Abstract
Plant diurnal oscillation is a 24-hour period based variation. The correlation between diurnal genes and biological pathways was widely revealed by microarray analysis in different species. Rice (Oryza sativa) is the major food staple for about half of the world's population. The rice flag leaf is essential in providing photosynthates to the grain filling. However, there is still no comprehensive view about the diurnal transcriptome for rice leaves. In this study, we applied rice microarray to monitor the rhythmically expressed genes in rice seedling and flag leaves. We developed a new computational analysis approach and identified 6,266 (10.96%) diurnal probe sets in seedling leaves, 13,773 (24.08%) diurnal probe sets in flag leaves. About 65% of overall transcription factors were identified as flag leaf preferred. In seedling leaves, the peak of phase distribution was from 2:00am to 4:00am, whereas in flag leaves, the peak was from 8:00pm to 2:00am. The diurnal phase distribution analysis of gene ontology (GO) and cis-element enrichment indicated that, some important processes were waken by the light, such as photosynthesis and abiotic stimulus, while some genes related to the nuclear and ribosome involved processes were active mostly during the switch time of light to dark. The starch and sucrose metabolism pathway genes also showed diurnal phase. We conducted comparison analysis between Arabidopsis and rice leaf transcriptome throughout the diurnal cycle. In summary, our analysis approach is feasible for relatively unbiased identification of diurnal transcripts, efficiently detecting some special periodic patterns with non-sinusoidal periodic patterns. Compared to the rice flag leaves, the gene transcription levels of seedling leaves were relatively limited to the diurnal rhythm. Our comprehensive microarray analysis of seedling and flag leaves of rice provided an overview of the rice diurnal transcriptome and indicated some diurnal regulated biological processes and key functional pathways in rice.
Introduction
A diurnal cycle is defined as a pattern that recurs over a 24-hr period. Plant diurnal oscillation is universal for plants and coordinates many biological pathways related to extracellular or intracellular signals, adapting the plants to daily alternation and maintaining a balance between metabolic reactions during light and darkness, especially for fluctuations of the carbon balance [1], [2], [3], [4], [5], [6], [7], [8], [9], [10]. During the light period, carbon fixation in leaves leads to sucrose synthesis through photosynthesis and the starch produced accumulates in the leaves. During darkness, the starch is degraded and so starch content decreases. Rice (Oryza sativa) is the major food staple for about half of the world's population and it is also a model monocot for studies of crop plants with relatively smaller genomes, due to the completion of its genome sequence. The rice genes related to starch synthesis are essential to improving grain quality (such as eating and cooking quality). In rice, flag leaves play important role in providing photosynthates to the filling grain.
The diurnal cycle also coordinates the opening and closing of stomata, affecting transpiration and changing the water potential in leaves. The opening of stomata facilitates carbon dioxide uptake for photosynthesis during the light [11], [12], [13]. The light regulation is essential in the metabolic and physiological functions of plants during the diurnal periods. Light is also crucial to entrain the endogenous circadian clock, ensuring the precise cyclic expression of circadian-regulated genes during the day. The significant correlation between diurnally oscillating genes during the diurnal cycle and growth hormone-responsive genes was revealed mostly through microarray-based transcriptome analysis [14], [15]. The plant growth-hormone pathways tightly interact with light signalling and the diurnal cycle in the control of plant growth.
Microarray experiments are normally used to collect large-scale time-series data, monitor genome-wide gene expression, profile the changes in transcripts, and identify novel genes regulated in the diurnal cycling and circadian clock during a 24-hr period [16]. The first version of an Affymetrix Arabidopsis microarray (containing 11,521 Arabidopsis ESTs) was applied to study the circadian clock-regulated key pathways in Arabidopsis by Harmer in 2000 [17]. In 2001, Ellen Wisman's group used another platform of microarray and found about 11% of the Arabidopsis genes showed a diurnal expression pattern and about 2% with a circadian rhythm [18]. Then the ATH1 Arabidopsis whole-genome array was also used to analyze the transcriptome throughout the diurnal cycle for clues concerning the diurnal and circadian-regulated starch metabolism in Arabidopsis leaves [1]. The transcriptome analysis for the diurnal changes of the starch metabolism-related genes indicated that there was transcriptional and posttranscriptional regulation of starch metabolism in Arabidopsis leaves. Recently, large microarray datasets related to the plant diurnal cycle (e.g. diurnal and circadian microarray data for Arabidopsis [1], [15], [18], [19], maize [20], [21], [22], barley [23] and soybean [24]) were published and some are available in public databases. In addition, the database of DIURNAL project (http://diurnal.cgrb.oregonstate.edu/) provides web-based data-mining tools that are multiple user-friendly for searching the diurnal and circadian microarray data of Arabidopsis, rice and poplar [19], [25].
During statistical analysis of diurnal microarray data, there are many challenges to correctly identify the subset of genes with a clear diurnal signature. A variety of methods have been developed to extract the cycling-expressed genes from microarray data for diurnal, circadian rhythm or cell cycle research. The computing methods mainly fall into two categories: frequency-domain and time-domain analyses [26].
The frequency-domain method is generally based on classical spectral analysis (e.g. Fourier transform and periodogram). After applying those methods, microarray expression profiles are transformed into frequency domains, then the rhythmic genes will yield the spectra that have a well-defined peak in the frequency domain. For diurnal or circadian-related genes, their periodogram will have a peak near a 24-hr period. Several prior studies have adapted these spectral methods to analyze biological data, e.g. FFT-NLLS [27], average periodogram [28] and Lomb–Scargle periodogram [29].
The time-domain method is an alternative method to analyze a time series using a pattern matching technique. To detect periodic patterns, the theoretical model is usually sinusoid. Two different methods are commonly used to measure the similarity between the models and the real data: nonlinear least-square curve fitting and cross correlation (CC). These algorithms assign to each time-series the properties of the model to which it is most similar. There are several programs available to perform the computation, including Cosinor [30], CORRCOS [17] and COSOPT [31]. Recently, we developed a new algorithm called ARSER to analyze diurnal or circadian expression data by combining the time-domain and frequency-domain analyses [32]. Testing by synthetic and real experimental data showed that it efficiently identified periodicity in short time-series.
Recently, little is known about the possible mechanism related to the rice diurnal cycle involved in metabolism, cellular function and growth. Global transcriptome analysis of the rice diurnal-cycle is also limited. In our study, we used the GeneChip Rice Genome Array representing 51,279 transcripts to monitor the expression profiles of rice flag leaves and seedling leaves during diurnal cycling. We employed the ARSER and CC methods together to analyze our microarray datasets. We also compared the predicted diurnal genes between rice and Arabidopsis. To elucidate the biological process involved in the rice day–night cycle, several approaches are being used, such as gene ontology (GO) enrichment analysis, MapMan, and cis-element analysis for the genes with similar diurnal phases. The present study might give some interesting insight into the light–dark diurnal cycle in plants.
Results
Experimental design and data quality
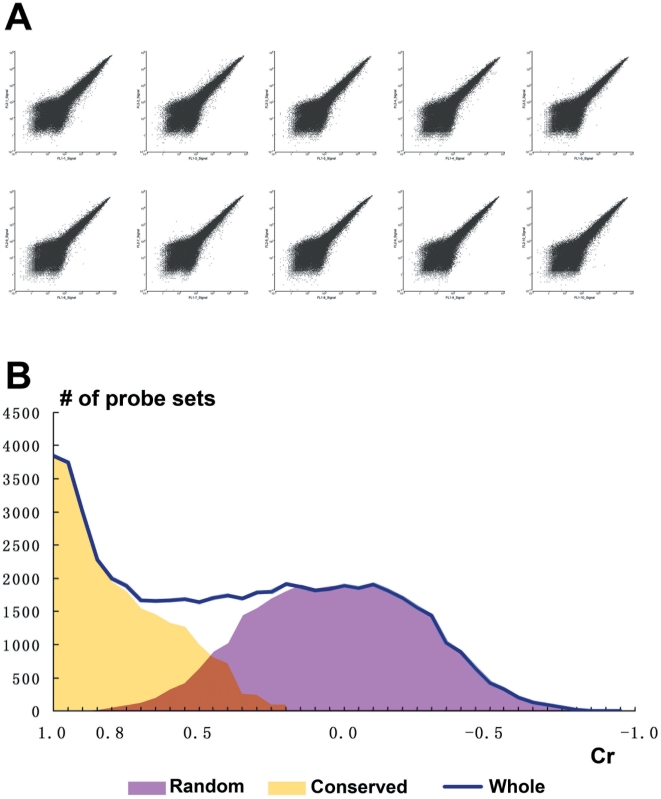
Rice flag leaf and seedling leaf samples were collected every 4 hr in a period of 36 hr with two biological replicates. The expression profile for each sample was obtained by Affymetrix Rice Genome GeneChip analysis. Detailed pair-wise scatter plots of biological replicates were generated for flag leaves (Figure 1A) and for seedling leaves (Figure S1A). For biological replicates of each time point, nearly all of probe sets were fallen along the diagonal of plots, indicating no major variation. The correlation coefficients and false-positive rates of each pair of biological replicate samples were calculated by GCOS (Table 1). All correlations were >0.95, while false-positive rates were <5%. In summary, the data quality was satisfactory for identifying genes with diurnal patterns.
