Abstract
The CREB binding protein (CBP) is a human transcriptional co-activator consisting of several conserved functional modules, which interacts with distinct transcription factors including nuclear receptors, CREB, and STAT proteins. Despite the importance of CBP in transcriptional regulation, many questions regarding the role of its particular domains in CBP functions remain unanswered. Therefore, developing small molecules capable of selectively modulating a single domain of CBP is of invaluable aid at unraveling its prominent activities. Here we report the design, synthesis and biological evaluation of conformationally restricted peptides as novel modulators for the acetyl-lysine binding bromodomain (BRD) of CBP. Utilizing a target structure–guided and computer-aided rational design approach, we developed a series of cyclic peptides with affinity for CBP BRD significantly greater than those of its biological ligands, including lysine-acetylated histones and tumor suppressor p53. The best cyclopeptide of the series exhibited a Kd of 8.0 µM, representing a 24-fold improvement in affinity over that of the linear lysine 382-acetylated p53 peptide. This lead peptide is highly selective for CBP BRD over BRDs from other transcriptional proteins. Cell-based functional assays carried out in colorectal carcinoma HCT116 cells further demonstrated the efficacy of this compound to modulate p53 stability and function in response to DNA damage. Our results strongly argue that these CBP modulators can effectively inhibit p53 transcriptional activity by blocking p53K382ac binding to CBP BRD and promoting p53 instability by changes of its post-translational modification states, a different mechanism to that of the p53 inhibitors reported to date.
Human transcriptional co-activator CREB binding protein (CBP) functions to physically bridge many DNA-binding transcription factors to the basal transcription machinery1. Despite its importance as a master nuclear integrator of transcriptional responses, many questions about CBP functions and regulation remain unanswered2,3. Thus, chemical probes able to modulate its specific domains are of great interest as such molecules would constitute powerful tools to systematically elucidate the biological functions of CBP with respect to endogenous proteins in cells.
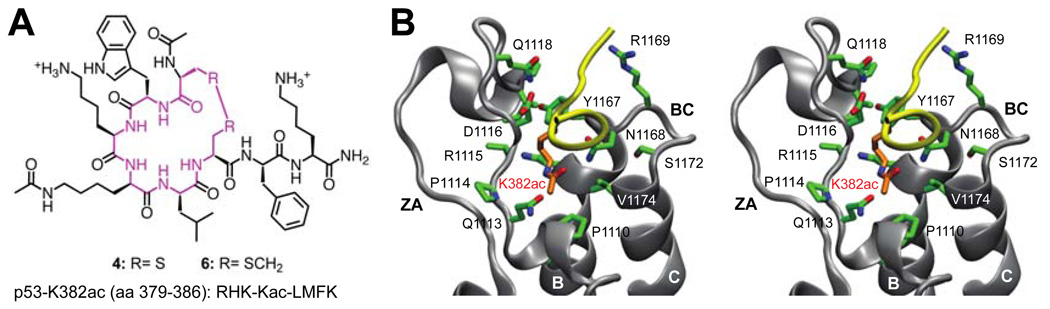
Recent studies show that upon DNA damage CBP is recruited by the tumor suppressor p53 to modify chromatin and aid transcription activation of p53 target genes. This co-activator recruitment process is facilitated by the bromodomain (BRD) of CBP binding to p53 at the C-terminal acetylated lysine 382 (K382ac)4,5. The molecular basis of this CBP BRD/p53 recognition was defined by the three-dimensional solution structure of CBP BRD bound to a lysine 382-acetylated p53 peptide (p53-K382ac)5. Using this complex structure and following a target-structure guided design, we have identified two cyclic peptides that selectively inhibit CBP’s acetylated p53 binding activity in cells under stress conditions. These cyclopeptide ligands represent the most potent CBP BRD chemical ligands reported to date (Fig. 1A).
Figure 1.
