Abstract
Genetic individuality is the foundation of personalized medicine, yet its determinants are currently poorly understood. One issue is the difference between monozygotic twins that are assumed identical and have been extensively used in genetic studies for decades [1]. Here, we report genome-wide alterations in two nuclear families each with a pair of monozygotic twins discordant for schizophrenia evaluated by the Affymetrix 6.0 human SNP array. The data analysis includes characterization of copy number variations (CNVs) and single nucleotide polymorphism (SNPs). The results have identified genomic differences between twin pairs and a set of new provisional schizophrenia genes. Samples were found to have between 35 and 65 CNVs per individual. The majority of CNVs (∼80%) represented gains. In addition, ∼10% of the CNVs were de novo (not present in parents), of these, 30% arose during parental meiosis and 70% arose during developmental mitosis. We also observed SNPs in the twins that were absent from both parents. These constituted 0.12% of all SNPs seen in the twins. In 65% of cases these SNPs arose during meiosis compared to 35% during mitosis. The developmental mitotic origin of most CNVs that may lead to MZ twin discordance may also cause tissue differences within individuals during a single pregnancy and generate a high frequency of mosaics in the population. The results argue for enduring genome-wide changes during cellular transmission, often ignored in most genetic analyses.
Introduction
Genome-wide constancy and change underlies evolution and familial inheritance but remains ill-defined. An assessment of changes as the genome is passed on from one generation (meiosis) and developmental cycle (mitosis) to the next is needed. It directly contributes to the sum of genetic individuality. At present, these inquiries are difficult [2], and require the development of new quantitative methods to assess genome-wide changes and their significance. This report assesses two common measures of genomic variation: copy number variations and (CNVs) and single nucleotide polymorphism (SNPs) across a generation and between monozygotic twins in two exceptional families. The results offer novel insight into meiotic and mitotic sources of variation, which results in genetic individuality between MZ twins. This individuality may account for discordance in monozygotic twins for a variety of diseases including schizophrenia.
CNVs are structural variants that are both frequent and relevant and may range in size in humans from 1 Kb to several Mb [3]. Given their impact on physiology and function, CNVs have a major influence on evolution and gene expression and on normal and disease related variation [3]. CNVs include duplications and deletions leading to a departure from the classic view that all autosomal genes are present in two copies, with one allele inherited from each parent. The majority of CNVs are copy number polymorphisms (CNPs), existing in a frequency that is greater than 1% and transmitted across generations. However, a small proportion of CNVs are novel events. CNVs may account for a major fraction (∼12%) of the genome, but appear to concentrate in some genomic regions depending on the sequence features [4], [5]. Unlike CNVs, SNPs are relatively small changes, usually involving replacement of a nucleotide with another. SNPs are common and distributed across the entire human genome. Individual SNPs mark a unique genomic location, and are usually neutral in nature. In other cases, they may change amino-acids, cause protein truncation or affect expression. They are easily detected, and have been extensively exploited in genetic analysis including the cloning of disease causing genes, individual identification and establishment of genetic relatedness.
Studies on these two genome-wide variations (SNPs and CNVs) have greatly enhanced our understanding of evolution and genetic individuality. They are also helping to elucidate the cause of genetic, and genomic disorders including schizophrenia [6]. A number of SNPs appear to be linked to this complex neuro-developmental disease, which has a heritability estimate of 80%. However, results of linkage studies have not been consistently reproducible [7], [8]. Individuals affected with schizophrenia (SCZ) have shown an elevated incidence of CNVs [9] and a few rare CNVs appear to have a major effect on the development of SCZ [10]. However, these CNVs account for only a small fraction of schizophrenia cases [11] and the challenge of identifying common genetic cause(s) of SCZ remains. The search for genes in SCZ currently relies on large number of patients and matched controls. The limited progress using these approaches emphasizes the need to pursue alternative approaches. Future studies may benefit from inclusion of two features. The first is a genome-wide comparison of the parents and their progeny affected by SCZ and the second is the assessment of genomes of monozygotic twins (that show ∼52% discordance for SCZ) [12], [13]. The current study reports genome-wide CNV and SNP results on two exceptional families that include monozygotic twins discordant for schizophrenia (Figure 1, Table 1).
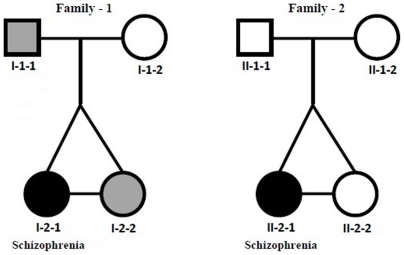
Figure 1. Pedigree of two families with monozygotic twins discordant for schizophrenia.
Members of the family one are indicated with (I-) and members of the family two are indicated with (II-). The designations included in this figure are followed in subsequent figures and tables.
Table 1. Demography and Clinical History.
| Family 1 | Family 2 | |||||||
| I-1-1 | I-1-2 | I-2-1 | I-2-2 | II-1-1 | II-1-2 | II-2-1 | II-2-2 | |
| Age (yr.) at assessment | 82 | 74 | 53 | 53 | N/A | N/A | 43 | 43 |
| Sex | Male | Female | Female | Female | Male | Female | Female | Female |
| Declared Race | Afro-American | Caucasian | ||||||
| Psychiatric features | Compulsive Personality Disorder | N/A | Schizophrenia, Paranoid Type, onset age 22 | Bipolar I Disorder, onset age 52 | Major depression and panic disorder for 6 months after cardiac surgery, onset age 73 | N/A | Schizoaffective Disorder, onset age 27 | Single episode of Major Depression, fully remitted, onset age 18 |
Demography and Clinical History of monozygotic (MZ) twins discordant for Schizophrenia (SCZ). Family one is indicated with (I), family two is indicated with (II). N/A = Not Applicable.
Results and Discussion
Familial Distribution of CNVs
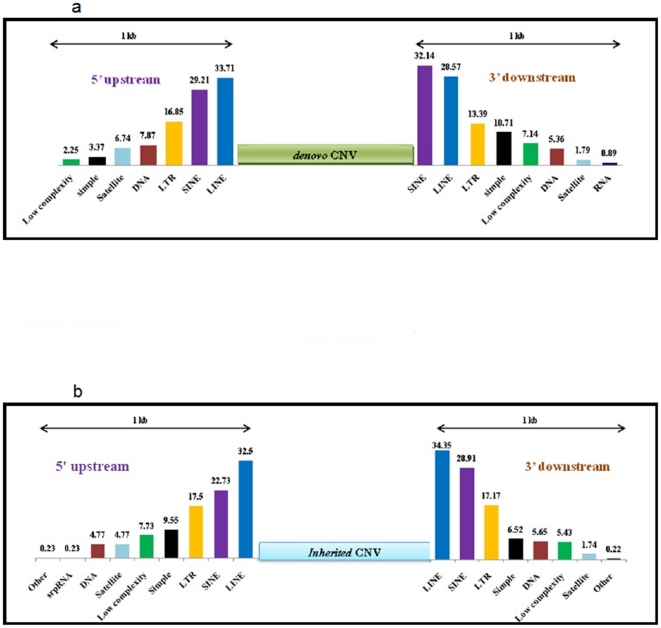
The number of CNVs per individual ranged from 35 to 65, with the exception of one individual who is described more fully later (Table 2). This is similar to the number of CNVs per subject reported from most other studies that have used Affymetrix 6.0 Human SNP arrays [14]. The range is also comparable with the number of CNVs found in Venter's genome (62) based on his complete genome sequence [15]. The exception in our study was the father in family 2 (II-1-1) who was found to harbour a rare chromosome 13q deletion containing 40 CNVs at a single genomic location. Although this finding is beyond the scope of this report, it is important to note that II-1-1 underwent chemotherapy treatment and that the samples utilized in this study were obtained towards the end of that treatment. Most CNVs identified were in the range of 100 to 200 Kb, consistent with the size distribution of CNVs reported in the literature [14]. The majority of CNVs observed (Table 3) were copy number gains (78.5%) and ∼10% of the CNVs identified are not listed in the Database of Genomic Variants (http://projects.tcag.ca/variation/) accessed on 8.2.2010. Further, the chromosomal distribution of CNVs was comparable across individuals with the exception of the father in family 2 who had consistently higher CNVs affecting most chromosomes (Table 4). Of the CNVs identified, >50 per cent overlapped RefSeq genes. The identified genes are frequently associated with metabolic pathways such as starch and sucrose metabolism as well as pathways involved in the metabolism of amino acids, for example, , phenylalanine, histidine and tyrosine (AMY2A,AMY1A,ALDH1L1,PSMC1). Structurally, >67% of the CNVs identified were flanked at both the 5′and 3′ end or at just the 5′ (>7%) or 3′ (>8%) end with a set of common repeats, represented by short interspersed nucleotide elements (SINEs), long interspersed nucleotide elements (LINEs), long terminal repeats (LTRs) and low copy repeats (LCRs) near the breakpoints. The majority of the deletion breakpoints had 1–30 bp of microhomology, whereas a small fraction of deletion breakpoints contained inserted sequences. The co-occurrence of microhomology and inserted sequence suggests that both recombination and replication based mutational mechanisms are operational in CNV generation. Recent studies have identified short DNA motifs that both determine the location of meiotic crossover hotspots and are significantly enriched at the breakpoints of recurrent non-allelic homologous recombination (NAHR) syndromes [16]. We found evidence for this mechanism in a subset of the breakpoint events (data not shown). This was true for the de novo (Figure 2a) as well as inherited (Figure 2b) CNVs. Such sequences may represent genomic architecture that is prone to genome instability by a predisposition to genomic rearrangements via non-homologous end joining (NHEJ), template switching and/or non-allelic homologous recombination (NAHR).
