Abstract
Therapeutics that discriminate between the genetic makeup of normal cells and tumour cells are valuable for treating and understanding cancer. Small molecules with oncogene-selective lethality may reveal novel functions of oncoproteins and enable the creation of more selective drugs1. Here we describe the mechanism of action of the selective anti-tumour agent erastin, involving the RAS–RAF–MEK signalling pathway functioning in cell proliferation, differentiation and survival. Erastin exhibits greater lethality in human tumour cells harbouring mutations in the oncogenes HRAS, KRAS or BRAF. Using affinity purification and mass spectrometry, we discovered that erastin acts through mitochondrial voltage-dependent anion channels (VDACs)—a novel target for anti-cancer drugs. We show that erastin treatment of cells harbouring oncogenic RAS causes the appearance of oxidative species and subsequent death through an oxidative, non-apoptotic mechanism. RNA-interference-mediated knockdown of VDAC2 or VDAC3 caused resistance to erastin, implicating these two VDAC isoforms in the mechanism of action of erastin. Moreover, using purified mitochondria expressing a single VDAC isoform, we found that erastin alters the permeability of the outer mitochondrial membrane. Finally, using a radiolabelled analogue and a filter-binding assay, we show that erastin binds directly to VDAC2. These results demonstrate that ligands to VDAC proteins can induce non-apoptotic cell death selectively in some tumour cells harbouring activating mutations in the RAS–RAF–MEK pathway.
In a screen of about 24,000 compounds, we discovered erastin, which induces rapid death in engineered human tumour cells (BJ-TERT/LT/ST/RASV12 cells, ref. 2) with oncogenic v-Ha-ras Harvey rat sarcoma viral oncogene homologue (HRAS)V12, but not in isogenic, non-tumorigenic cells lacking oncogenic RAS (BJ-TERT/LT/ST cells) (Fig. 1a, Supplementary Fig. 1 and ref. 3). This cell death was not dependent on the rate of cell division, nor was it idiosyncratic to these cells (Fig. 1a and Supplementary Fig. 2), because cell lines engineered in a similar way responded similarly.
Figure 1. Erastin activates a rapid, oxidative, non-apoptotic cell death process.
a, HRASV12-expressing cell lines are sensitive to erastin, whereas isogenic lines lacking HRASV12 are resistant, as determined by Trypan blue exclusion; the graph is a representative outcome of multiple independent experiments. b, Transmission electron microscopy images (×20,000) of BJ-TERT/LT/ST/RASV12 mitochondria after cells were treated with nothing, erastin (37 μM for 10 h) or staurosporine (STS, 1 μM for 5 h). c, Phase-contrast photograph of BJ-TERT/LT/ST/RASV12 cells 24 h after 9 μM erastin treatment indicates that nuclei are intact after cell death. d, Anti-oxidants suppress erastin-induced death in BJ-TERT/LT/ST/RASV12 cells. BHT, butylated hydroxytoluene; DMSO, dimethylsulphoxide. e, Level of intracellular oxidative species in BJ-TERT/LT/ST/RASV12 (black line) or BJ-TERT (grey line) cells treated with 4.6 μM erastin. y axis, percentage of cells exhibiting dichlorofluorescein (DCF) fluorescence within region of interest, M1, as measured by flow cytometry; error bars, one standard deviation, n = 2. f, PARP1 cleavage is not seen during erastin-induced cell death in A-673, HT-1080 and HeLa cells. NT, no treatment. g, STS, but not erastin, induces cytochrome c (cyt c) release from BJ-TERT/LT/ST/RASV12 mitochondria; mitochondrial fraction, mito; cytosolic fraction, cyto. h, Pro-caspase-3 is not cleaved in response to erastin.
We found that erastin-treated cells did not display changes in nuclear morphology characteristic of apoptosis (Fig. 1c, ref. 3). However, imaging by electron microscopy did reveal changes in mitochondrial morphology, such as loss of structural integrity (Fig. 1b). These mitochondrial morphological changes were not observed in response to staurosporine, hydrogen peroxide or rapamycin—compounds that induce apoptosis, necrosis and autophagy, respectively (Fig. 1b and data not shown).
