Abstract
Human ether-á-go-go–related gene (hERG) potassium channels have voltage-dependent closing (deactivation) kinetics that are unusually slow. A Per-Arnt-Sim (PAS) domain in the cytoplasmic N-terminal region of hERG regulates slow deactivation by making a direct interaction with another part of the hERG channel. The mechanism for slow deactivation is unclear, however, because the other regions of the channel that participate in regulation of deactivation are not known. To identify other functional determinants of slow deactivation, we generated hERG channels with deletions of the cytoplasmic C-terminal regions. We report that hERG channels with deletions of the cyclic nucleotide–binding domain (CNBD) had accelerated deactivation kinetics that were similar to those seen in hERG channels lacking the PAS domain. Channels with dual deletions of the PAS domain and the CNBD did not show further acceleration in deactivation, indicating that the PAS domain and the CNBD regulate deactivation by a convergent mechanism. A recombinant PAS domain that we previously showed could directly regulate PAS domain–deleted channels did not regulate channels with dual deletions of the PAS domain and CNBD, suggesting that the PAS domain did not interact with CNBD-deleted channels. Biochemical protein interaction assays showed that glutathione S-transferase (GST)–PAS (but not GST) bound to a CNBD-containing fusion protein. Coexpression of PAS domain–deleted subunits (with intact C-terminal regions) and CNBD-deleted subunits (with intact N-terminal regions) resulted in channels with partially restored slow deactivation kinetics, suggesting regulatory intersubunit interactions between PAS domains and CNBDs. Together, these data suggest that the mechanism for regulation of slow deactivation in hERG channels is an interaction between the N-terminal PAS domain and the C-terminal CNBD.
INTRODUCTION
The human ether-á-go-go–related gene (ERG [hERG]) encodes a voltage-activated potassium channel, which is the primary component of the cardiac delayed rectifier potassium current and helps to repolarize the ventricular action potential (Warmke and Ganetzky, 1994; Sanguinetti et al., 1995; Trudeau et al., 1995). Two types of potentially lethal cardiac arrhythmias are associated with hERG: inherited mutations in the hERG gene underlie type II long QT syndrome, and drug inhibition of hERG channels underlies acquired long QT syndrome (Curran et al., 1995; Sanguinetti et al., 1995). hERG channels are also expressed in the brain, although their role is less clearly defined (Wymore et al., 1997; Saganich et al., 2001; Guasti et al., 2005). Mammalian ERG plays a role in spike frequency adaptation in cultured neurons, and ERG is involved in membrane excitability in medial vestibular nucleus neurons and firing frequency adaptation in Purkinje cells (Chiesa et al., 1997; Sacco et al., 2003; Pessia et al., 2008). A primate-specific ERG isoform is associated with schizophrenia (Huffaker et al., 2009). Together, these data suggest a role for hERG in neuronal function and mental illness in addition to its well-defined role in the heart.
hERG channel gating is characterized by slow closing (deactivation; Sanguinetti et al., 1995; Trudeau et al., 1995), but the mechanism of deactivation is not fully understood. Previous data suggest that the N-terminal Per-Arnt-Sim (PAS) domain (amino acids 1–135) regulates deactivation gating (Morais Cabral et al., 1998; Wang et al., 1998; Viloria et al., 2000) because channels with deletions of most of the N-terminal region or deletions of just the PAS domain have deactivation rates up to 10-fold faster than those for wild-type hERG (Schönherr and Heinemann, 1996; Spector et al., 1996; Wang et al., 1998). Channels with point mutations in the PAS domain also have rapid deactivation kinetics (Morais Cabral et al., 1998; Chen et al., 1999; Gustina and Trudeau, 2009). The PAS domain is proposed to form a stable, direct interaction with the hERG channel, as indicated by the ability of the PAS domain to regulate channel gating when expressed as a purified protein or a separate gene fragment (Morais Cabral et al., 1998; Gustina and Trudeau, 2009). Förster resonance energy transfer experiments show that soluble PAS domains are in close proximity to the channel at the plasma membrane (Gustina and Trudeau, 2009). The data strongly suggest that the PAS domain interacts directly with other parts of the channel to regulate deactivation gating, but the identity of these other regions is not known.
hERG is a member of the ether-á-go-go family of potassium channels, which is homologous to the cyclic nucleotide–gated (CNG) and hyperpolarization-activated cyclic nucleotide–modulated (HCN) families of cation channels (Warmke and Ganetzky, 1994; Zagotta et al., 2003). hERG, CNG, and HCN channels possess large intracellular C-terminal regions that contain cyclic nucleotide–binding domains (CNBDs; Kaupp et al., 1989; Kaupp, 1991; Warmke and Ganetzky, 1994; Morais Cabral et al., 1998; Zagotta et al., 2003). Cyclic nucleotides bind to and regulate gating in CNG and HCN channels; in contrast, cyclic nucleotides bind with low affinity but do not regulate hERG channels (Brelidze et al., 2009). In HCN channels, the C-linker and CNBD have been shown to form a tetramer that occupies the intracellular space beneath the channel pore (Zagotta et al., 2003). A similar structure has been proposed for hERG and would position the CNBD to interact with other regions of the channel (Miranda et al., 2008; Al-Owais et al., 2009).
In this study, we focus on directly identifying the site of interaction in the hERG channel for the PAS domain. We created targeted deletions of the C-terminal regions and found that channels with deletions of the CNBD have rapid deactivation similar to channels with deletions of the N-terminal PAS domain. Channels with CNBD deletions also had increased rates of activation. The CNBD-deleted channels lack regulation by a genetically encoded PAS domain fragment, suggesting that the PAS domain is not able to interact with channels lacking the CNBD. Protein biochemistry experiments demonstrate direct binding of the PAS domain to a CNBD-containing C-terminal region protein. Coexpression of PAS domain–deleted subunits (with intact C-terminal regions) and CNBD-deleted subunits (with intact N-terminal regions) resulted in channels with partially restored slow deactivation kinetics, suggesting regulatory intersubunit interactions between PAS domains and CNBDs. Together, our results show that the N-terminal PAS domain interacts with the C-terminal CNBD and regulates deactivation gating in hERG channels. We found that the CNBD-deleted channels also had more rapid activation gating, indicating a separate role for the CNBD in regulation of activation gating.