A. Structure of cyclopeptides developed in this study, and a linear p53-K382ac peptide. B. Stereoview of the representative structure of the CBP BRD/p53-K382ac complex for the 50 ns MD simulations
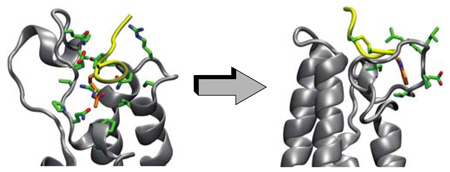
Our rational ligand design began with performing molecular dynamics (MD) simulations on the NMR structure of the CBP BRD bound to p53-K382ac (PDB id: 1JSP). In this complex the p53-K382ac peptide lies across a pocket formed between the ZA and BC loops in the CBP BRD and adopts a β-turn-like conformation with the K382ac being at the beginning of the turn5 (Fig. 1B). This β turn-like conformation of the p53 peptide is a distinctive feature of the CBP BRD/p53-K382ac recognition as compared to other BRD structures and likely plays a pivotal role in ligand specificity and affinity4,5.
The MD simulations showed that the two ends of the octapeptide were considerably more flexible than K382ac and its flanking residues, which are anchored in the binding pocket of the CBP BRD (Suppl. Fig. 1A). We computed the distance distributions curves between Cβ atoms of residues R379 and H380 on the N-terminal side of K382ac and the residues on the C-terminal side of the β-turn (L383, M384 and F385, Suppl. Fig. 1B). The results suggested that the β turn-like conformation could be stabilized by cyclizing the linear peptide through residues M384 and either R379 or H380, and by means of a linker two or three atoms long. Since none of these residues showed important contributions to the binding energy, we reasoned that they could be replaced by cysteines that could then anchor cyclization of the peptide.
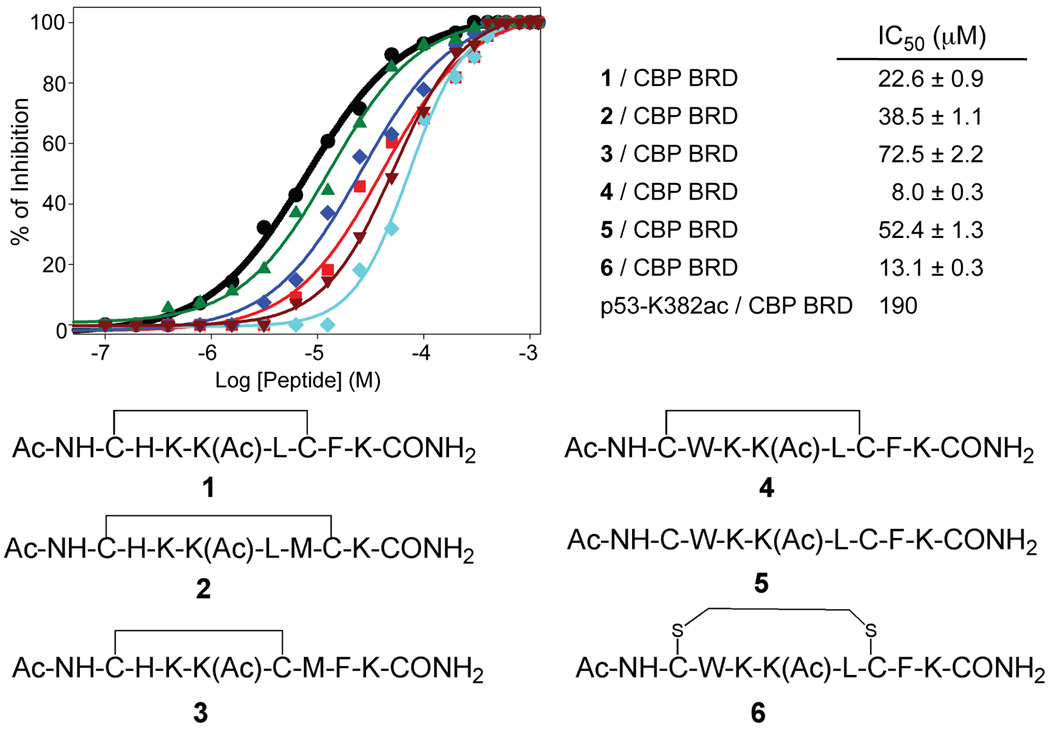
The synthesis of these two cyclic peptides was carried out on solid phase (Suppl. Scheme S1). To explore the molecular determinants of CBP BRD/p53-K382ac binding as well as to validate our binding model, we prepared another 4 cyclopeptides, which resulted from the combination of linking through either R379 or H380 and F385 or L383. The affinities of the six cyclic peptides (a–f) for CBP BRD were next evaluated in vitro using a competition fluorescent polarization (FP) assay. The cyclization of p53-K382ac between positions H380 and F385, M384 or L383 (d–f), resulted in a major loss of the p53 binding indicating that H380 is essential for the CBP BRD/p53 interaction (Suppl. Table 1). This result is in agreement with our previous study using mutant p53 peptides5. Conversely, incorporation of conformational constrains through R379 and F385, M384 or L383 led to a 5-, 9- or 2-fold increase (b, a, c) in binding affinity, respectively, over that of the linear p53-K382ac peptide (Fig. 2). The best peptide (a or 1) exhibited an IC50 of 22.6 µM. This result is fully aligned with the predictions made in our molecular modeling, thereby validating our model.