Given that erastin is a new compound found in a cell-based screen, we had no insight into its mechanism of action. We used a two-pronged approach to define the mechanism, involving, first, a suppressor screen to identify annotated compounds that prevent erastin-induced cell death and, second, an affinity purification approach to identify proteins that mediate the activity of erastin. First, we performed a suppressor screen using a library of about 2,000 biologically active compounds4 and found that antioxidants (α-tocopherol, butylated hydroxytoluene and β-carotene) prevent erastin-induced death (Fig. 1d, Supplementary Fig. 3). Moreover, we detected generation of an oxidizing species in response to erastin treatment in BJ-TERT/LT/ST/RASV12 cells, but not in isogenic BJ-TERT cells (Fig. 1e, Supplementary Fig. 4). Finally, we found that erastin-induced death in the HT-1080 fibrosarcoma cell line was also suppressed by antioxidants (Supplementary Fig. 5).
To characterize the mode of erastin-induced cell death, we looked for features of well characterized death pathways. We determined that the oxidizing species generated in BJ-TERT/LT/ST/RASV12 cells in the presence of erastin emanate from mitochondria (see Supplementary Discussion), consistent with the perturbation in mitochondrial morphology. We discovered that the oxidizing species do not cause poly(ADP ribose) polymerase 1 (PARP1) cleavage (Fig. 1f, Supplementary Fig. 6), cytochrome c release from mitochondria (Fig. 1g), or pro-caspase-3 cleavage (Fig. 1h)3,5,6—all of which are hallmarks of apoptosis, a stereotypical form of cell death activated by many anti-tumour agents3,7–10 as well as by staurosporine (Fig. 1f–h). Moreover, we looked for these hallmarks in four different cell lines sensitive to erastin; none showed activation of these markers (Fig. 1f, h). Other canonical hallmarks of apoptosis were similarly absent (see Supplementary Information). In summary, these initial studies revealed that erastin induces rapid, oxidative, non-apoptotic death in tumour cells with oncogenic HRAS.
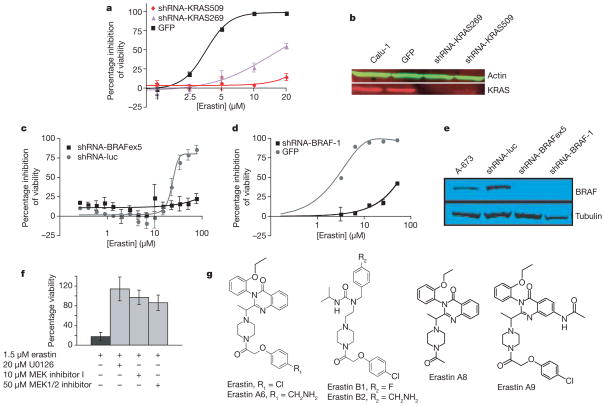
To define the genetic basis of the selective lethality of erastin, we used a lentiviral-based RNA interference system11. We originally discovered erastin in a screen for oncogenic-HRAS-selective lethal compounds. However, v-Ki-ras2 Kirsten rat sarcoma viral oncogene homologue (KRAS) is more frequently mutated in human cancers than HRAS12. Selective toxicity in mutant-KRAS-expressing cell lines would broaden the applicability of erastin as a therapeutic. Thus, we tested whether erastin was selectively lethal to tumour cells harbouring oncogenic KRAS, and whether this lethality could be arrested by knockdown of KRAS. We found that a lung carcinoma cell line (Calu-1) with an activating mutation in KRAS was sensitive to erastin (half-maximal inhibitory concentration, IC50 = 4 μM; Fig. 2a); when infected with lentiviral constructs expressing two different short hairpin RNAs (shRNAs) targeting KRAS (Supplementary Table 4), these cells exhibited resistance to erastin (Fig. 2a, b).