Table 2. Distribution of CNV among family members according to size.
| CNV Size | Family 1 | Family 2 | ||||||
| I-1-1 | I-1-2 | I-2-1 | I-2-2 | II-1-1 | II-1-2 | II-2-1 | II-2-2 | |
| < = 100 kb | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 1 |
| >100 to 200 kb | 17 | 18 | 15 | 20 | 119 | 50 | 24 | 24 |
| >200 to 300 kb | 11 | 6 | 4 | 10 | 25 | 6 | 13 | 9 |
| >300 to 400 kb | 5 | 5 | 6 | 5 | 11 | 3 | 1 | 4 |
| >400 to 500 kb | 2 | 0 | 2 | 2 | 6 | 1 | 1 | 2 |
| >500 to 1000 kb | 6 | 2 | 4 | 7 | 7 | 2 | 5 | 2 |
| >1 to 10 Mb | 9 | 4 | 5 | 3 | 5 | 2 | 4 | 5 |
| >10 to 20 Mb | 5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| >20 Mb | 3 | 0 | 0 | 0 | 2 | 0 | 2 | 2 |
| Total | 58 | 35 | 36 | 47 | 177 | 64 | 50 | 49 |
Numerical values in each cell of the table indicate how many CNVs of that particular size range were observed in that particular individual.
Table 3. Identity of copy number variants across individual family members.
| CNVs | Family 1 | Family 2 | ||||||
| I-1-1 | I-1-2 | I-2-1 | I-2-2 | II-1-1 | II-1-2 | II-2-1 | II-2-2 | |
| No. of Loss | 21 | 6 | 5 | 6 | 52 | 11 | 6 | 4 |
| No. of Gain | 37 | 29 | 31 | 41 | 125 | 53 | 44 | 45 |
| Novel (absent in DGV) | 1 | 1 | 0 | 2 | 42 | 6 | 1 | 0 |
| Present in DGV | 57 | 34 | 36 | 45 | 135 | 58 | 49 | 49 |
| Total (for the individual) | 58 | 35 | 36 | 47 | 177 | 64 | 50 | 49 |
Frequency of CNVs which are losses (deletion) or gains (duplication) and characterization as present or absent from The Database of Genomic Variants (DGV).
Table 4. Chromosome wise distribution of CNV.
| Chr No. | Family 1 | Family 2 | ||||||
| I-1-1 | I-1-2 | I-2-1 | I-2-2 | II-1-1 | II-1-2 | II-2-1 | II-2-2 | |
| 1 | 4 | 2 | 2 | 2 | 11 | 2 | 6 | 6 |
| 2 | 4 | 2 | 5 | 5 | 11 | 2 | 2 | 3 |
| 3 | 1 | 4 | 2 | 3 | 7 | 4 | 4 | 2 |
| 4 | 4 | 3 | 2 | 4 | 8 | 1 | 3 | 2 |
| 5 | 0 | 0 | 0 | 0 | 12 | 0 | 2 | 1 |
| 6 | 0 | 0 | 0 | 0 | 10 | 2 | 0 | 0 |
| 7 | 2 | 6 | 3 | 4 | 10 | 5 | 3 | 4 |
| 8 | 1 | 0 | 3 | 3 | 7 | 3 | 3 | 3 |
| 9 | 1 | 1 | 2 | 3 | 4 | 4 | 3 | 4 |
| 10 | 2 | 1 | 1 | 1 | 3 | 5 | 1 | 1 |
| 11 | 3 | 1 | 2 | 1 | 5 | 1 | 2 | 2 |
| 12 | 0 | 1 | 0 | 1 | 4 | 3 | 0 | 1 |
| 13 | 0 | 0 | 0 | 0 | 40 | 1 | 1 | 0 |
| 14 | 4 | 6 | 3 | 5 | 4 | 6 | 5 | 3 |
| 15 | 4 | 3 | 1 | 3 | 2 | 6 | 6 | 8 |
| 16 | 2 | 1 | 2 | 3 | 9 | 1 | 1 | 1 |
| 17 | 4 | 1 | 2 | 3 | 8 | 2 | 2 | 1 |
| 18 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 |
| 19 | 0 | 1 | 0 | 0 | 7 | 3 | 2 | 1 |
| 20 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| 21 | 1 | 2 | 2 | 3 | 2 | 3 | 2 | 1 |
| 22 | 3 | 0 | 3 | 2 | 3 | 1 | 1 | 1 |
| X | 18 | 0 | 1 | 1 | 9 | 9 | 1 | 2 |
| Total | 58 | 35 | 36 | 47 | 177 | 64 | 50 | 49 |
Chromosome specific distribution of de novo (present in twin(s) and not in parents) and inherited (present in at least one parent) CNVs in family 1 and family 2.
Chr. No = Chromosome number.
Figure 2. Distribution of repeat elements 1 kb upstream (5′) and 1 kb downstream (3′) of the de novo (2a) and inherited (2b) CNVs across eight individuals.
These include LINE (blue), SINE (purple), LTR (yellow), Satellite (sky blue), simple repeats (black) and low complexity repeats (green) with numerical values on top of the bars representing percentage of that repeat.
Familial vs de novo Origin of CNVs
A novel feature of the data included in this report is that we are able to classify observed CNVs into two groups based on their absence or presence in one of the parents. CNVs that were found in one or both twins and not seen in either parent, were classified as de novo. If a de novo CNV was present in both twins, it was considered to have originated during parental meiosis and when present in only one of the two twins, it was assumed to have originated in mitosis during development. This classification allowed us to identify 14 and 26 de novo CNVs in family 1 (Table 5) and family 2 (Table 6) respectively. The table includes genomic locations as well as individual specific break points which allow for the assessment of regions of overlap with the Database of Genomic Variants (Toronto, Ontario). Mitotic origin of CNVs was ∼3 times higher than CNVs generated during parental meiosis. Of the mitotic de novo CNVs identified two (loss at 14q32.11 as well as loss at 8q11.21) were specific to the schizophrenia patient in family 1 and one (gain at 19q13.41) was specific to the patient in family 2. Such results are novel in the literature. Further, it is enticing to ask the question, do the genes disturbed by CNVs contribute to the development of their disease symptoms? Although the answers to such questions are of paramount importance, the results available do not offer a direct assessment of such questions. Nonetheless, it is appropriate to entertain the discussion that the known features of these genes are or are not compatible with disturbances observed in schizophrenia, which is discussed below.