MATERIALS AND METHODS
Molecular biology
The hERG, hERG ΔN, and N1–135 expression clones were previously described (Trudeau et al., 1995, 1999; Wang et al., 1998; Gustina and Trudeau, 2009). C-terminal region deletions were created using overlap extension PCR with custom-made primers. All constructs contained S620T, a point mutation that has been shown to increase channel expression and remove inactivation without affecting PAS domain regulation of gating (Herzberg et al., 1998; Wang et al., 1998; Viloria et al., 2000; Gustina and Trudeau, 2009). All constructs were verified by standard fluorescence-based DNA sequencing. For expression in Xenopus laevis oocytes, mRNAs were made with the mMessage mMachine kit (Applied Biosystems).
Preparation and injection of oocytes
Oocytes were collected from female frogs (Xenopus; Xenopus Express) anesthetized by a 30–45-min exposure to 0.3% tricaine (Sigma-Aldrich). All animal procedures were approved by the University of Maryland School of Medicine Animal Care and Use Committee. The follicular membranes were removed from oocytes by treatment with collagenase B (Sigma-Aldrich), and stage IV oocytes were sorted for injection. For electrophysiological recordings, Xenopus oocytes were pressure injected with the same volume of RNA (52 nl) for each experiment and incubated for 3–10 d at 16°C in ND96 with 50 µg/ml gentamicin (Sigma-Aldrich).
Electrophysiology and analysis
Recordings from whole oocytes were performed at room temperature with a two-electrode voltage-clamp (OC-725C; Warner Instruments) connected to an analog to digital converter (ITC-18; Instrutech). Glass recording microelectrodes had resistances of 1–2 MΩ and were filled with 3-M KCl. The bath (external) solution contained 4-mM KCl, 94-mM NaCl, 1-mM MgCl2, 0.3-mM CaCl2, and 5-mM HEPES, pH 7.4. Data were recorded using Patchmaster software (HEKA) and analyzed using Igor Pro software (Wavemetrics). The holding potential was −80 mV throughout. For clarity, capacitance transients were manually removed from current traces.
The I-V relationship was determined by normalizing peak outward current after 1-s depolarizations from −100 to 80 mV in 20-mV increments to the peak outward current for that cell. The steady-state voltage dependence of activation (G-V) was determined by normalizing the instantaneous tail currents at −50 mV after 1-s steps from −100 to 40 mV in 20-mV increments to the peak instantaneous tail current at −50 mV. These data were fit with a Boltzman function (y = 1/[1 + e[(V1/2 − V)/k]]) in which V1/2 is the half-maximal activation potential and k is the slope factor. For the time to half-maximal activation, we manually determined the time after the depolarizing step at which the current reached half of its maximal value (based on the peak current at the end of the 1-s depolarization). Current relaxations with repolarizing voltage steps (deactivation) were fit with an exponential function (y = Ae(–t/τ))in which t is time and τ is the time constant of deactivation. A single exponential was found to provide the best fit for the data. Averaged data are presented as the mean ± SEM. n represents the number of cells. Statistical analyses were performed using one-way ANOVA with Tukey’s test for pairwise comparisons. A value of P < 0.01 was considered statistically significant.
Protein biochemistry
Glutathione S-transferase (GST) N1–135 or GST-only constructs were transformed in BL21 (DE3) competent cells (Agilent Technologies). Bacterial cultures were prepared and incubated at 37°C with shaking until the exponential growth phase was reached. Cultures were cooled at 30°C for 30 min. Protein expression was induced using 0.4-mM isopropyl-β-d-thiogalactoside (Research Products International), and cultures were incubated at 30°C overnight with shaking. Bacteria were harvested with centrifugation at 4,000 rpm for 10 min, resuspended in buffer S (50-mM Tris, pH 8, 150-mM NaCl, 25-mM imidazole, 0.5% CHAPS, and 0.25% Tween 20; Sigma-Aldrich), and lysed with sonication, and the resulting samples were centrifuged at 13,000 rpm for 20 min to remove cellular debris. The supernatants were applied to preequilibrated glutathione Sepharose 4 Fast Flow beads (GE Healthcare) and incubated with rotation at 4°C for 30 min. The beads were washed three times with buffer S to remove unbound proteins. Bound protein concentration was assessed using spectrophotometry.
H6 hERG 666–872 Flag was transformed in M15 (pPREP4) competent cells (VWR Scientific Products). Bacterial cultures were prepared and incubated at 37°C with shaking until the exponential growth phase was reached. Cultures were cooled at 18°C for 30 min. Protein expression was induced using 1-mM isopropyl-β-d-thiogalactoside, and cultures were incubated at 18°C overnight with shaking. Bacteria were harvested via centrifugation at 4,000 rpm for 10 min, resuspended in buffer S, and lysed with sonication, and the resulting samples were centrifuged at 13,000 rpm for 20 min to remove cellular debris. The supernatant was applied to a nickel column (HiTrap Chelating HP; GE Healthcare) to bind the H6 hERG 666–872 Flag protein. The purified protein was eluted from the column by buffer S with 500-mM imidazole.
Purified H6 hERG 666–872 Flag was combined with purified GST N1–135 or GST only conjugated to glutathione Sepharose beads in buffer S containing 2% CHAPS. Samples were incubated at 4°C overnight with rotation. Samples were washed to remove any unbound proteins. Bound proteins were then stripped off the beads and run on a 4–15% Tris-HCl gel (Criterion Precast; Bio-Rad Laboratories) using SDS-PAGE at 150 V for 90 min. To detect H6 hERG 666–872 Flag, proteins were blotted onto nitrocellulose membrane at 100 V for 45 min at 4°C. The blot was developed with the Hoefer Processor Plus (GE Healthcare) at 4°C using mouse anti-Flag primary antibody at 1:2,000 (Sigma-Aldrich) and goat anti–mouse horseradish peroxidase–conjugated secondary antibody at 1:1,000 (Thermo Fisher Scientific). Blots were developed using SuperSignal West Femto (Thermo Fisher Scientific) and imaged with a chemiluminescence imager (ChemiDoc XRS; Bio-Rad Laboratories). The gel was stained with Coomassie brilliant blue (Bio-Rad Laboratories) to visualize the GST-tagged proteins.
Online supplemental material
The steady-state voltage dependence of activation (G-V), V1/2, and k (slope) values for all constructs used in this study can be found in Table S1. Online supplemental material is available at http://www.jgp.org/cgi/content/full/jgp.201010582/DC1.