Figure 2. Erastin lethality is dependent on the RAS–RAF–MEK pathway.
a, Calu-1 cells infected with lentivirus containing shRNAs targeting KRAS were resistant to erastin-induced lethality as quantified by way of comparison with green fluorescent protein (GFP) control in Trypan blue exclusion assay. Sequences of shRNAs are indicated by starting nucleotide in the KRAS mRNA coding sequence. b, KRAS knockdown was confirmed by western blot analysis. c, d, A-673 cells were infected with lentivirus expressing indicated shRNAs or a control GFP plasmid. Cells were treated with erastin for 24 h; percentage inhibition of viability (y axis) was measured using c, Alamar blue and d, Trypan blue. Luc, luciferase; BRAFex5, exon 5 of BRAF transcript. e, BRAF knockdown was confirmed by western blot analysis. f, 48 h treatment with MEK inhibitors (U0126, Sigma; MEK inhibitor 1, Calbiochem; MEK1/2 inhibitor, Calbiochem) prevents erastin-induced lethality in BJ-TERT/LT/ST/RASV12 cells; percentage viability (y axis) was determined using Trypan blue. g, Structures of erastin and related analogues. All error bars in Fig. 2 represent one standard deviation; n =2 or 3.
RAS proteins are known to modulate many downstream pathways, and genetic lesions in these pathways are associated with many cancer types12. To clarify the connection between erastin and oncogenic RAS signalling, we sought evidence that erastin acts in a manner that is specific to cells with activated RAS–RAF–MEK signalling (Supplementary Table 1 and Supplementary Discussion). One cell line with moderate sensitivity to erastin was A-673 (Supplementary Table 1), containing an activating V600E mutation in v-raf murine sarcoma viral oncogene homologue B1 (BRAF)—a direct target of RAS13. To determine whether the activating mutation in BRAF influences erastin sensitivity, we created two different shRNAs targeting BRAF messenger RNA (Supplementary Table 4). We found that A-673 cells containing either of these constructs were resistant to erastin (Fig. 2c–e). Moreover, co-expression of a non-targetable V600E mutant BRAF partially restored sensitivity of these cells to erastin (Supplementary Fig. 7).
To confirm that activated RAS–RAF–MEK signalling renders other tumour cells sensitive to erastin, we examined the effect of mitogen-activated protein kinase (MAPK)/extracellular signal-regulated kinase (ERK) kinase 1/2 (MEK1/2) inhibitors on erastin sensitivity. All three inhibitors caused erastin resistance in both BJ-TERT/LT/ST/RASV12 and HT-1080 cells, with activating mutations in HRAS and NRAS, respectively (Fig. 2f and Supplementary Fig. 8; Supplementary Table 2 shows the effect of these MEK inhibitors alone on cell viability). Finally, we found a modest correlation between ERK1/2 phosphorylation status and erastin sensitivity in 12 sarcoma cell lines (Table 1). In summary, KRAS and BRAF knockdown, MEK inhibition, and analysis of ERK1/2 phosphorylation status together suggest that erastin contains a degree of selectivity for cells in which the RAS–RAF–MEK pathway is constitutively activated.
Table 1.
Correlation between erastin sensitivity and phospho-ERK level
| Human cell line | Erastin sensitivity (%) | Phospho-ERK1/2 |
|---|---|---|
| A-673 | 54 | 0.66 |
| BJ-TERT | 22 | 0.09 |
| BJ-TERT/LT/ST/RASV12 | 100 | 0.92 |
| EWS 502 | 42 | 0.12 |
| HL-60 | 0 | 0.10 |
| HT-1080 | 98 | 0.53 |
| SK-ES-1 | 96 | 0.25 |
| SK-N-MC | 95 | 0.05 |
| SW 872 | 0 | 0.12 |
| TC 32 | 88 | 0.12 |
| TC 71 | 92 | 0.23 |
| U-937 | 73 | 0.27 |
The maximum percentage cell death induced by erastin in each cell line is shown, along with the level of phospho-ERK1/2 as quantified by western blot (arbitrary units). The correlation is 0.41.