Table 5. de novo CNVs in Family 1.
| Sl. No | Location | Family 1 | Status | Meiosis | Mitosis | Novel | Genes (Overlapping or Nearby) | SD | |||||
| I-2-1 | Size (kb) | Breakpoints | I-2-2 | Size (kb) | Breakpoints | ||||||||
| 1 | 1p36.13 | Yes | 112 | 16724089…16835888 | Gain | Yes | NBPF1, NBPF10 | 1 | |||||
| 2 | 2p25.3 | Yes | 152 | 1407209…1559511 | Yes | 152 | 1407209…1559511 | Gain | Yes | TPO | 0 | ||
| 3 | 2p11.2 | Yes | 1147 | 89862331…91008912 | Yes | 1159 | 89850279…91008912 | Loss | Yes | 0 | |||
| 4 | 4q28.3 | Yes | 191 | 132801221…132992517 | Gain | Yes | 1 | ||||||
| 5 | 7q11.21 | Yes | 118 | 64706066…64823721 | Yes | 118 | 64704377…64822216 | Loss | Yes | 1 | |||
| 6 | 8p23.1 | Yes | 126 | 7847289…7973253 | Loss | Yes | 1 | ||||||
| 7 | 8q11.1 | Yes | 336 | 47045602…47381308 | Yes | 250 | 47131383…47381308 | Gain | Yes | 0 | |||
| 8 | 8q11.21 | Yes | 154 | 48178242…48332398 | Loss | Yes | Yes | KIAA0146 | 0 | ||||
| 9 | 9p11.2 | Yes | 569 | 45361389…45929992 | Gain | Yes | FAM27A | 1 | |||||
| 10 | 9p13.1 | Yes | 141 | 38777481…38918566 | Gain | Yes | 1 | ||||||
| 11 | 9q12 | Yes | 861 | 65412415…66273526 | Gain | Yes | 1 | ||||||
| 12 | 12p13.31 | Yes | 196 | 8303317…8499801 | Gain | Yes | CLEC6A | 1 | |||||
| 13 | 14q32.11 | Yes | 103 | 89780137…89883415 | Loss | Yes | Yes | PSMC1, C14orf102 | 0 | ||||
| 14 | 21q11.2 | Yes | 119 | 13891136…14009908 | Gain | Yes | ANKRD21, LOC441956 | 1 | |||||
| 15 | Xp11.23 | Yes | 149 | 47917899…48066856 | Yes | 149 | 47917899…48066856 | Gain | Yes | SSX5, SSX1, SSX9 | 0 | ||
Table 6. de novo CNVs in Family 2.
| Sl. No | Location | Family 2 | Status | Meiosis | Mitosis | Novel | Genes (Overlapping or Nearby) | SD | |||||
| II-2-1 | Size (kb) | Breakpoints | II-2-2 | Size (kb) | Breakpoints | ||||||||
| 1 | 1q21.1 | Yes | 120 | 143867807…143987616 | Gain | Yes | NOTCH2NL | 0 | |||||
| 2 | 1q21.1 | Yes | 104 | 147353175…147456930 | Yes | 104 | 147353175…147456930 | Loss | Yes | 0 | |||
| 3 | 1q43 | Yes | 119 | 241230453…241349107 | Gain | Yes | 0 | ||||||
| 4 | 3q21.2 | Yes | 155 | 126958012…127112518 | Gain | Yes | 1 | ||||||
| 5 | 4p11 | Yes | 299 | 48986100…49285347 | Gain | Yes | 0 | ||||||
| 6 | 5p15.33 | Yes | 101 | 770367…871743 | Yes | 107 | 770367…877436 | Gain | Yes | ZDHHC11 | 1 | ||
| 7 | 5p13.3 | Yes | 151 | 34119387…34269887 | Gain | Yes | 1 | ||||||
| 8 | 7q11.21 | Yes | 202 | 61761008…61962936 | Gain | Yes | 0 | ||||||
| 9 | 7q11.21 | Yes | 116 | 64588316…64704125 | Yes | 125 | 64579322…64704125 | Gain | Yes | 1 | |||
| 10 | 7q35 | Yes | 100 | 142956516…143056637 | Gain | Yes | LOC441294, FAM139A | 1 | |||||
| 11 | 8p23.1 | Yes | 220 | 12071704…12291845 | Yes | 220 | 12071704…12291845 | Gain | Yes | FAM86B1, DEFB130 | 0 | ||
| 12 | 9p12 | Yes | 2720 | 41465094…44184864 | Yes | 1901 | 42249132…44149779 | Gain | Yes | ANKRD20A2, ANKRD20A3, FOXD4L4, FOXD4L2 | 1 | ||
| 13 | 9q12 | Yes | 250 | 67416254…67665974 | Gain | Yes | ANKRD20A1, ANKRD20A3 | 1 | |||||
| 14 | 11q13.2 | Yes | 267 | 67239223…67505822 | Yes | 139 | 67239223…67378031 | Gain | Yes | 1 | |||
| 15 | 12p13.31 | Yes | 189 | 8310909…8499801 | Gain | Yes | 1 | ||||||
| 16 | 13q11 | Yes | 208 | 18138676…18346383 | Gain | Yes | 1 | ||||||
| 17 | 14q11.1 | Yes | 601 | 18072112…18672662 | Yes | 601 | 18072112…18672662 | Gain | Yes | OR11H12, ACTBL1 | 1 | ||
| 18 | 15q11.1 | Yes | 106 | 18276329…18382609 | Gain | Yes | 1 | ||||||
| 19 | 15q11.2 | Yes | 203 | 19882763…20085783 | Yes | 221 | 19864583…20085783 | Gain | Yes | OR4M2, OR4N4, LOC650137 | 1 | ||
| 20 | 15q13.1 | Yes | 227 | 26808083…27035216 | Gain | Yes | APBA2 | 0 | |||||
| 21 | 15q13.2 | Yes | 110 | 28452853…28563274 | Gain | Yes | CHRFAM7A | 1 | |||||
| 22 | 17p11.1 | Yes | 199 | 22127012…22326425 | Gain | Yes | 0 | ||||||
| 23 | 19q13.41 | Yes | 109 | 58847652…58957090 | Gain | Yes | Yes | ZNF331, DPRX | 0 | ||||
| 24 | 20q11.1 | Yes | 118 | 28147331…28264860 | Gain | Yes | 1 | ||||||
| 25 | 21p11.2 | Yes | 3480 | 10106540…13586186 | Yes | 3814 | 9758730…13572586 | Gain | Yes | BAGE2, BAGE4, BAGE | 0 | ||
Identity of de novo CNVs found in Family 1 (5a) and Family 2 (5b) and the gene regions (overlapping or nearby). De novo CNVs are defined as those that are present in either or both twins but not found in parents. SD displays the percentage of overlap with segmental duplications, ‘0’ indicates no overlap between the CNV and segmental duplication and ‘1’ indicates 90–100% overlap. The table includes genomic locations as well as twin specific breakpoints which allow for the assessment of regions of overlap with the Database of Genomic Variants (Toronto, Ontario). SI No. = Serial number. Novel indicates a CNV which is not present in The Database of Genomic Variants (DGV).