RESULTS
Channels with deletions in the C-terminal CNBD have an increased rate of activation and rapid deactivation
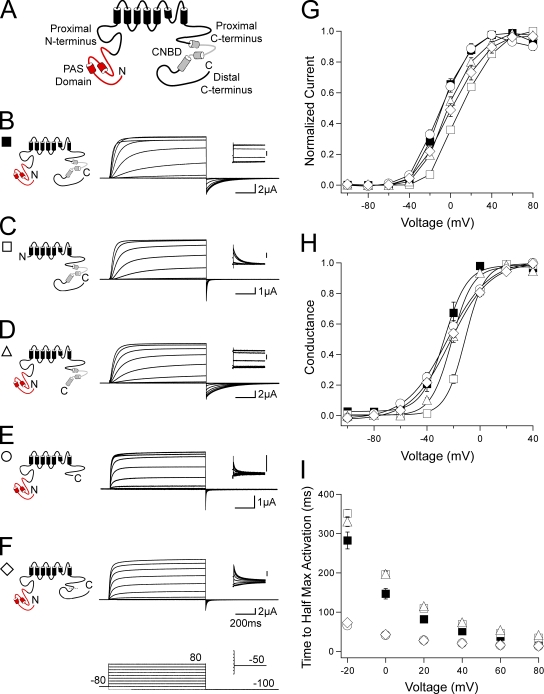
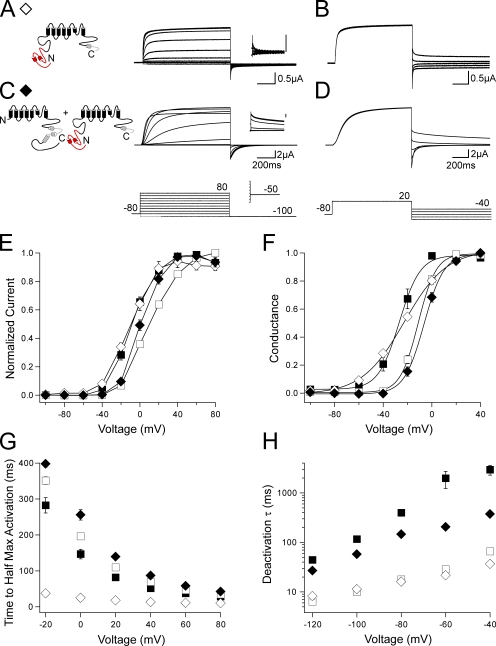
To examine the role of the C-terminal regions in gating of hERG channels, we made a family of channels lacking the C-terminal CNBD and/or the distal C-terminal region (Fig. 1 A). Channels were expressed in oocytes, and currents were recorded using a two-electrode voltage-clamp (see Materials and methods). All channels in this study contained a serine 620 to threonine (S620T) point mutation in the pore region, which removes inactivation gating and enhances channel expression, to more directly measure activation and deactivation gating (Herzberg et al., 1998; Wang et al., 1998). To elicit a family of outward currents, channels were activated by a series of pulses from −100 to 80 mV in 20-mV steps followed by a repolarizing pulse to −100 mV (Fig. 1). Channels with a deletion of most of the N-terminal region (amino acids 2–354), hERG ΔN (Fig. 1, C, G, and H; and Table S1), demonstrated a rightward shift in the I-V relationship and a significant (P < 0.001, ANOVA) right shift in the G-V curve when compared with hERG (Fig. 1, B, G, and H; and Table S1). Channels with a deletion of only the distal C-terminal region (amino acids 873–1,159), hERG Δdistal C, had no measurable differences in the I-V, G-V, or time to half-maximal activation compared with hERG (Fig. 1, D and G–I; and Table S1). In channels with the CNBD (amino acids 749–872) deleted in addition to the distal C-terminal region deletion, hERG ΔCNBD/distal C (Fig. 1 E), the I-V and G-V were not measurably shifted (Fig. 1, G and H; and Table S1); however, we observed a significant (P < 0.01 at all voltages, ANOVA) and marked decrease in the time to half-maximal activation (Fig. 1 I). Channels with a deletion of only the CNBD, hERG ΔCNBD, also demonstrated a significant (P < 0.01 at all voltages, ANOVA) decrease in the time to half-maximal activation with no measurable changes to the I-V and G-V (Fig. 1, F–I). These results suggest that the CNBD, but not the distal C-terminal region, is involved in activation gating. These recordings also show that channels lacking the CNBD (Fig. 1, E and F) had a very rapidly deactivating tail current at repolarization to −100 mV, similar to that in hERG ΔN channels but unlike that in wild-type hERG or hERG Δdistal C. These results suggest that the CNBD also plays a role in regulation of deactivation gating.
Figure 1.
hERG activation gating is altered by C-terminal CNBD deletion. (A) Schematic of the hERG K+ channel with relevant N- and C-terminal domains indicated. (B–F) Channel schematics and two-electrode voltage-clamp recordings of a family of currents from hERG (B), hERG ΔN (C), hERG Δdistal C (D), hERG ΔCNBD/distal C (E), and hERG ΔCNBD (F). Currents were elicited using the pulse protocols indicated. Insets showing tail currents at −50 mV are 200 ms in duration; inset scale bars are 0.5 μA. (G) I-V relationship for B–F. The currents at the end of each depolarizing pulse were normalized to the peak current for that cell and plotted versus voltage. n ≥ 4 for each. (H) The steady-state voltage dependence of activation (G-V) for B–F. The instantaneous tail currents at −50 mV were normalized to the peak instantaneous tail current for that cell and plotted versus voltage. n ≥ 5 for each. Data were fit with a Boltzmann function to determine the V1/2 and k (slope) values. (I) Plot of the time to half-maximal activation versus voltage for B–F. n ≥ 4 for each. For G–I, the values are plotted as the means ± SEM. The error bars are within the points if not visible.
Deactivation was further investigated by recording a family of tail currents. Channels were activated with a pulse to 20 mV followed by repolarizing pulses between −120 and −40 mV in 20-mV increments (Fig. 2). The tail currents were fit with a single exponential function to derive the time constant of deactivation (see Materials and methods). As previously reported (Schönherr and Heinemann, 1996; Spector et al., 1996; Morais Cabral et al., 1998; Wang et al., 1998; Viloria et al., 2000), deletion of the N-terminal region in hERG ΔN caused a significant (P < 0.001 at all voltages, ANOVA) speeding of deactivation compared with hERG (Fig. 2, A, B, and F). The deactivation kinetics of hERG Δdistal C (Fig. 2, C and F) were not measurably different from those of hERG. Channels in which the CNBD was deleted, hERG ΔCNBD/distal C (Fig. 2, D and F) and hERG ΔCNBD (Fig. 2, E and F), had rapid deactivation kinetics that were not different from those of hERG ΔN, indicating that the CNBD was necessary for slow deactivation. These results show that channels with deletion of the CNBD have fast deactivation kinetics, like channels with deletion of the N-terminal region, over a range of repolarizing voltages and suggest that the N-terminal region and the CNBD may regulate deactivation gating by a convergent mechanism.