Figure 2.
Structure and binding affinity to CBP BRD of the synthesized peptide ligands.
The molecular determinants of the CBP BRD/1 interaction were further studied by MD simulation (Suppl. Material). In distinction from the CBP BRD/p53-K382ac binding6, in this cyclic octapeptide, H380 was found to be sitting in a different site of the acetyl-lysine-binding pocket interacting with key residues such as Tyr1167, Asn1168 and Arg1169 in the BC loop of CBP BRD (Suppl. Figure 1D). As such, we envisioned that replacing H380 by a tryptophan could enhance peptide binding. This hypothesis was supported by further MD simulations studies of the CBP BRD/4 complex (Suppl. Material).
We then synthesized the cyclic H380W peptide 4 and determined its binding affinity using the same FP competition assay described above. As predicted by our molecular modeling studies, this peptide showed improved affinity over the histidine-substituted one (1), with an IC50 of 8.0 ± 0.3 µM, thus proving to be the most active peptide of the series (Fig. 2). This result represents a 24-fold improvement in affinity as compared to that of the linear p53-K382ac peptide. We next studied the contribution of the conformational effect on the affinity of 4. To this end, we measured the binding to CBP BRD of its linear analog (5), using the FP competition assay. Indeed, the weaker IC50 obtained for this linear peptide confirmed that the cyclization does make an important contribution to the binding affinity with a free energy change of −1.1 kcal/mol (ΔΔG = ΔGcyclic−ΔGlinear) (Fig. 2). Therefore, the overall improvement observed in the affinity of this cyclopeptide is the result of two combined effects: the modification made to the peptide sequence, and the stabilization of the bioactive conformation by means of the cyclization.
To understand the molecular basis of CBP BRD/4 recognition, we performed NMR titration of the BRD binding to both 4 and the linear p53-K382ac peptide. Comparison of 2D 1H-15N HSQC (heteronuclear single quantum coherence) spectra of CPB BRD with and without a peptide (molar ratio of 1:3) revealed that binding of the cyclized peptide 4 led to more extended chemical shift changes of protein NMR resonances than that of the linear p53 peptide5, thus confirming the stronger nature of the former interaction (Suppl. Fig. 2). Overall, a similar set of protein residues exhibited the major perturbations upon peptide binding, confirming the similarity of the binding mode of the two peptides.
