Abstract
The accumulation of protein aggregates in neurons appears to be a basic feature of neurodegenerative disease. In huntington disease (HD), a progressive and ultimately fatal neurodegenerative disorder caused by an expansion of the polyglutamine repeat within the protein huntingtin (Htt), the immediate proximal cause of disease is well understood. However, the cellular mechanisms which modulate the rate at which fragments of Htt containing polyglutamine accumulate in neurons is a central issue in the development of approaches to modulate the rate and extent of neuronal loss in this disease. We have recently found that Htt is phosphorylated by the kinase IKK on serine (s) 13, activating its phosphorylation on S16 and its acetylation and poly-SUMOylation, modifications that modulate its clearance by the proteasome and lysosome in cells.1 In the discussion here I suggest that Htt may have a normal function in the lysosomal mechanism of selective macroautophagy involved in its own degradation which may share some similarity with the yeast cytoplasm to vacuole targeting (Cvt) pathway. Pharmacologic activation of this pathway may be useful early in disease progression to treat HD and other neurodegenerative diseases characterized by the accumulation of disease proteins.
Key words: Huntington disease, Huntingtin, polyglutamine, autophagy, IKK
An age-related reduction in protein clearance mechanisms has been implicated in the pathogenesis of neurodegenerative diseases including the polyglutamine (polyQ) repeat diseases, Alzheimer disease (AD), Parkinson disease (PD) and Amyotrophic Lateral Sclerosis (ALS). These diseases are each associated with the accumulation of insoluble protein aggregates in diseased neurons. Huntington Disease (HD), caused by an expansion of the polyQ repeat in the protein Huntingtin (Htt), is one such disease of aging in which mutant Htt inclusions form in striatal and cortical neurons as disease progresses. Clarification of the mechanisms of Htt clearance is paramount to finding therapeutic targets to treat HD that may be broadly useful in the treatment of these currently incurable neurodegenerative diseases.
The Kinase IKK Regulates Htt Degradation
In an effort to better understand the factors which influence mutant Htt degradation, an important focus has been the changes in pathways of mutant Htt clearance which occur as a consequence of cellular stress. Cellular stress can be generated as the direct consequence of mutant Htt expression or by inflammatory stimuli, oxidative stress, DNA damage or any combination of these factors. No matter what the combination of stimuli, activation of the downstream signaling IKK kinase complex appears to be a key regulator of Htt degradation. When IKK is activated, the pattern and control of mutant Htt degradation is altered. We have focused on the details of the cellular response to IKK activation and the consequences on the rate and extent of degradation of mutant Htt.1
In cell models of HD, we find that expanded polyQ mutant Htt is less efficiently phosphorylated and cleared upon activation of IKK than unexpanded Htt, resulting in mutant Htt accumulation and reduced degradation by the proteasome and lysosome.1 IKK is activated in cell culture and mouse models of HD with expression of mutant Htt.2 HD patients exhibit innate immune activation over a decade before overt neurological manifestation of disease3 consistent with a compensatory activation of IKK in vivo to help remove mutant Htt from the cell.
Taking a biochemical approach to study the effects of IKK activation in HD, we have found that IKK directly modifies Htt when activated, phosphorylating Htt on serine (S) 13, inducing its subsequent phosphorylation on S16 and acetylation on lysine (K) 9. These modifications occur on a transient species of Htt that is degraded by both the proteasome and the lysosome in the presence of activated IKK.
A central element to this process now appears to be the recently demonstrated activation of the lysosomal clearance mechanism associated with autophagy by IKK.4 Consistent with this mechanism, we find that degradation of phosphorylated Htt involves lysosomal-associated membrane protein LAMP-2A and the heat shock protein Hsc70.1 Phosphorylation of Htt on S16 creates a consensus Hsc70 interaction motif. Further, mimicking Htt phosphorylation reduces its abundance in cell culture but increases its interaction with Hsc70 in vitro, suggesting that phosphorylation may activate Htt recognition by a chaperone complex required for selective protein degradation.
In addition, we have shown that the autophagy protein Atg7, essential for autophagosome formation, regulates clearance of phosphorylated Htt which implies that vesicles may mediate phosphorylated Htt autophagic degradation. S13/S16 phosphorylated and K9 acetylated wildtype (wt) Htt is present in mouse brain and appears to be relatively insoluble, consistent with oligomerization of the species of Htt being cleared by the cell. This species is also phosphorylated on threonine (T) 3; phosphorylation of T3 appears to increase Htt aggregation.1,5
IKK Regulates Htt Post-Translational Modification and Cellular Localization
Ubiquitination, SUMOylation and acetylation are post-translational modifications that have been previously demonstrated to regulate protein degradation, and these modifications can be modulated upstream by phosphorylation (reviewed in ref. 1). We find that IKK-activated phosphorylation of Htt S13/S16 regulates multiple post-translational modifications of Htt including ubiquitination, SUMOylation and acetylation and induces its nuclear localization, all events that may be involved in its clearance.1 The order in which these modifications occur and their specific functions in Htt degradation have yet to be determined. We previously reported that Htt amino acids 1–17 can function as a cytoplasmic retention sequence.6 I suggest that phosphorylation of S13S16 modifies the structure of this putative cytoplasmic retention sequence to increase nuclear localization and activate Htt poly-SUMOylation.
SUMOylation has been reported to target proteins to subnuclear structures called PML bodies, which contain the acetyltransferase CBP/p300.7 CBP/p300 interacts directly with and is phosphorylated by IKK in the nucleus, activating its acetyltransferase activity.8 We previously found that Htt also interacts directly with this acetyltransferase,9,10 which has been implicated in the acetylation of Htt on K444, a modification that increases mutant Htt clearance by macroautophagy.11 Clearance of K444-acetylated Htt has been suggested to be mediated by p62/SQSTM1, a protein that shuttles between the cytoplasm and nucleus and recruits polyubiquitinated proteins to PML bodies.11,12
PML bodies may also be the site of Htt caspase cleavage initiated by IKK,13 as caspase cleavage of proteins has been demonstrated to occur within this sub-nuclear structure.14 The PML body is a site of proteasomal degradation of poly-SUMOylated proteins, a process dependent on the SUMO-binding ubiquitin E3 ligase RNF4,15,16 and polyQ aggregates colocalize with PML bodies.17 I propose that Htt is phosphorylated and forms a complex with IKK in the cytoplasm, which then increases the extent of its nuclear localization. In this view, Htt is then poly-SUMOylated, targeting it to PML bodies, acetylated on lysines 9 and 444, cleaved by caspases, and either degraded by the proteasome in an SUMO/RNF4-dependent fashion or targeted to the cytoplasm where it is cleared by macroautophagy.
Mutant Htt Post-Translational Modification Modulates Its Toxicity
The post-translational modifications described above appear to influence degradation of the Htt protein. With aging, both proteasomal and lysosomal function are reduced.18–21 We find that inhibition of the proteasome or the lysosome results in an accumulation of phosphorylated and acetylated Htt and Htt fragments. It has been previously shown that a build-up of toxic amino-terminal mutant Htt fragments in the nucleus is a marker of HD pathogenesis, and that IKK may facilitate formation of these fragments.13,22
We find that mimicking phosphorylation of mutant Htt exon 1 protein (Httex1p) in an acute striatal slice culture model or full-length mutant Htt in BACHD transgenic mice reduces its toxicity,1,23 implying that mutant Htt clearance is activated in these phosphomimetic models. In contrast, mimicking phosphorylation of S13 and S16 in a Drosophila model of HD increases mutant Httex1p toxicity and its accumulation into aggregates (Marsh JL, personal communication). This difference could reflect the absence of lysosomal degradation of phosphorylated human mutant Httex1p since lysosomal clearance mechanisms are not closely conserved in Drosophila. For instance, there is no structural homolog of LAMP-2A and the amino-terminal domain of Drosophila Htt does not contain S13 or S16. Therefore, this Drosophila model could mimic patients already having impaired lysosomal clearance upon aging, at which point induction of the IKK kinase by innate immune activation no longer is protective due to nuclear accumulation of a modified form of Htt that is toxic. Consistent with the idea that IKK could have both beneficial and toxic effects depending upon the cell's ability to clear post-translationally modified Htt, inhibition of IKK activity can reduce mutant Htt toxicity and the protein RIP-2, an activator of IKK, has been implicated in enhancement of HD pathogenesis.2,24
In keeping with the idea that modifications may have different consequences depending on the cellular milieu, SUMOylation can be both protective and pathogenic in expanded polyQ-repeat diseases, which may reflect that poly-SUMOylation of Htt increases its clearance while undegraded SUMOylated Htt is toxic.6,25–27 Acetylation may also control pathogenicity of Htt as both inhibition and activation of specific histone deacetylases can reduce Htt-mediated toxicity.9,28–30 Therapies to treat HD may thus be challenging to develop if activation of Htt phosphorylation, SUMOylation and acetylation early on is protective, but later in the course of disease may result in the formation of an accumulating, toxic species. Therefore, significant care must be taken to match potential therapeutics for HD to the stage of disease progression reached by the patient.
Selective Autophagy, Conserved Across Phylogeny, Has Been Well Defined in Yeast
Selective lysosomal protein degradation mechanisms are essential to the viability of the cell to maximize efficiency of cellular function. These mechanisms have been particularly well defined in yeast. The yeast vacuole is the equivalent of the mammalian lysosome. Genetic screens in Saccharomyces cerevisiae have led to the identification of several mechanisms in which select proteins or organelles are directed to be degraded by the vacuole. For example, under glucose and nutrient-rich conditions, yeast gluconeogenic enzymes such as fructose-1,6-bisphosphatase (FBPase) and cytoplasmic malate dehydrogenase (MDH2) become unnecessary. Using biochemical and genetic approaches, the vacuole import and degradation (Vid) pathway has been defined, which selectively targets these enzymes for degradation by the proteasome, or by the vacuole via Vid vesicles.31 The yeast heat shock protein Ssa2 is required for import of FBPase into Vid vesicles, which cluster at actin patches and merge with endocytic vesicles as they are forming on the plasma membrane. FBPase is degraded following vacuolar fusion with the endosome.32–34
Another selective pathway of vacuolar targeting exists in yeast under glucose-rich conditions: the cytoplasm to vacuole targeting (Cvt) pathway. In this pathway, the target protein can be recognized by yeast heat shock proteins Ssa1 and Ssa2,35 oligomerizes and interacts with the Cvt receptor protein Atg19, which binds Atg11, a scaffolding protein that interacts with actin. Actin and the actin-binding complex Arp2/3 are required for the Cvt pathway and are suggested to act as a track to guide cargos to the site of vesicle formation through interaction with Atg11.36 The Atg19/Atg11-coated oligomers are surrounded by the Cvt vesicle which then fuses with the vacuolar membrane, dependent on Vam3, an integral vacuolar membrane t-SNARE protein. Atg19 binds to Atg8-phosphatidylethanolamine localized to the forming Cvt vesicle, an event that may trigger completion of the vesicle.37 While this targeting pathway has been considered biosynthetic, and was defined for resident vacuolar proteins aminopeptidase I (Ape1) and α-mannosidase (Ams1), it may also target proteins for vacuolar degradation.38
By both the Vid and Cvt pathways, select proteins are targeted to vesicles that ultimately fuse with the endosome/vacuole when yeast are grown in glucoserich media. Perixosomes, unnecessary for energy generation under nutrient-rich conditions in yeast can become selectively degraded by another autophagic mechanism requiring Atg11, pexophagy.39 Pexophagy and the Vid and Cvt pathways allow selective targeting of proteins and organelles to the vacuole under nutrientrich conditions, regulating vacuolar degradation to maximize cellular efficiency.