De novo CNVs and Schizophrenia
The genes overlapping disease specific de novo CNVs in family 1 included PSMC1 (proteasome 26S subunit, ATPase, 1) and C14orf102 (chromosome 14 open reading frame 102 gene) on 14q32.11 and KIAA0146 on 8q11.21. PSMC1 (MIM 602706) is an ATP-dependent protease [17] that may include protein ubiquitination in response to DNA damage [18]. It is composed of a 20S catalytic proteasome and 2 PA700 regulatory modules and contains an AAA (ATPases associated with diverse cellular activities) domain [17]. The human and mouse proteins are 99% identical [19] and may play a significant role in ubiquitin-mediated proteasomal proteolysis in the molecular pathogenesis of neurological diseases such as spinocerebellar ataxia type 7 (SCA7). Also, several studies (for review, see [20], [21]), have indicated that the genes related to ubiquitination are altered in the brains of patients with schizophrenia. Further, this CNV also affects another gene (C14orf102; chromosome 14 open reading frame 102) which is conserved across phyla and highly expressed in the brain (Affymetrix GNF Expression Atlas 2 Data). The other CNV affected in this patient of family 1 represents a loss at 8q11.21, that contains the still uncharacterized gene, KIAA0146, which is expressed in the brain, may contain a CAG repeat and is conserved in chimpanzee, dog, cow, mouse, rat, chicken, and zebra fish. It is a transcription factor with CCAAT enhancer binding protein (CEBP) function [22]. Further the gene is highly expressed in the brain and hippocampus that may implicate it in mental disorders (www.genecards.org). Although we cannot rule out a role for these three genes (PSMC1, C14orf102 and KIAA0146) in schizophrenia, such conclusions would be premature. Only a follow up study will establish if any of the three genes directly contribute to the development of schizophrenia in the patient from family 1. A similar analysis of CNVs in family 2 has identified a 109 kb gain at 19q13.41 that is specific to the schizophrenia patient in family 2. Translocations involving 19q13 are a frequent finding in follicular adenomas of the thyroid and may represent the most frequent type of structural aberration in human epithelial tumors [23]. The CNV identified in this region contains two genes; DPRX1 and ZNF331. DPRX1 (divergent-paired related homeobox) is a member of the DPRX homeobox gene family, contains a single conserved homeodomain and may function as a putative transcription factor. It may bind a promoter or enhancer sequence or interact with a DNA binding transcription factor and is involved in early embryonic development and cell differentiation [24]. The drosophila homologue of the DPRX1 gene (dPrx5; Drosophila peroxiredoxin 5) confers protection against oxidative stress, apoptosis and also promotes longevity [25]. The next gene, ZNF331(zinc finger protein 331) affected by this CNV is also involved in DNA-dependent regulation of transcription as a transcriptional repressor [26]. Interestingly, it is one of the imprinted genes that exhibits monoallelic expression in a parent-of-origin specific manner [27]. Imprinted genes are important for development and behaviour and disruption of their expression is associated with many human disorders [28]. In conclusion the three genes affected in the schizophrenia patient in family 1 (PSMC1, C14orf102, KIAA0146) and the two genes affected in the patient of family 2 (DPRX1 and ZNF331) could not be excluded from their potential involvement in the development of schizophrenia in the two patients. If applicable, the biological systems affected in the two patients is hypothesized to be different. The patient in family one is hypothesized to have a ubiquitin-mediated proteasomal proteolysis while the patient of family 2 could have errors in regulatory mechanisms affecting gene regulation. Such conclusions must remain hypothetical until proven by independent supporting evidence.
De novo changes may lead to mosaicism
The genotypes generated by the Affymetrix 6.0 array have also allowed us to establish that ∼0.12% (1086 and 1022 in twin pair 1 and 2 respectively; 11 substitutions shared by both pairs) of the SNPs in the twins represented de novo substitutions, but unlike CNVs, (that primarily originated during ontogeny in mitosis) most (63–65%) originated during parental meiosis. These results suggest that DNA replication fidelity at the level of single base pairs (SNPs) vs replication forks (CNVs) is differentially exercised during meiosis and mitosis. The single base pairing is much more stringent in mitosis (evolved to produce identical daughter cells), compared to meiosis where errors can facilitate potentially beneficial variations. In contrast, CNVs which affect the phenotype may be advantageous when occurring during mitosis and selected for during development. Thus, cell type specific CNVs may play a role in growth and development, offering advantageous variability. This would mean that most individuals are mosaics [29]: a hypothesis that is difficult to assess and evaluate. It is likely that the ratio of mosaic cells may be maintained throughout the differentiated (ectoderm, mesoderm, endoderm, etc) tissues over the lifetime [30], [31]; an exception being when other factors are directly influencing DNA stability. Such a mechanism may generate genomic differences and differential mosaicism in most or all individuals. If this is the case, it will complicate traditional genetic analysis that assumes stability of the genome with rare exceptions.
We have been able to establish genome-wide (CNVs and SNPs) discordance for MZ twin pairs. Also, given that the twins are discordant for schizophrenia, it is possible to assign provisional CNVs (and genes) as well as substitutions (SNPs) that may be associated with the disease status of the affected twins in family 1 and family 2 (Table 7,8). Similarly, we identified substitutions (SNPs) that were different between the affected and unaffected member of the two sets of twins including their distribution along the chromosomes, introns and exons and the predicted effect on the gene product. Identity of de novo CNVs found in Family 1 (Table 5) and Family 2 (Table 6) and the gene regions which they overlap was reported. De novo CNVs are defined as those that are present in either twin but not found in parents. In the tables, SD indicates the percentage of overlap between segmental duplications and the CNVs, ‘0’ means there is no overlap between CNV and segmental duplication and ‘1’ means 90–100% overlap.
Table 7. Inherited CNVs in Family 1.
| Sl. No | Location | Family 1 | Status | Novel | Genes (Overlapping or Nearby) | SD | |||||||||||
| I-1-1 | Size(kb) | Breakpoints | I-1-2 | Size(kb) | Breakpoints | I-2-1 | Size(kb) | Breakpoints | I-2-2 | Size(kb) | Breakpoints | ||||||
| 1 | 1p36.33 | Yes | 167 | 51586…218557 | Yes | 707 | 51586…758644 | Yes | 707 | 51586…758644 | Gain | OR4F5, OR4F3, OR4F16, OR4F29 | 1 | ||||