Next, we studied the specificity of the CBP-BRD/4 association (most potent cyclopeptide) over other BRDs by FP, using a fluorescein-labeled analog of it (Fl-4), which was synthesized as described in the Suppl. material. The binding affinity of this fluorescent cyclopeptide for CBP-BRD, as determined in a direct in vitro FP assay (Suppl. Fig. 3A), was almost identical to the IC50 value previously calculated for 4. This result validates our in vitro competition FP data. The selectivity of this peptide was assessed against BRDs from co-activator PCAF (p300-CBP associated factor) and a BET family protein BRD4 (BRD-containing protein 4). Neither PCAF BRD nor the BRD4 BRDs showed significant interactions with the cyclopeptide Fl-4, thus demonstrating the specificity of the CBP BRD/4 association (Suppl. Fig. 3A).
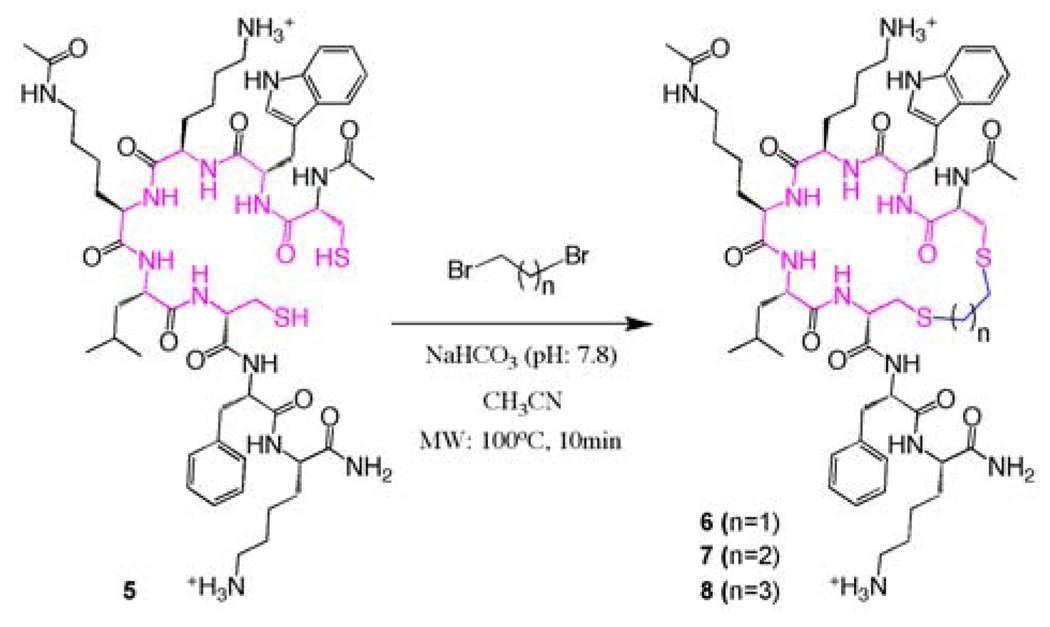
While cyclic peptides are considerably more stable than their linear counterparts, disulfide bridges could be reduced inside cells. Since our goal was to test this cyclic peptide in a functional assay, we next focused our efforts in preparing more stable cyclic analogs of 4. Specifically, we reasoned that cyclization through a thioether-like linker would provide improved stability while retaining the binding properties of the parent cyclic peptide. Thus, we synthesized bis-thioether linked analogs of 4 by using a previously reported chemoselective strategy6 with a modification. This procedure involved treating linear unprotected dicysteine-containing peptides with polybromobenzyl derivatives in CH3CN/NH4HCO3 solution at pH 7.8. In this study, we used instead, dibromoalkyl analogs as alkylating agents. At first, the previously reported methodology did not produce the expected cyclic peptides, and very little reaction was observed after 24 hours stirring at room temperature. However, microwave irradiation at 100°C for 10 min afforded the desired cyclic bis-thioether peptides in almost quantitative yields (Scheme 1).
Scheme 1.