In contrast, under starvation conditions, alternate vacuolar targeting pathways are activated in yeast. In glucose-poor media, macroautophagy is induced to generate energy through vacuolar digestion of cytoplasmic proteins and organelles, freeing necessary amino acids that are then transported out of the vacuole to the cytoplasm for use by the cell. Starvation-induced macroautophagy is generally a nonspecific mechanism of vacuolar protein clearance where autophagosomes form and engulf organelles and cytoplasmic contents, but in some physiological conditions, unnecessary proteins or dysfunctional organelles are selectively degraded in the vacuole by macroautophagy.37,40 Also under starvation conditions, nuclear proteins can be targeted to the vacuole by piecemeal microautophagy of the nucleus (PMN), during which small nuclear envelope blebs pinch off and fuse with the vacuole, requiring Atg11 and Vam3.41
Autophagic Pathways in Mammalian Cells
Autophagic pathways used in mammalian cells appear to show significant similarities to those used in yeast. Many of the key proteins in yeast autophagic pathways have orthologs in mammals judged by both sequence and functional similarity. For example, Atg5, Atg7, Atg8 (LC3) and Atg12, required for vesicle formation in starvation-induced macroautophagy, PMN and the Cvt pathway, have clear orthologs in mammals.37 Macroautophagy, stimulated by starvation conditions and by inhibition of mTOR complex 1 (mTORC1) with rapamycin, has been extensively defined in mammalian cells, while PMN has not yet been directly demonstrated, although a process similar to PMN has been defined in Bloom's syndrome where nuclear microvesicles are released into the cytoplasm.42
Signals such as nutrient availability in mammalian cells appear to be involved in the induction of autophagy and activity of autophagy components.43 Under nutrient-rich conditions, activation of insulinsignaling pathways, Akt, mTORC1 and S6 kinase increase mutant Htt fragment clearance by autophagy in mammalian cells despite the fact that these pathways generally inhibit starvation-induced macroautophagy.44 Similarly, Akt, IKK and Cdk5, kinases active in nutrient-rich conditions, directly phosphorylate mutant Htt and reduce its toxicity.1,45–47 mTORC1, downstream of Akt, can activate the IKK complex while rapamycin, an inhibitor of mTORC1, can inhibit IKK activation48 and activate starvationinduced macroautophagy.37 We find that the phosphorylated/acetylated species of wt Htt has reduced solubility, suggesting this clearance intermediate may oligomerize like substrates for the Cvt pathway.1 Under starvation conditions, the transcription factor FOXO3a activates macroautophagy49 but under nutrient-rich conditions, both Akt and IKK directly phosphorylate FOXO3a and target it for degradation50 thereby inhibiting nonspecific macroautophagy.49
It is possible, therefore, that the mechanism of IKK-mediated selective autophagic clearance of Htt1 may be similar to the vegetative yeast Cvt or Vid pathways rather than to starvation-induced macroautophagy. Since in yeast, oligomerized proteins normally selectively targeted to Cvt vesicles in glucose-rich conditions may be delivered to the vacuole by macroautophagy under starvation conditions,51 nonspecific macroautophagosomal targeting of Htt to the lysosome induced by mTORC1 inhibition52 may be compensatory for a reduction in function of a mammalian Cvt-like macroautophagic pathway.
Since starvation conditions increase lysosome numbers and levels of LAMP-2A,53 periodic nutrient limitation may later bolster lysosomal activity under nutrient-rich conditions, as has been demonstrated for the yeast FBPase protein. Yeast cells that are glucose-starved for short periods largely degrade FBPase by the proteasome upon introduction of glucose to the media, while cells starved for longer periods primarily utilize the vacuole upon transfer to glucose.31 Periodic fasting, thought for centuries to be healthful, may generally boost lysosome numbers and function and later improve lysosomal clearance of proteins under nutrient-rich conditions.
Atg proteins required for non-selective macroautophagy are conserved from yeast to man, while those involved in selective autophagy like the Cvt pathway or pexophagy show poor conservation, suggesting that specialized selective autophagy genes have evolved.39 In particular, close structural homologs for the proteins essential for the yeast Cvt pathway, Vac8, Atg11, Atg20, Atg23 and Atg27 have not been identified in mammals.
Atg19, the substrate receptor protein for the Cvt pathway, is functionally similar to mammalian proteins p62/SQSTM1 and NBR1 because they all interact with oligomerized cargo, directly bind Atg8/LC3 and are degraded by the mechanism of selective autophagy that they regulate.54,55 These considerations suggest that a version of the Cvt pathway may exist in mammals.
Atg20 is a sorting nexin family member required for the Cvt pathway and endosomal sorting which interacts directly with Atg11.56,57 I find that yeast Atg20 shares some structural similarity with human Optineurin (Suppl. Fig. 1), a protein that can regulate inflammatory pathways and membrane trafficking,58,59 interact directly with Htt and is mutated in the neurodegenerative diseases ALS and glaucoma.60,61 Another sorting nexin that interacts with Atg20, Atg24/SNX4, shares some similarity with Huntingtin-associated protein-1 (HAP-1, Suppl. Fig. 2), proteins that both interact with the largest subunit of the yeast/human molecular motor dynactin complex, NIP100/p150Glued respectively.62,63
Three proteins in the Atg1 kinase complex, involved in the energetic toggle between starvation-induced macroautophagy and the Cvt pathway, yeast Atg1, Atg13 and Atg17, have been proposed to be Ulk1/Ulk2, mAtg13 and FIP200 in mammalian cells.39,64,65 While Ulk1/Ulk2 are strongly homologous to the C. elegans Atg1-like protein Unc-51, the putative mammalian Atg13 and Atg17 functional homologs share little similarity to the yeast proteins by sequence. In fact, FIP200, a protein that promotes mTORC1 activation,66 is a member of the NCBI Pfam: Family: ATG11 (PF10377) and is structurally similar to S. Pombe (Taz1-interacting factor 1) and C. elegans (WBGene00020334) Atg11s. Yeast Atg11 and Atg17 both act as scaffolds for PAS (phagophore assembly site or preautophagosomal structure) organization as part of the Atg1 kinase complex, Atg11 under nutrient-rich conditions and Atg17 under starvation conditions.67 I find that yeast Atg17 may show similarity to the recently reported mAtg13-binding protein, Atg101,67 (Suppl. Fig. 3), and suggest that FIP200/Atg101 may be the mammalian counterparts of Atg11/Atg17 respectively (Table 1). Yeast Vam3 is a vacuolar integral membrane t-SNARE protein involved in fusion of Cvt, PMN and starvation-induced macroautophagy vesicles with the vacuole.41,68 Vam3 plays a role in vacuolar assembly and trafficking, and interacts genetically with NIP100, part of the dynactin complex.69 Vam3 may have some functional similarity to mammalian LAMP-2, an integral lysosomal membrane protein involved in autophagy, vesicular trafficking and lysosome biogenesis.70
Table 1.
Yeast Cvt pathway proteins and their hypothetical human functional homologs
| Yeast protein | Function | Interactors | Human protein | Function | Interactors |
| Atg1 | Kinase required for vesicle and PAS formation | Atg1, Atg8, Atg11, Atg13, Atg14, Atg17, Atg18, Atg21, Atg29, Slx5 | ULK1/2 | Kinase that regulates autophagy156 | mAtg13 |
| Atg8 | Vesicle component, phagophore expansion, vesicle fusion | Atg1, Atg3, Atg4, Atg7, Atg19, Atg32, Vam3 | LC3 | Recruited to the phagophore membrane and remains associated with completed vesicle102 | p62/SQSTM1 |
| Atg11 | Directs receptor-bound cargo to the PAS for packaging into vesicles; required for recruiting other proteins to the PAS | Atg1, Atg9, Atg11, Atg12, Atg17, Atg19, Atg20, Atg29, Atg32 | Huntingtin aa 1743–3144 | Regulation of early endosome motility89 | HAP40/Rab5 complex |
| FIP200 | Regulation of cell growth, proliferation, cell migration, apoptosis, microtubule dynamics, stress response156,157 | mAtg13, ULK1/2, PIASy, p53, FAK, TSC1, Pyk2, ASK1 stathmin, TRAF2 | |||
| Atg13 | Regulatory subunit of Atg1 complex, required for vesicle formation, involved in Atg9/23/27 cycling | Vac8, Atg1, Atg17 | mAtg13 | Plays an essential role in autophagosome formation, phosphorylated by ULK and mTOR156,158 | ULK1/2, FIP200, Atg101 |
| Atg17 | Scaffold protein responsible for PAS site organization, stimulates Atg1 kinase activity | Atg1, Atg9, Atg11, Atg12, Atg13, Atg17, Atg24, Atg29, NIP100 | Atg101 | Important for stability and phosphorylation of ULK and mAtg13158 | mAtg13 |
| Atg19 | Cvt pathway receptor protein delivers cargo to PAS | Atg8, Atg11, Ubi4 | p62/SQSTM1 NBR1 | Shuttle proteins transport ubiquitinated proteins to lysosome54,55,93,102,159 | LC3, Huntingtin, ubiquitin |
| Atg20 | Sorting nexin for Cvt pathway and endosomal sorting | Atg11, Atg24 | Optineurin | Membrane trafficking, cellular morphogenesis59 | Huntingtin, Rab8 |
| Atg23 | Peripheral membrane protein required for Atg9/Atg27 shuttling/membrane trafficking | Atg9, Atg27 | Huntingtin aa 1–586 | Contains polyQ repeat, vesicle/membrane trafficking, localizes to microtubules59,77,85,160 | HAP-1, Optineurin, p53, Sp1, Akt, CBP, SH3GL3, RasGaP, PSD-95, HIP-1, HYP-J, PACSIN, actin |
| Atg24 | Sorting nexin involved in Cvt pathway and in retrieval of late-Golgi SNAREs from post-Golgi endosomes to the trans-Golgi network | Atg17, Atg20, Cog6, NIP100 | HAP-1 | Adaptor protein, mediates vesicle trafficking62,161 | Huntingtin (171–230) PCM-1, kinesin-like protein, neurofilament M, p150glued |
| Vac8 | Vacuole inheritance, the Cvt pathway, mitophagy, PMN, vacuole/vacuole fusion | Atg13, Vam3, Act1 | Huntingtin aa 807–1653 | DNA-damage response signal protein45 | Cdk5 |
| Vam3 | Integral vacuolar membrane protein required for vacuolar assembly and protein trafficking, Golgi to vacuole transport, vesicular fusion, PMN | Atg8, Vac8, Vam3, Vam7 | LAMP-2A | Integral lysosomal membrane protein involved in vesicular trafficking and lysosome biogenesis70,150,162 | UCH-L1/Hsc70/Hsp90 complex |
| NIP100 | Large subunit of dynactin complex | Atg24 | p150glued | Large subunit of dynactin complex62 | HAP-1 |
Select direct interactions listed. Yeast protein data obtained from the Saccharomyces Genome Database: www.yeastgenome.org.