For the second prong of our approach to defining the mechanism of action of erastin (affinity-based target identification), we synthesized erastin analogues that could be linked to a solid-phase resin for biochemical purification of potential targets. We found that replacement of the p-chloro substituent in erastin with the aminomethyl group necessary for linkage to a resin resulted in an analogue (erastin A6, Fig. 2g) that, though less potent, retained the ability to kill BJ-TERT/LT/ST/RASV12 cells, but not BJ-TERT cells (Supplementary Fig. 9). We also identified a suitable analogue (erastin B2, Supplementary Information) that lacked activity (Fig. 2g and Supplementary Fig. 9), and thus could serve as a negative control for target purification.
We immobilized erastin A6 and erastin B2 on solid-phase resin and sought proteins that interact with A6, but not B2. Using BJ-TERT/LT/ST/RASV12 cell lysates, we identified all three isoforms of the human mitochondrial voltage-dependent anion channels (VDAC1, VDAC2 and VDAC3) on the A6 resin, but only VDAC1 on the B2 resin (Supplementary Table 3, 7–14). Using BJ-TERT cell lysates, we identified a small amount of VDAC1 on the A6 resin, but none of the VDACs on the B2 resin. Thus, it appears that erastin A6 isolates VDAC2 and VDAC3 more efficiently than does erastin B2. Furthermore, the finding that erastin pulls down a mitochondrial protein (VDAC) is consistent with observations that erastin induces mitochondria-driven oxidative death.
VDACs (also known as eukaryotic porins) are membrane-spanning channels that facilitate transmembrane transport of ions and metabolites14,15, most notably across the outer mitochondrial membrane16. It has been demonstrated that VDACs are gated by membrane voltage, at least in vitro (Supplementary Discussion): in the closed state, ions, but not small molecule metabolites, can penetrate VDAC pores17; in the open state, both ions and metabolites pass through VDAC channels. In addition, the closed state is cation-selective, whereas the open state is anion-selective.
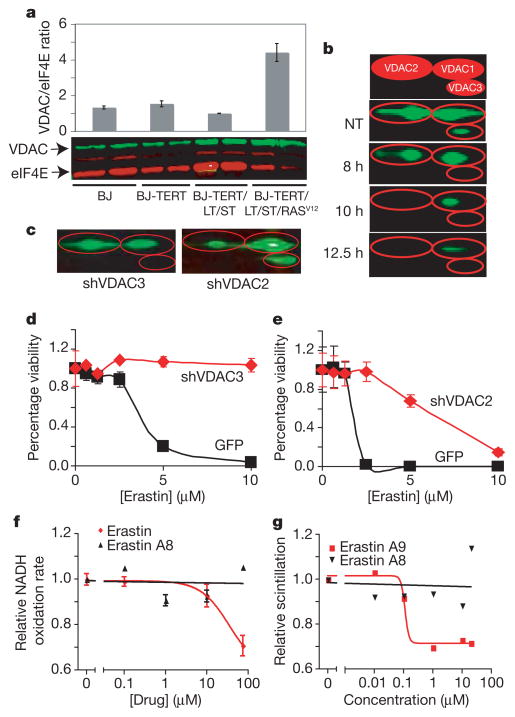
Having identified VDACs as a potential target of erastin, we sought evidence that altered expression of VDACs contributes to altered erastin sensitivity. To determine whether VDACs are upregulated in response to oncogenic HRAS, we measured VDAC abundance in the BJ cell series (see ref. 2 and Supplementary Discussion for engineering and genetics of BJ cell series). In BJ-TERT/LT/ST/RASV12 cells, the total amount of VDAC protein is increased (Fig. 3a and Supplementary Fig. 10). These results suggest that erastin acts by a gain-of-function mechanism, and that cells with more VDAC protein are more sensitive to erastin, although other aspects of cellular physiology may also be relevant.
