risk for alcohol dependence. Am J Med Genet. 1998;81(3):207–15. [PubMed] [Google Scholar]
- 10.Wolff PH. Ethnic differences in alcohol sensitivity. Science. 1972;175(20):449–50. doi: 10.1126/science.175.4020.449. [DOI] [PubMed] [Google Scholar]
- 11.Goedde HW, Agarwal DP, Harada S. Genetic studies on alcohol-metabolizing enzymes: detection of isozymes in human hair roots. Enzyme. 1980;25(4):281–6. doi: 10.1159/000459265. [DOI] [PubMed] [Google Scholar]
- 12.Harada S, Agarwal DP, Goedde HW, et al. Possible protective role against alcoholism for aldehyde dehydrogenase isozyme deficiency in Japan. Lancet. 1982;2(8302):827. doi: 10.1016/s0140-6736(82)92722-2. [DOI] [PubMed] [Google Scholar]
- 13.Edenberg HJ. The genetics of alcohol metabolism: role of alcohol dehydrogenase and aldehyde dehydrogenase variants. Alcohol Res Health. 2007;30(1):5–13. [PMC free article] [PubMed] [Google Scholar]
- 14.Oota H, Pakstis AJ, Bonne-Tamir B, et al. The evolution and population genetics of the ALDH2 locus: random genetic drift, selection, and low levels of recombination. Ann Hum Genet. 2004;68(Pt 2):93–109. doi: 10.1046/j.1529-8817.2003.00060.x. [DOI] [PubMed] [Google Scholar]
- 15.Chen CC, Lu RB, Chen YC, et al. Interaction between the functional polymorphisms of the alcohol-metabolism genes in protection against alcoholism. Am J Hum Genet. 1999;65(3):795–807. doi: 10.1086/302540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Luczak SE, Glatt SJ, Wall TL. Meta-analyses of ALDH2 and ADH1B with alcohol dependence in Asians. Psychol Bull. 2006;132(4):607–21. doi: 10.1037/0033-2909.132.4.607. [DOI] [PubMed] [Google Scholar]
- 17.Thomasson HR, Edenberg HJ, Crabb DW, et al. Alcohol and aldehyde dehydrogenase genotypes and alcoholism in Chinese men. Am J Hum Genet. 1991;48(4):677–81. [PMC free article] [PubMed] [Google Scholar]
- 18.Higuchi S, Matsushita S, Imazeki H, et al. Aldehyde dehydrogenase genotypes in Japanese alcoholics. Lancet. 1994;343(8899):741–2. doi: 10.1016/s0140-6736(94)91629-2. [DOI] [PubMed] [Google Scholar]
- 19.Whitfield JB. Alcohol dehydrogenase and alcohol dependence: variation in genotype-associated risk between populations. Am J Hum Genet. 2002;71(5):1247–50. doi: 10.1086/344287. author reply: 1250–1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Edenberg HJ, Dick DM, Xuei X, et al. Variations in GABRA2, encoding the alpha 2 subunit of the GABA(A) receptor, are associated with alcohol dependence and with brain oscillations. Am J Hum Genet. 2004;74(4):705–14. doi: 10.1086/383283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Agrawal A, Edenberg HJ, Foroud T, et al. Association of GABRA2 with drug dependence in the collaborative study of the genetics of alcoholism sample. Behav Genet. 2006;36(5):640–50. doi: 10.1007/s10519-006-9069-4. [DOI] [PubMed] [Google Scholar]
- 22.Covault J, Gelernter J, Hesselbrock V, et al. Allelic and haplotypic association of GABRA2 with alcohol dependence. Am J Med Genet B Neuropsychiatr Genet. 2004;129(1):104–9. doi: 10.1002/ajmg.b.30091. [DOI] [PubMed] [Google Scholar]
- 23.Fehr C, Sander T, Tadic A, et al. Confirmation of association of the GABRA2 gene with alcohol dependence by subtype-specific analysis. Psychiatr Genet. 2006;16(1):9–17. doi: 10.1097/01.ypg.0000185027.89816.d9. [DOI] [PubMed] [Google Scholar]