Htt Has Similarities to Several Proteins Required for Selective Autophagy in Yeast
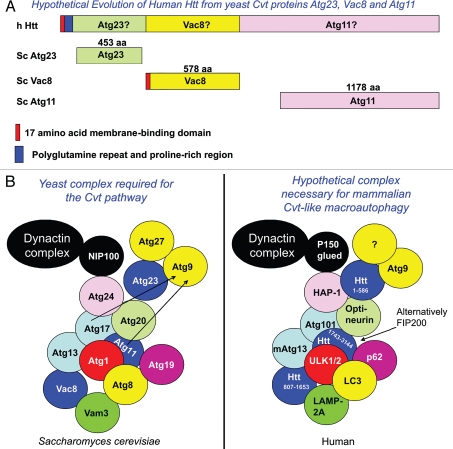
The Huntingtin protein may play a role in the mechanism of protein degradation involved in its own clearance. Conditional knockout of Htt in the central nervous system results in an accumulation of neuropil protein aggregates containing ubiquitin and p62/SQSTM1 (Zeitlin S, personal communication), demonstrating a loss of function in autophagy similar to that observed with neuronal knockdown of mammalian autophagy proteins Atg5, Atg7 and FIP200.71–73 Conversely, deletion of the polyQ stretch within wt mouse Htt stimulates autophagy via an mTOR-independent pathway in vivo,74 suggesting that wt Htt has a function in cellular autophagy. Htt may play a role in autophagic cargo recognition as this process has been recently demonstrated to be impaired in HD, where autophagosomes form normally and are eliminated by lysosomes, but fail to efficiently trap cytosolic cargo in their lumen.75 Based on these considerations I propose the hypothesis that Htt is a protein integral to the function of a Cvt-like mammalian macroautophagic pathway and may have evolved through fusion of proteins essential for the yeast Cvt pathway (Fig. 1 and Table 1).
Figure 1.
Htt may have evolved from Cvt proteins Atg23, Vac8 and Atg11. (A) The yeast Vac8 protein contains 11 armadillo repeats which can be described as ancestral HEAT repeats. Throughout evolution, Vac8 may have fused with yeast protein Atg23 on the amino-terminus and Atg11 on the C-terminus. These yeast proteins are essential for the cytoplasm to vacuole targeting (Cvt) selective autophagic pathway, which is activated by TORC1 in yeast grown under nutrient-rich conditions. The cartoon is not drawn to scale. (B) A protein complex essential for the yeast Cvt pathway may be similar to a hypothetical human complex required for selective macroautophagy under nutrient-rich conditions.
Yeast Atg23 is a peripheral-membrane protein that co-localizes with Atg9 and Atg27, integral membrane proteins essential for autophagic vesicle formation at the PAS.76 These three proteins may be required for bringing membrane to forming vesicles. Atg9 is needed for both nutrient-rich and starvation-induced vesicle formation, while Atg23 and Atg27 largely function in the Cvt pathway but may aid in the formation of starvation-induced macroautophagic vesicles. Atg23 interacts directly with Atg9 and Atg27 at the perivacuolar PAS and punctate non-PAS peripheral cytoplasmic locations and cycles between the periphery and the PAS. Atg23 and Atg11 together are required for movement of Atg9 towards the PAS, while Atg1 and Atg13 aid in retrograde movement of Atg23 away from the PAS.
I find that the amino-terminal 586 amino acid caspase-6 fragment of Htt has some structural similarity to yeast Atg23 (Suppl. Fig. 4). Like Atg23, the Htt protein also plays a role in vesicular trafficking.77,78 Htt localizes to microtubules and is associated with proteins of the molecular motor machinery including dynein and HAP-1, that interact with p150Glued and kinesin-1. The first 480 amino acids of wt Htt can activate vesicular movement in cells in which full-length Htt expression is knocked down; this ability becomes impaired with expansion of the polyQ repeat. This suggests that Htt's role in vesicular trafficking is contained within its amino-terminal 480 amino acids that are similar to yeast Atg23.
The yeast Vac8 phosphoprotein is essential for multiple vacuolar processes including vacuole inheritance, the Cvt pathway, pexophagy, PMN and vacuole/vacuole fusion and co-sediments with actin filaments.79–81 Vac8 contains 11 armadillo repeats that can be described as ancestral HEAT repeats, spanning its entire sequence.82 Vac8 is both myristoylated and palmitoylated, modifications required for its targeting to membranes via its first 18 amino acids which contain a Src kinase SH4 domain.81,83 Similar to the yeast Vac8 protein, the Htt protein contains 12 HEAT repeats between amino acids 745 and 1710,22 can be palmitoylated,84 associates with actin,85 and interacts with membranes via its first 17 amino acids.86,87 Both Vac8 and Htt regulate longevity.74,88 I find that the Vac8 protein shares some structural similarity with Htt amino acids 807–1,652 (Suppl. Fig. 5).
Within the C-terminal end of Htt, between amino acids 1,815 and 3,144, there exists a domain required for interaction with Hap40/Rab5 and regulation of early endosome motility.89 This domain contains weak similarity to the yeast Atg11 protein (Suppl. Fig. 6), essential for the Cvt pathway, PMN, pexophagy and selective mitochondrial degradation by autophagy: mitophagy.90 Starting at amino acid 3,037, Htt may also contain a highly conserved consensus WXXL LC3-binding domain preceded by an aspartic acid residue.91 Atg11 is an adapter protein necessary for cargo loading that oligomerizes and interacts directly with Cvt receptor protein Atg19.57 Like Atg11, Htt has been described as a protein scaffold92 and we have found that the phosphorylated/acetylated wt Htt species may have a tendency to aggregate.1 Htt aggregates interact with p62/SQSTM1,93 a protein with similarity to Atg19, which has also been found to be required for targeting of a high molecular weight species of K444-acetylated Htt for lysosomal degradation.11 p62/SQSTM1 can regulate activation of the IKK complex through multiple mechanisms including oxidative stress94,95 and may therefore induce Htt post-translational modification and caspase cleavage, activating Atg23 and Atg11 for functioning in a mammalian Cvt-like pathway in which the amino-terminal Atg23-like fragment ultimately oligomerizes and is cleared.1
Like the C-terminal domain of Htt, FIP200 may also be a mammalian Atg11 protein by structural comparison and is similar to Htt amino acids 1,743–3,144 (Suppl. Fig. 7), but may specifically affect the cerebellum, as neuronal knockdown of FIP200 leads to cerebellar degeneration.73 A loss of Htt function may particularly affect the striatum which degenerates in HD. The differential function of Atg11-like proteins in brain regions may cause the neuronal specificity of degeneration found in many neurodegenerative diseases. The recent observation of a lack of cargo recognition in HD cells75 may reflect a loss of Atg11-like function.
Some functional and structural similarities may exist between Htt and yeast Atg23, Vac8 and Atg11, all proteins required for the yeast Cvt pathway. Vac8 and Atg11 are also essential for PMN and pexophagy and Atg11 for mitophagy.41,90 These proteins may have fused throughout evolution to create Htt (Fig. 1A), which may be a protein whose function is integral for mammalian versions of the Cvt and PMN pathways, pexophagy and mitophagy essential for neuronal homeostasis in the striatum. Evolutionary fusion of yeast protein interactors (Slx5/Slx8) to form one functional mammalian protein (RNF4) has been previously described for proteins involved in a poly-SUMOylation/ubiquitination clearance pathway,96 which may be tied to this mechanism of selective protein degradation. Fusion has also been proposed for Saccharomyces cerevisiae Atg29 and Atg31 proteins to form Atg28 in Pichia pastoris.97
Similar to the function of heat shock proteins Ssa1 and Ssa2 in recognition of proteins targeted to the vacuole in the yeast Cvt pathway, human Hsc70 may regulate lysosomal targeting of proteins selectively bound for degradation under nutrient-rich conditions. The Hsc70 consensus binding sequence is a pentapeptide with a glutamine on either end, a large hydrophobic residue (isoleucine, leucine, valine or phenylalanine), a positively charged residue (lysine or arginine), a negatively charged residue (aspartate or glutamate) and a repeated basic or hydrophobic amino acid.98 This sequence shares some overlap with the consensus SUMOylation four amino acid motif containing a large hydrophobic residue followed by the lysine to which SUMO is conjugated, any amino acid and ending in a glutamate.99 We find that phosphorylation of Htt S16 may create a consensus Hsc70 binding sequence with phosphorylated serine mimicking aspartate/glutamate.1 Similarly, acetylation of lysine can mimic glutamine to potentially create an Hsc70-binding pentapeptide. SUMOylation and acetylation, activated by protein phosphorylation, may be linked to target protein interaction with Hsc70 in the induction of selective protein degradation by the proteasome and by the lysosome.
Mutant Htt Activates the IKK Kinase Complex Impacting Transcription
Short-lived nuclear proteins, in addition to long-lived cytoplasmic proteins, may be selectively targeted for lysosomal as well as proteasomal degradation, as the MEF2D transcription factor was demonstrated to be exported from the nucleus and cleared by the lysosome, dependent on LAMP-2A function.100,101 p62/SQSTM1 has been shown to activate both proteasomal and lysosomal protein clearance and these two mechanisms may be intimately related.102–104 We previously found that truncated mutant Htt (exon 1 protein, Httex1p) expression represses p53-activated transcription.10 If the Htt protein physically plays a role in transcription factor clearance by p62/SQSTM1-mediated selective autophagy and/or proteasomal degradation, then activation of a transcription factor's degradation may effectively result in a repression of transcription mediated by the factor. Expression of mutant expanded polyQ Htt results in activation of the IKK complex in cell culture and in vivo.2 I hypothesize that the repression of p53-mediated transcription we previously reported may reflect expanded polyQ Httex1p activating IKK and wt Htt to in turn induce clearance of p53, which itself is a target of IKK.105
As this protein clearance mechanism becomes compromised in HD, it may result in accumulation of active p53, effectively enhancing neurodegeneration as previously described in HD transgenic mice.106 Similarly, there is an early reduction in Sp1 and CREB-mediated transcription in cells expressing mutant Htt, while these transcription factors accumulate in HD transgenic mice in parallel with pathogenesis.107–114 In addition, the transcriptional repressor REST accumulates in the nucleus with mutant Htt expression and represses transcription.115 I propose that expression of mutant Htt activates IKK, which induces wt Htt to increase clearance of active, acetylated transcription factors including p53, Sp1, CREB and REST, but that these factors all accumulate when protein clearance mechanisms fail, dysregulating transcription, which may precipitate HD pathogenesis.
Activation of IKK by mutant Htt expression may also have direct effects on Htt post-translational modification and the efficacy of drugs and proteins to induce Htt clearance. Mutant expanded polyQ “specific” Htt SUMOylation27 or acetylation11 may be driven by endogenous IKK activation in the presence of mutant Htt; these modifications may occur even more efficiently on the wt protein with independent activation of the kinase. Similarly, drugs found only to increase mutant Htt clearance by “mTOR independent autophagy”116 may actually work to enhance wt Htt clearance in the presence of IKK activation. Recently, the Alfy protein was found to be a scaffold between p62/SQSTM1-bound proteins and the molecular machinery that builds autophagosomes.117 Alfy was shown to enhance aggregated mutant Httex1p clearance while having no effect on wt soluble Httex1p. IKK increases levels of insoluble phosphorylated wt Htt;1 wt Htt oligomerization, activated by IKK, may normally regulate its Alfy-mediated lysosomal degradation, which is not induced without activation of the kinase.