Figure 1. Expression profiles between the biological replicate samples of flag leaves.
A: Pair-wise scatter plots for the raw probe-set intensity data across all time points. B: The number distribution of all probe sets based on the correlation coefficient (Cr) across the time series between the biological Os.7890.1.S1_x_at samples
Table 1. Comparison of the expression profiles between the biological replicate samples.
| Sample # | Time point (hr) | Seedling leaves | Flag leaves | Comments | ||
| correlation | False Positive Rate (%) | correlation | False Positive Rate (%) | |||
| 1 | 8 | 0.98 | 0.85 | 0.98 | 3.82 | Day 1 8:00am, the end of night |
| 2 | 12 | 0.99 | 1.26 | 0.98 | 3.05 | |
| 3 | 16 | 0.99 | 1.18 | 0.98 | 2.21 | |
| 4 | 20 | 0.99 | 0.92 | 0.98 | 2.08 | Day 1 8:00pm, the start of night |
| 5 | 24 | 0.98 | 0.95 | 0.96 | 4.71 | Day 2 0:00am |
| 6 | 28 | 0.98 | 1.00 | 0.96 | 3.64 | |
| 7 | 32 | 0.98 | 1.00 | 0.99 | 1.62 | Day 2 8:00am, the end of night |
| 8 | 36 | 0.99 | 1.20 | 0.99 | 1.72 | |
| 9 | 40 | 0.96 | 4.09 | 0.99 | 1.50 | |
| 10 | 44 | 0.99 | 1.57 | 0.98 | 2.55 | Day 2 8:00pm, the start of night |
In addition, we calculated the correlation coefficient of each probe set (Cr) across the time-series of two sets of biological replicates in flag leaves and seedling leaves (Figure 1B and Figure S1B, respectively). As we had assumed, there were two forms among the expression patterns of each probe set along the detection time points: one was consistent and similar between biological replicates, with the Cr of replicate time-series tending to 1; the other was random, occasionally matched in biological replicates, and the average of Cr tended to 0. The number distribution of Cr (flag or seedling leaves) could be divided into two parts during analysis: the random part, close to a normal distribution, shown in purple in Figure 1B and Figure S1B; and the conserved part, shown in yellow, which represented the probe sets with consistent expression pattern.
In the meanwhile, we performed real-time RT-PCR to validate microarray results of flag leaves (Figure S2A) and seedling leaves (Figure S2B) in seven time points from 12 hr to 36 hr. Several genes were selected, including light-harvesting chlorophyll a/b binding protein (LOC_Os03g39610), starch and sucrose metabolism pathway related proteins such as glycogen/starch synthases (LOC_Os07g22930), 4-alpha-glucanotransferase (LOC_Os07g46790), and Glucan water dikinase (LOC_Os06g30310).The real-time RT-PCR results mostly matched the microarray expression patterns.
Algorithms selected for diurnal pattern identification
To identify the genes with periodic expression patterns, we assumed that the expression profile of a gene exhibiting a diurnal pattern approximated a cosine wave with a period of nearly 24 hr. Several methods or algorithms are available for diurnal pattern identification, and two were selected: ARSER and CC. ARSER is a newly developed algorithm to analyze diurnal or circadian expression data by combining the time-domain and frequency-domain analyses, and was shown to be efficient in identifying periodicity in short time-series [32]. CC calculates the Pearson's correlation between a rhythmically-expressed gene and a theoretical cosine wave with a defined 24-hr period, and is a typical pattern matching technique [17].
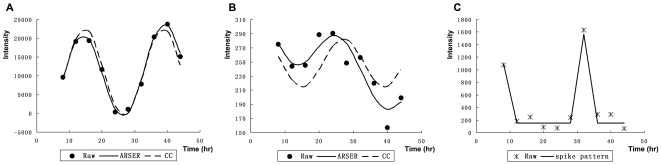
Figure 2 shows some probe set examples with a diurnal pattern identified. Figure 2A presents the Os.7890.1.S1_x_at (CAB gene) expression value during 8–44 hr in 4 hr spaces with its predicted cosine-curve model using ARSER (AR) and CC methods. The calculation showed that the two methods almost agreed. The ARSER method gave a better fit than CC for the probe set for Os.15803.1.S1_at (Figure 2B). The probe set for Os.22928.2.S1_at showed a typical spike diurnal pattern (Figure 2C), which was only recognized by the ARSER method.
Figure 2. Examples for diurnal probe sets for different pattern identification algorithms.
The scatter plot is a raw expression value of probe sets for 8–44 h, with sampling every 4 h; the solid line represents the cosine-curve model with ARSER (AR) simulation; the dash line represent the cosine-curve model with cross-correlation (CC) simulation. A: Os.7890.1.S1_x_at, seedling leaves, replicate sample 1. B: Os.15803.1.S1_at, seedling leaves, replicate sample 2. C: Os.22928.2.S1_at, flag leaves, replicate sample 1
The rice diurnal pattern genes in flag and seedling leaves
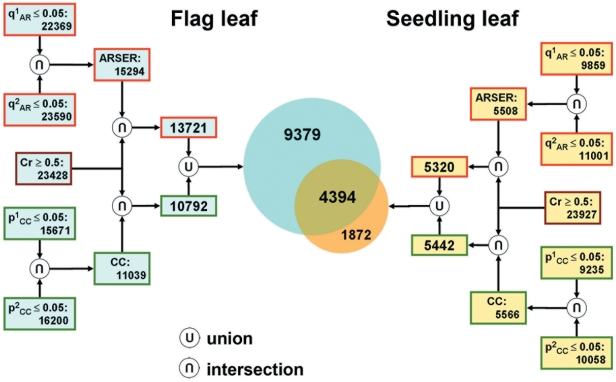
The cosine-curve model of each probe set was calculated by the ARSER and CC algorithms, for every replicate sample series of flag and seedling leaves. To obtain more reliable diurnal pattern genes, we considered the p- or q-value of the model curve and the Cr between biological replicate sample series for each probe set. If the p- or q-values were all <0.05 in biological replicates and the Cr >0.5, then we used the combined results of the two algorithms. With the combinative criteria (Figure 3), 6,266 probe sets (10.96%) were identified with diurnal patterns in seedling leaves and 13,773 (24.08%) in flag leaves. The detailed information of these probe sets included raw intensity, cosine-curve model parameters, and gene annotation were shown in File S1. Within these diurnal pattern probe sets, there were 4,394 probe sets identified with diurnal patterns in both seedling and flag leaves, and 9,379 showed diurnal patterns that were preferred in flag leaves (Figure 3).
Figure 3. Identify the probe sets with diurnal pattern between flag leaves and seedling leaves.
The orange shape indicates the diurnal-pattern probe sets only in seedling leaves, the light blue shape indicates the diurnal-pattern probe sets only in flag leaves, and the brown shape indicates the diurnal-pattern probe sets both in seedling and flag leaves.
The phase distribution of rice diurnal pattern genes in seedling and flag leaves
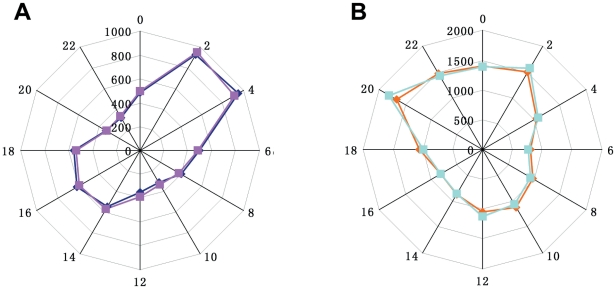
To further analyze the rice diurnal-pattern genes in seedling and flag leaves, all these genes were disassembled based on their phase of the cosine-curve model. The distribution of these probe sets in seedling and flag leaves are shown in Figure 4A and 4B, respectively. The distribution in two biological replicates was almost identical. For seedling leaves, the phases were during 2:00 am to 4:00 am, whereas in flag leaves the phases were during 8:00 pm to 2:00 am.