Cyclization to bis-thioether linked peptides. [NaHCO3 (20 mM, pH = 7.8) / CH3CN (7:1)].
The affinity of the bis-thioether cyclic peptides 6–8 for CBP BRD was measured in the FP competition assay, of which 7 and 8 showed weak affinities (IC50 > 90 µM), whereas 6 was almost as potent as the parent peptide 4 (Fig. 2). These results further highlight the importance of the conformational effect on the binding of these cyclopeptides to CBP BRD.
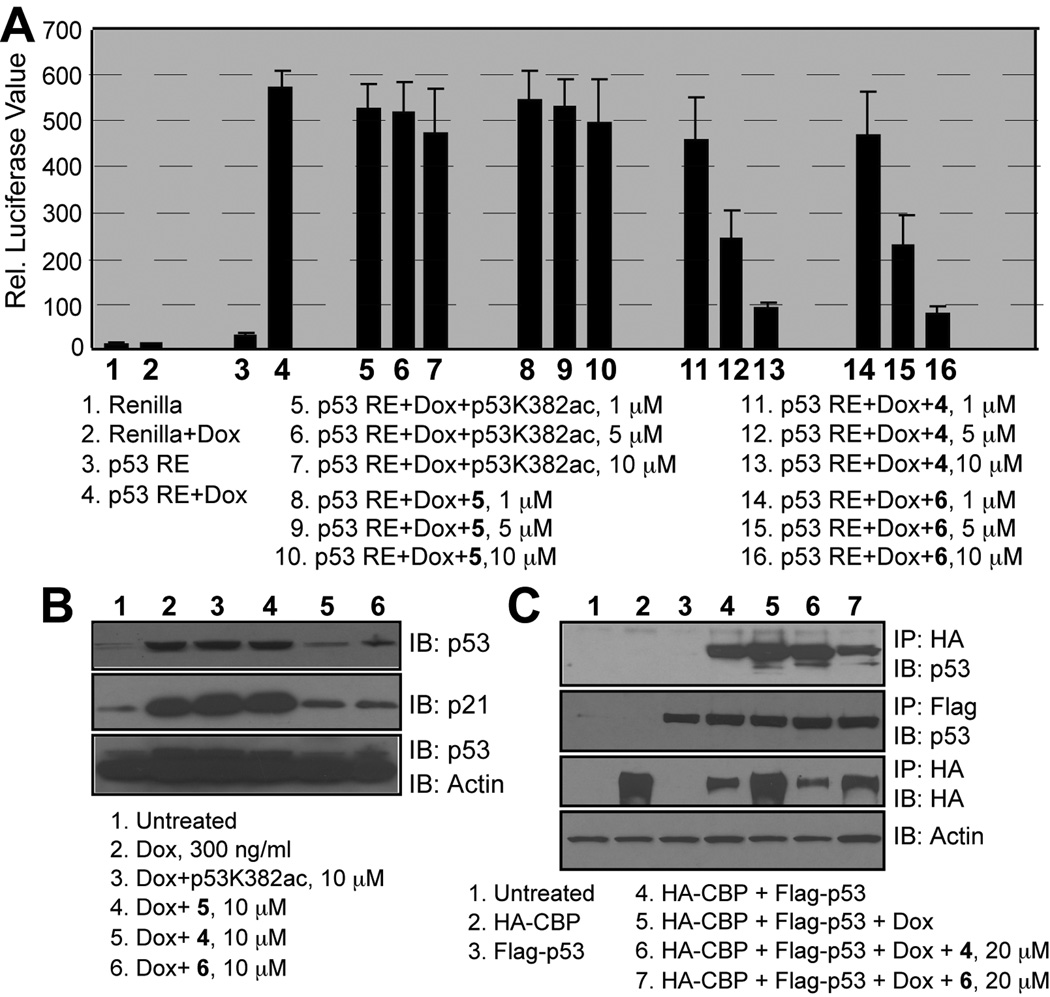
Finally, we characterized the lead peptide’s ability to modulate p53 functions in cells, including p53- induced p21 activation, p53 stability, and p53 association with CBP upon stimulation. For p53-based p21-luciferase assay, colorectal carcinoma HCT116 cells were co-transfected with p21 promoter in tandem with luciferase and renilla luciferase plasmids. Doxorubicin treatment of the HCT116 cells caused activation of p21 reflected by luciferase activity (Fig. 3A). This p53-induced p21 activation showed a dose-dependent inhibition by the cyclic peptides 4 and 6, but not by the linear peptides. We next investigated effects of these peptides on the level of endogenous p53 and p21 proteins in HCT116 cells. The HCT116 cells were incubated with the peptide ligands for 12 hours prior to the treatment with doxorubicin (300 ng/ml) for next 24 hrs. The western blots showed that whereas the linear peptides were almost inactive, the cyclic peptides 4 and 6 effectively reduced the protein level of endogenous p53 and p21 (Fig. 3B). These results indicated that inhibition of lysine-acetylated p53 association with CBP results in a decreased stability of p53, likely due to deacetylation and ubiquitination-mediated protein degradation, leading to a reduced activation of p21. We then assessed how these peptides inhibit p53/CBP interaction by over-expressing HA-CBP and Flag-p53 in human embryonic kidney (HEK) 293T cells. The HA-CBP pull-down and western blot analyses revealed that consistent with its enhanced stability of cyclization, 6 showed much better ability in inhibiting doxorubicin-induced p53/CBP binding than that of 4 (Fig. 3C). The apparently less profound inhibitory effects of 6 or 4 in this experiment than those seen in the p53 luciferase study could be due to differences in the peptide cell permeability with different cell lines, and also readout sensitivity of the two assays.