| 2 | 1q21.1 | Yes | 765 | 147303136…148068045 | Yes | 734 | 147311699…148045353 | Yes | 577 | 147381253…147958358 | Gain | PPIAL4, FCGR1A, HIST2H2BF | 1 | ||||
| 3 | 2p11.2 | Yes | 338 | 88917155…89254935 | Yes | 322 | 88914734…89236978 | Yes | 325 | 88917155…89242149 | Yes | 327 | 88914734…89242149 | Gain | 1 | ||
| 4 | 2p11.1 | Yes | 138 | 91017077…91154841 | Yes | 143 | 91017077…91160399 | Yes | 137 | 91017077…91154463 | Gain | 1 | |||||
| 5 | 2q21.2 | Yes | 236 | 132597824…132833718 | Yes | 222 | 132597824…132819911 | Yes | 222 | 132597824…132819911 | Gain | 1 | |||||
| 6 | 3p12.3 | Yes | 306 | 75677859…75984129 | Yes | 380 | 75597086…75977210 | Yes | 182 | 75583442…75764996 | Yes | 402 | 75582277…75984129 | Gain | 1 | ||
| 7 | 3q21.2 | Yes | 132 | 126907150…127039328 | Yes | 185 | 126907150…127091652 | Gain | 1 | ||||||||
| 8 | 3q21.3 | Yes | 170 | 131198515…131368353 | Yes | 166 | 131213377…131379054 | Yes | 176 | 131214431…131389948 | Gain | 1 | |||||
| 9 | 4p16.2 | Yes | 196 | 4040542…4236511 | Yes | 363 | 3873500…4236511 | Yes | 366 | 3870638…4236511 | Gain | 0 | |||||
| 10 | 4p11 | Yes | 436 | 48849363…49285347 | Yes | 497 | 48788531…49285347 | Yes | 497 | 48788531…49285347 | Gain | 1 | |||||
| 11 | 4q35.2 | Yes | 232 | 191021837…191254119 | Yes | 195 | 191059369…191254119 | Yes | 223 | 191031042…191254119 | Gain | FRG1, TUBB4Q, FRG2, DUX4 | 0 | ||||
| 12 | 7p11.1 | Yes | 231 | 57523223…57753919 | Yes | 101 | 57640100…57741512 | Yes | 315 | 57640100…57954861 | Yes | 117 | 57640100…57757406 | Gain | 1 | ||
| 13 | 7q11.21 | Yes | 112 | 61365830…61477958 | Yes | 111 | 61365830…61476918 | Gain | 0 | ||||||||
| 14 | 7q11.21 | Yes | 253 | 64320173…64573380 | Yes | 415 | 64204380…64619667 | Yes | 385 | 64204380…64589253 | Loss | ZNF92 | 0 | ||||
| 15 | 8p23.1 | Yes | 694 | 7209579…7903560 | Yes | 215 | 7027251…7242508 | Yes | 270 | 7021193…7291135 | Loss | DEFB103A, DEFB103B, SPAG11B, DEFB104B, DEFB104A, DEFB106B, DEFB106A, DEFB105B, DEFB105A, DEFB107B, DEFB107A, SPAG11A, DEFB4 | 2 | ||||
| 16 | 9q12 | Yes | 690 | 68115006…68805366 | Yes | 1141 | 68115006…69256300 | Yes | 694 | 68115006…68809437 | Gain | FOXD4L6, CBWD6, ANKRD20A4, CCDC29 | 1 | ||||
| 17 | 10q11.1 | Yes | 183 | 41972779…42155347 | Yes | 105 | 41934430…42039743 | Yes | 183 | 41972779…42155347 | Yes | 239 | 41934430…42173117 | Gain | 1 | ||
| 18 | 11p15.4 | Yes | 175 | 3405799…3580813 | Yes | 131 | 3430789…3561991 | Yes | 139 | 3430789…3569305 | Yes | 156 | 3406002…3561991 | Gain | 1 | ||
| 19 | 11q13.2 | Yes | 227 | 67239223…67466368 | Yes | 193 | 67273413…67466368 | Gain | 1 | ||||||||
| 20 | 14q11.1 | Yes | 1322 | 18138794…19460382 | Yes | 1103 | 18072112…19175240 | Yes | 705 | 18072112…18776746 | Yes | 705 | 18072112…18776746 | Gain | OR11H12, ACTBL1, OR4Q3, OR4M1, OR4N2, OR4K5 | 0 | |
| 21 | 14q32.33 | Yes | 126 | 105265510…105391419 | Yes | 167 | 105100670…105268160 | Yes | 632 | 105190672…105822317 | Yes | 181 | 105149735…105331052 | Gain | 0 | ||
| Yes | 156 | 105413825…105569826 | Yes | 213 | 105289618…105502685 | Yes | 178 | 105827891…106005581 | Yes | 261 | 105341035…105601720 | Gain | 0 | ||||
| Yes | 205 | 105612786…105818132 | Yes | 279 | 105508896…105788389 | Yes | 280 | 105612786…105892769 | Gain | 0 | |||||||
| 22 | 15q11.1 | Yes | 471 | 18370252…18841457 | Yes | 178 | 18522238…18700540 | Yes | 1223 | 18845990…20068512 | Yes | 562 | 18276329…18838423 | Gain | LOC283755, POTE15, OR4M2 | 1 | |
| Yes | 1177 | 18845990…20022565 | Yes | 344 | 18845990…19189673 | Yes | 1078 | 18845990…19923712 | Gain | OR4N4, LOC650137 | 1 | ||||||
| Yes | 264 | 19303160…19566863 | Gain | 1 | |||||||||||||
| 23 | 15q11.2 | Yes | 189 | 22026287…22214843 | Yes | 174 | 22026287…22200408 | Gain | 1 | ||||||||
| 24 | 16p11.2 | Yes | 1217 | 32303108…33520394 | Yes | 1297 | 32538757…33836128 | Yes | 1142 | 32538757…33680554 | Yes | 249 | 32538757…32787273 | Gain | LOC729355, TP53TG3 | 1 | |
| Yes | 752 | 32910319…33662480 | Gain | 1 | |||||||||||||
| 25 | 16p11.2 | Yes | 250 | 34374795…34624994 | Yes | 249 | 34375533…34624994 | Yes | 249 | 34375533…34624994 | Gain | 1 | |||||
| 26 | 17p11.2 | Yes | 140 | 20559979…20700133 | Yes | 164 | 20538867…20703365 | Gain | 1 | ||||||||
| 27 | 17q21.31 | Yes | 229 | 41521621…41750183 | Yes | 123 | 41521621…41644356 | Yes | 123 | 41521621…41644356 | Gain | KIAA1267, LRRC37A | 0 | ||||
| 28 | 17q21.31 | Yes | 351 | 41756820…42107467 | Yes | 296 | 41811739…42107467 | Yes | 392 | 41700624…42092926 | Yes | 302 | 41700624…42002447 | Gain/Loss | ARL17, LRRC37A2, NSF | 1 | |
| 29 | 21p11.2 | Yes | 204 | 9758730…9962501 | Yes | 204 | 9758730…9962501 | Yes | 204 | 9758730…9962501 | Gain | TPTE | 0 | ||||
| 30 | 21p11.1 | Yes | 3411 | 10106540…13517603 | Yes | 3419 | 10106540…13525448 | Yes | 3419 | 10106540…13525448 | Yes | 3477 | 10106540…13583117 | Gain | BAGE2, BAGE4, BAGE | 2 | |
| 31 | 22q11.1 | Yes | 339 | 14435171…14774593 | Yes | 320 | 14435207…14754960 | Yes | 320 | 14435207…14754960 | Gain | ACTBL1 | 1 | ||||
| 32 | 22q11.21 | Yes | 124 | 20051708…20175282 | Yes | 136 | 20145854…20281562 | Yes | 136 | 20145854…20281562 | Gain | HIC2, UBE2L3 | 1 | ||||
| 33 | 22q11.22 | Yes | 240 | 21292462…21532509 | Yes | 127 | 21327799…21454509 | Gain | GGTL4 | 0 | |||||||
Table 8. Inherited CNVs in Family 2.
| Sl. No | Location | Family 2 | Status | Novel | Genes (Overlapping or Nearby) | SD | |||||||||||
| II-1-1 | Size(kb) | Breakpoints | II-1-2 | Size(kb) | Breakpoints | II-2-1 | Size(kb) | Breakpoints | II-2-2 | Size(kb) | Breakpoints | ||||||
| 1 | 1p36.33 | Yes | 707 | 51586…758644 | Yes | 537 | 218557…755132 | Gain | OR4F5, OR4F3, OR4F16, OR4F29 | 1 | |||||||
| 2 | 1p36.13 | Yes | 117 | 16718622…16835888 | Yes | 167 | 16718622…16885360 | Yes | 345 | 16718622…17063437 | Gain | NBPF1, NBPF10 | 1 | ||||
| 3 | 1p21.1 | Yes | 130 | 103910749…104041200 | Yes | 127 | 103931691…104058426 | Gain | AMY2B, AMY2A, AMY1A, AMY1C, AMY1B | 1 | |||||||
| 4 | 1p11.2 | Yes | 21680 | 121045307…142725034 | Yes | 21725 | 121045307…142770353 | Yes | 21725 | 121045307…142770353 | Gain | 0 | |||||
| 5 | 1q23.3 | Yes | 121 | 159775403…159896554 | Yes | 116 | 159780383…159896554 | Yes | 121 | 159775403…159896554 | Loss | FCGR3A, FCGR2C, FCGR3B | 1 | ||||
| 6 | 2p11.2 | Yes | 401 | 88925215…89326446 | Yes | 759 | 88914227…89673147 | Yes | 935 | 88926972…89861763 | Yes | 450 | 88914734…89365010 | Gain | 1 | ||
| 7 | 2p11.1 | Yes | 160 | 91017077…91176948 | Yes | 268 | 91017077…91285520 | Yes | 1275 | 89879561…91154463 | Gain | 1 | |||||
| 8 | 2q21.2 | Yes | 260 | 132593436…132853218 | Yes | 183 | 132597824…132780848 | Yes | 222 | 132597824…132819911 | Gain | 1 | |||||
| 9 | 3p12.3 | Yes | 260 | 75583442…75843060 | Yes | 226 | 75538978…75764996 | Yes | 260 | 75583442…75843060 | Yes | 182 | 75583442…75764996 | Gain | 1 | ||
| 10 | 3q12.2 | Yes | 108 | 101822746…101930873 | Yes | 102 | 101822746…101925168 | Gain | GPR128, TFG | 0 | |||||||
| 11 | 3q21.3 | Yes | 141 | 131198817…131339424 | Yes | 195 | 131194669…131389948 | Yes | 199 | 131198515…131397648 | Gain | 1 | |||||