Induction of Selective Protein Clearance Pathways Early in Disease Progression May Slow Neurodegeneration
I propose that activating selective protein clearance pathways in presymptomatic or early phases of disease progression may be generally useful to treat neurodegenerative disease. In later stages, when LAMP-2A levels and lysosome and proteasome function are reduced, activation of these pathways may create acetylated, SUMOylated and caspase-cleaved protein clearance intermediates that accumulate and are toxic to the cell. Evidence for both protective and pathologic outcomes involving activation of the signaling cascades that may induce selective clearance pathways in models of neurodegenerative disease provide support for my thesis. Signal transduction pathways that may induce selective clearance mechanisms include insulin signaling, DNA damage, oxidative stress and inflammation. These stimuli activate Akt, Cdk5 and IKK,48,118–122 kinases that have been found to phosphorylate neurodegenerative disease-causing proteins and influence their toxicity and clearance.
The pro-survival kinase Akt, activated by insulin signaling,122 can modulate neurodegeneration in multiple disease models and can function to activate IKK.48 Akt phosphorylates S776 of Ataxin-1, the disease protein in the polyQ disorder Spinocerebellar Ataxia type 1 and enhances its toxicity in cell culture, transgenic Drosophila and mice.123,124 Similarly, an inactivation of insulin signaling/Akt in mouse models of AD rescues behavioral deficits.125–127 Conversely, Akt phosphorylation of the androgen receptor, mutated in Spinal and Bulbar Muscular Atrophy, also a polyQ disorder, increases its clearance and reduces its toxicity in vivo and in vitro.128 Akt phosphorylates Htt at S421 which has been shown to be neuroprotective and to reduce inclusion formation in cell culture.129 Activation of Akt is also associated with increased Htt clearance by macroautophagy.44
IKK activation has been implicated in neurodestructive outcomes in HD, AD and PD2,13,24,130,131 while immuno-activities and inflammatory processes may also be neuroprotective.132 We find that Htt phosphorylation by IKK can both reduce and increase its toxicity, which may depend on the ability of the cell to clear the modified species.1
Cdk5, which may activate Akt in neurons, is a protein kinase involved in the maturation and maintenance of the central nervous system with roles in the regulation of the cell cycle, migration, survival and neuronal integration.121,133 Cdk5 phosphorylates the tau protein, which intraneuronally accumulates in a temporal pattern that parallels pathogenesis of AD and Frontotemporal Dementias; upregulation of Cdk5 activity has been generally observed to enhance AD pathogenesis.134 However, we have found that increases in the levels of the Cdk5 coactivator p25, previously shown to correlate with improved learning and memory,135,136 are associated with reduced abundance of phosphorylated tau in brain and improved phenotype of a mouse model of AD137 implicating a role for Cdk5 in a phosphorylation-driven mechanism of tau clearance. Cdk5 phosphorylates Htt serines 434, 1181 and 1201 and reduces mutant Htt-mediated toxicity and aggregation in cell culture.45,46 Phosphorylation of both Htt and tau by Cdk5 may similarly regulate their clearance, and accumulation of the toxic phosphorylated protein may reflect a loss of age-related protein degradation mechanisms. Cdk5 may therefore be a third protein kinase, similar to Akt and IKK, which may regulate selective protein clearance that becomes impaired during aging.
Recently proteins have been found to be acetylated by acetyltransferases under nutrient-rich conditions and deacetylated by Class III histone deacetylases with starvation; the level of protein acetylation may reflect energy status and regulate unneeded protein degradation.138,139 p62/SQSTM1 levels are increased by acetyltransferase p300/CBP, while starvation-induced macroautophagy requires Sirt1, a Class III histone deacetylase.43,140 I hypothesize that IKK may induce Htt acetylation at K9 and K4441,11 within its Atg23-like domain, activating its cleavage, function in vesicle trafficking and degradation as well as inducing the activity of the carboxy-terminal Htt Atg11-like fragment in selective autophagy. p62/SQSTM1-mediated selective autophagy may be increased by inhibition of Class III histone deacetylases with nicotinamide, a treatment which we have found reduces levels of phosphorylated tau and slows neurodegeneration in a mouse model of AD.137 This treatment works early in disease progression but has no effect in older animals, which may reflect a loss in protein clearance mechanisms with age.
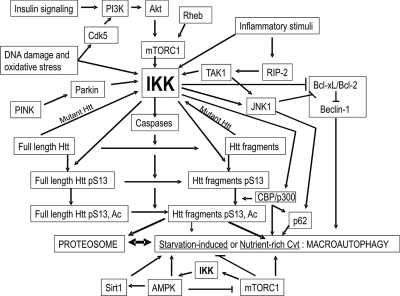
I propose that activation of a Cvt-like mechanism of mammalian macroautophagy may be very useful therapeutically to treat neurodegenerative diseases of aging (Fig. 2). This mechanism may previously have been defined as “mTOR Independent Autophagy”,141 “Chaperone-Assisted Selective Autophagy (CASA)”,142 or “Quality Control Autophagy.”143 Activation of this protein clearance mechanism may occur with inflammatory stimuli, insulin signaling, DNA damage or oxidative stress activating Cdk5, Akt, mTORC1 and ultimately IKK.48,121,144 IKK activation to induce this Cvt-like pathway may be enhanced by two proteins mutated in PD, PINK1 and parkin which have recently been demonstrated to activate mitophagy.145–147 IKK activates CBP/p300 and JNK1 increasing levels of p62/SQSTM1,4,8,43,148,149 and reducing the interaction of Bcl-xL/Bcl-2 with Beclin-14,13,148 thereby potentially inducing Cvt-like macroautophagy. The complex interacting with the substrate protein may include Htt, BAG-3, p62/SQSTM1, CHIP, Hsc70 and HspB8.1,142 Function of this complex may be impaired with expansion of the Htt polyQ repeat, as mutant Htt is not as efficiently phosphorylated by IKK,1 Cdk5,46 and Akt,47 all of which may activate the complex's function. Alfy may assist in bridging this complex to the molecular machinery that builds autophagosomes.117 F-actin and cortactin regulated by HDAC6 may facilitate fusion of the vesicle to the lysosome143 and LAMP-2A may be required for optimal association of lysosomes with dynein/dynactin to activate vesicular trafficking which is essential for vesicle/lysosome fusion.70,150 All of these proteins may be therapeutic targets for the activation of Cvt-like selective macroautophagy. The polyphenol resveratrol, previously shown to induce autophagy, may therapeutically activate this Cvt-like mechanism via induction of JNK1-mediated p62/SQSTM1 expression and caspase activation,148 consistent with a possible role for resveratrol in the upstream activation of IKK. Proteins involved in vesicle expansion, completion and nucleation are equivalent for both nutrient-rich and starvation-induced autophagic pathways, and therefore may not be useful to specifically increase Cvt-like selective macroautophagy. If this mechanism becomes overwhelmed as in later stages of disease when LAMP-2A levels are reduced,151 starvation/AMPK/rapamycin/Sirt1-induced macroautophagy may compensate by clearing the oligomerized substrate protein complexes which may have accumulated to form inclusions. At this point, activation of starvation-induced macroautophagy and Sirt1 may also be protective against the further progression of the disease.
Figure 2.
Model: The kinase IKK may activate Htt degradation by a mammalian version of the yeast Cvt pathway. The kinase IKK, activated by inflammatory stimuli, insulin signaling, DNA damage and oxidative stress pathways,48,118–120,153,154 Rheb155 and by PINK/parkin,146,147 phosphorylates Htt S13 and activates Htt acetylation by CBP/p300 and caspase cleavage,13 targeting the amino-terminal fragments of Htt for degradation by the proteasome and lysosome.1 IKK reduces interaction of Bcl-xL/Bcl-2 with Beclin-1,4,13,148 thereby priming activation of both starvation-induced and the proposed mammalian Cvt macroautophagic pathways. IKK activates JNK1 and CBP/p300 to increase levels of p62/SQSTM1,4,8,43,148,149 required for the Cvt pathway and AMPK4 which may then activate starvation-induced macroautophagy as a compensatory feedback mechanism. Mutant Htt expression activates IKK.2 The proposed mammalian Cvt pathway, activated by CBP/p300 or inhibition of Sirt1 by nicotinamide, may be involved in vesicle-mediated degradation of phosphorylated/acetylated Htt N-terminal fragments by the lysosome. With aging and mutant Htt expression, the proteasome and this Cvt-like mechanism of autophagy may become impaired, and this loss of function may be compensated for by activation of starvation/rapamycin/AMPK/Sirt1-induced macroautophagy.
Conclusions
Htt may share some functional and structural similarities with three yeast proteins, Atg23, Vac8 and Atg11, required for selective vacuolar targeting in the yeast Cvt pathway. Htt may therefore play a functional and regulatory role in a selective protein clearance mechanism ultimately involved in its own processing. Phosphorylation of Htt by the IKK complex activates its poly-SUMOylation and acetylation; I propose that these modifications may regulate a normal role for Htt in a selective mechanism of protein clearance requiring both the proteasome and the lysosome, involving protein oligomerization. This mechanism may be activated by Akt/Cdk5/mTORC1/IKK kinases, and have some similarities to the yeast Cvt pathway, which functions under glucose/nutrient-rich conditions, when starvation-induced macroautophagy is inhibited, and in which oligomers are selectively targeted to the vacuole. If the process becomes inefficient, mutant Htt may accumulate in inclusion bodies, which can be cleared by starvation-induced macroautophagy52 potentially reflecting a compensatory response. This mammalian Cvt-like autophagic clearance pathway may decline with age and become compromised by expansion of the polyQ repeat within Htt, precipitating neurodegeneration. This mechanism may be particularly important in the brain as it depends on glucose as an energy substrate.152 Therapeutically activating this selective protein clearance mechanism may be a valuable approach to the early treatment of many inclusion-associated diseases like HD and the polyQ-repeat diseases, PD, AD and ALS and to extend lifespan. It will be of great interest to carry out the challenging experimental work that will be required to investigate and potentially validate this approach to therapeutic intervention in these disorders.
Acknowledgements
I would like to thank Drs. David Housman, Leslie M. Thompson, Scott O. Zeitlin, Ralph Bradshaw, J. Lawrence Marsh, Kara Neely, Maya Koike and Daniel A. Keys for helpful discussion and critical reading of the manuscript. I am very grateful to the Hereditary Disease Foundation and the Fox Family Foundation for their support of our work, as well as the CHDI Foundation and NIH (NS52789).