- 24.Soyka M, Preuss UW, Hesselbrock V, et al. GABA-A2 receptor subunit gene (GABRA2) polymorphisms and risk for alcohol dependence. J Psychiatr Res. 2008;42(3):184–91. doi: 10.1016/j.jpsychires.2006.11.006. [DOI] [PubMed] [Google Scholar]
- 25.Glowa JR, Crawley J, Suzdak PD, et al. Ethanol and the GABA receptor complex: studies with the partial inverse benzodiazepine receptor agonist Ro 15-4513. Pharmacol Biochem Behav. 1988;31(3):767–72. doi: 10.1016/0091-3057(88)90263-8. [DOI] [PubMed] [Google Scholar]
- 26.Boyle AE, Segal R, Smith BR, et al. Bidirectional effects of GABAergic agonists and antagonists on maintenance of voluntary ethanol intake in rats. Pharmacol Biochem Behav. 1993;46(1):179–82. doi: 10.1016/0091-3057(93)90338-t. [DOI] [PubMed] [Google Scholar]
- 27.Treutlein J, Cichon S, Ridinger M, et al. Genome-wide association study of alcohol dependence. Arch Gen Psychiatry. 2009;66(7):773–84. doi: 10.1001/archgenpsychiatry.2009.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hashibe M, McKay JD, Curado MP, et al. Multiple ADH genes are associated with upper aerodigestive cancers. Nat Genet. 2008;40(6):707–9. doi: 10.1038/ng.151. [DOI] [PubMed] [Google Scholar]
- 29.Vink JM, Willemsen G, Boomsma DI. Heritability of smoking initiation and nicotine dependence. Behav Genet. 2005;35(4):397–406. doi: 10.1007/s10519-004-1327-8. [DOI] [PubMed] [Google Scholar]
- 30.Bierut LJ, Madden PA, Breslau N, et al. Novel genes identified in a high-density genome wide association study for nicotine dependence. Hum Mol Genet. 2007;16(1):24–35. doi: 10.1093/hmg/ddl441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Saccone SF, Hinrichs AL, Saccone NL, et al. Cholinergic nicotinic receptor genes implicated in a nicotine dependence association study targeting 348 candidate genes with 3713 SNPs. Hum Mol Genet. 2007;16(1):36–49. doi: 10.1093/hmg/ddl438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Saccone N, Wang JC, Breslau N, et al. The CHRNA5-CHRNA3-CHRNB4 nicotinic receptor subunit gene cluster affects risk for nicotine dependence in AfricanAmericans and in European-Americans. Cancer Res. 2009;69(17):6848–56. doi: 10.1158/0008-5472.CAN-09-0786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Spitz MR, Amos CI, Dong Q, et al. The CHRNA5-A3 region on chromosome 15q24-25.1 is a risk factor both for nicotine dependence and for lung cancer. J Natl Cancer Inst. 2008;100(21):1552–6. doi: 10.1093/jnci/djn363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thorgeirsson TE, Geller F, Sulem P, et al. A variant associated with nicotine dependence, lung cancer and peripheral arterial disease. Nature. 2008;452(7187):638–42. doi: 10.1038/nature06846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Weiss RB, Baker TB, Cannon DS, et al. A candidate gene approach identifies the CHRNA5-A3-B4 region as a risk factor for age-dependent nicotine addiction. PLoS Genet. 2008;4(7):e1000125. doi: 10.1371/journal.pgen.1000125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Berrettini W, Yuan X, Tozzi F, et al. Alpha-5/alpha-3 nicotinic receptor subunit alleles increase risk for heavy smoking. Mol Psychiatry. 2008;13(4):368–73. doi: 10.1038/sj.mp.4002154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bierut LJ, Stitzel JA, Wang JC, et al. Variants in nicotinic receptors and risk for nicotine dependence. Am J Psychiatry. 