Abstract
Lrs4 and Csm1, components of the monopolin complex, localize to the rDNA where they regulate rDNA maintenance and segregation. During meiosis, the complex also associates with kinetochores to bring about sister kinetochore co-orientation, an essential aspect of meiosis I chromosome segregation. We show here that the Lrs4-Csm1 complex associates with kinetochores during mitosis. This kinetochore localization is observed during anaphase and depends on the on the Mitotic Exit Network, a signaling cascade essential for the completion of mitosis. Furthermore, we find that Lrs4 and Csm1 are important for chromosome segregation fidelity. Our results reveal a previously unanticipated function for Lrs4-Csm1 in mitotic chromosome segregation.
Key words: mitosis, monopolin, Lrs4, Csm1, kinetochore, Mitotic Exit Network, chromosome segregation
Introduction
During mitosis, each pair of sister chromatids must be accurately attached to the mitotic spindle during metaphase and segregated during anaphase, such that the resultant daughter cells inherit a copy of each chromosome. To ensure proper chromosomes segregation, the kinetochores of each pair of sister chromatids attach to microtubules emanating from opposite poles of the cell, in a process known as bi-orientation. In S. cerevisiae, chromosomes remain attached to microtubules for the duration of the cell cycle, save for a brief disassociation during DNA synthesis.1 Despite the static nature of much of the complex, regulatory proteins dynamically localize to kinetochores. For example, during metaphase components of the spindle assembly checkpoint associate with unattached kinetochores hindering chromosome segregation until all chromatid pairs have made the proper attachments (reviewed in ref. 2). In higher eukaryotes, kinetochore proteins have been identified that selectively associate with kinetochores during anaphase to facilitate the loading of the centromere-specific histone H3 variant Cenp-A.3–5 We show here that the Lrs4-Csm1 protein complex, previously shown to play a key role in rDNA maintenance and kinetochore function during meiosis I, localizes to mitotic kinetochores during anaphase, where they appear to ensure chromosome segregation fidelity.
During the mitotic cell cycle, Lrs4 and Csm1 form a complex that is required for linking sister chromatids at the rDNA locus. Due to its highly repetitive nature and its size, the rDNA locus requires special mechanisms and special timing to ensure the maintenance of chromosome fidelity during mitosis.6 Lrs4 and Csm1 reside in the nucleolus where they form a complex called the RENT (Regulator of nucleolar silencing and telophase exit) complex, which binds to the replication fork barrier site (RFB) within the non-transcribed region NTS1 in the ribosomal DNA repeats.7,8 This complex promotes rDNA segregation, mediates transcriptional silencing and prevents unequal sister chromatid exchange within the repetitive rDNA array.7,8 Lrs4 and Csm1 also bind to and recruit condensins to the rDNA, where they participate in rDNA segregation,9 as well as Heh1 and Nur1, nuclear envelope proteins, which are required to maintain rDNA repeat stability.10 By linking RENT complex components anchored at NTS1 regions of the rDNA to both condensin complexes and nuclear envelope proteins, Lrs4 and Csm1 “clamp” sister chromatids in frame, restricting their movement and thereby preventing recombination events that would result in expansions or contractions of the rDNA array.
During the meiotic divisions, Lrs4 and Csm1 play a key role in chromosome segregation. Meiosis is a specialized cell division where one round of DNA synthesis is followed by two rounds of chromosome segregation. To coordinate the first round of chromosome segregation, each pair of sister chromatids remains attached at their kinetochores and are segregated towards the same pole while the homologous pair of sister chromatids is segregated towards opposite poles. This process requires that the kinetochores of a pair of sister chromatids orient themselves towards the same pole (co-orientation). In the second round of chromosomal division, the kinetochores of each pair of sister chromatids bi-orient and are segregated away from each other. During meiosis I, Lrs4 and Csm1 are components of the quadripartite monopolin complex that links sister chromatids. In addition to Lrs4 and Csm1, the complex contains the meiosis-specific Mam1 protein and the protein kinase Hrr25.11–13 In essence, the monopolin complex fuses or clamps adjacent kinetochores in a cohesin-independent mechanism,14 thereby ensuring that only one microtubule attaches to each unique pair of sister chromatids15 (reviewed in ref. 16). In the absence of the monopolin complex during meiosis I, the kinetochores of a sister chromatid pair become active and are capable of making attachments towards opposite poles, resulting in the mis-segregation of chromosomes and inviable spores.11–13,17
Here we investigate a potential role for Lrs4 and Csm1 at kinetochores during mitosis. We find that the two proteins localize to kinetochores during anaphase. This association is precipitated by a signaling pathway known as the Mitotic Exit Network (MEN). The MEN triggers exit from mitosis by promoting the release of the protein phosphatase Cdc14 from the nucleolus. It also promotes the dissociation of Lrs4 and Csm1 from the rDNA and their dispersal throughout the nucleus.7 Consistent with a role of Lrs4 and Csm1 at kinetochores, we find that cells lacking either protein exhibit increased plasmid loss rates. Our results indicate that the Lrs4-Csm1 complex is important for ensuring accurate chromosome segregation.