Figure 3.
A. Inhibition of p53-induced p21 luciferase expression by the peptide ligands; B. Effects of the p53 peptides on endogenous p53 and p21 in HCT116 cells after doxorubicin treatment. C. Inhibition of overexpressed HA-CBP and Flag-p53 interactions in HEK293T cells by the peptides, 4 and 6.
The mechanism of action of our peptidic ligands in p53 inhibition differs from that of the p53 inhibitors known to date that may act by blocking mitochondrial trans-localization of p53 in transcription-dependent or independent pathways8,9. Because excess of p53 activity has been reportedly linked to diseases such as neurodegenerative disorders (Alzheimer’s disease, Parkinson’s disease and multiple sclerosis)9,10, infectious9 and autoimmune diseases9, chemical ligands capable of temporal and selective inhibition of p53 are of great interest. Further, recent studies have catapulted p53 at the center of stem cell biology showing that disruption of p53/p21 pathway enhances efficiency of reprogramming of somatic cells into induced pluripotent stem (iPS) cells by over-expression of transcription factors Oct4, Sox2, Klf4 and c-Myc11–17. In these studies, p53 inhibition was achieved by using siRNA gene knockdown approach. The p53 inhibitors would be an attractive technology for facilitating iPS production through inhibiting endogenous p53 in a temporal and revisable manner. Therefore our study opens doors for future investigations that examine the utility of this new class of p53 inhibitors in disease models in which a temporal down-regulation of p53 may be beneficial, as well as for studies of the role of p53 acetylation in transcriptional activity in different biological systems.
Supplementary Material
Acknowledgments
This work was supported by the NCI’s Research Supplements to Promote Diversity in Health-Related Research Program (G.G.-N.) and grants from the National Institutes of Health (M.-M.Z. and S.M.).
Footnotes
Supporting Information Available: Schemes S1 to S3, Figures S1 to S4, Table S1, experimental procedures for the peptide synthesis, computational protocols, binding and cell-based assays. This material is available free of charge via the internet at http://pubs.acs.org.