Figure 4. The phase distribution of the diurnal pattern probe sets in seedling leaves and flag leaves.
A. The phase distribution of diurnal-pattern probe sets in seedling leaves: the dark blue indicates seedling leaf sample replicate 1, and the purple indicates seedling leaf sample replicate 2. B.The phase distribution of diurnal-pattern probe sets in flag leaves: the yellow indicates flag leaf sample replicate 1, and the light blue indicates flag leaf sample replicate 2
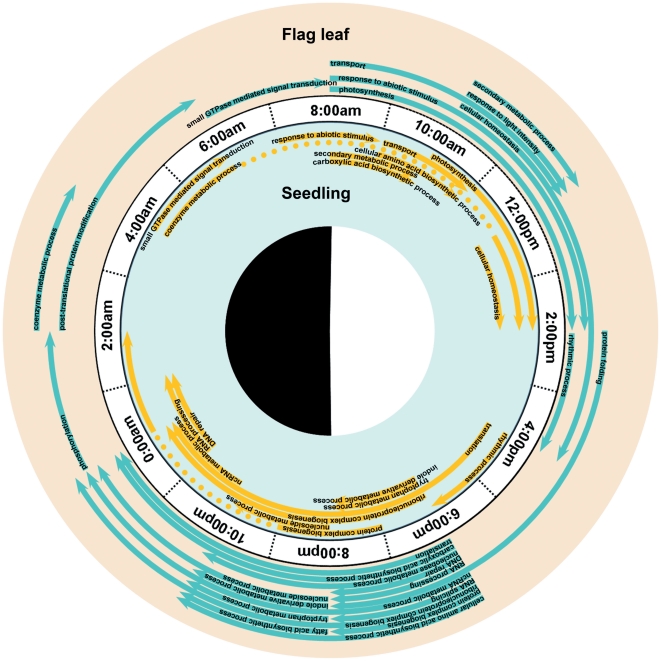
To elucidate the biological process of the rice diurnal-pattern genes involved, we employed gene ontology (GO) enrichment analysis [33] on the probe sets within each phase-period (Figure 5). In the enriched GO term distribution, along with the diurnal pattern phase, there were several interesting findings. Some important processes were induced by the light, such as photosynthesis, response to abiotic stimulus, transporters, and secondary metabolic processes. Some other biological processes related to the nucleus and ribosomes were active during the night: the transcriptional regulation-related processes such as RNA processing, RNA splicing and DNA repair, were enriched in the later afternoon and evening; circadian rhythm was enriched in late afternoon; and small GTPase-mediated signal transduction was highlighted in early morning. Some flag leaves' specifically enriched GO terms were also shown in Figure 5, such as fatty acid biosynthesis processes in the early evening, phosphorylation at midnight, and post-translational protein modification from night to early dawn.
Figure 5. The Gene Ontology (GO) distribution along with the phase of the diurnal pattern probe-sets in seedling leaves and flag leaves.
The middle circle with time stamp indicates the day–night cycle, inside for GO term distribution of seedling leaves and outside for flag leaves. Each line arc indicates the start point and end point (with arrow sign) of one GO term, the dashed arc indicates the temporary break of the GO term. The name of each GO term is shown on the arc.
Further, we predicted common motifs in the promoter regions (2 kb, 5′-upstream of the ATG) of the genes with the same diurnal pattern phase. The possible diurnal cis-regulatory elements were identified using the published Web-based program ELEMENT [25] (http://element.cgrb.oregonstate.edu/) for rice genes distributed over the different phases of the entire day (Table 2). There were several diurnal-related cis-elements with significant presence in seedling and flag leaves, including Morning Element (ME, *CCACAC*), Evening Element (EE, *TATC*), SORLIP2AT (*GGCCC* or *GGGCC*), DOFCOREZM (*AAAG*), midnight enriched telobox (TBX, *AACCCT*), Element II of Arabidopsis PCNA-2 (*GGCCCA*, *TGGGCC*, or *AGCCCA*), AtSR1 (CaMBP, *CGCG*), ARR1AT, EECCRCAH1 (*AAAATC*), and TATABOX5. In flag leaves, the ME combined with the G-box (CACGTG) was significantly enriched in morning-expressed genes. Some important motifs were only present in the promoters of the flag leaves' preferentially expressed genes, such as GCCcore, CGACG element, SURECOREATSULTR11 (*GAGAC*, G-box/ABRE (*CACGTG*), BIHD1OS and WRKY71OS (*ATGTCA*).
Table 2. Common motifs in the promoter regions of diurnal pattern genes in phases.
| Element | Phase | 0 | 2 | 4 | 6 | 8 | 10 | 12 | 14 | 16 | 18 | 20 | 22 | Annotation |
| *TATC* | Flag leaf | √ | √ | √ | √ | √ | √ | √ | Evening Element (EE) | |||||
| Seedling | √ | √ | √ | √ | ||||||||||
| *GGCCCA* | Flag leaf | √ | √ | √ | √ | √ | √ | √ | √ | √ | √ | Element II ofArabidopsis PCNA-2 | ||
| Seedling | √ | √ | √ | √ | ||||||||||
| *TGGGCC* | Flag leaf | √ | √ | √ | √ | √ | √ | √ | √ | Element II ofArabidopsis PCNA-2 | ||||
| Seedling | √ | √ | ||||||||||||
| *AGCCCA* | Flag leaf | √ | √ | √ | Element II ofArabidopsis PCNA-2 | |||||||||
| Seedling | ||||||||||||||
| *GCCCA* | Flag leaf | √ | √ | √ | √ | √ | √ | √ | √ | √ | √ | overrepresented inthe OxPhos promoter | ||
| Seedling | √ | √ | √ | √ | ||||||||||
| *GGCCC* | Flag leaf | √ | √ | √ | √ | √ | √ | √ | √ | √ | SORLIP2AT | |||
| Seedling | √ | √ | √ | √ | ||||||||||
| *GGGCC* | Flag leaf | √ | √ | √ | √ | √ | √ | √ | √ | √ | √ | SORLIP2AT | ||
| Seedling | √ | √ | √ | √ | ||||||||||
| *GCCGCC* | Flag leaf | √ | √ | GCCCORE | ||||||||||
| Seedling | ||||||||||||||
| *AACCCT* | Flag leaf | √ | √ | √ | TBX,overrepresentedaround midnight | |||||||||
| Seedling | √ | |||||||||||||
| *GATAA* | Flag leaf | √ | √ | √ | √ | √ | √ | I-box; rbcSt; | ||||||
| Seedling | √ | √ | ||||||||||||
| *CAAAAC* | Flag leaf | √ | √ | √ | √ | ANAERO4CONSENSUS | ||||||||
| Seedling | ||||||||||||||
| *CGCTT* | Flag leaf | √ | WINPSTPIIIK | |||||||||||
| Seedling | √ | |||||||||||||
| *AAAG* | Flag leaf | √ | √ | √ | √ | √ | √ | √ | √ | √ | √ | √ | DOFCOREZM | |
| Seedling | √ | √ | √ | √ | √ | √ | √ | √ | ||||||
| *CCACAC* | Flag leaf | √ | √ | √ | morning element (ME)RBCSBOX2PS | |||||||||
| Seedling | √ | √ | ||||||||||||
| *CACGTG* | Flag leaf | √ | √ | √ | √ | √ | G-box/ABRE | |||||||
| Seedling | ||||||||||||||
| *ACGTG* | Flag leaf | √ | √ | √ | √ | √ | √ | ACGTATERD1 | ||||||
| Seedling | √ | |||||||||||||
| *GCCAC* | Flag leaf | √ | √ | SORLIP1AT | ||||||||||
| Seedling | √ | √ | √ | |||||||||||
| *CGCG* | Flag leaf | √ | √ | √ | √ | √ | √ | AtSR1(CaMBP) | ||||||
| Seedling | √ | √ | √ | |||||||||||
| *CGACG* | Flag leaf | √ | √ | √ | CGACG element | |||||||||
| Seedling | ||||||||||||||
| *GAAAAA* | Flag leaf | √ | √ | √ | √ | √ | √ | √ | GT1CONSENSUS andGT1GMSCAM4 | |||||
| Seedling | √ | √ | √ | √ | √ | |||||||||
| *GAGAC* | Flag leaf | √ | √ | √ | √ | SURECOREATSULTR11 | ||||||||
| Seedling | ||||||||||||||
| *AAATAA* | Flag leaf | √ | √ | TATABOX5 | ||||||||||
| Seedling | √ | √ | ||||||||||||
| *AAAATC* | Flag leaf | √ | √ | √ | √ | ARR1AT and EECCRCAH1 | ||||||||
| Seedling | √ | √ | ||||||||||||
| *ATGTCA* | Flag leaf | √ | √ | BIHD1OS and WRKY71OS | ||||||||||
| Seedling |
Sign √ indicates the element significantly present in the promoters of the genes shown diurnal pattern in proper phase.