| 12 | 4p16.2 | Yes | 407 | 3870638…4278016 | Yes | 180 | 3964803…4144453 | Yes | 366 | 3870638…4236511 | Yes | 357 | 3870638…4227503 | Gain | OTOP1 | 0 | |
| 13 | 4q35.2 | Yes | 200 | 191053845…191254119 | Yes | 226 | 191028537…191254119 | Yes | 158 | 191052245…191210542 | Gain | FRG1, TUBB4Q, FRG2, DUX4 | 0 | ||||
| 14 | 7p22.1 | Yes | 157 | 6838697…6995298 | Yes | 155 | 6840798…6995298 | Gain | 1 | ||||||||
| 15 | 7p11.1 | Yes | 117 | 57640100…57757406 | Yes | 114 | 57640100…57753919 | Yes | 149 | 57604989…57753919 | Yes | 123 | 57597399…57720623 | Gain | 1 | ||
| 16 | 8p23.1 | Yes | 203 | 12415742…12618442 | Yes | 199 | 12415742…12614748 | Yes | 136 | 12415742…12551430 | Gain | 1 | |||||
| 17 | 8p11.23 | Yes | 139 | 39349470…39488053 | Yes | 151 | 39354748…39506110 | Yes | 133 | 39354748…39488053 | Yes | 151 | 39354748…39506110 | Loss | 0 | ||
| 18 | 9p11.2 | Yes | 21937 | 44336683…66273526 | Yes | 21015 | 45258754…66273526 | Yes | 21015 | 45258754…66273526 | Gain | FAM27A, FAM75A7 | 0 | ||||
| 9q12 | Yes | 861 | 68352238…69213455 | Yes | 103 | 68115006…68218485 | Yes | 1180 | 68076544…69256300 | Yes | 1099 | 68115006…69213671 | Gain | FOXD4L6, CBWD6, ANKRD20A4, CCDC29 | 1 | ||
| Yes | 457 | 68352238…68809437 | Gain | ||||||||||||||
| 19 | 10q11.1 | Yes | 129 | 41974796…42103488 | Yes | 236 | 41934430…42170853 | Yes | 105 | 41934430…42039743 | Yes | 239 | 41934430…42173117 | Gain | 1 | ||
| 20 | 11p15.4 | Yes | 206 | 3383178…3588946 | Yes | 205 | 3376078…3580813 | Yes | 131 | 3430789…3561991 | Gain | 1 | |||||
| 21 | 14q11.2 | Yes | 186 | 21602854…21788783 | Yes | 172 | 21625813…21787161 | Loss | 0 | ||||||||
| 22 | 14q11.2 | Yes | 226 | 21804698…22030660 | Yes | 226 | 21804698…22030660 | Loss | 0 | ||||||||
| 23 | 14q32.33 | Yes | 386 | 105190672…105576359 | Yes | 253 | 105345270…105597999 | Yes | 411 | 105190672…105601397 | Yes | 336 | 105265510…105601397 | Gain | 0 | ||
| 24 | 14q32.33 | Yes | 137 | 105760582…105897672 | Yes | 173 | 105645593…105818132 | Yes | 150 | 105638133…105788389 | Yes | 182 | 105640496…105822317 | Gain | 0 | ||
| 25 | 15q11.2 | Yes | 156 | 18682380…18838423 | Yes | 183 | 18655531…18838423 | Yes | 156 | 18682380…18838423 | Gain | LOC283755, POTE15 | 1 | ||||
| Yes | 1067 | 18861808…19928521 | Yes | 572 | 18850029…19422452 | Yes | 344 | 18845990…19189673 | Gain | OR4M2, OR4N4, LOC650137 | 1 | ||||||
| Yes | 624 | 19207088…19835514 | Gain | 1 | |||||||||||||
| 26 | 15q25.3 | Yes | 161 | 83524791…83685356 | Yes | 228 | 83524791…83752853 | Yes | 228 | 83524791…83752450 | Gain | AKAP12 | 0 | ||||
| 27 | 15q25.3 | Yes | 123 | 83784507…83907801 | Yes | 157 | 83784507…83941483 | Yes | 159 | 83790259…83949305 | Gain | AKAP13 | 0 | ||||
| 28 | 16p11.2 | Yes | 118 | 31882658…32000323 | Yes | 1131 | 32531735…33662480 | Yes | 1377 | 32303108…33680554 | Gain | LOC729355, TP53TG3 | 1 | ||||
| Yes | 294 | 32088275…32382422 | Gain | 1 | |||||||||||||
| Yes | 185 | 32538757…32723310 | Gain | 1 | |||||||||||||
| Yes | 474 | 32962147…33436245 | Gain | 1 | |||||||||||||
| Yes | 211 | 33451476…33662480 | Gain | 1 | |||||||||||||
| 29 | 17q21.31 | Yes | 586 | 41521621…42107467 | Yes | 198 | 41521621…41719935 | Yes | 586 | 41521621…42107467 | Yes | 407 | 41700624…42107467 | Gain | KIAA1267, LRRC37A, ARL17, LRRC37A2, NSF | 1 | |
| 30 | 18p11.21 | Yes | 1545 | 15262486…16807594 | Yes | 130 | 15218647…15348836 | Gain | ROCK1 | 1 | |||||||
| 31 | 19q13.31 | Yes | 116 | 47991257…48107552 | Yes | 133 | 47991257…48123857 | Yes | 235 | 47986218…48221228 | Loss | PSG1, PSG6, PSG7, PSG11 | 1 | ||||
| 32 | 21p11.2 | Yes | 204 | 9758730…9962501 | Yes | 204 | 9758730…9962501 | Yes | 204 | 9758730…9962501 | Gain | TPTE | 0 | ||||
| 33 | 22q11.22 | Yes | 148 | 21300127…21448190 | Yes | 200 | 21298324…21498767 | Yes | 178 | 21298324…21476564 | Gain | GGTL4 | 0 | ||||
| 34 | Xp11.23 | Yes | 112 | 47917899…48029446 | Yes | 269 | 47917899…48186708 | Yes | 184 | 47917899…48102337 | Yes | 257 | 47935225…48192383 | Gain | SSX5, SSX1, SSX9, SSX3 | 1 | |
| 35 | Xq13.1 | Yes | 185 | 71869375…72054837 | Yes | 185 | 71869375…72054837 | Gain | Yes | DMRTC1 | 1 | ||||||
Identity of inherited CNVs found in Family 1 (7), Family 2 (8) and the gene regions which they overlap. Inherited CNVs are those which are present in either or both parents and transmitted to either or both twins. All size is in kb. SD indicates the percentage of overlap between segmental duplications and CNVs. ‘0’ means there is no overlap between CNV and segmental duplication, ‘1’ means 90–100% and ‘2’ means 50–90% overlap. Parental CNVs not transmitted to offspring were not included in Table 5– 8 so the total number of CNVs present in Table 2– 4 was not same as Table 5– 8. The table includes genomic locations as well as individual specific break points which allow for the assessment of regions of overlap with the Database of Genomic Variants (Toronto, Ontario). SI No. = Serial number. Novel indicates a CNV which is not present in The Database of Genomic Variants (DGV).
We also analyzed genes that overlapped de novo CNVs (gains and losses) in order to assess their potential effect on physiology and function starting with GO ontology annotation (http://www.geneontology.org). Interestingly, the majority of genes belonged to transcription, DNA replication, transport, and cell signalling pathways, including ‘binding’ or ‘catalytic’ functions. A number of these genes are expressed in the brain, some with potential to affect neurophysiology, neurodevelopment and function and a set of them are known to show altered expression in schizophrenia (www.schizophreniaforum.org). Also of significance is the observation that the FAM19A5 protein encoded by the FAM19A5 gene (22q13.32) belongs to the TAFA protein family which are predominantly expressed in the brain, and are postulated to function as brain-specific chemokines or neurokines, that act as regulators of immune and nervous cells [32]. This finding adds to the existing speculation about the role of the Major Histocompatability Loci (MHC) and infection in SCZ. Functional analysis of this gene and upstream regulatory elements for characteristic patterns of nucleosome occupancy changes associated with enhancers could yield novel insights into the role of this gene in psychiatric disorders. IPA analysis of gene networks of CNVs and SNPs converged on cell cycle, cellular growth and proliferation. Genes involved in genetic disorders such as hematological disease, immunological, inflammatory and developmental disorders were overrepresented. These results support the hypothesis that schizophrenia is a “developmental disorder” at the molecular level. Interestingly, a recent co-expression network analysis of microarray-based brain gene expression data revealed perturbations in developmental processes in schizophrenia [33]. However, given that these results are based on only two twin pairs, and schizophrenia is highly heterogeneous, the results on disease causations cannot be generalized. Also, we have offered other explanations for twin discordance that may involve epigenetic changes [34].