Footnotes
Previously published online: www.landesbioscience.com/journals/cc/article/12718
Supplementary Material
References
- 1.Thompson LM, Aiken CT, Kaltenbach LS, Agrawal N, Illes K, Khoshnan A, et al. IKK phosphorylates Huntingtin and targets it for degradation by the proteasome and lysosome. J Cell Biol. 2009;187:1083–1099. doi: 10.1083/jcb.200909067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Khoshnan A, Ko J, Watkin EE, Paige LA, Reinhart PH, Patterson PH. Activation of the IkappaB kinase complex and nuclear factor-kappaB contributes to mutant huntingtin neurotoxicity. J Neurosci. 2004;24:7999–8008. doi: 10.1523/JNEUROSCI.2675-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bjorkqvist M, Wild EJ, Thiele J, Silvestroni A, Andre R, Lahiri N, et al. A novel pathogenic pathway of immune activation detectable before clinical onset in Huntington's disease. J Exp Med. 2008;205:1869–1877. doi: 10.1084/jem.20080178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Criollo A, Senovilla L, Authier H, Maiuri MC, Morselli E, Vitale I, et al. The IKK complex contributes to the induction of autophagy. EMBO J. 2010;29:619–631. doi: 10.1038/emboj.2009.364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aiken CT, Steffan JS, Guerrero CM, Khashwji H, Lukacsovich T, Simmons D, et al. Phosphorylation of threonine 3: implications for Huntingtin aggregation and neurotoxicity. J Biol Chem. 2009;284:29427–29436. doi: 10.1074/jbc.M109.013193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Steffan JS, Agrawal N, Pallos J, Rockabrand E, Trotman LC, Slepko N, et al. SUMO modification of Huntingtin and Huntington's disease pathology. Science. 2004;304:100–104. doi: 10.1126/science.1092194. [DOI] [PubMed] [Google Scholar]
- 7.Van Damme E,, Laukens K,, Dang TH, Van Ostade X. A manually curated network of the PML nuclear body interactome reveals an important role for PMLNBs in SUMOylation dynamics. Int J Biol Sci. 2010;6:51–67. doi: 10.7150/ijbs.6.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang WC,, Ju TK, Hung MC, Chen CC. Phosphorylation of CBP by IKKalpha promotes cell growth by switching the binding preference of CBP from p53 to NFkappaB. Mol Cell. 2007;26:75–87. doi: 10.1016/j.molcel.2007.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Steffan JS, Bodai L, Pallos J, Poelman M, McCampbell A, Apostol BL, et al. Histone deacetylase inhibitors arrest polyglutamine-dependent neurodegeneration in Drosophila. Nature. 2001;413:739–743. doi: 10.1038/35099568. [DOI] [PubMed] [Google Scholar]
- 10.Steffan JS, Kazantsev A, Spasic-Boskovic O, Greenwald M, Zhu YZ, Gohler H, et al. The Huntington's disease protein interacts with p53 and CREB-binding protein and represses transcription. Proc Natl Acad Sci USA. 2000;97:6763–6768. doi: 10.1073/pnas.100110097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jeong H, Then F, Melia TJJ, Mazzulli JR, Cui L, Savas JN, et al. Acetylation Targets Mutant Huntingtin to Autophagosomes for Degradation. Cell. 2009;137:60–72. doi: 10.1016/j.cell.2009.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pankiv S, Lamark T, Bruun JA, Overvatn A, Bjorkoy G, Johansen T. Nucleocytoplasmic shuttling of p62/SQSTM1 and its role in recruitment of nuclear polyubiquitinated proteins to promyelocytic leukemia bodies. J Biol Chem. 2010;285:5941–5953. doi: 10.1074/jbc.M109.039925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Khoshnan A, Ko J, Tescu S, Brundin P, Patterson PH. IKKalpha and IKKbeta regulation of DNA damage-induced cleavage of huntingtin. PLoS One. 2009:45768. doi: 10.1371/journal.pone.0005768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tan JA, Sun Y, Song J, Chen Y, Krontiris TG, Durrin LK. SUMO conjugation to the matrix attachment region-binding protein, special AT-rich sequence-binding protein-1 (SATB1), targets SATB1 to promyelocytic nuclear bodies where it undergoes caspase cleavage. J Biol Chem. 2008;283:18124–18134. doi: 10.1074/jbc.M800512200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lallemand-Breitenbach V, Jeanne M, Benhenda S, Nasr R, Lei M, Peres L, et al. Arsenic degrades PML or PML-RARalpha through a SUMO-triggered RNF4/ubiquitin-mediated pathway. Nat Cell Biol. 2008;10:547–555. doi: 10.1038/ncb1717. [DOI] [PubMed] [Google Scholar]
- 16.Tatham MH, Geoffroy MC, Shen L, Plechanovova A, Hattersley N, Jaffray EG, et al. RNF4 is a poly-SUMO-specific E3 ubiquitin ligase required for arsenic-induced PML degradation. Nat Cell Biol. 2008;10:538–546. doi: 10.1038/ncb1716. [DOI] [PubMed] [Google Scholar]
- 17.Janer A, Martin E, Muriel MP, Latouche M, Fujigasaki H, Ruberg M, et al. PML clastosomes prevent nuclear accumulation of mutant ataxin-7 and other polyglutamine proteins. J Cell Biol. 2006;174:65–76. doi: 10.1083/jcb.200511045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chondrogianni N, Gonos ES. Proteasome activation as a novel antiaging strategy. IUBMB Life. 2008;60:651–655. doi: 10.1002/iub.99. [DOI] [PubMed] [Google Scholar]
- 19.Cuervo AM, Bergamini E, Brunk UT, Droge W, Ffrench M, Terman A. Autophagy and aging: the importance of maintaining “clean” cells. Autophagy. 2005;1:131–140. doi: 10.4161/auto.1.3.2017. [DOI] [PubMed] [Google Scholar]
- 20.Martinez-Vicente M, Cuervo AM. Autophagy and neurodegeneration: when the cleaning crew goes on strike. Lancet Neurol. 2007;6:352–361. doi: 10.1016/S1474-4422(07)70076-5. [DOI] [PubMed] [Google Scholar]
- 21.Tonoki A, Kuranaga E, Tomioka T, Hamazaki J, Murata S, Tanaka K, et al. Genetic evidence linking age-dependent attenuation of the 26S proteasome with the aging process. Mol Cell Biol. 2009;29:1095–1106. doi: 10.1128/MCB.01227-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Warby SC, Doty CN, Graham RK, Carroll JB, Yang YZ, Singaraja RR, et al. Activated caspase-6 and caspase-6-cleaved fragments of huntingtin specifically colocalize in the nucleus. Hum Mol Genet. 2008;17:2390–2404. doi: 10.1093/hmg/ddn139. [DOI] [PubMed] [Google Scholar]
- 23.Gu X, Greiner ER, Mishra R, Kodali R, Osmand A, Finkbeiner S, et al. Serines 13 and 16 are critical determinants of full-length human mutant Huntingtin induced disease pathogenesis in HD mice. Neuron. 2009;64:828–840. doi: 10.1016/j.neuron.2009.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang X, Wang H, Figueroa BE, Zhang WH, Huo C, Guan Y, et al. Dysregulation of receptor interacting protein-2 and caspase recruitment domain only protein mediates aberrant caspase-1 activation in Huntington's disease. J Neurosci. 2005;25:11645–11654. doi: 10.1523/JNEUROSCI.4181-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chan HY, Warrick JM, Andriola I, Merry D, Bonini NM. Genetic modulation of polyglutamine toxicity by protein conjugation pathways in Drosophila. Hum Mol Genet. 2002;11:2895–2904. doi: 10.1093/hmg/11.23.2895. [DOI] [PubMed] [Google Scholar]
- 26.Janer A, Werner A, Takahashi-Fujigasaki J, Daret A, Fujigasaki H, Takada K, et al. SUMOylation attenuates the aggregation propensity and cellular toxicity of the polyglutamine expanded ataxin-7. Hum Mol Genet. 2010;19:181–195. doi: 10.1093/hmg/ddp478. [DOI] [PubMed] [Google Scholar]
- 27.Subramaniam S, Sixt KM, Barrow R, Snyder SH. Rhes, a striatal specific protein, mediates mutant-huntingtin cytotoxicity. Science. 2009;324:1327–1330. doi: 10.1126/science.1172871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hockly E, Richon VM, Woodman B, Smith DL, Zhou X, Rosa E, et al. Suberoylanilide hydroxamic acid, a histone deacetylase inhibitor, ameliorates motor deficits in a mouse model of Huntington's disease. Proc Natl Acad Sci USA. 2003;100:2041–2046. doi: 10.1073/pnas.0437870100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pallos J, Bodai L, Lukacsovich T, Purcell JM, Steffan JS, Thompson LM, et al. Inhibition of specific HDACs and sirtuins suppresses pathogenesis in a Drosophila model of Huntington's disease. Hum Mol Genet. 2008;17:3767–3775. doi: 10.1093/hmg/ddn273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Parker JA, Arango M, Abderrahmane S, Lambert E, Tourette C, Catoire H, et al. Resveratrol rescues mutant polyglutamine cytotoxicity in nematode and mammalian neurons. Nat Genet. 2005;37:349–350. doi: 10.1038/ng1534. [DOI] [PubMed] [Google Scholar]
- 31.Hung GC, Brown CR, Wolfe AB, Liu J, Chiang HL. Degradation of the gluconeogenic enzymes fructose-1,6-bisphosphatase and malate dehydrogenase is mediated by distinct proteolytic pathways and signaling events. J Biol Chem. 2004;279:49138–49150. doi: 10.1074/jbc.M404544200. [DOI] [PubMed] [Google Scholar]
- 32.Brown CR, McCann JA, Chiang HL. The heat shock protein Ssa2p is required for import of fructose-1, 6-bisphosphatase into Vid vesicles. J Cell Biol. 2000;150:65–76. doi: 10.1083/jcb.150.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brown CR, Wolfe AB, Cui D, Chiang HL. The vacuolar import and degradation pathway merges with the endocytic pathway to deliver fructose-1,6-bisphosphatase to the vacuole for degradation. J Biol Chem. 2008;283:26116–26127. doi: 10.1074/jbc.M709922200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brown CR, Dunton D, Chiang HL. The vacuole import and degradation pathway utilizes early steps of endocytosis and actin polymerization to deliver cargo proteins to the vacuole for degradation. J Biol Chem. 2010;285:1516–1528. doi: 10.1074/jbc.M109.028241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Silles E, Mazon MJ, Gevaert K, Goethals M, Vandekerckhove J, Leber R, et al. Targeting of aminopeptidase I to the yeast vacuole is mediated by Ssa1p, a cytosolic member of the 70 kDa stress protein family. J Biol Chem. 2000;275:34054–34059. doi: 10.1074/jbc.M003514200. [DOI] [PubMed] [Google Scholar]
- 36.Reggiori F, Monastyrska I, Shintani T, Klionsky DJ. The actin cytoskeleton is required for selective types of autophagy, but not nonspecific autophagy, in the yeast Saccharomyces cerevisiae. Mol Biol Cell. 2005;16:5843–5856. doi: 10.1091/mbc.E05-07-0629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Klionsky DJ. The molecular machinery of autophagy: unanswered questions. J Cell Sci. 2005;118:7–18. doi: 10.1242/jcs.01620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kageyama T, Suzuki K, Ohsumi Y. Lap3 is a selective target of autophagy in yeast, Saccharomyces cerevisiae. Biochem Biophys Res Commun. 2009;378:551–557. doi: 10.1016/j.bbrc.2008.11.084. [DOI] [PubMed] [Google Scholar]