Rice transcription factor gene families with diurnal patterns in flag and seedling leaves
There were large numbers of transcription factor genes with diurnal patterns in both seedling and flag leaves. Several gene families of transcription factors, such as EIL, ZIM and ZF-HD, showed a flag-leaf-preferred diurnal pattern. About 65% of overall transcription factors were flag leaf preferred (Table 3). There were 20 families with a higher proportion of flag-leaf-preferred diurnal pattern, five were >90%, eight were within 80 to <90%, another four within 70 to <80%, and three within 65 to <70%.
Table 3. The rice transcription factors families showing flag leaf prefer diurnal patterns.
| TF family | Seedling | Flag leaf | Flag leaf prefer | % |
| zf-HD | 0 | 8 | 8 | 100.00 |
| EIL | 0 | 7 | 7 | 100.00 |
| ZIM | 1 | 6 | 6 | 100.00 |
| SNF2 | 5 | 18 | 17 | 94.44 |
| ABI3VP1 | 1 | 10 | 9 | 90.00 |
| TCP | 3 | 8 | 7 | 87.50 |
| SET | 5 | 19 | 16 | 84.21 |
| MYB | 13 | 24 | 20 | 83.33 |
| Alfin-like | 1 | 6 | 5 | 83.33 |
| Jumonji | 3 | 6 | 5 | 83.33 |
| WRKY | 10 | 26 | 21 | 80.77 |
| NAC | 13 | 20 | 16 | 80.00 |
| PHD | 6 | 20 | 16 | 80.00 |
| ARF | 7 | 19 | 15 | 78.95 |
| AUX/IAA | 6 | 17 | 13 | 76.47 |
| C2H2 | 20 | 30 | 22 | 73.33 |
| HB | 17 | 41 | 29 | 70.73 |
| C3H | 19 | 40 | 27 | 67.50 |
| MADS | 9 | 12 | 8 | 66.67 |
| TUB | 4 | 9 | 6 | 66.67 |
| AP2-EREBP | 20 | 40 | 26 | 65.00 |
| HSF | 7 | 17 | 11 | 64.71 |
| bHLH | 21 | 28 | 18 | 64.29 |
| CCAAT-HAP | 7 | 14 | 9 | 64.29 |
| MYB-related | 16 | 37 | 23 | 62.16 |
| Trihelix | 5 | 13 | 8 | 61.54 |
| GRAS | 10 | 15 | 9 | 60.00 |
| OVERALL | 383 | 747 | 490 | 65.60 |
Diurnal patterns of starch and sucrose metabolism related genes in seedling and flag leaves
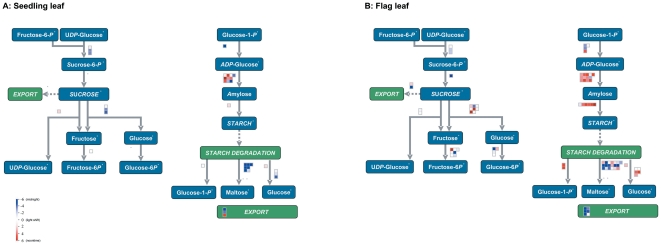
The rice genes related to starch synthesis are essential to improving grain quality (such as eating and cooking quality). We searched the possible rice orthologs for the Arabidopsis starch-related genes, and made a rice gene-expression profile for the diurnal changes of sucrose metabolism-related enzymes in seedling and flag leaves, which were further displayed by MapMan (Version 3.0). We employed BLAST to map the probe sets to the BINs of MapMan, giving detailed mapping information of every probe set with a diurnal pattern (File S2). The phase distribution of the diurnal pattern probe-sets related to starch and sucrose biosynthesis and degradation pathways in seedling and flag leaves are shown in Figure 6A and 6B, respectively. There were similarities and a diversity of diurnal patterns between rice flag leaves and seedling leaves. The enzymes involved in the starch synthesis pathway mainly showed a daytime diurnal phase both in seedling and flag leaves. Overall, more probe sets with diurnal patterns occurred in flag leaves compared to seedling leaves: e.g. in the starch synthesis pathway, the majority of related genes showed a diurnal pattern during day time, both in seedling and flag leaves, but the genes encoding ADP-glucose pyrophosphorylases (AGPases) and starch-branching enzymes had significant diurnal patterns in flag leaves. Furthermore, the pathways derived from Tian et al. [34] showed diurnal patterns of individual enzymes in the starch synthesis pathway (Figure S3).
Figure 6. The diurnal pattern of key genes in starch and sucrose related pathway.
The phase of each probe set was redefined as its relation to the time point of light shift (the start or the end of night). Red, blue and white squares to indicate the noon, midnight and light shift phase of the probe set, respectively.
Comparative analysis of rice and Arabidopsis diurnal pattern genes
There were similarities and differences between Arabidopsis and rice transcriptome analyses throughout the diurnal cycle. Recently, a large body of Arabidopsis expression profiling data related to diurnal or circadian rhythms was made publicly available. We selected the raw data (NCBI's Gene Expression Omnibus (GEO) accession number: GSE3416) from the study by Blasing [1] on diurnal gene expression of 5-6-week-old rosette leaves in Arabidopsis thaliana Col-0, and recalculated the diurnal pattern genes using the same approach, i.e. combining ARSER and CC. The detailed diurnal pattern Arabidopsis genes of GSE3416 are also listed in File S1. Thus, it is possible to compare the Arabidopsis diurnal pattern genes with those we identified in rice seedling leaves and flag leaves. Using the regular BLAST method, a close homolog between rice and Arabidopsis genes was mapped. Most diurnal-pattern genes in seedling and flag leaves had the Arabidopsis homolog (Table 4), and about half of these had a diurnal expression pattern. However, there were a large number of genes preferentially expressed with a diurnal pattern in rice flag leaves and seedling leaves that were not found in Arabidopsis leaves.
Table 4. Comparison between rice diurnal pattern genes in seedling leaves and flag leaves with Arabidopsis genes.
| Probe set ID | Match to Arabidopsis | Probe set ID (with Arabidopsis diurnal gene) | % | |
| Diurnal in flag leaf | 13773 | 11916 | 5868 | 49.24% |
| Diurnal in seedling | 6266 | 5515 | 2792 | 50.63% |
| Overlap | 4394 | 3942 | 2116 | 53.68% |
| Flag leaf prefer | 9379 | 7974 | 3752 | 47.05% |
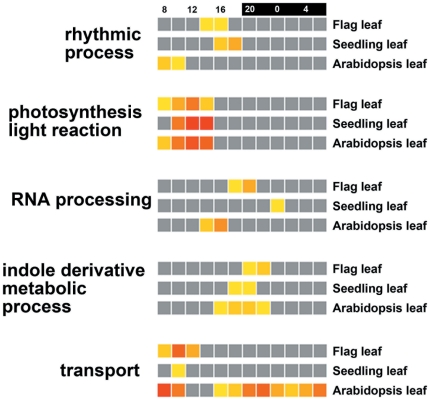
Some phase specific diurnal cis-elements were compared between rice and Arabidopsis. The morning element, G-box, Evening Element, TBX and Element II of PCNA-2 were conserved and enriched in similar phases across rice and Arabidopsis. We also compared the GO term distribution between Arabidopsis and rice diurnal-pattern genes (Figure 7). Several processes such as photosynthesis and indole-derivative metabolic processes were similar between rice and Arabidopsis; however, many (e.g. rhythmic processes and RNA processing) were in different phases between the species.
Figure 7. Selected GO term distribution along with the phase of the diurnal pattern between Arabidopsis and rice leaves.
The comparison is displayed in HTML table mode for rice flag leaf, rice seedling leaf, and Arabidopsis leaf. The color blocks (yellow-to-red) represent the level of each term at a certain time-point. The greyscale represents non-significant enrichment.