Figure 3. Erastin compounds act through VDACs.
a, VDAC/eukaryotic initiation factor 4E (eIF4E) protein ratio in engineered BJ-derived cells as quantified using western blot. b, BJ-TERT/LT/ST/RASV12 cells were treated with erastin and harvested at indicated time points for quantitative two-dimensional western blot. The top panel is an illustration of VDAC isoforms separated by two-dimensional gel electrophoresis. c, Isoform-specific knockdown of VDAC in HT-1080 cells infected with virus expressing either VDAC3- or VDAC2-targeted shRNA plasmid (shVDAC3 or shVDAC2) was confirmed using two-dimensional protein gels. d, e, These cells were then treated with erastin dilutions, and viability relative to no treatment (y axis) was determined using Trypan blue exclusion and compared to an identical process using a GFP control plasmid. f, Rate of NADH oxidation, normalized to no treatment, in mitochondria purified from VDAC-knockout yeast expressing murine VDAC2 was determined in the presence of erastin (red) or an inactive analogue, erastin A8 (black). g, Direct binding to VDAC2 was assayed using tritium-labelled erastin A9 in competition with unlabelled erastin A9 (red) or erastin A8 (black). All error bars in Fig. 3 represent one standard deviation, n =2 or 3.
Next, using two-dimensional gel electrophoresis to evaluate protein expression in BJ-TERT/LT/ST/RASV12 cells (Supplementary Fig. 11), we found that, after 8 h of erastin treatment, VDAC3 was no longer detectable, and, after 10 h, VDAC2 became undetectable (Fig. 3b). Doxorubicin and camptothecin are examples of agents that act by a gain-of-function mechanism on their molecular targets, topoisomerase IIα and topoisomerase I, respectively18. It may be that a cellular response to erastin is downregulation of VDAC2/3 after lethal oxidative species have been generated, as occurs with camptothecin and topoisomerase I following DNA damage. The fact that VDAC1 is still present at later time points suggests that the loss of VDAC2/3 is not simply caused by loss of mitochondria.
To test the gain-of-function hypothesis, we reduced VDAC protein levels using lentiviral shRNA expression11. We created multiple shRNA constructs targeting each VDAC isoform (Supplementary Table 4 and Supplementary Fig. 12) and tested their effects on cell sensitivity to erastin (Supplementary Figs 13–15). We found that knockdown of VDAC3 caused significant resistance to erastin (Fig. 3c, d and Supplementary Fig. 15). We also observed some degree of erastin resistance when VDAC2 was knocked down (Fig. 3c, e and Supplementary Fig. 14). In contrast, overexpression of VDAC3 alone in BJ-TERT cells yielded no increase in sensitivity to erastin (Supplementary Fig. 16), suggesting that VDAC3, and to some degree VDAC2, is necessary but not sufficient for sensitivity to erastin and that other downstream features of RAS–RAF–MEK signalling are also needed. Overall, these results are consistent with a gain-of-function mechanism involving erastin and VDAC2/3. Furthermore, this effect is specific to erastin, and not to other lethal compounds: VDAC2-deficient embryonic stem cells have been shown to be more sensitive, not less sensitive, to staurosporine and etoposide19.
To test the hypothesis that erastin alters mitochondrial outer membrane permeability, we monitored the function of VDACs using purified mitochondria from yeast engineered to express a single mouse VDAC isoform in place of yeast VDAC20. A previous report demonstrated that the rate of NADH uptake through the outer membrane of such mitochondria is dependent on the specific VDAC expressed in yeast. We found that erastin treatment yielded a decrease in the rate of NADH oxidation, suggesting a reduced permeability of these mitochondria to NADH when mouse VDAC1 or VDAC2 were expressed (Fig. 3f and Supplementary Fig. 17). We found little NADH oxidation in mitochondria expressing VDAC3, suggesting minimal intrinsic membrane permeability (Supplementary Fig. 17); this is consistent with previous reports that VDAC3 does not gate well in vitro20. Finally, we found that an inactive analogue of erastin (erastin A8, Fig. 2g and Supplementary Fig. 9) had no such effect (Fig. 3f and Supplementary Fig. 17). These results suggest that erastin affects VDAC gating, possibly switching its ion selectivity and allowing cationic species into mitochondria.