References
- 1.Kwok RP, Lundblad JR, Chrivia JC, Richards JP, Bachinger HP, Brennan RG, Roberts SG, Green MR, Goodman RH. Nature. 1994;370(6486):177–178. doi: 10.1038/370223a0. [DOI] [PubMed] [Google Scholar]
- 2.Goodman RH, Smolik. S. Genes Dev. 2000;14:1553–1577. [PubMed] [Google Scholar]
- 3.Chan HM, Thangue NB. J. Cell Sci. 2001;114:2363–2373. doi: 10.1242/jcs.114.13.2363. [DOI] [PubMed] [Google Scholar]
- 4.Zeng L, Zhang Q, Gerona-Navarro G, Moshkina N, Zhou MM. Structure. 2008;16:643–652. doi: 10.1016/j.str.2008.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mujtaba S, He Y, Zeng L, Yan S, Plotnikova O, Sachchidanand, Sanchez R, Zeleznik-Le NJ, Ronai Z, Zhou MM. Mol Cell. 2004;13:251–263. doi: 10.1016/s1097-2765(03)00528-8. [DOI] [PubMed] [Google Scholar]
- 6.Timmerman P, Beld J, Puijk WC, Meloen RH. ChemBioChem. 2005;6:821–824. doi: 10.1002/cbic.200400374. [DOI] [PubMed] [Google Scholar]
- 7.Sachchidanand, Resnick-Silverman L, Yan S, Mutjaba S, Liu WJ, Zeng L, Manfredi JJ, Zhou MM. Chemistry & Biology. 2006;13:81–90. doi: 10.1016/j.chembiol.2005.10.014. [DOI] [PubMed] [Google Scholar]
- 8.Strom E, Sathe S, Komarov PG, Chernova OB, Pavlovska I, Shyshynova I, Bosykh DA, Burdelya LG, Macklis RM, Skaliter R, Komarova EA, Gudkov AV. Nat. Chem. Biol. 2006;2(9):474–479. doi: 10.1038/nchembio809. [DOI] [PubMed] [Google Scholar]
- 9.Nayak SK, Panesar PS, Kumar H. Curr Med Chem. 2009;16(21):2627–2640. doi: 10.2174/092986709788681976. [DOI] [PubMed] [Google Scholar]
- 10.Li J, Ghiani C, Kim JY, Liu A, Sandoval J, DeVellis J, Casaccia-Bonnefil P. J Neurosci. 2008;28:6118–6127. doi: 10.1523/JNEUROSCI.0184-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Banito A, Rashid ST, Acosta JC, Li S, Pereira CF, Geti I, Pinho S, Silva JC, Azuara V, Walsh M, Vallier L, Gil J. Genes Dev. 2009;23:2134–2139. doi: 10.1101/gad.1811609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hanna J, Saha K, Pando B, van Zon J, Lengner CJ, Creyghton MP, van Oudenaarden A, Jaenisch R. Nature. 2009;462:595–601. doi: 10.1038/nature08592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hong H, Takahashi K, Ichisaka T, Aoi T, Kanagawa O, Nakagawa M, Okita K, Yamanaka S. Nature. 2009;460:1132–1135. doi: 10.1038/nature08235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kawamura T, Suzuki J, Wang YV, Menendez S, Morera LB, Raya A, Wahl GM, Belmonte JC. Nature. 2009;460:1140–1144. doi: 10.1038/nature08311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li H, Collado M, Villasante A, Strati K, Ortega S, Canamero M, Blasco MA, Serrano M. Nature. 2009;460:1136–1139. doi: 10.1038/nature08290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marion RM, Strati K, Li H, Murga M, Blanco R, Ortega S, Fernandez-Capetillo O, Serrano M, Blasco MA. Nature. 2009;460:1149–1153. doi: 10.1038/nature08287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Utikal J, Polo JM, Stadtfeld M, Maherali N, Kulalert W, Walsh RM, Khalil A, Rheinwald JG, Hochedlinger K. Nature. 2009;460:1145–1148. doi: 10.1038/nature08285. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.