Figure 2.
C-terminal CNBD deletion affects hERG deactivation. (A–E) Channel schematics and two-electrode voltage-clamp recordings of a family of tail currents from hERG (A), hERG ΔN (B), hERG Δdistal C (C), hERG ΔCNBD/distal C (D), and hERG ΔCNBD (E). Currents were elicited using the pulse protocol indicated. (F) Plot of the time constants (τ) of deactivation derived from a single exponential fit (see Materials and methods) to the tail currents in A–E. The values are plotted as the means ± SEM. The error bars are within the points if not visible. n ≥ 5 for each.
N-terminal region deletion does not further alter gating in channels with a CNBD deletion
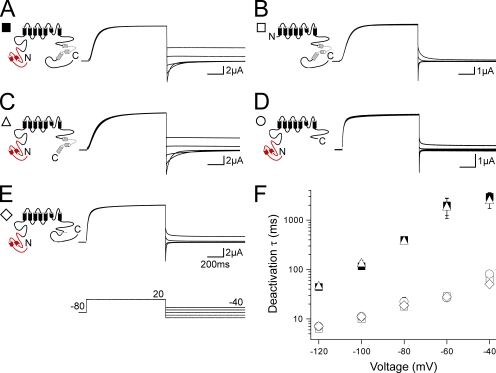
We next asked whether gating in channels with deletions of both the N- and C-terminal regions was additive to or independent of C-terminal region–only deletions. We recorded families of outward currents and tail currents from N-terminal region–deleted channels that also had deletions of the distal C terminus, the CNBD, or both the CNBD and distal C-terminal region (Fig. 3). hERG ΔN Δdistal C channels (Fig. 3, A and B) had a rightward shift in the I-V relationship and a significant (P < 0.001, ANOVA) right shift in the G-V curve when compared with hERG Δdistal C channels (Fig. 3, G–I; and Table S1). The rightward shifts in the I-V and G-V were surprisingly greater than the shifts observed with hERG ΔN alone (Fig. 3, G–I, open squares). hERG ΔN Δdistal C also had fast deactivation (Fig. 3 J) that was not measurably different from hERG ΔN, indicating that N-terminal region deletion speeds deactivation gating irrespective of the presence of the distal C-terminal region. hERG ΔN ΔCNBD/distal C (Fig. 3, C, D, and G–J) and hERG ΔN ΔCNBD (Fig. 3, E–J) showed no measurable differences compared with hERG ΔCNBD/distal C and hERG ΔCNBD, indicating that there was no additional effect of the N-terminal deletions on channel gating in channels lacking the CNBD. These results suggest that the N-terminal region and the CNBD exert their effects on channel deactivation via a convergent mechanism.
Figure 3.
N-terminal region deletion does not further alter gating in C-terminal–deleted channels that include a CNBD deletion. (A, C, and E) Channel schematics and two-electrode voltage-clamp recordings of a family of currents from hERG ΔN Δdistal C (A), hERG ΔN ΔCNBD/distal C (C), and hERG ΔN ΔCNBD (E). Currents were elicited using the pulse protocols indicated. Insets showing tail currents at −50 mV are 200 ms in duration; inset scale bars are 0.5 μA. (B, D, and F) Two-electrode voltage-clamp recordings of a family of tail currents from the constructs in A, C, and E. Currents were elicited using the pulse protocol indicated. (G) Normalized I-V relationship for A, C, and E. n ≥ 6 for each. (H) Normalized G-V relationship for A, C, and E. Data were fit with a Boltzmann function to determine the V1/2 and k (slope) values. n ≥ 6 for each. (I) Plot of the time to half-maximal activation for A, C, and E. n ≥ 6 for each. (J) Plot of the time constants (τ) of deactivation derived from a single exponential fit to the tail currents in B, D, and F. n ≥ 5 for each. For G–J, the values are plotted as the means ± SEM. The error bars are within the points if not visible. hERG (closed squares) and hERG ΔN (open squares) are included on all plots for reference.
Coexpression of a genetically encoded PAS domain fragment modifies gating in distal C-terminal region–deleted channels but has no effect on CNBD-deleted channels
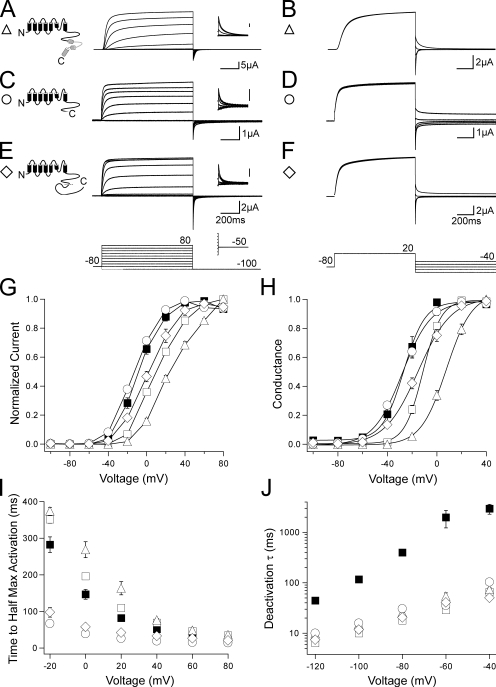
To more directly test the ability of the N-terminal region, specifically the PAS domain (amino acids 1–135), to regulate gating in channels with C-terminal region deletions, we probed channels with a genetically encoded PAS domain fragment (N1–135). N1–135 was shown to noncovalently regulate channel deactivation gating in channels lacking the N-terminal region (Gustina and Trudeau, 2009). We coexpressed N1–135 with channels containing both N- and C-terminal region deletions and recorded families of outward currents and tail currents. hERG ΔN Δdistal C + N1–135 (Fig. 4, A and B) showed a left shift in the I-V toward the values obtained for hERG Δdistal C and hERG, although no change was seen in the G-V curve from hERG ΔN Δdistal C (Fig. 4, G and H; and Table S1). When N1–135 was coexpressed with hERG ΔN Δdistal C, the deactivation rate was significantly (P < 0.01 at all voltages, ANOVA) slowed to a rate similar to that of hERG Δdistal C and hERG (Fig. 4 J). These results suggest that N1–135 interacted with hERG ΔN Δdistal C channels to regulate gating, meaning that the presence of the distal C-terminal region was not required for the PAS domain to regulate gating. Coexpression of N1–135 with hERG ΔN ΔCNBD/distal C and hERG ΔN ΔCNBD yielded no measurable changes in the I-V, G-V, time to half-maximal activation, or deactivation kinetics (Fig. 4, C–J; and Table S1), indicating that N1–135 is unable to regulate gating in channels that are lacking the CNBD. These results suggest the PAS domain may interact with the CNBD.