2008;165(9):1163–71. doi: 10.1176/appi.ajp.2008.07111711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang JC, Cruchaga C, Saccone N, et al. Risk for nicotine dependence and lung cancer is conferred by mRNA expression levels and amino acid change in CHRNA5. Hum Mol Genet. 2009;18(16):3125–35. doi: 10.1093/hmg/ddp231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ehringer MA, Clegg HV, Collins AC, et al. Association of the neuronal nicotinic receptor beta2 subunit gene (CHRNB2) with subjective responses to alcohol and nicotine. Am J Med Genet B Neuropsychiatr Genet. 2007;144(5):596–604. doi: 10.1002/ajmg.b.30464. [DOI] [PubMed] [Google Scholar]
- 40.Ehringer MA, McQueen MB, Hoft NR, et al. Association of CHRN genes with “dizziness” to tobacco. Am J Med Genet B Neuropsychiatr Genet. 2010;153B(2):600–9. doi: 10.1002/ajmg.b.31027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sherva R, Wilhelmsen K, Pomerleau CS, et al. Association of a single nucleotide polymorphism in neuronal acetylcholine receptor subunit alpha 5 (CHRNA5) with smoking status and with ‘pleasurable buzz’ during early experimentation with smoking. Addiction. 2008;103(9):1544–52. doi: 10.1111/j.1360-0443.2008.02279.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Breitling LP, Dahmen N, Mittelstrass K, et al. Smoking cessation and variations in nicotinic acetylcholine receptor subunits alpha-5, alpha-3, and beta-4 genes. Biol Psychiatry. 2009;65(8):691–5. doi: 10.1016/j.biopsych.2008.10.004. [DOI] [PubMed] [Google Scholar]
- 43.Baker TB, Weiss RB, Bolt D, et al. Human neuronal acetylcholine receptor A5-A3-B4 haplotypes are associated with multiple nicotine dependence phenotypes. Nicotine Tob Res. 2009;11(7):785–96. doi: 10.1093/ntr/ntp064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Freathy RM, Ring SM, Shields B, et al. A common genetic variant in the 15q24 nicotinic acetylcholine receptor gene cluster (CHRNA5-CHRNA3-CHRNB4) is associated with a reduced ability of women to quit smoking in pregnancy. Hum Mol Genet. 2009;18(15):2922–7. doi: 10.1093/hmg/ddp216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hung RJ, McKay JD, Gaborieau V, et al. A susceptibility locus for lung cancer maps to nicotinic acetylcholine receptor subunit genes on 15q25. Nature. 2008;452(7187):633–7. doi: 10.1038/nature06885. [DOI] [PubMed] [Google Scholar]
- 46.Amos CI, Wu X, Broderick P, et al. Genome-wide association scan of tag SNPs identifies a susceptibility locus for lung cancer at 15q25.1. Nat Genet. 2008;40(5):616–22. doi: 10.1038/ng.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pillai SG, Ge D, Zhu G, et al. A genome-wide association study in chronic obstructive pulmonary disease (COPD): identification of two major susceptibility loci. PLoS Genet. 2009;5(3):e1000421. doi: 10.1371/journal.pgen.1000421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Le Marchand L, Derby KS, Murphy SE, et al. Smokers with the CHRNA lung cancer-associated variants are exposed to higher levels of nicotine equivalents and a carcinogenic tobacco-specific nitrosamine. Cancer Res. 2008;68(22):9137–40. doi: 10.1158/0008-5472.CAN-08-2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Substance Abuse and Mental Health Services Administration, Office of Applied Studies . Results from the 2007 National Survey on Drug Use and Health: National Findings. Rockville, MD: [Accessed October 16, 2009]. 2008. NSDUH Series H-34, DHHS Publication No. SMA 08-4343. Available at: http://www.oas.samhsa.gov/NSDUH/2k7NSDUH/2k7results.cfm. [Google Scholar]