Discussion
Identification of diurnal pattern genes in rice seedling and flag leaves
A variety of methods have been developed to extract the cycling-expressed genes from microarray data for circadian rhythm or diurnal research. The computational methods mainly fall into two categories: traditional time-series spectral analysis and cosine-based pattern matching methods. The advantages of the traditional spectral method are that they have been widely used to analyze time-series, and many programs are available for their calculation; however, their limitation is that they only perform well for long time-series. For microarray data, which usually have only a few points (due to high costs), the traditional spectral methods do not efficiently detect the exact diurnal or circadian rhythm period [35]. The cosine-based pattern matching methods are mathematically convenient with a reasonably good description of well-defined properties; however, they may not efficiently identify non-sinusoidal periodic patterns. Thus it is essential to use a suitable approach to identify the rhythmically-expressed genes during our analysis of diurnal microarray data.
The ARSER method [32] combines the time-domain and frequency-domain analyses and was shown efficient in identifying the periodicity in short time-series. Comparing with other cycling algorithms, ARSER can handle noise in the expression data and identify periodic patterns from limited sample-sizes and low numbers of replications of short time-series. Particularly, unlike cosine-curve-based algorithms, ARSER can identify both non-sinusoidal and sinusoidal patterns. For example, two transcripts with sinusoidal expression pattern can be identified as periodic by both ARSER and CC methods (Figure 2A and 2B), while one transcript with a non-sinusoidal (spike) expression pattern was identified as periodic by ARSER (Figure 2C). ARSER determines the statistically significant periodic transcripts by FDR q-values, which are calculated based on the distribution of p-values. The period range for computing will also impact on the selection of gene sets during the analysis of microarray data. By setting appropriate values for parameters, we could get more significant results using ARSER.
The combined use of ARSER and CC methods showed some special periodic patterns that were non-sinusoidal periodic patterns, such as probe set Os.22928.2.S1_at (Figure 2C) with a periodic expression value and a spike-shaped pattern according to HAYSTACK (http://haystack.cgrb.oregonstate.edu/). This approach appears to be efficient in analyzing short time-series compared with the spectral method and has a simpler computational procedure than for nonlinear curve fitting.
Combining the mathematical model and the reproducibility in biological replicates, we globally defined 6,266 (10.96%) diurnal probe-sets in seedling leaves and 13,773 (24.08%) in flag leaves. Transcriptome analysis showed that gene transcription levels in flag leaves were mostly of diurnal rhythm. Recently, a maize custom high-density 105 K Agilent microarray was conducted to investigate the diurnal expression patterns between the leaf and developing ears; about 22.7% (10,037) of expressed transcripts exhibited a diurnal cycling pattern in leaves, but only 0.39% (47) in developing ears [22]. These results indicated that the diurnal rhythms are related to developmental stages and tissue specificity, revealing a ‘third-dimension’ of diurnal rhythm regulation.
Interactions between diurnal patterns and plant hormone signalling regulation in rice leaves
Light is a key environmental cue, and interactions among light, plant hormones and the circadian clock appear to control the diurnal patterns of plant growth. We analyzed the rice transcription factor families with flag-leaf-preferred diurnal patterns, and the results suggested that ethylene may also affect the diurnal pattern in rice flag leaves (Table 3). For example, four ethylene-insensitive3-like (EIL) genes (LOC_Os07g48630, LOC_Os03g20780, LOC_Os03g20790 and LOC_Os09g31400) were identified as diurnal only in flag leaves but not seedling leaves. EIL may be involved in the ethylene signal-transduction pathway. Additionally, we also found that TIFY family proteins (recently discovered to play a critical role in repression of jasmonate signalling [36], [37], [38], [39], [40]), might also affect the rice diurnal pattern. Among 20 rice TIFY genes, only OsTIFY11a (OsJAZ9, LOC_Os03g08310) showed a diurnal pattern in seedling leaves, while another five showed a diurnal pattern in flag leaves, including OsTIFY3 (OsJAZ1, LOC_Os04g55920), OsTIFY6a (OsJAZ3, LOC_Os08g33160), OsTIFY6b (OsJAZ4, LOC_Os09g23660), OsTIFY10b (OsJAZ7, LOC_Os07g42370) and OsTIFY10c (OsJAZ8, LOC_Os09g26780).
Rice diurnal microarray analysis showed that some abscisic acid (ABA)-dependent transcription factor genes (e.g. ABI3VP1 and OsNAC5) had a significant diurnal pattern in rice flag leaves (Table 3). Common motifs such as CACGTG (G-box/ABRE), ACGTG (ABRELATERD1) and CACG (NAC core motif), were identified in the promoter regions (2 kb 5′-upstream from the ATG) of the genes with similar diurnal pattern peaks in the day time (from phase 8 to phase 18). In Arabidopsis, a large number of ABA-responsive and/or methyl jasmonate (MeJA)-responsive genes were identified with oscillation expression diurnally and robustly during the light–dark cycle [14], [15]. Plant stomatal movements are rhythmic and ABA can regulate the diurnal oscillator period [41], [42]. From motif analysis (Table 2), we also found that CGCG Box (calmodulin-binding) was enriched in the promoter regions of the diurnal genes with expression peaks from morning to noon (phases 6, 8, 10, 12 and 14). This may be related to the possible dynamic change of the concentration of internal calcium which oscillates diurnally, peaking during the day and dropping at night.
Auxin is a key regulator of plant growth and development, and auxin signal transduction can be regulated by the circadian clock in Arabidopsis [15], [43]. The plant sensitivity to auxin was observed to vary according to the time of day. Through microarray analysis, several AUX/IAA and ARF genes showed significant diurnal patterns in rice flag and seedling leaves. Through promoter analysis for the rice diurnal genes, we found that common motif ATGTCA/TGTCA [SURECOREATSULTR11, the core of sulfur-responsive element (SURE) which contains auxin response factor (ARF) binding sequence] was significantly enriched in the promoter regions of the diurnal genes in flag leaves, with expression peaks from morning to noon (phases 8, 10 and 12), while in seedling leaves the expression peaks were in the evening (phase 22).
From the diurnal genes in flag and seedling leaves, some gibberellin-mediated signalling and metabolic-related genes were enriched in seedling leaves, including ent-kaurene synthase A (LOC_Os04g09900 and LOC_Os02g36210), ent-kaurene synthase B (LOC_Os02g36140, LOC_Os04g10060, LOC_Os11g28530 and LOC_Os12g30824) and ent-kaurene oxidase (LOC_Os06g37364). Among the seven gibberellin-related genes, two (LOC_Os12g30824 and LOC_Os06g37364) also peaked at midnight in flag leaves.
Starch metabolism in rice diurnal cycling
During the diurnal cycle, starch is stored in leaves in a pattern such that starch content increases during periods of light and decreases during darkness [44], [45], [46]. Starch is one of the most important compounds synthesized by plants, and higher starch levels in leaves can lead to increased biomass. The carbon contained in leaves can be converted to ethanol for use as a bio-fuel and an alternate energy source. The genes related to starch synthesis are also essential for improving grain quality (e.g. eating and cooking quality). Our microarray-based diurnal gene identification showed similarities and diversity of diurnal patterns for starch metabolism-related genes between rice flag and seedling leaves (Figure 6 and Figure S3). The enzymes involved in the starch synthesis pathway mainly showed a daytime diurnal phase, both in seedling and flag leaves; whereas there were more genes of the starch synthesis enzymes with a diurnal pattern only in flag leaves (e.g. AGPase and SBE1). Our results showed no AGPases with a diurnal pattern in seedling leaves, but LOC_Os09g12660 (the small subunit of AGPase) had a diurnal phase at 4:00pm in flag leaves. AGPase catalyzes the reaction generating the sugar nucleotide ADP-glucose and inorganic pyrophosphate from glucose 1-phosphate and ATP, which is the first step of starch biosynthesis. AGPase is considered as a major enzyme controlling starch synthesis [47]. Starch branching enzyme (SBE) acts on glucose polymers through α-1,6-glucosidic bonds to form branches on the α-1,4-linked glucose backbone, which is also a key enzyme in the starch biosynthesis pathway [48], [49], [50]. In sorghum endosperm, three SBE genes showed a diurnal rhythm in gene expression levels during a 24-h cycle [51]. Our result showed that four rice probe-sets matched LOC_Os06g51084 (SBE1) with a diurnal pattern only in flag leaves. The starch metabolism genes with a diurnal pattern in flag leaves may play special roles during graining filling, beneficial to grain quantity and quality.