Figure 4.
N1–135 coexpression has no effect on channels that have a CNBD deletion but does modify gating in distal C-terminal region–deleted channels. (A, C, and E) Channel schematics and two-electrode voltage-clamp recordings of a family of currents from hERG ΔN Δdistal C + N1–135 (A), hERG ΔN ΔCNBD/distal C + N1–135 (C), and hERG ΔN ΔCNBD + N1–135 (E). Currents were elicited using the pulse protocols indicated. Insets showing tail currents at −50 mV are 200 ms in duration; inset scale bars are 0.5 μA. (B, D, and F) Two-electrode voltage-clamp recordings of a family of tail currents from the constructs in A, C, and E. Currents were elicited using the pulse protocol indicated. (G) Normalized I-V relationship for A, C, and E. n ≥ 5 for each. (H) Normalized G-V relationship for A, C, and E. Data were fit with a Boltzmann function to determine the V1/2 and k (slope) values. n ≥ 6 for each. (I) Plot of the time to half-maximal activation for A, C, and E. n ≥ 5 for each. (J) Plot of the time constants (τ) of deactivation derived from a single exponential fit to the tail currents in B, D, and F. n ≥ 5 for each. For G–J, the values are plotted as the means ± SEM. The error bars are within the points if not visible. hERG (closed squares) and hERG ΔN (open squares) are included on all plots for reference.
A biochemical interaction assay indicates binding of the PAS domain to the CNBD
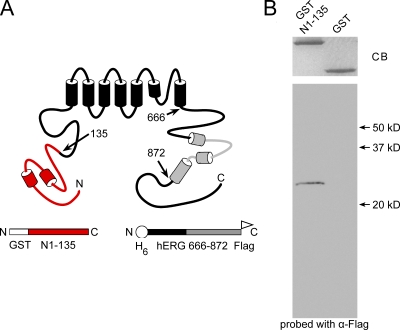
To directly test for an interaction between the PAS domain and the CNBD, we performed a protein interaction assay. First, we fused the PAS domain to GST (GST N1–135; Fig. 5 A). Then, we added polyhistidine and Flag tags to a C-terminal region comprised of the C-linker region and the CNBD (H6 hERG 666–872 Flag; Fig. 5 A). (We included the C-linker region because similar C-linker–CNBD proteins from HCN2 and hERG were soluble [Zagotta et al., 2003; Brelidze et al., 2009].) GST N1–135 or GST (as a negative control) was conjugated to glutathione beads. H6 hERG 666–872 Flag proteins were purified using a nickel column (see Materials and methods). To test for interactions, H6 hERG 666–872 Flag was added to the bead-bound GST N1–135 or GST proteins and allowed to incubate overnight. The next day, the beads were washed to remove any unbound fusion proteins, and hERG 666–872 fusion proteins that were associated with bead-bound GST N1–135 were analyzed by SDS-PAGE and identified with a Western blot. As seen in Fig. 5 B, GST N1–135 and GST were present in relatively equivalent amounts as indicated by Coomassie blue staining; however, the CNBD-containing protein hERG 666–872 was only detected when GST N1–135 was present. These results indicate a direct and specific interaction of the PAS domain with the C-terminal CNBD and/or C-linker region.
Figure 5.
Biochemical interaction of hERG 1–135 with the CNBD. (A) Schematic of the hERG channel and the constructs used for the biochemical interaction assay. (B) Coomassie stain (CB, Coomassie blue) and Western blot of GST N1–135 and GST-only control probed with α-Flag to detect bound hERG H6 C-linker/CNBD-Flag.
Evidence for an intersubunit interaction between N- and C-terminal regions
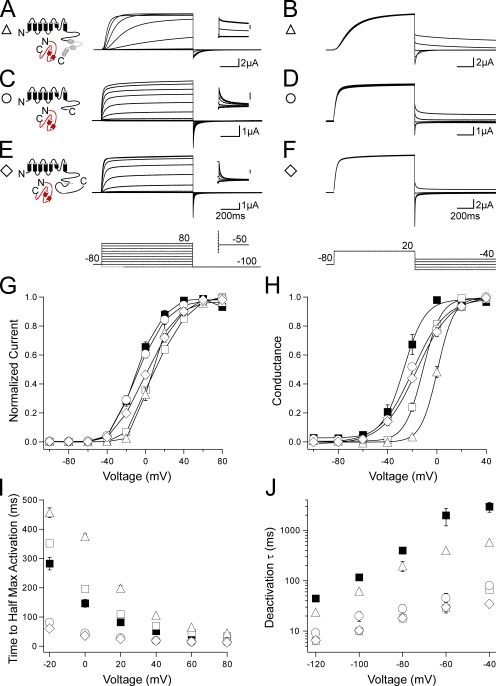
We next hypothesized that functional interactions between PAS domains and CNBDs could occur between different subunits. To test this, we first generated and characterized a channel lacking the distal C-terminal region and the majority of the CNBD (residues 815–1,159), hERG ΔpCNBD/distal C (Fig. 6, A and B). This channel demonstrated a shallow G-V curve, faster activation as indicated by a significantly (P < 0.001 at all voltages, ANOVA) decreased time to half-maximal activation, and a significantly (P < 0.01 at all voltages, ANOVA) faster deactivation rate compared with hERG (Fig. 6, E–H; and Table S1). The channel also had rapid deactivation that was not different from the rate of deactivation of hERG ΔN, suggesting that partial deletion of the CNBD is sufficient to disrupt PAS domain–dependent regulation of the channel. When hERG ΔpCNBD/distal C was coexpressed with hERG ΔN, the G-V curve was right shifted toward that of hERG ΔN, and the time to half-maximal activation was significantly (P < 0.001 at all voltages, ANOVA) increased, indicating slower activation than that of hERG ΔN (Fig. 6, C, E, F, and G; and Table S1). Surprisingly, hERG ΔN + hERG ΔpCNBD/distal C demonstrated a significant (P < 0.001 at all voltages, ANOVA) slowing of deactivation similar to that seen with wild-type hERG (Fig. 6, D and H). Independently, hERG ΔN and hERG ΔpCNBD/distal C channels have fast deactivation kinetics, but, when coexpressed, they have slow deactivation. These results are not explained by an additive effect of two separate populations of channels. Rather, these results suggest N-terminal region modulation of gating of the adjacent subunits, which lack a functional N-terminal region in heterotetrameric channels. This could occur through an intersubunit interaction between the N-terminal region of hERG ΔpCNBD/distal C and the C-terminal region of hERG ΔN.