- 39.Meijer WH, van der Klei IJ, Veenhuis M, Kiel JA. ATG genes involved in non-selective autophagy are conserved from yeast to man, but the selective Cvt and pexophagy pathways also require organism-specific genes. Autophagy. 2007;3:106–116. doi: 10.4161/auto.3595. [DOI] [PubMed] [Google Scholar]
- 40.Mijaljica D, Prescott M, Klionsky DJ, Devenish RJ. Autophagy and vacuole homeostasis: a case for self-degradation? Autophagy. 2007;3:417–421. doi: 10.4161/auto.4441. [DOI] [PubMed] [Google Scholar]
- 41.Krick R, Muehe Y, Prick T, Bremer S, Schlotterhose P, Eskelinen EL, et al. Piecemeal microautophagy of the nucleus requires the core macroautophagy genes. Mol Biol Cell. 2008;19:4492–4505. doi: 10.1091/mbc.E08-04-0363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mijaljica D, Prescott M, Devenish RJ. Nibbling within the nucleus: turnover of nuclear contents. Cell Mol Life Sci. 2007;64:581–588. doi: 10.1007/s00018-007-6395-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee IH, Finkel T. Regulation of autophagy by the p300 acetyltransferase. J Biol Chem. 2009;284:6322–6328. doi: 10.1074/jbc.M807135200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yamamoto A, Cremona ML, Rothman JE. Autophagy-mediated clearance of huntingtin aggregates triggered by the insulin-signaling pathway. J Cell Biol. 2006;172:719–731. doi: 10.1083/jcb.200510065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Anne SL, Saudou F, Humbert S. Phosphorylation of huntingtin by cyclin-dependent kinase 5 is induced by DNA damage and regulates wild-type and mutant huntingtin toxicity in neurons. J Neurosci. 2007;27:7318–7328. doi: 10.1523/JNEUROSCI.1831-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Luo S, Vacher C, Davies JE, Rubinsztein DC. Cdk5 phosphorylation of huntingtin reduces its cleavage by caspases: implications for mutant huntingtin toxicity. J Cell Biol. 2005;169:647–656. doi: 10.1083/jcb.200412071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Warby SC, Chan EY, Metzler M, Gan L, Singaraja RR, Crocker SF, et al. Huntingtin phosphorylation on serine 421 is significantly reduced in the striatum and by polyglutamine expansion in vivo. Hum Mol Genet. 2005;14:1569–1577. doi: 10.1093/hmg/ddi165. [DOI] [PubMed] [Google Scholar]
- 48.Dan HC, Cooper MJ, Cogswell PC, Duncan JA, Ting JP, Baldwin AS. Akt-dependent regulation of NF-{kappa}B is controlled by mTOR and Raptor in association with IKK. Genes Dev. 2008;22:1490–1500. doi: 10.1101/gad.1662308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhao J, Brault JJ, Schild A, Cao P, Sandri M, Schiaffino S, et al. FoxO3 coordinately activates protein degradation by the autophagic/lysosomal and proteasomal pathways in atrophying muscle cells. Cell Metab. 2007;6:472–483. doi: 10.1016/j.cmet.2007.11.004. [DOI] [PubMed] [Google Scholar]
- 50.Hu MC, Lee DF, Xia W, Golfman LS, Ou-Yang F, Yang JY, et al. IkappaB kinase promotes tumorigenesis through inhibition of forkhead FOXO3a. Cell. 2004;117:225–237. doi: 10.1016/s0092-8674(04)00302-2. [DOI] [PubMed] [Google Scholar]
- 51.Wang CW, Klionsky DJ. The molecular mechanism of autophagy. Mol Med. 2003;9:65–76. [PMC free article] [PubMed] [Google Scholar]
- 52.Ravikumar B, Vacher C, Berger Z, Davies JE, Luo S, Oroz LG, et al. Inhibition of mTOR induces autophagy and reduces toxicity of polyglutamine expansions in fly and mouse models of Huntington disease. Nat Genet. 2004;36:585–595. doi: 10.1038/ng1362. [DOI] [PubMed] [Google Scholar]
- 53.Finn PF, Dice JF. Proteolytic and lipolytic responses to starvation. Nutrition. 2006;22:830–844. doi: 10.1016/j.nut.2006.04.008. [DOI] [PubMed] [Google Scholar]
- 54.Chang CY, Huang WP. Atg19 mediates a dual interaction cargo sorting mechanism in selective autophagy. Mol Biol Cell. 2007;18:919–929. doi: 10.1091/mbc.E06-08-0683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lynch-Day MA, Klionsky DJ. The Cvt pathway as a model for selective autophagy. FEBS Lett. 2010;584:1359–1366. doi: 10.1016/j.febslet.2010.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nice DC, Sato TK, Stromhaug PE, Emr SD, Klionsky DJ. Cooperative binding of the cytoplasm to vacuole targeting pathway proteins, Cvt13 and Cvt20, to phosphatidylinositol 3-phosphate at the pre-autophagosomal structure is required for selective autophagy. J Biol Chem. 2002;277:30198–31207. doi: 10.1074/jbc.M204736200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yorimitsu T, Klionsky DJ. Atg11 links cargo to the vesicle-forming machinery in the cytoplasm to vacuole targeting pathway. Mol Biol Cell. 2005;16:1593–1605. doi: 10.1091/mbc.E04-11-1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhu G, Wu CJ, Zhao Y, Ashwell JD. Optineurin negatively regulates TNFalpha-induced NFkappaB activation by competing with NEMO for ubiquitinated RIP. Curr Biol. 2007;17:1438–1443. doi: 10.1016/j.cub.2007.07.041. [DOI] [PubMed] [Google Scholar]
- 59.Hattula K, Peranen J. FIP-2, a coiled-coil protein, links Huntingtin to Rab8 and modulates cellular morphogenesis. Curr Biol. 2000;10:1603–1606. doi: 10.1016/s0960-9822(00)00864-2. [DOI] [PubMed] [Google Scholar]
- 60.Maruyama H, Morino H, Ito H, Izumi Y, Kato H, Watanabe Y, et al. Mutations of optineurin in amyotrophic lateral sclerosis. Nature. 2010;465:223–226. doi: 10.1038/nature08971. [DOI] [PubMed] [Google Scholar]
- 61.Fuse N. Genetic bases for glaucoma. Tohoku J Exp Med. 2010;221:1–10. doi: 10.1620/tjem.221.1. [DOI] [PubMed] [Google Scholar]
- 62.Engelender S, Sharp AH, Colomer V, Tokito MK, Lanahan A, Worley P, et al. Huntingtin-associated protein 1 (HAP1) interacts with the p150Glued subunit of dynactin. Hum Mol Genet. 1997;6:2205–2212. doi: 10.1093/hmg/6.13.2205. [DOI] [PubMed] [Google Scholar]
- 63.Vollert CS, Uetz P. The phox homology (PX) domain protein interaction network in yeast. Mol Cell Proteomics. 2004;3:1053–1064. doi: 10.1074/mcp.M400081-MCP200. [DOI] [PubMed] [Google Scholar]
- 64.Chan EY, Longatti A, McKnight NC, Tooze SA. Kinase-inactivated ULK proteins inhibit autophagy via their conserved C-terminal domains using an Atg13-independent mechanism. Mol Cell Biol. 2009;29:157–171. doi: 10.1128/MCB.01082-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hara T, Takamura A, Kishi C, Iemura S, Natsume T, Guan JL, et al. FIP200, a ULK-interacting protein, is required for autophagosome formation in mammalian cells. J Cell Biol. 2008;181:497–510. doi: 10.1083/jcb.200712064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gan B, Guan JL. FIP200, a key signaling node to coordinately regulate various cellular processes. Cell Signal. 2008;20:787–794. doi: 10.1016/j.cellsig.2007.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Suzuki K, Ohsumi Y. Current knowledge of the pre-autophagosomal structure (PAS) FEBS Lett. 2010;584:1280–1286. doi: 10.1016/j.febslet.2010.02.001. [DOI] [PubMed] [Google Scholar]
- 68.Darsow T, Rieder SE, Emr SD. A multispecificity syntaxin homologue, Vam3p, essential for autophagic and biosynthetic protein transport to the vacuole. J Cell Biol. 1997;138:517–529. doi: 10.1083/jcb.138.3.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Costanzo M, Baryshnikova A, Bellay J, Kim Y, Spear ED, Sevier CS, et al. The genetic landscape of a cell. Science. 327:425–431. doi: 10.1126/science.1180823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Saftig P, Beertsen W, Eskelinen EL. LAMP-2: a control step for phagosome and autophagosome maturation. Autophagy. 2008;4:510–512. doi: 10.4161/auto.5724. [DOI] [PubMed] [Google Scholar]
- 71.Hara T, Nakamura K, Matsui M, Yamamoto A, Nakahara Y, Suzuki-Migishima R, et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441:885–889. doi: 10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- 72.Komatsu M, Waguri S, Chiba T, Murata S, Iwata J, Tanida I, et al. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 2006;441:880–884. doi: 10.1038/nature04723. [DOI] [PubMed] [Google Scholar]
- 73.Liang CC, Wang C, Peng X, Gan B, Guan JL. Neural-specific deletion of FIP200 leads to cerebellar degeneration caused by increased neuronal death and axon degeneration. J Biol Chem. 2010;285:3499–3509. doi: 10.1074/jbc.M109.072389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zheng S, Clabough EB, Sarkar S, Futter M, Rubinsztein DC, Zeitlin SO. Deletion of the huntingtin polyglutamine stretch enhances neuronal autophagy and longevity in mice. PLoS Genet. 2010;6:1000838. doi: 10.1371/journal.pgen.1000838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Martinez-Vicente M, Talloczy Z, Wong E, Tang G, Koga H, Kaushik S, et al. Cargo recognition failure is responsible for inefficient autophagy in Huntington's disease. Nat Neurosci. 2010;13:567–576. doi: 10.1038/nn.2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Legakis JE, Yen WL, Klionsky DJ. A cycling protein complex required for selective autophagy. Autophagy. 2007;3:422–432. doi: 10.4161/auto.4129. [DOI] [PubMed] [Google Scholar]
- 77.Colin E, Zala D, Liot G, Rangone H, Borrell-Pages M, Li XJ, et al. Huntingtin phosphorylation acts as a molecular switch for anterograde/retrograde transport in neurons. EMBO J. 2008;27:2124–2134. doi: 10.1038/emboj.2008.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zala D, Colin E, Rangone H, Liot G, Humbert S, Saudou F. Phosphorylation of mutant huntingtin at S421 restores anterograde and retrograde transport in neurons. Hum Mol Genet. 2008;17:3837–3846. doi: 10.1093/hmg/ddn281. [DOI] [PubMed] [Google Scholar]
- 79.Wang YX, Catlett NL, Weisman LS. Vac8p, a vacuolar protein with armadillo repeats, functions in both vacuole inheritance and protein targeting from the cytoplasm to vacuole. J Cell Biol. 1998;140:1063–1074. doi: 10.1083/jcb.140.5.1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Fry MR, Thomson JM, Tomasini AJ, Dunn WA., Jr Early and late molecular events of glucose-induced pexophagy in Pichia pastoris require Vac8. Autophagy. 2006;2:280–288. doi: 10.4161/auto.3164. [DOI] [PubMed] [Google Scholar]