It is not surprising that genomic studies have begun to use monozygotic twins. In fact a number of them have identified copy number variations [35] and epigenetic [36]–[38] differences between them; an exception to these results is a recent study by Baranzini et al [39]. They studied three pairs of monozygotic twins discordant for Multiple Sclerosis (MS) and found no difference that could account for the disease causation. The results may be viewed as not surprising for a number of reasons. First, MS is known to have significant environmental components including sunlight and viruses, among others, [40] and the concordance rate in monozygotic twins is only ∼30%. Second, they assessed the CD4+ lymphocytes only that may or may not represent the causative cell type. Also, they sequenced the genome of CD4+ cells from a single pair corresponding to 21.7 and 22.5-fold coverage representing 99.6% and 99.5% of the NCBI human reference genome, which may or may not be effective. Only additional genomic and epigenomic studies on MZ twins will offer insights into the dynamics of genomic stability and change, that forms the focus of this report.
In summary, the present study adds to the recent effort in human genetics to define the phenomenon of constancy and change using inheritance and origin of genome-wide CNVs and SNPs. The results demonstrate that CNVs often result from mitosis during early development facilitated by flanking repeats. They may lead to CNV differences among different tissue and make most individuals mosaics. The described approach expands the search for disease related genetic changes, indicates the time of their occurrence and begins to interrogate the mechanisms involved.
Materials and Methods
This research was approved by the Committee on Research Involving Human Subjects at the University of Western Ontario. The families and patients were identified, recruited and clinically assessed by Dr. Richard O'Reilly (Psychiatrist) and all participants (Figure 1) gave informed consent and provided blood and buccal cells for this research. All subjects were interviewed using the Structured Clinical Interview for DSM IV and the SCID II (for personality disorders) and their medical records collected and reviewed. Diagnoses and demographic information are listed in Table 1. DNA was extracted from the collected white blood cells using the perfect pure DNA blood kit (5prime.com) following the manufacturer's protocol. Subsequent microarray analysis was performed using the Affymetrix Genome-Wide Human SNP Array 6.0 at the London Regional Genomics Centre (LRGC) following manufacturer's protocol and stringent quality control measures. Briefly, 5 µg of genomic DNA was labelled and hybridized to Affymetrix SNP 6.0 arrays. CNVs called by both Affymetrix Genotyping Console 4.0 and Partek® Genotyping Suite™ software suites were retained for analysis. In both cases, the CNVs were identified by continuity of markers on a segment. Two CNVs that overlapped by >50% in the two methods of data analysis were given the same identity. Every measure was undertaken to avoid inclusion of false positives including correction for segmental duplications. We found evidence of CNVs associated with segmental duplications which agrees with previous studies [41]. The CNVs identified were further assessed by comparison to the Database of Genomic Variants (http://projects.tcag.ca/variation/) and annotated with gene symbols by importing the annotation file from the UCSC genome browser (NCBI36/hg 18). A CNV that was present in both members of the twin pair and not in either of their two parents was considered to be meiotic de novo (originated during gamete formation), while a CNV that was present in one of the two twins and not present in either parent was considered to be mitotic de novo (originated during development). Further, a CNV present in the SCZ affected twin only (as compared to the two parents and unaffected member of the pair or the database) was classified as “provisional de novo CNV” for this disease. Novel CNVs discovered in this study were validated for predicted CNVs by Real Time PCR analysis with an internal control (RNAseP gene) using TaqMan detection chemistry and the ABI Prism 7300 Sequence Detection System (Applied Biosystems, http://www.appliedbiosystems.org). The copy number of the test locus in each case was defined as 2T−ΔΔC where ΔCT is the difference in threshold cycle number for the test and reference loci.
Additional CNV analysis focused on two aspects. The first deals with identification of putative repeat elements in the flanking regions of CNVs; within a 1 kb region upstream and downstream of the CNV breakpoint which could promote breakage, deletion and duplication. The identification of repeat elements was carried out using repeat masker (http://www.repeatmasker.org/). Secondly, a probable mechanism associated with sequence-specific susceptibility to CNVs was queried. This data was used to test models related to the origin of CNVs. Previously reported candidates for CNV mechanisms include Non-Allelic Homologous Recombination (NAHR), Non-Homologous End Joining (NHEJ), Fork Stalling and Template Switching (FoSTeS) and Microhomology-Mediated Break-Induced Replication (MMBIR) [42]. The second line of investigation involved functional characterization of genes by matching of the identified genes with the Schizophrenia Gene Database (http://www.schizophreniaforum.org/res/sczgene/default.asp) as well as their assessment by GO ontology (http://www.geneontology.org/). The genes identified were also subjected to IPA analysis (www.ingenuity.com) that identified the nature of gene interactions and the pathways involved.
The use of Affymetrix 6.0 Human SNP array also allowed us to assess the transmission of a total of 909622 SNPs that are contained on the array. It allowed us to identify SNPs in the twins that were not present in either of the two parents; considered to be de novo. The origin of the de novo SNPs was assumed to be parental meiosis if both twins carried the novel nucleotide. In contrast, the origin of the de novo SNPs was assumed to be somatic development (mitosis) if only one of the two twins carried the novel nucleotide. We were able to assign novel substitutions to different categories including their potential effect on the gene and gene product, as well as pathways that may be affected.
Acknowledgments
We thank members of the two families who participated in this research.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: Work was supported by the Canadian Institutes of Health Research (CIHR) http://www.cihr-irsc.gc.ca/ Grant number: R2258A15; Ontario Mental Health Foundation (OMHF) http://www.omhf.on.ca/ Grant number: R2258A14; and the Schizophrenia Society of Ontario (SSO) http://www.schizophrenia.on.ca/ Grant number: R2258A10. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Fisher RA. The Genesis of Twins. Genetics. 1919;4:489–499. doi: 10.1093/genetics/4.5.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Venter JC. Multiple personal genomes await. Nature. 2010;464:676–677. doi: 10.1038/464676a. [DOI] [PubMed] [Google Scholar]
- 3.Lee C, Scherer SW. The clinical context of copy number variation in the human genome. Expert Rev Mol Med. 2010;12:e8. doi: 10.1017/S1462399410001390. [DOI] [PubMed] [Google Scholar]
- 4.Iafrate AJ, Feuk L, Rivera MN, Listewnik ML, Donahoe PK, et al. Detection of large-scale variation in the human genome. Nat Genet. 2004;36:949–951. doi: 10.1038/ng1416. [DOI] [PubMed] [Google Scholar]
- 5.Sebat J, Lakshmi B, Troge J, Alexander J, Young J, et al. Large-scale copy number polymorphism in the human genome. Science. 2004;305:525–528. doi: 10.1126/science.1098918. [DOI] [PubMed] [Google Scholar]
- 6.Stankiewicz P, Lupski JR. Structural variation in the human genome and its role in disease. Annu Rev Med. 2010;61:437–455. doi: 10.1146/annurev-med-100708-204735. [DOI] [PubMed] [Google Scholar]
- 7.Walsh T, McClellan JM, McCarthy SE, Addington AM, Pierce SB, et al. Rare structural variants disrupt multiple genes in neurodevelopmental pathways in schizophrenia. Science. 2008;320:539–543. doi: 10.1126/science.1155174. [DOI] [PubMed] [Google Scholar]