Figure 6.
Coexpression of N-terminal region–deleted hERG with C-terminal region–deleted hERG partially compensates for gating alterations caused by either N- or C-terminal region deletion. (A and C) Channel schematics and two-electrode voltage-clamp recordings of a family of currents from hERG ΔpCNBD/distal C (A) and hERG ΔN + hERG ΔpCNBD/distal C (C). Currents were elicited using the pulse protocols indicated. Insets showing tail currents at −50 mV are 200 ms in duration; inset scale bars are 0.5 μA. (B and D) Two-electrode voltage-clamp recordings of a family of tail currents from the constructs in A and C. Currents were elicited using the pulse protocol indicated. (E) Normalized I-V relationship for A and C. n ≥ 5 for each. (F) Normalized G-V relationship for A and C. Data were fit with a Boltzmann function to determine the V1/2 and k (slope) values. n ≥ 6 for each. (G) Plot of the time to half-maximal activation for A and C. n ≥ 5 for each. (H) Plot of the time constants (τ) of deactivation derived from a single exponential fit to the tail currents in B and D. n ≥ 5 for each. For E–H, the values are the means ± SEM. The error bars are within the points if not visible. hERG (closed squares) and hERG ΔN (open squares) are included on all plots for reference.
DISCUSSION
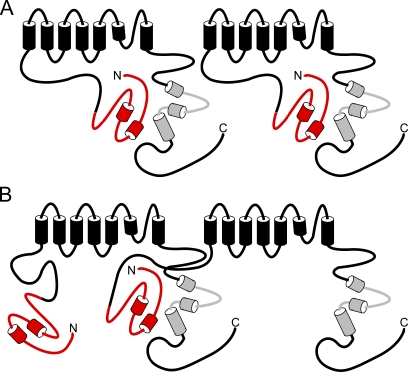
In this study, we present evidence that an interaction between the N-terminal PAS domain and the C-terminal CNBD is the mechanism for regulation of hERG channel deactivation gating. We showed that channels with deletions of the CNBD have fast deactivation (similar to that seen in channels with N-terminal region deletions). The CNBD-deleted channels lack regulation by a genetically encoded PAS domain fragment, although channels with distal C-terminal region deletions maintain PAS domain modulation. Protein biochemistry experiments demonstrate direct binding of purified N-terminal PAS and C-terminal CNBD–containing proteins. A model we propose to explain our results is that the PAS domain forms a stable interaction with the CNBD either within individual subunits of the channel (Fig. 7 A) or between adjacent subunits of the channel (Fig. 7 B). If the CNBD is not present in the channel, our data show that the PAS domain is not sufficient to regulate channel gating. We propose that, when the CNBD is deleted, the PAS domain is not properly positioned to regulate deactivation gating.
Figure 7.
Model of N- and C-terminal region interactions in the hERG K+ channel. (A) Interaction of the N-terminal PAS domain with the C-terminal CNBD within the same subunit of the hERG channel. (B) Interaction of the PAS domain with the CNBD on adjacent subunits of the hERG channel.
We have also found evidence for intersubunit interaction between N- and C-terminal regions in hERG channels. hERG ΔN homomeric channels and hERG ΔpCNBD/distal C homomeric channels had fast deactivation kinetics; however, coexpression of hERG ΔN and hERG ΔpCNBD/distal C subunits resulted in channels that had slow deactivation kinetics. These results strongly suggest the formation of heterotetrameric channels, as two independent populations of fast deactivating channels could not demonstrate slow deactivation gating. These results also indicate an intersubunit interaction between the functional N-terminal region on one subunit with a functional C-terminal region on another subunit within a heterotetramer (Fig. 7 B). (We also noted that the mean current amplitude of hERG ΔN channels [1.58 ± 0.33 μA; n = 3] and hERG ΔpCNBD/distal C channels [0.86 ± 0.14 μA; n = 5] was modest, but when these channels were coexpressed, we measured a marked enhancement of the current [11.61 ± 0.57 μA; n = 7], consistent with subunit heterotetramerization.) When individual subunits possess only either N-terminal or C-terminal regions, we propose that the N- and C-terminal regions make an intersubunit interaction. However, this does not exclude the possibility that wild-type subunits, which possess both N- and C-terminal regions, could form intrasubunit N- and C-terminal interactions (Fig. 7 A). Perhaps interdomain and intersubunit interactions between N-terminal and C-terminal regions are a general regulatory mechanism in channels with CNBDs because the N-terminal region interacts with the C-terminal region in rod and olfactory CNG channels (Varnum and Zagotta, 1997; Trudeau and Zagotta, 2002).
Our data indicate that both the PAS domain and the CNBD are required for regulation of deactivation.Because channels with deletions of the proximal N-terminal region (located between the PAS domain and the transmembrane S1 domain) are regulated by recombinant PAS domains, the proximal N-terminal region does not appear to play a central role in deactivation gating. Other studies have proposed that the S4–S5 linker interacts with the PAS domain and regulates deactivation gating (Morais Cabral et al., 1998; Wang et al., 2000; Li et al., 2010). In our study, channels (hERG ΔCNBD and hERG ΔCNBD/distal C) with intact PAS domains and intact S4–S5 linkers (but deleted CNBDs) had accelerated deactivation, indicating that PAS domains and S4–S5 linkers are not sufficient for regulation of deactivation gating and suggesting that CNBDs are required for regulation of deactivation. However, our results do not rule out the possibility that the S4–S5 linker may play a necessary role in slow deactivation. It is possible that interactions between the S4–S5 linker and (a) the PAS domain, (b) just the first ∼26 amino acids of the PAS domain, (c) the C-linker/CNBD, or (d) a PAS domain and C-linker/CNBD complex may contribute to regulation of deactivation gating. Determining these possible interactions and their putative role in regulation of deactivation will require further investigation.
We found that channels with CNBD deletions (Fig. 1, E, F, and H; Fig. 6, A and F; and Table S1) had a twofold increase in the slope (k) values of the G-V curves compared with that in wild-type hERG (Fig. 1, B and H; and Table S1). We interpret these findings to mean that the CNBD regulates gating transitions in the activation and deactivation pathways. In heteromeric channels formed from N-deleted and CNBD-deleted subunits, the slope of the G-V was restored to a value similar to that of wild-type channels (Fig. 6, C and F; and Table S1). This result suggests that an interaction between PAS domains and CNBDs is necessary for shaping the voltage dependence of activation and deactivation in hERG channels.