Having demonstrated interactions between erastin and VDAC using affinity-based target identification and VDAC functional assays, we explored the direct binding of erastin to VDACs. Using modified versions of previously reported protocols, we isolated VDAC2 from Escherichia coli for use in a competition binding experiment21,22 using a radiolabelled analogue (erastin A9). The results demonstrate that the cold RAS-selective lethal erastin A9 (IC50 = 1.9 μM, Supplementary Fig. 9), unlike inactive erastin A8, directly binds to VDAC2 (dissociation constant, Kd = 112 nM; Fig. 3g), in the process competing off radiolabelled erastin A9.
The data presented herein are consistent with a model in which erastin interacts with VDAC proteins to induce mitochondrial dysfunction, release of oxidative species and, ultimately, non-apoptotic, oxidative cell death. This process has a degree of selectivity for cells with activated RAS–RAF–MEK signalling. These results demonstrate that it is feasible to discover oncogene-selective compounds and to use them to clarify oncogene-related cell death mechanisms.
METHODS SUMMARY
VDAC identification using pull-down assay
Erastin A6 and B2 were linked to a solid-phase resin, which was then incubated with BJ-TERT/LT/ST/RASV12 or BJ-TERT cell lysate and washed before protein was eluted and then digested as described23. Reverse-phase high-performance liquid chromatography was performed before samples were analysed using both a 4700 Proteomics Analyser matrix-assisted laser desorption/ionization-time of flight (TOF)/TOF (TOF/TOF; Applied Biosystems) and a Q Trap (AB/MDS Sciex). Tandem mass spectrometry (MS/MS) data was then compared against either the corrected NCBInr protein sequence database or the HumanNR database (http://www.ncbi.nlm.nih.gov/). GPS Explorer (Applied Biosystems) was used for submitting data acquired from the TOF/TOF for database searching.
Knockdown using lentiviral shRNAs
HT-1080 and Calu-1 cells were infected with lentiviruses expressing shRNAs targeting VDAC and KRAS, respectively (Supplementary Table 4). A-673 cells were infected with retroviruses24 expressing shRNAs against either BRAF or luciferase (Supplementary Table 4).
NADH oxidation assay
NADH oxidation in the mitochondria was measured by resuspending mitochondria isolated from yeast in 25 μM NADH, with or without erastin, and monitoring absorbance at λ =340 nm (ref. 20). As a control for mitochondrial intactness, parallel assays were run using hypotonically shocked mitochondria.
VDAC2 binding assay
VDAC2 protein was isolated from E. coli using a modified version of a previously described protocol22. To assay direct binding of erastin analogues, 40 μg of purified VDAC2 was incubated in the presence of 20 μM radiolabelled erastin A9 and serial dilutions of unlabelled erastin A9 or erastin A8. The mixture was then deposited onto a binding filter using vacuum filtration. After rinsing, radioactivity was detected on a liquid scintillation counter (LKB Wallac 1,211 RACKBETA).
Full protocols
For detailed methods, which additionally include cell culture, western blotting, cell viability assays, PARP cleavage, cytochrome c release, oxidative species detection, transmission electron microscopy, VDAC3 overexpression, PCR with reverse transcription and quantitative PCR, see Methods.
Full Methods and any associated references are available in the online version of the paper at www.nature.com/nature.