Possible ribosome and chromatin related transcriptional regulation during light–dark diurnal cycle
Enriched GO term analysis for the assigned diurnal genes (Figure 5) showed that the genes related to the nuclear and ribosome-involved processes were active mostly during the period of light–dark change, which is similar to the GO phase distribution of diurnal transcripts in maize leaves [22]. This may be related to light-caused DNA mutations and DNA repair during the day and night. We also found that SNF2 family genes were significantly expressed with a diurnal pattern in flag leaves (Table 3). SNF2 is the catalytic subunit of the SWI/SNF chromatin remodelling complex and, SNF2-family genes play important roles in transcriptional regulation, maintenance of chromosome integrity and DNA repair [52], [53], [54], [55]. The Arabidopsis SWI2/SNF2 chromatin remodelling gene-family was reported to be involved in DNA damage response and recombination [53]. The diurnally-regulated SNF2 family genes may be related to changes in chromatin structure at the core of the diurnal oscillator, which may provide a clue concerning the regulation of diurnal progression by chromatin dynamics.
Materials and Methods
Plant materials
Seeds of rice (Oryza sativa subsp. japonica var. Nipponbare) were surface-sterilized in 5% (w/v) sodium hypochlorite for 20 min and then washed in distilled water three or four times, then germinated in water for 2 d at room temperature and 1 d at 37°C. The seedlings were transferred to water-saturated Whatman filter paper and grown in a greenhouse (28/25°C and 12/12 h of light/dark, and 83% relative humidity). After about 17 d, seedling leaves were harvested every 4 hours.
For flag leaf samples, the rice plants were grown in the field under natural conditions, within the May–October growing season, on an experimental farm in Zhejiang, China. Three-month-old rice plants were entrained into the greenhouse under the regular condition (32/30°C and 12/12 h of light/dark). Flag leaves were harvested every 4 hours.
RNA isolation and Affymetrix GeneChip experiments
All leaf samples were homogenized in liquid nitrogen before isolation of RNA. Total RNA was isolated using TRIZOL® reagent (Invitrogen, CA, USA) and purified using Qiagen RNeasy columns (Qiagen, Hilden, Germany). For each sample, 8 µg of total RNA was used for making biotin-labelled cRNA targets; cDNA and cRNA synthesis; cRNA fragmentation, hybridization, washing and staining; and scanning, following the GeneChip Standard Protocol (Eukaryotic Target Preparation). In this experiment, a Poly-A RNA Control Kit and a One-Cycle cDNA Synthesis kit were applied. Affymetrix rice genome arrays were used for hybridizations.
Real-time RT-PCR
Reverse transcription was performed using M-MLV kit (Invitrogen). We heated 10 µl samples containing 2 µg of total RNA, and 20 pmol of random hexamers (Invitrogen) at 70°C for 10 minutes to denature the RNA, and then chilled the samples on ice for 2 min. We added reaction buffer and M-MLV enzyme to a total volume of 20 µl containing 500 µM dNTPs, 50 mM Tris–HCl (pH 8.3), 75 mM KCl, 3 mM MgCl2, 5 mM dithiothreitol, 200 units of M-MLV, and 20 pmol random hexamers. The samples were then heated at 37°C for 1 h. The cDNA samples were diluted to 8 ng/µl for real-time RT-PCR analysis.
For real-time RT-PCR, triplicate assays were performed on 1 µl of each cDNA dilution using the SYBR Green Master Mix (Applied Biosystems, PN 4309155) with an ABI 7900 sequence detection system according to the manufacture's protocol (Applied Biosystems). The gene-specific primers were designed by using PRIMER3 (http://frodo.wi.mit.edu/primer3/input.htm). The amplification of 18S rRNA was used as an internal control to normalize all data (forward primer, 5′-CGGCTACCACATCCAAGGAA-3′; reverse primer, 5′- TGTCACTACCTCCCCGTGTCA-3′). The primer sets of four selected genes were listed below: LOC_Os03g39610 (5′-ATGTTCTCCATGTTCGGCTTCT-3′ and 5′-GTATGCCCAGGCGTTGTTG-3′), LOC_Os07g22930 (5′-GACGTCACAGCGGTTGCTT-3′ and 5′-TTCTTGGAATGCCGGTGTGT-3′), LOC_Os07g46790 (5′-TGAAAACGAGGAGTGGTTGAAA-3′ and 5′-GGCCCCATTGGCTGTGA-3′), and LOC_Os06g30310 (5′-TTACAGATCAGGGATTCCAAAAATC-3′ and 5′-ATAAAGCTCCTCAATGGCATGAC-3′). The relative quantification method (DDCT) was used to evaluate quantitative variation between replicates examined.
Transcriptome data analysis and diurnal pattern identification
The signal intensity for each probe set on the GeneChip microarray was extracted by Affymetrix GCOS software and the TGT (target mean value) was scaled as 500 for each chip. Pair-wise scatter plots of replicate samples were generated by Partek Genomics Suite (Version 6.3). For each probe set, we calculated the correlation coefficient (Cr) of two sets of biological replicates across the time series.
Two methods were applied to identify diurnal pattern probe sets: ARSER and cross-correlation (CC). ARSER employs autoregressive spectral estimation to predict the periodicity of an expression pattern from the frequency spectrum and then models the rhythmic patterns using a harmonic regression model to fit the time-series [32]. There are four steps during the ARSER method: the ‘detrending process’, performs a data pre-processing strategy that removes any linear trend from the time-series; then autoregressive spectral analysis calculates the power spectral density of the time-series; further harmonic analysis provides the estimates of the parameters that describe the rhythmic patterns; and finally, false-discovery rate q-values are calculated for multiple comparisons. The CC method was used to calculate the Pearson's correlation between a rhythmically-expressed gene and a theoretical cosine wave with defined phase [17]. The brief calculation process follows. First, we used the cosine curve (Equation 1) with phases of 0–24 hr to prepare 60 test cosine-curves of 24-hr periodicity. The time span was 36 hr long with one and half cycle and interval between adjacent phases equal to 0.4 hr. Second, we calculated the Cα of the best-fitting cosine curve for each expression profile and the phase of the best-fitting cosine curve was defined as the phase of the related probe sets. Third, we used a random Monte Carlo simulation to determine the statistical significance p-value: we randomly produced 100,000 expression profiles, and calculated the maximum Cβ for each of them. We then counted the number of times that a random expression profile showed Cβ greater than a specified value and defined the p-value as the number divided by the number of all random expression profiles [56].
| (1) |
Gene annotation, Gene Ontology analysis, and pathway analysis
The consensus sequence of each probe set was compared by BLAST (Basic Local Alignment and Search Tool) against the TIGR Rice Genome version 5 to map the probe set ID to the locus ID in the rice genome. The cut-off e-value was set as 1e–20. Within the 57,195 designed probe sets in the Affymetrix rice genome array, there are 52,697 probe-sets mapped to rice genes in TIGR rice pseudomolecules.
The promoter sequences were extracted from MSU Rice Genome Annotation Website (http://rice.plantbiology.msu.edu/) and ELEMENT (http://element.cgrb.oregonstate.edu/). ELEMENT and Place (http://www.dna.affrc.go.jp/PLACE/) [57] were used for motif search and cis-element identification.
The GO category enrichment analysis was applied by in-house agriGO analysis service [33], the Singular Enrichment Analysis (SEA) and Cross comparison of SEA (SEACOMPARE) tools with default parameters for the Affymetrix rice genome array used for analysis.
MapMan (http://gabi.rzpd.de/projects/MapMan) was used for key regulation group analysis. The starch biosynthesis pathway was adopted from that of Tian [34] and the corresponding MapMan pathways were created through the mapping files.
Supporting Information
Expression profiles between the biological replicate samples of seedling leaves. A: Pair-wise scatter plots for the raw probe-set intensity data across all time points. B: The number distribution of all probe sets based on the correlation coefficient (Cr) across the time series between the biological replicate samples (including file name, file format, name and URL link of appropriate viewer if format is unusual).
(TIF)
Real-time RT-PCR validation for selected genes. Four genes (LOC_Os03g39610, LOC_Os07g22930, LOC_Os07g46790, and LOC_Os06g30310) were selected for real-time RT-PCR to validate the expression patterns of flag leaves (A) and seedling leaves (B) in seven time points from 12 hr to 36 hr. The purple and yellow bars represent the expression level in microarrys, and the blue bars represent the relative intensity of real-time RT-PCR from independent biological replicates.
(TIF)
The diurnal pattern of the key genes in the starch biosynthesis pathway. The phase of each probe set was redefined as its relation to the time point of light shift (the start or the end of night). Red, blue and white squares to indicate the noon, midnight and light shift phase of the probe set, respectively.
(TIF)
Raw tables for diurnal probe-sets list.