- 8.Need AC, Ge D, Weale ME, Maia J, Feng S, et al. A genome-wide investigation of SNPs and CNVs in schizophrenia. PLoS Genet. 2009;5:e1000373. doi: 10.1371/journal.pgen.1000373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Singh SM, Castellani CA, O'Reilly RL. Copy number variation showers in schizophrenia: an emerging hypothesis. Mol Psychiatry. 2009;14:356–358. doi: 10.1038/mp.2008.149. [DOI] [PubMed] [Google Scholar]
- 10.Grozeva D, Kirov G, Ivanov D, Jones IR, Jones L, et al. Rare copy number variants: a point of rarity in genetic risk for bipolar disorder and schizophrenia. Arch Gen Psychiatry. 2010;67:318–327. doi: 10.1001/archgenpsychiatry.2010.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kirov G. The role of copy number variation in schizophrenia. Expert Rev Neurother. 2010;10:25–32. doi: 10.1586/ern.09.133. [DOI] [PubMed] [Google Scholar]
- 12.Gottesman II, Bertelsen A. Confirming unexpressed genotypes for schizophrenia. Risks in the offspring of Fischer's Danish identical and fraternal discordant twins. Arch Gen Psychiatry. 1989;46:867–872. doi: 10.1001/archpsyc.1989.01810100009002. [DOI] [PubMed] [Google Scholar]
- 13.McGuffin P, Asherson P, Owen M, Farmer A. The strength of the genetic effect. Is there room for an environmental influence in the aetiology of schizophrenia? Br J Psychiatry. 1994;164:593–599. doi: 10.1192/bjp.164.5.593. [DOI] [PubMed] [Google Scholar]
- 14.Ku CS, Pawitan Y, Sim X, Ong RT, Seielstad M, et al. Genomic copy number variations in three Southeast Asian populations. Hum Mutat. 2010;31:851–857. doi: 10.1002/humu.21287. [DOI] [PubMed] [Google Scholar]
- 15.Levy S, Sutton G, Ng PC, Feuk L, Halpern AL, et al. The diploid genome sequence of an individual human. PLoS Biol. 2007;5:e254. doi: 10.1371/journal.pbio.0050254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Myers S, Freeman C, Auton A, Donnelly P, McVean G. A common sequence motif associated with recombination hot spots and genome instability in humans. Nat Genet. 2008;40:1124–1129. doi: 10.1038/ng.213. [DOI] [PubMed] [Google Scholar]
- 17.Tanahashi N, Suzuki M, Fujiwara T, Takahashi E, Shimbara N, et al. Chromosomal localization and immunological analysis of a family of human 26S proteasomal ATPases. Biochem Biophys Res Commun. 1998;243:229–232. doi: 10.1006/bbrc.1997.7892. [DOI] [PubMed] [Google Scholar]
- 18.Huen MS, Chen J. The DNA damage response pathways: at the crossroad of protein modifications. Cell Res. 2008;18:8–16. doi: 10.1038/cr.2007.109. [DOI] [PubMed] [Google Scholar]
- 19.Hoyle J, Fisher EM. Genomic organization and mapping of the mouse P26s4 ATPase gene: a member of the remarkably conserved AAA gene family. Genomics. 1996;31:115–118. doi: 10.1006/geno.1996.0017. [DOI] [PubMed] [Google Scholar]
- 20.Konradi C. Gene expression microarray studies in polygenic psychiatric disorders: applications and data analysis. Brain Res Brain Res Rev. 2005;50:142–155. doi: 10.1016/j.brainresrev.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 21.Mirnics K, Levitt P, Lewis DA. Critical appraisal of DNA microarrays in psychiatric genomics. Biol Psychiatry. 2006;60:163–176. doi: 10.1016/j.biopsych.2006.02.003. [DOI] [PubMed] [Google Scholar]
- 22.Lee YW, Eum SY, Chen KC, Hennig B, Toborek M. Gene expression profile in interleukin-4-stimulated human vascular endothelial cells. Mol Med. 2004;10:19–27. doi: 10.2119/2004-00024.lee. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Belge G, Roque L, Soares J, Bruckmann S, Thode B, et al. Cytogenetic investigations of 340 thyroid hyperplasias and adenomas revealing correlations between cytogenetic findings and histology. Cancer Genet Cytogenet. 1998;101:42–48. doi: 10.1016/s0165-4608(97)00057-5. [DOI] [PubMed] [Google Scholar]
- 24.Booth HA, Holland PW. Annotation, nomenclature and evolution of four novel homeobox genes expressed in the human germ line. Gene. 2007;387:7–14. doi: 10.1016/j.gene.2006.07.034. [DOI] [PubMed] [Google Scholar]
- 25.Michalak K, Orr WC, Radyuk SN. Drosophila peroxiredoxin 5 is the second gene in a dicistronic operon. Biochem Biophys Res Commun. 2008;368:273–278. doi: 10.1016/j.bbrc.2008.01.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu H, Zhang S, Qiu W, Zhang G, Xia Q, et al. Isolation, characterization, and mapping of a novel human KRAB zinc finger protein encoding gene ZNF463. Biochim Biophys Acta. 2001;1518:190–193. doi: 10.1016/s0167-4781(01)00172-5. [DOI] [PubMed] [Google Scholar]
- 27.Daelemans C, Ritchie ME, Smits G, Abu-Amero S, Sudbery IM, et al. High-throughput analysis of candidate imprinted genes and allele-specific gene expression in the human term placenta. BMC Genet. 2010;11:25. doi: 10.1186/1471-2156-11-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meiboom M, Murua EH, Pentimalli F, Fusco A, Belge G, et al. A 3.4-kbp transcript of ZNF331 is solely expressed in follicular thyroid adenomas. Cytogenet Genome Res. 2003;101:113–117. doi: 10.1159/000074165. [DOI] [PubMed] [Google Scholar]
- 29.Piotrowski A, Bruder CE, Andersson R, Diaz dS, Menzel U, et al. Somatic mosaicism for copy number variation in differentiated human tissues. Hum Mutat. 2008;29:1118–1124. doi: 10.1002/humu.20815. [DOI] [PubMed] [Google Scholar]
- 30.Mkrtchyan H, Gross M, Hinreiner S, Polytiko A, Manvelyan M, et al. Early embryonic chromosome instability results in stable mosaic pattern in human tissues. PLoS One. 2010;5:e9591. doi: 10.1371/journal.pone.0009591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vanneste E, Voet T, Le Caignec C, Ampe M, Konings P, et al. Chromosome instability is common in human cleavage-stage embryos. Nat Med. 2009;15:577–583. doi: 10.1038/nm.1924. [DOI] [PubMed] [Google Scholar]
- 32.Schwartz M. The emergence of a new science of the mind: immunology benefits the mind. Mol Psychiatry. 2010;15:337–338. doi: 10.1038/mp.2010.22. [DOI] [PubMed] [Google Scholar]
- 33.Torkamani A, Dean B, Schork NJ, Thomas EA. Coexpression network analysis of neural tissue reveals perturbations in developmental processes in schizophrenia. Genome Res. 2010;20:403–412. doi: 10.1101/gr.101956.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Singh SM, O'Reilly R. (Epi)genomics and neurodevelopment in schizophrenia: monozygotic twins discordant for schizophrenia augment the search for disease-related (epi)genomic alterations. Genome. 2009;52:8–19. doi: 10.1139/G08-095. [DOI] [PubMed] [Google Scholar]
- 35.Bruder CE, Piotrowski A, Gijsbers AA, Andersson R, Erickson S, et al. Phenotypically concordant and discordant monozygotic twins display different DNA copy-number-variation profiles. Am J Hum Genet. 2008;82:763–771. doi: 10.1016/j.ajhg.2007.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ollikainen M, Smith KR, Joo EJ, Ng HK, Andronikos R, et al. DNA methylation analysis of multiple tissues from newborn twins reveals both genetic and intrauterine components to variation in the human neonatal epigenome. Hum Mol Genet. 2010 doi: 10.1093/hmg/ddq336. [DOI] [PubMed] [Google Scholar]
- 37.Fraga MF, Ballestar E, Paz MF, Ropero S, Setien F, et al. Epigenetic differences arise during the lifetime of monozygotic twins. Proc Natl Acad Sci U S A. 2005;102:10604–10609. doi: 10.1073/pnas.0500398102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kaminsky ZA, Tang T, Wang SC, Ptak C, Oh GH, et al. DNA methylation profiles in monozygotic and dizygotic twins. Nat Genet. 2009;41:240–245. doi: 10.1038/ng.286. [DOI] [PubMed] [Google Scholar]
- 39.Baranzini SE, Mudge J, van Velkinburgh JC, Khankhanian P, Khrebtukova I, et al. Genome, epigenome and RNA sequences of monozygotic twins discordant for multiple sclerosis. Nature. 2010;464:1351–1356. doi: 10.1038/nature08990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Milo R, Kahana E. Multiple sclerosis: geoepidemiology, genetics and the environment. Autoimmun Rev. 2010;9:A387–A394. doi: 10.1016/j.autrev.2009.11.010. [DOI] [PubMed] [Google Scholar]
- 41.Hastings PJ, Lupski JR, Rosenberg SM, Ira G. Mechanisms of change in gene copy number. Nat Rev Genet. 2009;10:551–564. doi: 10.1038/nrg2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang F, Khajavi M, Connolly AM, Towne CF, Batish SD, et al. The DNA replication FoSTeS/MMBIR mechanism can generate genomic, genic and exonic complex rearrangements in humans. Nat Genet. 2009;41:849–853. doi: 10.1038/ng.399. [DOI] [PMC free article] [PubMed] [Google Scholar]