- 81.Tang F, Peng Y, Nau JJ, Kauffman EJ, Weisman LS. Vac8p, an armadillo repeat protein, coordinates vacuole inheritance with multiple vacuolar processes. Traffic. 2006;7:1368–1377. doi: 10.1111/j.1600-0854.2006.00458.x. [DOI] [PubMed] [Google Scholar]
- 82.Andrade MA, Petosa C, O'Donoghue SI, Muller CW, Bork P. Comparison of ARM and HEAT protein repeats. J Mol Biol. 2001;309:1–18. doi: 10.1006/jmbi.2001.4624. [DOI] [PubMed] [Google Scholar]
- 83.Subramanian K, Dietrich LE, Hou H, LaGrassa TJ, Meiringer CT, Ungermann C. Palmitoylation determines the function of Vac8 at the yeast vacuole. J Cell Sci. 2006;119:2477–2485. doi: 10.1242/jcs.02972. [DOI] [PubMed] [Google Scholar]
- 84.Yanai A, Huang K, Kang R, Singaraja RR, Arstikaitis P, Gan L, et al. Palmitoylation of huntingtin by HIP14 is essential for its trafficking and function. Nat Neurosci. 2006;9:824–831. doi: 10.1038/nn1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Angeli S, Shao J, Diamond MI. F-actin binding regions on the androgen receptor and huntingtin increase aggregation and alter aggregate characteristics. PLoS One. 2010;5:9053. doi: 10.1371/journal.pone.0009053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Rockabrand E, Slepko N, Pantalone A, Nukala VN, Kazantsev A, Marsh JL, et al. The first 17 amino acids of Huntingtin modulate its sub-cellular localization, aggregation and effects on calcium homeostasis. Hum Mol Genet. 2007;16:61–77. doi: 10.1093/hmg/ddl440. [DOI] [PubMed] [Google Scholar]
- 87.Atwal RS, Xia J, Pinchev D, Taylor J, Epand RM, Truant R. Huntingtin Has a Membrane Association Signal that Can Modulate Huntingtin Aggregation, Nuclear Entry and Toxicity. Hum Mol Genet. 2007;16:2600–2615. doi: 10.1093/hmg/ddm217. [DOI] [PubMed] [Google Scholar]
- 88.Tang F, Watkins JW, Bermudez M, Gray R, Gaban A, Portie K, et al. A life-span extending form of autophagy employs the vacuole-vacuole fusion machinery. Autophagy. 2008;4:874–886. doi: 10.4161/auto.6556. [DOI] [PubMed] [Google Scholar]
- 89.Pal A, Severin F, Lommer B, Shevchenko A, Zerial M. Huntingtin-HAP40 complex is a novel Rab5 effector that regulates early endosome motility and is upregulated in Huntington's disease. J Cell Biol. 2006;172:605–618. doi: 10.1083/jcb.200509091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kraft C, Reggiori F, Peter M. Selective types of autophagy in yeast. Biochim Biophys Acta. 2009;1793:1404–1412. doi: 10.1016/j.bbamcr.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 91.Noda NN, Kumeta H, Nakatogawa H, Satoo K, Adachi W, Ishii J, et al. Structural basis of target recognition by Atg8/LC3 during selective autophagy. Genes Cells. 2008;13:1211–1218. doi: 10.1111/j.1365-2443.2008.01238.x. [DOI] [PubMed] [Google Scholar]
- 92.Atwal RS, Truant R. A stress sensitive ER membrane-association domain in Huntingtin protein defines a potential role for Huntingtin in the regulation of autophagy. Autophagy. 2008;4:91–93. doi: 10.4161/auto.5201. [DOI] [PubMed] [Google Scholar]
- 93.Bjorkoy G, Lamark T, Brech A, Outzen H, Perander M, Overvatn A, et al. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J Cell Biol. 2005;171:603–614. doi: 10.1083/jcb.200507002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ishii T, Yanagawa T, Kawane T, Yuki K, Seita J, Yoshida H, et al. Murine peritoneal macrophages induce a novel 60 kDa protein with structural similarity to a tyrosine kinase p56lck-associated protein in response to oxidative stress. Biochem Biophys Res Commun. 1996;226:456–460. doi: 10.1006/bbrc.1996.1377. [DOI] [PubMed] [Google Scholar]
- 95.Duran A, Serrano M, Leitges M, Flores JM, Picard S, Brown JP, et al. The atypical PKC-interacting protein p62 is an important mediator of RANK-activated osteoclastogenesis. Dev Cell. 2004;6:303–309. doi: 10.1016/s1534-5807(03)00403-9. [DOI] [PubMed] [Google Scholar]
- 96.Perry JJ, Tainer JA, Boddy MN. A SIM-ultaneous role for SUMO and ubiquitin. Trends Biochem Sci. 2008;33:201–208. doi: 10.1016/j.tibs.2008.02.001. [DOI] [PubMed] [Google Scholar]
- 97.Nazarko TY, Farre JC, Subramani S. Peroxisome size provides insights into the function of autophagy-related proteins. Mol Biol Cell. 2009;20:3828–3839. doi: 10.1091/mbc.E09-03-0221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Dice JF. Peptide sequences that target cytosolic proteins for lysosomal proteolysis. Trends Biochem Sci. 1990;15:305–309. doi: 10.1016/0968-0004(90)90019-8. [DOI] [PubMed] [Google Scholar]
- 99.Sampson DA, Wang M, Matunis MJ. The small ubiquitin-like modifier-1 (SUMO-1) consensus sequence mediates Ubc9 binding and is essential for SUMO-1 modification. J Biol Chem. 2001;276:21664–21669. doi: 10.1074/jbc.M100006200. [DOI] [PubMed] [Google Scholar]
- 100.Yang Q, She H, Gearing M, Colla E, Lee M, Shacka JJ, et al. Regulation of neuronal survival factor MEF2D by chaperone-mediated autophagy. Science. 2009;323:124–127. doi: 10.1126/science.1166088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Fuertes G, Martin De Llano JJ, Villarroya A, Rivett AJ, Knecht E. Changes in the proteolytic activities of proteasomes and lysosomes in human fibroblasts produced by serum withdrawal, amino-acid deprivation and confluent conditions. Biochem J. 2003;375:75–86. doi: 10.1042/BJ20030282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Komatsu M, Ichimura Y. Physiological significance of selective degradation of p62 by autophagy. FEBS Lett. 2010;584:1374–1378. doi: 10.1016/j.febslet.2010.02.017. [DOI] [PubMed] [Google Scholar]
- 103.Seibenhener ML, Babu JR, Geetha T, Wong HC, Krishna NR, Wooten MW. Sequestosome 1/p62 is a polyubiquitin chain binding protein involved in ubiquitin proteasome degradation. Mol Cell Biol. 2004;24:8055–8068. doi: 10.1128/MCB.24.18.8055-8068.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Lamark T, Johansen T. Autophagy: links with the proteasome. Curr Opin Cell Biol. 2010;22:192–198. doi: 10.1016/j.ceb.2009.11.002. [DOI] [PubMed] [Google Scholar]
- 105.Xia Y, Padre RC, De Mendoza TH, Bottero V, Tergaonkar VB, Verma IM. Phosphorylation of p53 by IkappaB kinase 2 promotes its degradation by beta-TrCP. Proc Natl Acad Sci USA. 2009;106:2629–2634. doi: 10.1073/pnas.0812256106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Bae BI, Xu H, Igarashi S, Fujimuro M, Agrawal N, Taya Y, et al. p53 mediates cellular dysfunction and behavioral abnormalities in Huntington's disease. Neuron. 2005;47:29–41. doi: 10.1016/j.neuron.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 107.Chen-Plotkin AS, Sadri-Vakili G, Yohrling GJ, Braveman MW, Benn CL, Glajch KE, et al. Decreased association of the transcription factor Sp1 with genes downregulated in Huntington's disease. Neurobiol Dis. 2006;22:233–241. doi: 10.1016/j.nbd.2005.11.001. [DOI] [PubMed] [Google Scholar]
- 108.Dunah AW, Jeong H, Griffin A, Kim YM, Standaert DG, Hersch SM, et al. Sp1 and TAFII130 transcriptional activity disrupted in early Huntington's disease. Science. 2002;296:2238–2243. doi: 10.1126/science.1072613. [DOI] [PubMed] [Google Scholar]
- 109.Li SH, Cheng AL, Zhou H, Lam S, Rao M, Li H, et al. Interaction of Huntington disease protein with transcriptional activator Sp1. Mol Cell Biol. 2002;22:1277–1287. doi: 10.1128/mcb.22.5.1277-1287.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Obrietan K, Hoyt KR. CRE-mediated transcription is increased in Huntington's disease transgenic mice. J Neurosci. 2004;24:791–796. doi: 10.1523/JNEUROSCI.3493-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Qiu Z, Norflus F, Singh B, Swindell MK, Buzescu R, Bejarano M, et al. Sp1 is upregulated in cellular and transgenic models of Huntington disease, and its reduction is neuroprotective. J Biol Chem. 2006;281:16672–16680. doi: 10.1074/jbc.M511648200. [DOI] [PubMed] [Google Scholar]
- 112.Shimohata T, Nakajima T, Yamada M, Uchida C, Onodera O, Naruse S, et al. Expanded polyglutamine stretches interact with TAFII130, interfering with CREB-dependent transcription. Nat Genet. 2000;26:29–36. doi: 10.1038/79139. [DOI] [PubMed] [Google Scholar]
- 113.Sugars KL, Brown R, Cook LJ, Swartz J, Rubinsztein DC. Decreased cAMP response element-mediated transcription: an early event in exon 1 and full-length cell models of Huntington's disease that contributes to polyglutamine pathogenesis. J Biol Chem. 2004;279:4988–4999. doi: 10.1074/jbc.M310226200. [DOI] [PubMed] [Google Scholar]
- 114.Wyttenbach A, Swartz J, Kita H, Thykjaer T, Carmichael J, Bradley J, et al. Polyglutamine expansions cause decreased CRE-mediated transcription and early gene expression changes prior to cell death in an inducible cell model of Huntington's disease. Hum Mol Genet. 2001;10:1829–1845. doi: 10.1093/hmg/10.17.1829. [DOI] [PubMed] [Google Scholar]
- 115.Zuccato C, Tartari M, Crotti A, Goffredo D, Valenza M, Conti L, et al. Huntingtin interacts with REST/NRSF to modulate the transcription of NRSE-controlled neuronal genes. Nat Genet. 2003;35:76–83. doi: 10.1038/ng1219. [DOI] [PubMed] [Google Scholar]
- 116.Williams A, Sarkar S, Cuddon P, Ttofi EK, Saiki S, Siddiqi FH, et al. Novel targets for Huntington's disease in an mTOR-independent autophagy pathway. Nat Chem Biol. 2008;4:295–305. doi: 10.1038/nchembio.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Filimonenko M, Isakson P, Finley KD, Anderson M, Jeong H, Melia TJ, et al. The selective macroautophagic degradation of aggregated proteins requires the PI3P-binding protein Alfy. Mol Cell. 2010;38:265–279. doi: 10.1016/j.molcel.2010.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Hayden MS, Ghosh S. Signaling to NFkappaB. Genes Dev. 2004;18:2195–2224. doi: 10.1101/gad.1228704. [DOI] [PubMed] [Google Scholar]
- 119.Gloire G, Legrand-Poels S, Piette J. NFkappaB activation by reactive oxygen species: fifteen years later. Biochem Pharmacol. 2006;72:1493–1505. doi: 10.1016/j.bcp.2006.04.011. [DOI] [PubMed] [Google Scholar]
- 120.Wu ZH, Shi Y, Tibbetts RS, Miyamoto S. Molecular linkage between the kinase ATM and NFkappaB signaling in response to genotoxic stimuli. Science. 2006;311:1141–1146. doi: 10.1126/science.1121513. [DOI] [PubMed] [Google Scholar]