Results
The monopolin complex colocalizes with Ndc80 during anaphase.
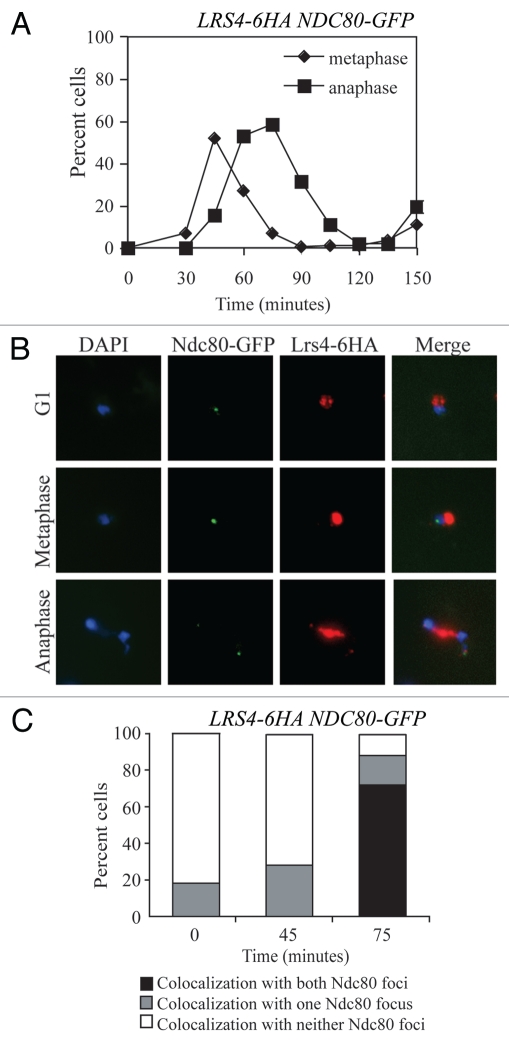
Our previous studies showed that during mitosis, Lrs4 and Csm1 become released from the nucleolus during anaphase.7 To determine whether, as in meiosis, the mitotic monopolin complex localizes to kinetochores after its release from the nucleolus, we analyzed Lrs4 localization on chromosome spreads prepared from cells progressing through the cell cycle in a synchronous manner. Because the location of Lrs4 and Csm1 are interdependent,12,18 we focused our localization studies on Lrs4. To identify kinetochores, cells carrying an epitope-tagged version of Lrs4 (Lrs4-6HA) also carried a GFP-tagged version of the kinetochore component Ndc80. We found that during anaphase, a portion of Lrs4 remained associated with the rDNA as judged by Lrs4-6HA localization between the DNA lobes of anaphase cells (note that the rDNA is one of the last genomic regions to segregate; Fig. 1B). Interestingly, Lrs4 was also found to co-localize with Ndc80-GFP (Fig. 1B and C). This localization was only detected during anaphase, when a fraction of Lrs4 is released from the nucleolus (Figs. 1B). Co-localization with kinetochore subunits during anaphase does not necessarily demonstrate that a protein associates with kinetochores, as spindle pole bodies (the yeast equivalents of centrosomes) and centromeres are closely associated during anaphase.19 However, we will demonstrate below that the co-localization of Lrs4 with Ndc80 indeed reflects kinetochore association of Lrs4 during anaphase.
Figure 1.
Lrs4 and Csm1 localize to kinetochores or spindle pole bodies during mitotic anaphase. (A–C) Wild-type cells carrying an Lrs4-6HA and an Ndc80-GFP fusion (A15126) were arrested in G1 using α-factor pheromone (5 µg/ml) and released into medium lacking the pheromone at 25°C. At the indicated times, samples were taken to determine the percentage of cells with metaphase (diamonds) and anaphase (squares) spindles (A) and the percentage of cells showing co-localization of Lrs4-6HA with both, one or neither Ndc80-GFP marked spindle pole body (C) 200 cells were counted per time-point. The micrographs in (B) show Ndc80-GFP (green) and Lrs4-6HA (red) localization on chromosome spreads at 0, 45 and 75 minutes after release from the G1 arrest. DNA is shown in blue. At least 50 cells were counted per timepoint.
Lrs4 and Csm1 are the only members of the RENT complex that localize to kinetochores.