Supplementary Material
Acknowledgments
We thank S. Flaherty and S. Dolma for supporting experiments, H. Widlund for supplying the BRAF shRNA construct, R. Becklin and J. Savage for help with the analysis of the pull-down data, P. Robbins for help with the pull-down experiments, K. Brown for assistance with transmission electron microscopy, and M. Colombini for supplying engineered yeast and for discussions. B.R.S. is supported by a Career Award at the Scientific Interface from the Burroughs Welcome Fund and by the National Cancer Institute. S.L.L. is supported by an NCI grant, an American Cancer Society Research Scholars Grant, the Terri Anna Perine Sarcoma Fund, a Primary Children’s Medical Center Foundation Innovative Research Grant, a Hope Street Kids grant and a Catalyst Grant from the University of Utah School of Medicine.
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author Contributions N.Y. designed and performed the RNAi and VDAC overexpression, quantitative PCR, erastin analogue viability and chemical characterization experiments. E.Z. performed two-dimensional western analysis, PARP-1 and pro-caspase-3 cleavage, and cytochrome c release experiments. E.Z. and N.Y. performed transmission electron microscopy experiments. A.J.B., D.J.F. and N.Y. performed the NADH oxidation and direct binding experiments. W.S.Y. characterized sensitivity to erastin in the BJ-derived cell series. A.J.W. performed the MEK1/2 inhibitor experiment. I.S. and A.J.B. synthesized erastin analogues. R.S. and S.L.L. provided BRAF shRNAs, analysis of BRAF knockdown and the phospho-ERK western analysis. J.M.P., J.J.B. and S.S. were responsible for setting up the technology platform to pull down proteins binding to small molecule compounds. M.v.R. and J.M.P. performed the pull-down experiments. J.J.B., J.M.P. and S.S. designed, reviewed and supervised the pull-down experiments, and contributed to the analysis of the data. B.R.S. conceived of and supervised the project, designed and analysed experiments, and performed the anti-oxidant studies. B.R.S. and N.Y. prepared the manuscript.
Author Information Reprints and permissions information is available at www.nature.com/reprints. The authors declare no competing financial interests. Correspondence and requests for materials should be addressed to B.R.S. (stockwell@biology.columbia.edu).
References
- 1.Kaelin WG. The concept of synthetic lethality in the context of anticancer therapy. Nature Rev Cancer. 2005;5:689–698. doi: 10.1038/nrc1691. [DOI] [PubMed] [Google Scholar]
- 2.Hahn WC, et al. Creation of human tumour cells with defined genetic elements. Nature. 1999;400:464–468. doi: 10.1038/22780. [DOI] [PubMed] [Google Scholar]
- 3.Dolma S, Lessnick SL, Hahn WC, Stockwell BR. Identification of genotype-selective antitumor agents using synthetic lethal chemical screening in engineered human tumor cells. Cancer Cell. 2003;3:285–296. doi: 10.1016/s1535-6108(03)00050-3. [DOI] [PubMed] [Google Scholar]
- 4.Root DE, Flaherty SP, Kelley BP, Stockwell BR. Biological mechanism profiling using an annotated compound library. Chem Biol. 2003;10:881–892. doi: 10.1016/j.chembiol.2003.08.009. [DOI] [PubMed] [Google Scholar]
- 5.Song Z, Steller H. Death by design: mechanism and control of apoptosis. Trends Cell Biol. 1999;9:M49–M52. [PubMed] [Google Scholar]
- 6.Majno G, Joris I. Apoptosis, oncosis, and necrosis. An overview of cell death. Am J Pathol. 1995;146:3–15. [PMC free article] [PubMed] [Google Scholar]
- 7.Wyllie AH. Glucocorticoid-induced thymocyte apoptosis is associated with endogenous endonuclease activation. Nature. 1980;284:555–556. doi: 10.1038/284555a0. [DOI] [PubMed] [Google Scholar]