We show that the distal C-terminal region is not apparently involved in hERG activation or deactivation gating, as channels lacking only this region, hERG Δdistal C, did not have activation or deactivation properties that were different from those of wild-type hERG channels. Additionally, channels lacking the CNBD and distal C terminus, hERG ΔCNBD/distal C, did not have activation or deactivation properties that were different from channels that lacked only the CNBD, hERG ΔCNBD. An interesting observation was noted when the N-terminal region was deleted in addition to the distal C-terminal region: hERG ΔN Δdistal C channels demonstrated a right shift in the V1/2 of the G-V and an increased time to half-maximal activation when compared with hERG Δdistal C, indicating a unique effect of N-terminal region deletion in these channels. Coexpression of N1–135 with hERG ΔN Δdistal C resulted in only a partial restoration of the V1/2 of the G-V to the values of hERG Δdistal C, suggesting that the V1/2 shifts in the G-V are not entirely mediated by the PAS domain. As hERG ΔN Δdistal C channels alone or coexpressed with N1–135 are lacking the proximal N terminus (amino acids 136–354), it is possible that the proximal N-terminal region plays a role in determining V1/2.
Our experiments show the usefulness of using hERG S620T to study activation and deactivation kinetics, especially in channels that may not otherwise be expressed in sufficient numbers at the cell surface to produce measurable currents. We observe that the channels with larger C-terminal deletions of 344 and 410 amino acids (hERG ΔpCNBD/distal C and hERG ΔCNBD/distal C, respectively) have smaller mean current amplitudes than the other constructs used in this study, indicating a possible reduction in channel expression. A previous study shows that channels with C-terminal deletions of ≥311 amino acids do not show functional expression (Aydar and Palmer, 2001). We presume that the difference between the previous study and our study was that we used the S620T mutation to foster the expression of functional channels with deletions of the CNBD and C-terminal regions.
In addition to binding the PAS domain and regulating deactivation, our data indicate a separate role for the hERG CNBD in channel activation. In response to depolarization, channels lacking the CNBD (hERG ΔCNBD/distal C, hERG ΔCNBD, or hERG ΔpCNBD/distal C) demonstrated a less sigmoidal (i.e., a more square shaped) current compared with the more sigmoidal current of channels with intact CNBDs. CNBD-deleted channels also had a significantly (P < 0.01 at all voltages, ANOVA) reduced time to half-maximal activation compared with that of wild-type hERG channels (Fig. 1). These results suggest that the CNBD may play a role in regulation of the gating transitions that precede channel opening. The PAS domain does not appear to regulate these transitions, meaning that the effect of the CNBD on channel activation is independent of its interaction with the PAS domain.
Acknowledgments
We thank L. Leung for preparation of oocytes and technical assistance, Dr. G. Robertson for helpful discussions, and E. Gianulis for comments on the manuscript.
This work was supported by a National Institutes of Health grant HL-083121 (to M.C. Trudeau), an American Heart Association Mid-Atlantic Affiliate Predoctoral Fellowship (to A.S. Gustina), and a gift from the Helen Pumphrey Denit Trust.
Kenton J. Swartz served as editor.
Note added in proof. While this article was under review, Muskett et al. (2010. J. Biol. Chem. doi:10.1074/jbc.M110.199364) showed that mutations within the first 26 amino acids of hERG and mutations in the CNBD disrupted deactivation kinetics and proposed a structural model in which the first 26 residues of PAS (based on the nuclear magnetic resonance solution structure) interacted with the CNBD (based on a homology model; Muskett et al., 2010). Their results support our findings that the PAS domain and the C-linker/CNBD interact.
Footnotes
Abbreviations used in this paper:
- CNBD
- cyclic nucleotide–binding domain
- CNG
- cyclic nucleotide gated
- ERG
- ether-á-go-go–related gene
- GST
- glutathione S-transferase
- HCN
- hyperpolarization-activated cyclic nucleotide modulated
- hERG
- human ERG
- PAS
- Per-Arnt-Sim
References
- Al-Owais M., Bracey K., Wray D. 2009. Role of intracellular domains in the function of the herg potassium channel. Eur. Biophys. J. 38:569–576 10.1007/s00249-009-0408-2 [DOI] [PubMed] [Google Scholar]
- Aydar E., Palmer C. 2001. Functional characterization of the C-terminus of the human ether-à-go-go-related gene K(+) channel (HERG). J. Physiol. 534:1–14 10.1111/j.1469-7793.2001.t01-3-00001.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brelidze T.I., Carlson A.E., Zagotta W.N. 2009. Absence of direct cyclic nucleotide modulation of mEAG1 and hERG1 channels revealed with fluorescence and electrophysiological methods. J. Biol. Chem. 284:27989–27997 10.1074/jbc.M109.016337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Zou A., Splawski I., Keating M.T., Sanguinetti M.C. 1999. Long QT syndrome-associated mutations in the Per-Arnt-Sim (PAS) domain of HERG potassium channels accelerate channel deactivation. J. Biol. Chem. 274:10113–10118 10.1074/jbc.274.15.10113 [DOI] [PubMed] [Google Scholar]
- Chiesa N., Rosati B., Arcangeli A., Olivotto M., Wanke E. 1997. A novel role for HERG K+ channels: spike-frequency adaptation. J. Physiol. 501:313–318 (published erratum appears in J. Physiol. (Lond). 1997 502:715) 10.1111/j.1469-7793.1997.313bn.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curran M.E., Splawski I., Timothy K.W., Vincent G.M., Green E.D., Keating M.T. 1995. A molecular basis for cardiac arrhythmia: HERG mutations cause long QT syndrome. Cell. 80:795–803 10.1016/0092-8674(95)90358-5 [DOI] [PubMed] [Google Scholar]
- Guasti L., Cilia E., Crociani O., Hofmann G., Polvani S., Becchetti A., Wanke E., Tempia F., Arcangeli A. 2005. Expression pattern of the ether-a-go-go-related (ERG) family proteins in the adult mouse central nervous system: evidence for coassembly of different subunits. J. Comp. Neurol. 491:157–174 10.1002/cne.20721 [DOI] [PubMed] [Google Scholar]