- 121.Li BS, Ma W, Jaffe H, Zheng Y, Takahashi S, Zhang L, et al. Cyclin-dependent kinase-5 is involved in neuregulin-dependent activation of phosphatidylinositol 3-kinase and Akt activity mediating neuronal survival. J Biol Chem. 2003;278:35702–35709. doi: 10.1074/jbc.M302004200. [DOI] [PubMed] [Google Scholar]
- 122.Jiang ZY, Zhou QL, Coleman KA, Chouinard M, Boese Q, Czech MP. Insulin signaling through Akt/protein kinase B analyzed by small interfering RNA-mediated gene silencing. Proc Natl Acad Sci USA. 2003;100:7569–7574. doi: 10.1073/pnas.1332633100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Emamian ES, Kaytor MD, Duvick LA, Zu T, Tousey SK, Zoghbi HY, et al. Serine 776 of ataxin-1 is critical for polyglutamine-induced disease in SCA1 transgenic mice. Neuron. 2003;38:375–387. doi: 10.1016/s0896-6273(03)00258-7. [DOI] [PubMed] [Google Scholar]
- 124.Chen HK, Fernandez-Funez P, Acevedo SF, Lam YC, Kaytor MD, Fernandez MH, et al. Interaction of Akt-phosphorylated ataxin-1 with 14-3-3 mediates neurodegeneration in spinocerebellar ataxia type 1. Cell. 2003;113:457–468. doi: 10.1016/s0092-8674(03)00349-0. [DOI] [PubMed] [Google Scholar]
- 125.Cohen E, Paulsson JF, Blinder P, Burstyn-Cohen T, Du D, Estepa G, et al. Reduced IGF-1 signaling delays age-associated proteotoxicity in mice. Cell. 2009;139:1157–1169. doi: 10.1016/j.cell.2009.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Freude S, Hettich MM, Schumann C, Stohr O, Koch L, Kohler C, et al. Neuronal IGF-1 resistance reduces Abeta accumulation and protects against premature death in a model of Alzheimer's disease. FASEB J. 2009;23:3315–3324. doi: 10.1096/fj.09-132043. [DOI] [PubMed] [Google Scholar]
- 127.Killick R, Scales G, Leroy K, Causevic M, Hooper C, Irvine EE, et al. Deletion of Irs2 reduces amyloid deposition and rescues behavioural deficits in APP transgenic mice. Biochem Biophys Res Commun. 2009;386:257–262. doi: 10.1016/j.bbrc.2009.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Palazzolo I, Stack C, Kong L, Musaro A, Adachi H, Katsuno M, et al. Overexpression of IGF-1 in muscle attenuates disease in a mouse model of spinal and bulbar muscular atrophy. Neuron. 2009;63:316–328. doi: 10.1016/j.neuron.2009.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Humbert S, Bryson EA, Cordelieres FP, Connors NC, Datta SR, Finkbeiner S, et al. The IGF-1/Akt pathway is neuroprotective in Huntington's disease and involves Huntingtin phosphorylation by Akt. Dev Cell. 2002;2:831–837. doi: 10.1016/s1534-5807(02)00188-0. [DOI] [PubMed] [Google Scholar]
- 130.Ghosh A, Roy A, Liu X, Kordower JH, Mufson EJ, Hartley DM, et al. Selective inhibition of NFkappaB activation prevents dopaminergic neuronal loss in a mouse model of Parkinson's disease. Proc Natl Acad Sci USA. 2007;104:18754–18759. doi: 10.1073/pnas.0704908104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Mattson MP, Meffert MK. Roles for NFkappaB in nerve cell survival, plasticity and disease. Cell Death Differ. 2006;13:852–860. doi: 10.1038/sj.cdd.4401837. [DOI] [PubMed] [Google Scholar]
- 132.Hanisch UK, Johnson TV, Kipnis J. Toll-like receptors: roles in neuroprotection? Trends Neurosci. 2008;31:176–182. doi: 10.1016/j.tins.2008.01.005. [DOI] [PubMed] [Google Scholar]
- 133.Jessberger S, Gage FH, Eisch AJ, Lagace DC. Making a neuron: Cdk5 in embryonic and adult neurogenesis. Trends Neurosci. 2009;32:575–582. doi: 10.1016/j.tins.2009.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Dhariwala FA, Rajadhyaksha MS. An unusual member of the Cdk family: Cdk5. Cell Mol Neurobiol. 2008;28:351–369. doi: 10.1007/s10571-007-9242-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Angelo M, Plattner F, Irvine EE, Giese KP. Improved reversal learning and altered fear conditioning in transgenic mice with regionally restricted p25 expression. Eur J Neurosci. 2003;18:423–431. doi: 10.1046/j.1460-9568.2003.02746.x. [DOI] [PubMed] [Google Scholar]
- 136.Fischer A, Sananbenesi F, Pang PT, Lu B, Tsai LH. Opposing roles of transient and prolonged expression of p25 in synaptic plasticity and hippocampus-dependent memory. Neuron. 2005;48:825–838. doi: 10.1016/j.neuron.2005.10.033. [DOI] [PubMed] [Google Scholar]
- 137.Green KN, Steffan JS, Martinez-Coria H, Sun X, Schreiber SS, Thompson LM, et al. Nicotinamide restores cognition in Alzheimer's disease transgenic mice via a mechanism involving sirtuin inhibition and selective reduction of Thr231-phosphotau. J Neurosci. 2008;28:11500–11510. doi: 10.1523/JNEUROSCI.3203-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Wang Q, Zhang Y, Yang C, Xiong H, Lin Y, Yao J, et al. Acetylation of metabolic enzymes coordinates carbon source utilization and metabolic flux. Science. 2010;327:1004–1007. doi: 10.1126/science.1179687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Zhao S, Xu W, Jiang W, Yu W, Lin Y, Zhang T, et al. Regulation of cellular metabolism by protein lysine acetylation. Science. 2010;327:1000–1004. doi: 10.1126/science.1179689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Lee IH, Cao L, Mostoslavsky R, Lombard DB, Liu J, Bruns NE, et al. A role for the NAD-dependent deacetylase Sirt1 in the regulation of autophagy. Proc Natl Acad Sci USA. 2008;105:3374–3379. doi: 10.1073/pnas.0712145105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Sarkar S, Floto RA, Berger Z, Imarisio S, Cordenier A, Pasco M, et al. Lithium induces autophagy by inhibiting inositol monophosphatase. J Cell Biol. 2005;170:1101–1111. doi: 10.1083/jcb.200504035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Arndt V, Dick N, Tawo R, Dreiseidler M, Wenzel D, Hesse M, et al. Chaperone-assisted selective autophagy is essential for muscle maintenance. Curr Biol. 2010;20:143–148. doi: 10.1016/j.cub.2009.11.022. [DOI] [PubMed] [Google Scholar]
- 143.Lee JY, Koga H, Kawaguchi Y, Tang W, Wong E, Gao YS, et al. HDAC6 controls autophagosome maturation essential for ubiquitin-selective quality-control autophagy. EMBO J. 2010;29:969–980. doi: 10.1038/emboj.2009.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Mabb AM, Miyamoto S. SUMO and NFkappaB ties. Cell Mol Life Sci. 2007;64:1979–1996. doi: 10.1007/s00018-007-7005-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Henn IH, Bouman L, Schlehe JS, Schlierf A, Schramm JE, Wegener E, et al. Parkin mediates neuroprotection through activation of IkappaB kinase/nuclear factor-kappaB signaling. J Neurosci. 2007;27:1868–1878. doi: 10.1523/JNEUROSCI.5537-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Sha D, Chin LS, Li L. Phosphorylation of parkin by Parkinson disease-linked kinase PINK1 activates parkin E3 ligase function and NFkappaB signaling. Hum Mol Genet. 2010;19:352–363. doi: 10.1093/hmg/ddp501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Vives-Bauza C, Zhou C, Huang Y, Cui M, de Vries RL, Kim J, et al. PINK1-dependent recruitment of Parkin to mitochondria in mitophagy. Proc Natl Acad Sci USA. 2010;107:378–383. doi: 10.1073/pnas.0911187107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Puissant A, Robert G, Fenouille N, Luciano F, Cassuto JP, Raynaud S, et al. Resveratrol promotes autophagic cell death in chronic myelogenous leukemia cells via JNK-mediated p62/SQSTM1 expression and AMPK activation. Cancer Res. 2010;70:1042–1052. doi: 10.1158/0008-5472.CAN-09-3537. [DOI] [PubMed] [Google Scholar]
- 149.Verma UN, Yamamoto Y, Prajapati S, Gaynor RB. Nuclear role of IkappaB Kinase-gamma/NFkappa B essential modulator (IKKgamma/NEMO) in NFkappaB-dependent gene expression. J Biol Chem. 2004;279:3509–3515. doi: 10.1074/jbc.M309300200. [DOI] [PubMed] [Google Scholar]
- 150.Koga H, Kaushik S, Cuervo AM. Altered lipid content inhibits autophagic vesicular fusion. FASEB J. 2010;24:3052–3065. doi: 10.1096/fj.09-144519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Cuervo AM, Dice JF. Age-related decline in chaperone-mediated autophagy. J Biol Chem. 2000;275:31505–31513. doi: 10.1074/jbc.M002102200. [DOI] [PubMed] [Google Scholar]
- 152.Fehm HL, Kern W, Peters A. The selfish brain: competition for energy resources. Prog Brain Res. 2006;153:129–140. doi: 10.1016/S0079-6123(06)53007-9. [DOI] [PubMed] [Google Scholar]
- 153.Laplante M, Sabatini DM. mTOR signaling at a glance. J Cell Sci. 2009;122:3589–3594. doi: 10.1242/jcs.051011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Hasegawa M, Fujimoto Y, Lucas PC, Nakano H, Fukase K, Nunez G, et al. A critical role of RICK/RIP2 polyubiquitination in Nod-induced NFkappaB activation. EMBO J. 2008;27:373–383. doi: 10.1038/sj.emboj.7601962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Huang J, Manning BD. The TSC1-TSC2 complex: a molecular switchboard controlling cell growth. Biochem J. 2008;412:179–190. doi: 10.1042/BJ20080281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Jung CH, Ro SH, Cao J, Otto NM, Kim DH. mTOR regulation of autophagy. FEBS Lett. 2010;584:1287–1295. doi: 10.1016/j.febslet.2010.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Hara T, Mizushima N. Role of ULK-FIP200 complex in mammalian autophagy: FIP200, a counterpart of yeast Atg17? Autophagy. 2009;5:85–87. doi: 10.4161/auto.5.1.7180. [DOI] [PubMed] [Google Scholar]
- 158.Hosokawa N, Sasaki T, Iemura S, Natsume T, Hara T, Mizushima N. Atg101, a novel mammalian autophagy protein interacting with Atg13. Autophagy. 2009;5:973–979. doi: 10.4161/auto.5.7.9296. [DOI] [PubMed] [Google Scholar]
- 159.Xie Z, Klionsky DJ. Autophagosome formation: core machinery and adaptations. Nat Cell Biol. 2007;9:1102–1109. doi: 10.1038/ncb1007-1102. [DOI] [PubMed] [Google Scholar]
- 160.Harjes P, Wanker EE. The hunt for huntingtin function: interaction partners tell many different stories. Trends Biochem Sci. 2003;28:425–433. doi: 10.1016/S0968-0004(03)00168-3. [DOI] [PubMed] [Google Scholar]
- 161.Bertaux F, Sharp AH, Ross CA, Lehrach H, Bates GP, Wanker E. HAP1-huntingtin interactions do not contribute to the molecular pathology in Huntington's disease transgenic mice. FEBS Lett. 1998;426:229–232. doi: 10.1016/s0014-5793(98)00352-4. [DOI] [PubMed] [Google Scholar]
- 162.Kabuta T, Furuta A, Aoki S, Furuta K, Wada K. Aberrant interaction between Parkinson disease-associated mutant UCH-L1 and the lysosomal receptor for chaperone-mediated autophagy. J Biol Chem. 2008;283:23731–23738. doi: 10.1074/jbc.M801918200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.