- 50.Bierut LJ, Dinwiddie SH, Begleiter H, et al. Familial transmission of substance dependence: alcohol, marijuana, cocaine, and habitual smoking: a report from the Collaborative Study on the Genetics of Alcoholism. Arch Gen Psychiatry. 1998;55(11):982–8. doi: 10.1001/archpsyc.55.11.982. [DOI] [PubMed] [Google Scholar]
- 51.Grucza RA, Wang JC, Stitzel JA, et al. A risk allele for nicotine dependence in CHRNA5 is a protective allele for cocaine dependence. Biol Psychiatry. 2008;64(11):922–9. doi: 10.1016/j.biopsych.2008.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang JC, Grucza R, Cruchaga C, et al. Genetic variation in the CHRNA5 gene affects mRNA levels and is associated with risk for alcohol dependence. Mol Psychiatry. 2009;14(5):501–10. doi: 10.1038/mp.2008.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Klungsoyr O, Nygard JF, Sorensen T, et al. Cigarette smoking and incidence of first depressive episode: an 11-year, population-based follow-up study. Am J Epidemiol. 2006;163(5):421–32. doi: 10.1093/aje/kwj058. [DOI] [PubMed] [Google Scholar]
- 54.Breslau N, Peterson EL, Schultz LR, et al. Major depression and stages of smoking. A longitudinal investigation. Arch Gen Psychiatry. 1998;55(2):161–6. doi: 10.1001/archpsyc.55.2.161. [DOI] [PubMed] [Google Scholar]
- 55.Brown RA, Lewinsohn PM, Seeley JR, et al. Cigarette smoking, major depression, and other psychiatric disorders among adolescents. J Am Acad Child Adolesc Psychiatry. 1996;35(12):1602–10. doi: 10.1097/00004583-199612000-00011. [DOI] [PubMed] [Google Scholar]
- 56.Kendler KS, Neale MC, MacLean CJ, et al. Smoking and major depression. A causal analysis. Arch Gen Psychiatry. 1993;50(1):36–43. doi: 10.1001/archpsyc.1993.01820130038007. [DOI] [PubMed] [Google Scholar]
- 57.Goodman E, Capitman J. Depressive symptoms and cigarette smoking among teens. Pediatrics. 2000;106(4):748–55. doi: 10.1542/peds.106.4.748. [DOI] [PubMed] [Google Scholar]
- 58.Kandel DB, Davies M, Karus D, et al. The consequences in young adulthood of adolescent drug involvement. An overview. Arch Gen Psychiatry. 1986;43(8):746–54. doi: 10.1001/archpsyc.1986.01800080032005. [DOI] [PubMed] [Google Scholar]
- 59.Wu LT, Anthony JC. Tobacco smoking and depressed mood in late childhood and early adolescence. Am J Public Health. 1999;89(12):1837–40. doi: 10.2105/ajph.89.12.1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Breslau N, Novak SP, Kessler RC. Daily smoking and the subsequent onset of psychiatric disorders. Psychol Med. 2004;34(2):323–33. doi: 10.1017/s0033291703008869. [DOI] [PubMed] [Google Scholar]
- 61.Lichtenstein P, Yip BH, Bjork C, et al. Common genetic determinants of schizophrenia and bipolar disorder in Swedish families: a population-based study. Lancet. 2009;373(9659):234–9. doi: 10.1016/S0140-6736(09)60072-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Edvardsen J, Torgersen S, Roysamb E, et al. Unipolar depressive disorders have a common genotype. J Affect Disord. 2009;117(1–2):30–41. doi: 10.1016/j.jad.2008.12.004. [DOI] [PubMed] [Google Scholar]
- 63.Johnson C, Drgon T, McMahon FJ, et al. Convergent genome wide association results for bipolar disorder and substance dependence. Am J Med Genet B Neuropsychiatr Genet. 2009;150(2):182–90. doi: 10.1002/ajmg.b.30900. [DOI] [PubMed] [Google Scholar]