- 8.Kerr JF, Wyllie AH, Currie AR. Apoptosis: a basic biological phenomenon with wide-ranging implications in tissue kinetics. Br J Cancer. 1972;26:239–257. doi: 10.1038/bjc.1972.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Martin SJ, et al. Early redistribution of plasma membrane phosphatidylserine is a general feature of apoptosis regardless of the initiating stimulus: inhibition by overexpression of Bcl-2 and Abl. J Exp Med. 1995;182:1545–1556. doi: 10.1084/jem.182.5.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yuan J, Shaham S, Ledoux S, Ellis HM, Horvitz HR. The C. elegans cell death gene ced-3 encodes a protein similar to mammalian interleukin-1 beta-converting enzyme. Cell. 1993;75:641–652. doi: 10.1016/0092-8674(93)90485-9. [DOI] [PubMed] [Google Scholar]
- 11.Moffat J, et al. A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high-content screen. Cell. 2006;124:1283–1298. doi: 10.1016/j.cell.2006.01.040. [DOI] [PubMed] [Google Scholar]
- 12.Downward J. Targeting RAS signalling pathways in cancer therapy. Nature Rev Cancer. 2003;3:11–22. doi: 10.1038/nrc969. [DOI] [PubMed] [Google Scholar]
- 13.Davies H, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 14.Graham BH, Craigen WJ. Genetic approaches to analyzing mitochondrial outer membrane permeability. Curr Top Dev Biol. 2004;59:87–118. doi: 10.1016/S0070-2153(04)59004-X. [DOI] [PubMed] [Google Scholar]
- 15.Baker MA, Lane DJ, Ly JD, De Pinto V, Lawen A. VDAC1 is a transplasma membrane NADH-ferricyanide reductase. J Biol Chem. 2004;279:4811–4819. doi: 10.1074/jbc.M311020200. [DOI] [PubMed] [Google Scholar]
- 16.Rostovtseva T, Colombini M. ATP flux is controlled by a voltage-gated channel from the mitochondrial outer membrane. J Biol Chem. 1996;271:28006–28008. doi: 10.1074/jbc.271.45.28006. [DOI] [PubMed] [Google Scholar]
- 17.Mannella CA. Minireview: on the structure and gating mechanism of the mitochondrial channel, VDAC. J Bioenerg Biomembr. 1997;29:525–531. doi: 10.1023/a:1022489832594. [DOI] [PubMed] [Google Scholar]
- 18.Beck WT, Danks MK. Mechanisms of resistance to drugs that inhibit DNA topoisomerases. Semin Cancer Biol. 1991;2:235–244. [PubMed] [Google Scholar]
- 19.Cheng EH, Sheiko TV, Fisher JK, Craigen WJ, Korsmeyer SJ. VDAC2 inhibits BAK activation and mitochondrial apoptosis. Science. 2003;301:513–517. doi: 10.1126/science.1083995. [DOI] [PubMed] [Google Scholar]
- 20.Xu X, Decker W, Sampson MJ, Craigen WJ, Colombini M. Mouse VDAC isoforms expressed in yeast: channel properties and their roles in mitochondrial outer membrane permeability. J Membr Biol. 1999;170:89–102. doi: 10.1007/s002329900540. [DOI] [PubMed] [Google Scholar]
- 21.Poyurovsky MV, et al. Nucleotide binding by the Mdm2 RING domain facilitates Arf-independent Mdm2 nucleolar localization. Mol Cell. 2003;12:875–887. doi: 10.1016/s1097-2765(03)00400-3. [DOI] [PubMed] [Google Scholar]
- 22.Koppel DA, et al. Bacterial expression and characterization of the mitochondrial outer membrane channel. Effects of n-terminal modifications. J Biol Chem. 1998;273:13794–13800. doi: 10.1074/jbc.273.22.13794. [DOI] [PubMed] [Google Scholar]
- 23.Zhen Y, et al. Development of an LC-MALDI method for the analysis of protein complexes. J Am Soc Mass Spectrom. 2004;15:803–822. doi: 10.1016/j.jasms.2004.02.004. [DOI] [PubMed] [Google Scholar]
- 24.Sage J, Miller AL, Perez-Mancera PA, Wysocki JM, Jacks T. Acute mutation of retinoblastoma gene function is sufficient for cell cycle re-entry. Nature. 2003;424:223–228. doi: 10.1038/nature01764. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.