(XLS)
Tables for diurnal probe sets related to sucrose and starch pathways.
(XLS)
Acknowledgments
We would like to acknowledge Hong Yan for assistance with the GeneChip experiment.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported by grants from the Ministry of Science and Technology of China (2005CB120804 and 90817006). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Blasing OE, Gibon Y, Gunther M, Hohne M, Morcuende R, et al. Sugars and circadian regulation make major contributions to the global regulation of diurnal gene expression in Arabidopsis. Plant Cell. 2005;17:3257–3281. doi: 10.1105/tpc.105.035261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stitt M, Bulpin PV, ap Rees T. Pathway of starch breakdown in photosynthetic tissues of Pisum sativum. Biochim Biophys Acta. 1978;544:200–214. doi: 10.1016/0304-4165(78)90223-4. [DOI] [PubMed] [Google Scholar]
- 3.Stitt M, Gerhardt R, Kurzel B, Heldt HW. A Role for Fructose 2,6-Bisphosphate in the Regulation of Sucrose Synthesis in Spinach Leaves. Plant Physiol. 1983;72:1139–1141. doi: 10.1104/pp.72.4.1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Matt P, Schurr U, Klein D, Krapp A, Stitt M. Growth of tobacco in short-day conditions leads to high starch, low sugars, altered diurnal changes in the Nia transcript and low nitrate reductase activity, and inhibition of amino acid synthesis. Planta. 1998;207:27–41. doi: 10.1007/s004250050452. [DOI] [PubMed] [Google Scholar]
- 5.Geigenberger P, Stitt M. Diurnal changes in sucrose, nucleotides, starch synthesis and AGPS transcript in growing potato tubers that are suppressed by decreased expression of sucrose phosphate synthase. Plant J. 2000;23:795–806. doi: 10.1046/j.1365-313x.2000.00848.x. [DOI] [PubMed] [Google Scholar]
- 6.Gibon Y, Blaesing OE, Hannemann J, Carillo P, Hohne M, et al. A Robot-based platform to measure multiple enzyme activities in Arabidopsis using a set of cycling assays: comparison of changes of enzyme activities and transcript levels during diurnal cycles and in prolonged darkness. Plant Cell. 2004;16:3304–3325. doi: 10.1105/tpc.104.025973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gibon Y, Blasing OE, Palacios-Rojas N, Pankovic D, Hendriks JH, et al. Adjustment of diurnal starch turnover to short days: depletion of sugar during the night leads to a temporary inhibition of carbohydrate utilization, accumulation of sugars and post-translational activation of ADP-glucose pyrophosphorylase in the following light period. Plant J. 2004;39:847–862. doi: 10.1111/j.1365-313X.2004.02173.x. [DOI] [PubMed] [Google Scholar]
- 8.Gibon Y, Usadel B, Blaesing OE, Kamlage B, Hoehne M, et al. Integration of metabolite with transcript and enzyme activity profiling during diurnal cycles in Arabidopsis rosettes. Genome Biol. 2006;7:R76. doi: 10.1186/gb-2006-7-8-r76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Steer BT. Control of diurnal variations in photosynthetic products: I. Carbon metabolism. Plant Physiol. 1974;54:758–761. doi: 10.1104/pp.54.5.758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Geiger DR, Servaites, C J. Diurnal Regulation of Photosynthetic Carbon Metabolism in C3 Plants. Annual Review of Plant Physiology and Plant Molecular Biology. 1994;45:235–256. [Google Scholar]
- 11.Dietrich P, Sanders D, Hedrich R. The role of ion channels in light-dependent stomatal opening. J Exp Bot. 2001;52:1959–1967. doi: 10.1093/jexbot/52.363.1959. [DOI] [PubMed] [Google Scholar]
- 12.Shimazaki K, Doi M, Assmann SM, Kinoshita T. Light regulation of stomatal movement. Annu Rev Plant Biol. 2007;58:219–247. doi: 10.1146/annurev.arplant.57.032905.105434. [DOI] [PubMed] [Google Scholar]
- 13.Roelfsema MR, Hedrich R. In the light of stomatal opening: new insights into ‘the Watergate’. New Phytol. 2005;167:665–691. doi: 10.1111/j.1469-8137.2005.01460.x. [DOI] [PubMed] [Google Scholar]
- 14.Mizuno T, Yamashino T. Comparative transcriptome of diurnally oscillating genes and hormone-responsive genes in Arabidopsis thaliana: insight into circadian clock-controlled daily responses to common ambient stresses in plants. Plant Cell Physiol. 2008;49:481–487. doi: 10.1093/pcp/pcn008. [DOI] [PubMed] [Google Scholar]
- 15.Michael TP, Breton G, Hazen SP, Priest H, Mockler TC, et al. A morning-specific phytohormone gene expression program underlying rhythmic plant growth. PLoS Biol. 2008;6:e225. doi: 10.1371/journal.pbio.0060225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Doherty CJ, Kay SA. Circadian Control of Global Gene Expression Patterns. The Annual Review of Genetics. 2010;44:419–444. doi: 10.1146/annurev-genet-102209-163432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harmer SL, Hogenesch JB, Straume M, Chang HS, Han B, et al. Orchestrated transcription of key pathways in Arabidopsis by the circadian clock. Science. 2000;290:2110–2113. doi: 10.1126/science.290.5499.2110. [DOI] [PubMed] [Google Scholar]
- 18.Schaffer R, Landgraf J, Accerbi M, Simon V, Larson M, et al. Microarray analysis of diurnal and circadian-regulated genes in Arabidopsis. Plant Cell. 2001;13:113–123. doi: 10.1105/tpc.13.1.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mockler TC, Michael TP, Priest HD, Shen R, Sullivan CM, et al. The DIURNAL project: DIURNAL and circadian expression profiling, model-based pattern matching, and promoter analysis. Cold Spring Harb Symp Quant Biol. 2007;72:353–363. doi: 10.1101/sqb.2007.72.006. [DOI] [PubMed] [Google Scholar]
- 20.Khan S, Rowe SC, Harmon FG. Coordination of the maize transcriptome by a conserved circadian clock. BMC Plant Biol. 2010;10:126. doi: 10.1186/1471-2229-10-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Usadel B, Poree F, Nagel A, Lohse M, Czedik-Eysenberg A, et al. A guide to using MapMan to visualize and compare Omics data in plants: a case study in the crop species, Maize. Plant Cell Environ. 2009;32:1211–1229. doi: 10.1111/j.1365-3040.2009.01978.x. [DOI] [PubMed] [Google Scholar]
- 22.Hayes KevinR, MB, Meng Xin, Simmons CarlR, Habben JeffreyE, Danilevskaya OlgaN. Maize Global Transcriptomics Reveals Pervasive Leaf Diurnal Rhythms but Rhythms in Developing Ears Are Largely Limited to the Core Oscillator. PLoS ONE. 2010;5:e12887. doi: 10.1371/journal.pone.0012887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mangelsen E, Wanke D, Kilian J, Sundberg E, Harter K, et al. Significance of light, sugar, and amino acid supply for diurnal gene regulation in developing barley caryopses. Plant Physiol. 2010;153:14–33. doi: 10.1104/pp.110.154856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hudson KA. The Circadian Clock-controlled Transcriptome of Developing Soybean Seeds. The Plant Genome. 2010;3:3–13. [Google Scholar]
- 25.Michael TP, Mockler TC, Breton G, McEntee C, Byer A, et al. Network discovery pipeline elucidates conserved time-of-day-specific cis-regulatory modules. PLoS Genet. 2008;4:e14. doi: 10.1371/journal.pgen.0040014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chudova D, Ihler A, Lin KK, Andersen B, Smyth P. Bayesian detection of non-sinusoidal periodic patterns in circadian expression data. Bioinformatics. 2009;25:3114–3120. doi: 10.1093/bioinformatics/btp547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Plautz JD, Straume M, Stanewsky R, Jamison CF, Brandes C, et al. Quantitative analysis of Drosophila period gene transcription in living animals. J Biol Rhythms. 1997;12:204–217. doi: 10.1177/074873049701200302. [DOI] [PubMed] [Google Scholar]
- 28.Wichert S, Fokianos K, Strimmer K. Identifying periodically expressed transcripts in microarray time series data. Bioinformatics. 2004;20:5–20. doi: 10.1093/bioinformatics/btg364. [DOI] [PubMed] [Google Scholar]
- 29.Glynn EF, Chen J, Mushegian AR. Detecting periodic patterns in unevenly spaced gene expression time series using Lomb-Scargle periodograms. Bioinformatics. 2006;22:310–316. doi: 10.1093/bioinformatics/bti789. [DOI] [PubMed] [Google Scholar]