- Gustina A.S., Trudeau M.C. 2009. A recombinant N-terminal domain fully restores deactivation gating in N-truncated and long QT syndrome mutant hERG potassium channels. Proc. Natl. Acad. Sci. USA. 106:13082–13087 10.1073/pnas.0900180106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzberg I.M., Trudeau M.C., Robertson G.A. 1998. Transfer of rapid inactivation and sensitivity to the class III antiarrhythmic drug E-4031 from HERG to M-eag channels. J. Physiol. 511:3–14 10.1111/j.1469-7793.1998.003bi.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huffaker S.J., Chen J., Nicodemus K.K., Sambataro F., Yang F., Mattay V., Lipska B.K., Hyde T.M., Song J., Rujescu D., et al. 2009. A primate-specific, brain isoform of KCNH2 affects cortical physiology, cognition, neuronal repolarization and risk of schizophrenia. Nat. Med. 15:509–518 10.1038/nm.1962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaupp U.B. 1991. The cyclic nucleotide-gated channels of vertebrate photoreceptors and olfactory epithelium. Trends Neurosci. 14:150–157 10.1016/0166-2236(91)90087-B [DOI] [PubMed] [Google Scholar]
- Kaupp U.B., Niidome T., Tanabe T., Terada S., Bönigk W., Stühmer W., Cook N.J., Kangawa K., Matsuo H., Hirose T., et al. 1989. Primary structure and functional expression from complementary DNA of the rod photoreceptor cyclic GMP-gated channel. Nature. 342:762–766 10.1038/342762a0 [DOI] [PubMed] [Google Scholar]
- Li Q., Gayen S., Chen A.S., Huang Q., Raida M., Kang C. 2010. NMR solution structure of the N-terminal domain of hERG and its interaction with the S4-S5 linker. Biochem. Biophys. Res. Commun. 403:126–132 10.1016/j.bbrc.2010.10.132 [DOI] [PubMed] [Google Scholar]
- Miranda P., Manso D.G., Barros F., Carretero L., Hughes T.E., Alonso-Ron C., Domínguez P., de la Peña P. 2008. FRET with multiply labeled HERG K(+) channels as a reporter of the in vivo coarse architecture of the cytoplasmic domains. Biochim. Biophys. Acta. 1783:1681–1699 10.1016/j.bbamcr.2008.06.009 [DOI] [PubMed] [Google Scholar]
- Morais Cabral J.H., Lee A., Cohen S.L., Chait B.T., Li M., Mackinnon R. 1998. Crystal structure and functional analysis of the HERG potassium channel N terminus: a eukaryotic PAS domain. Cell. 95:649–655 10.1016/S0092-8674(00)81635-9 [DOI] [PubMed] [Google Scholar]
- Pessia M., Servettini I., Panichi R., Guasti L., Grassi S., Arcangeli A., Wanke E., Pettorossi V.E. 2008. ERG voltage-gated K+ channels regulate excitability and discharge dynamics of the medial vestibular nucleus neurones. J. Physiol. 586:4877–4890 10.1113/jphysiol.2008.155762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacco T., Bruno A., Wanke E., Tempia F. 2003. Functional roles of an ERG current isolated in cerebellar Purkinje neurons. J. Neurophysiol. 90:1817–1828 10.1152/jn.00104.2003 [DOI] [PubMed] [Google Scholar]
- Saganich M.J., Machado E., Rudy B. 2001. Differential expression of genes encoding subthreshold-operating voltage-gated K+ channels in brain. J. Neurosci. 21:4609–4624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanguinetti M.C., Jiang C., Curran M.E., Keating M.T. 1995. A mechanistic link between an inherited and an acquired cardiac arrhythmia: HERG encodes the IKr potassium channel. Cell. 81:299–307 10.1016/0092-8674(95)90340-2 [DOI] [PubMed] [Google Scholar]
- Schönherr R., Heinemann S.H. 1996. Molecular determinants for activation and inactivation of HERG, a human inward rectifier potassium channel. J. Physiol. 493:635–642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spector P.S., Curran M.E., Zou A., Keating M.T., Sanguinetti M.C. 1996. Fast inactivation causes rectification of the IKr channel. J. Gen. Physiol. 107:611–619 10.1085/jgp.107.5.611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trudeau M.C., Zagotta W.N. 2002. An intersubunit interaction regulates trafficking of rod cyclic nucleotide-gated channels and is disrupted in an inherited form of blindness. Neuron. 34:197–207 10.1016/S0896-6273(02)00647-5 [DOI] [PubMed] [Google Scholar]
- Trudeau M.C., Warmke J.W., Ganetzky B., Robertson G.A. 1995. HERG, a human inward rectifier in the voltage-gated potassium channel family. Science. 269:92–95 10.1126/science.7604285 [DOI] [PubMed] [Google Scholar]
- Trudeau M.C., Titus S.A., Branchaw J.L., Ganetzky B., Robertson G.A. 1999. Functional analysis of a mouse brain Elk-type K+ channel. J. Neurosci. 19:2906–2918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varnum M.D., Zagotta W.N. 1997. Interdomain interactions underlying activation of cyclic nucleotide-gated channels. Science. 278:110–113 10.1126/science.278.5335.110 [DOI] [PubMed] [Google Scholar]
- Viloria C.G., Barros F., Giráldez T., Gómez-Varela D., de la Peña P. 2000. Differential effects of amino-terminal distal and proximal domains in the regulation of human erg K(+) channel gating. Biophys. J. 79:231–246 10.1016/S0006-3495(00)76286-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Trudeau M.C., Zappia A.M., Robertson G.A. 1998. Regulation of deactivation by an amino terminal domain in human ether-à-go-go-related gene potassium channels. J. Gen. Physiol. 112:637–647 (published erratum appears in J. Gen. Physiol. 1999. 113:359) 10.1085/jgp.112.5.637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Myers C.D., Robertson G.A. 2000. Dynamic control of deactivation gating by a soluble amino-terminal domain in HERG K+ channels. J. Gen. Physiol. 115:749–758 10.1085/jgp.115.6.749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warmke J.W., Ganetzky B. 1994. A family of potassium channel genes related to eag in Drosophila and mammals. Proc. Natl. Acad. Sci. USA. 91:3438–3442 10.1073/pnas.91.8.3438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wymore R.S., Gintant G.A., Wymore R.T., Dixon J.E., McKinnon D., Cohen I.S. 1997. Tissue and species distribution of mRNA for the IKr-like K+ channel, erg. Circ. Res. 80:261–268 [DOI] [PubMed] [Google Scholar]
- Zagotta W.N., Olivier N.B., Black K.D., Young E.C., Olson R., Gouaux E. 2003. Structural basis for modulation and agonist specificity of HCN pacemaker channels. Nature. 425:200–205 10.1038/nature01922 [DOI] [PubMed] [Google Scholar]