- 30.Tabata M, Takeshima T, Burioka N, Nomura T, Ishizaki K, et al. Cosinor analysis of heart rate variability in ambulatory migraineurs. Headache. 2000;40:457–463. doi: 10.1046/j.1526-4610.2000.00068.x. [DOI] [PubMed] [Google Scholar]
- 31.Straume M. DNA microarray time series analysis: automated statistical assessment of circadian rhythms in gene expression patterning. Methods Enzymol. 2004;383:149–166. doi: 10.1016/S0076-6879(04)83007-6. [DOI] [PubMed] [Google Scholar]
- 32.Yang R, Su Z. Analyzing circadian expression data by harmonic regression based on autoregressive spectral estimation. Bioinformatics. 2010;26:i168–174. doi: 10.1093/bioinformatics/btq189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Du Z, Zhou X, Ling Y, Zhang Z, Su Z. agriGO: a GO analysis toolkit for the agricultural community. Nucleic Acids Res. 2010;38(Suppl):W64–70. doi: 10.1093/nar/gkq310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tian Z, Qian Q, Liu Q, Yan M, Liu X, et al. Allelic diversities in rice starch biosynthesis lead to a diverse array of rice eating and cooking qualities. Proc Natl Acad Sci U S A. 2009;106:21760–21765. doi: 10.1073/pnas.0912396106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Langmead CJ, Yan AK, McClung CR, Donald BR. Phase-independent rhythmic analysis of genome-wide expression patterns. J Comput Biol. 2003;10:521–536. doi: 10.1089/10665270360688165. [DOI] [PubMed] [Google Scholar]
- 36.Chung HS, Howe GA. A critical role for the TIFY motif in repression of jasmonate signaling by a stabilized splice variant of the JASMONATE ZIM-domain protein JAZ10 in Arabidopsis. Plant Cell. 2009;21:131–145. doi: 10.1105/tpc.108.064097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mach J. Alternative splicing produces a JAZ protein that is not broken down in response to jasmonic acid. Plant Cell. 2009;21:14. doi: 10.1105/tpc.108.210111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Turner JG. Stress responses: JAZ players deliver fusion and rhythm. Curr Biol. 2007;17:R847–849. doi: 10.1016/j.cub.2007.08.014. [DOI] [PubMed] [Google Scholar]
- 39.Thines B, Katsir L, Melotto M, Niu Y, Mandaokar A, et al. JAZ repressor proteins are targets of the SCF(COI1) complex during jasmonate signalling. Nature. 2007;448:661–665. doi: 10.1038/nature05960. [DOI] [PubMed] [Google Scholar]
- 40.Chini A, Fonseca S, Fernandez G, Adie B, Chico JM, et al. The JAZ family of repressors is the missing link in jasmonate signalling. Nature. 2007;448:666–671. doi: 10.1038/nature06006. [DOI] [PubMed] [Google Scholar]
- 41.Lee KH, Piao HL, Kim HY, Choi SM, Jiang F, et al. Activation of glucosidase via stress-induced polymerization rapidly increases active pools of abscisic acid. Cell. 2006;126:1109–1120. doi: 10.1016/j.cell.2006.07.034. [DOI] [PubMed] [Google Scholar]
- 42.Tallman G. Are diurnal patterns of stomatal movement the result of alternating metabolism of endogenous guard cell ABA and accumulation of ABA delivered to the apoplast around guard cells by transpiration? J Exp Bot. 2004;55:1963–1976. doi: 10.1093/jxb/erh212. [DOI] [PubMed] [Google Scholar]
- 43.Covington MF, Harmer SL. The circadian clock regulates auxin signaling and responses in Arabidopsis. PLoS Biol. 2007;5:e222. doi: 10.1371/journal.pbio.0050222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zeeman SC, Smith SM, Smith AM. The diurnal metabolism of leaf starch. Biochem J. 2007;401:13–28. doi: 10.1042/BJ20061393. [DOI] [PubMed] [Google Scholar]
- 45.Lu Y, Gehan JP, Sharkey TD. Daylength and circadian effects on starch degradation and maltose metabolism. Plant Physiol. 2005;138:2280–2291. doi: 10.1104/pp.105.061903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Orzechowski S. Starch metabolism in leaves. Acta Biochim Pol. 2008;55:435–445. [PubMed] [Google Scholar]
- 47.Salamone PR, Greene TW, Kavakli IH, Okita TW. Isolation and characterization of a higher plant ADP-glucose pyrophosphorylase small subunit homotetramer. FEBS Lett. 2000;482:113–118. doi: 10.1016/s0014-5793(00)01985-2. [DOI] [PubMed] [Google Scholar]
- 48.James MG, Denyer K, Myers AM. Starch synthesis in the cereal endosperm. Curr Opin Plant Biol. 2003;6:215–222. doi: 10.1016/s1369-5266(03)00042-6. [DOI] [PubMed] [Google Scholar]
- 49.Hannah LC, James M. The complexities of starch biosynthesis in cereal endosperms. Curr Opin Biotechnol. 2008;19:160–165. doi: 10.1016/j.copbio.2008.02.013. [DOI] [PubMed] [Google Scholar]
- 50.Tetlow IJ, Beisel KG, Cameron S, Makhmoudova A, Liu F, et al. Analysis of protein complexes in wheat amyloplasts reveals functional interactions among starch biosynthetic enzymes. Plant Physiol. 2008;146:1878–1891. doi: 10.1104/pp.108.116244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mutisya J, Sun C, Rosenquist S, Baguma Y, Jansson C. Diurnal oscillation of SBE expression in sorghum endosperm. J Plant Physiol. 2009;166:428–434. doi: 10.1016/j.jplph.2008.06.017. [DOI] [PubMed] [Google Scholar]
- 52.Lans H, Marteijn JA, Schumacher B, Hoeijmakers JH, Jansen G, et al. Involvement of global genome repair, transcription coupled repair, and chromatin remodeling in UV DNA damage response changes during development. PLoS Genet. 2010;6:e1000941. doi: 10.1371/journal.pgen.1000941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shaked H, Avivi-Ragolsky N, Levy AA. Involvement of the Arabidopsis SWI2/SNF2 chromatin remodeling gene family in DNA damage response and recombination. Genetics. 2006;173:985–994. doi: 10.1534/genetics.105.051664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Park JH, Park EJ, Hur SK, Kim S, Kwon J. Mammalian SWI/SNF chromatin remodeling complexes are required to prevent apoptosis after DNA damage. DNA Repair (Amst) 2009;8:29–39. doi: 10.1016/j.dnarep.2008.08.011. [DOI] [PubMed] [Google Scholar]
- 55.Zhao Q, Wang QE, Ray A, Wani G, Han C, et al. Modulation of nucleotide excision repair by mammalian SWI/SNF chromatin-remodeling complex. J Biol Chem. 2009;284:30424–30432. doi: 10.1074/jbc.M109.044982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yamada R, Ueda HR. Microarrays: statistical methods for circadian rhythms. Methods Mol Biol. 2007;362:245–264. doi: 10.1007/978-1-59745-257-1_17. [DOI] [PubMed] [Google Scholar]
- 57.Higo K, Ugawa Y, Iwamoto M, Korenaga T. Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res. 1999;27:297–300. doi: 10.1093/nar/27.1.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Expression profiles between the biological replicate samples of seedling leaves. A: Pair-wise scatter plots for the raw probe-set intensity data across all time points. B: The number distribution of all probe sets based on the correlation coefficient (Cr) across the time series between the biological replicate samples (including file name, file format, name and URL link of appropriate viewer if format is unusual).
(TIF)
Real-time RT-PCR validation for selected genes. Four genes (LOC_Os03g39610, LOC_Os07g22930, LOC_Os07g46790, and LOC_Os06g30310) were selected for real-time RT-PCR to validate the expression patterns of flag leaves (A) and seedling leaves (B) in seven time points from 12 hr to 36 hr. The purple and yellow bars represent the expression level in microarrys, and the blue bars represent the relative intensity of real-time RT-PCR from independent biological replicates.
(TIF)
The diurnal pattern of the key genes in the starch biosynthesis pathway. The phase of each probe set was redefined as its relation to the time point of light shift (the start or the end of night). Red, blue and white squares to indicate the noon, midnight and light shift phase of the probe set, respectively.
(TIF)
Raw tables for diurnal probe-sets list.
(XLS)
Tables for diurnal probe sets related to sucrose and starch pathways.
(XLS)