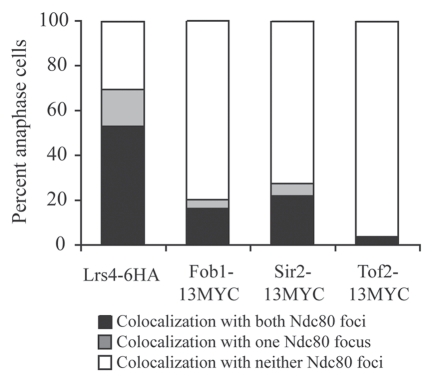
Lrs4 and Csm1 are not the only nucleolar proteins whose localization changes during anaphase. The RENT complex that contains Cdc14 and its inhibitor Cfi1/Net1; Sir2, a protein required for gene silencing; Tof2, a protein required for rDNA silencing and condensation; and the replication fork-binding protein Fob1, disassembles during anaphase, releasing Cdc14, Lrs4-Csm1 and Sir2 from the nucleolus.20–22 The condensin complex, which interacts with the Lrs4-Csm1 portion of the RENT complex, associates with chromosomes in early mitosis and becomes enriched in the nucleolus during early anaphase.6,9,23 During late anaphase, similar to the RENT complex, the condensin complex is released from the rDNA by the Mitotic Exit Network24 and becomes enriched at kinetochores.18,25,26
To test whether other RENT complex components associate with kinetochores during anaphase, we examined the localization of the RENT complex components Tof2, Sir2 and Fob1 in chromosome spreads of anaphase cells. Although Lrs4 co-localized with the kinetochore component Ndc80 in over half of anaphase cells, fewer than 20% of anaphase cells showed co-localization of Fob1, Sir2 and Tof2 with Ndc80 (Fig. 2). Despite evidence from fission yeast of Cdc14-homolog Clp1p localization to kinetochores during mitosis,27 we have not observed Cdc14-3HA localization at kinetochores using immunofluorescence (data not shown). Our results indicate that Lrs4 and Csm1 are the only members of the RENT complex that associate with kinetochores in budding yeast during anaphase at a detectable level.
Figure 2.
Kinetochore association during anaphase is not a general feature of RENT complex components. Cells carrying Ndc80-GFP and either Lrs4-6HA (A15127), Fob1-13MYC (A20431), Sir2-13MYC (A20432) or Tof2-13MYC (A20433) fusions were released from a pheromone-induced G1 arrest at 25°C. The percent of anaphase cells showing co-localization of the tagged proteins with Ndc80-GFP was determined. At least 50 cells were counted per strain.
The Mitotic Exit Network is required for Lrs4 association with kinetochores.
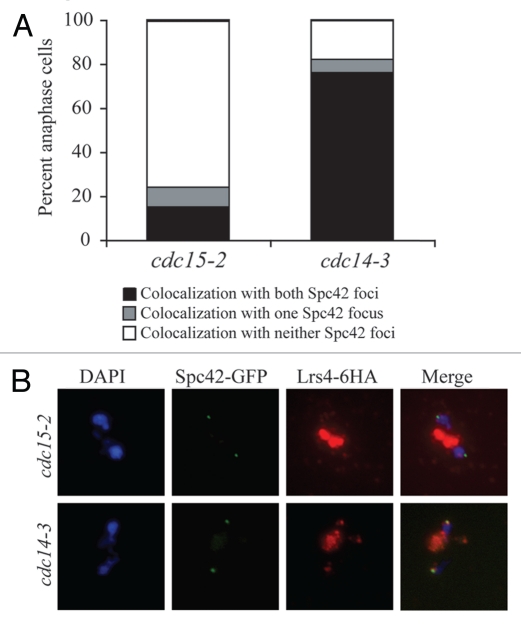
The Mitotic Exit Network (MEN) promotes exit from mitosis by releasing the protein phosphatase Cdc14 from the nucleolus during anaphase (reviewed in ref. 28). The MEN but not Cdc14 is also required for the release of Lrs4 and Csm1 from the nucleolus.7 As expected, the MEN was also needed for Lrs4-Csm1 to associate with kinetochores but CDC14 was not (Fig. 3). Whereas 80% of anaphase-arrested cells in cdc14-3 mutants showed co-localization between Lrs4-6HA and Spc42-GFP, only 20% showed co-localization in anaphase-arrested cdc15-2 MEN mutant cells (Fig. 3). Thus, the MEN is required for Lrs4-Csm1 release from the nucleolus and association with kinetochores.
Figure 3.
The Mitotic Exit Network is required for Lrs4-Csm1 association with kinetochores during anaphase. (A and B) cdc15-2 (A16755) and cdc14-3 (A16802) cells carrying an Spc42-GFP and a Lrs4-6HA fusion were released from a pheromone-induced G1 arrest at 37°C. Chromosome spreads were performed on samples taken 150 minutes after release and the percentage of cells with Lrs4-6HA co-localized with Ndc80-GFP was determined (A). The micrographs in (B) show Lrs4-6HA (red) and Spc42-GFP (green) localization in cdc14-3 and cdc15-2 mutants. At least 50 cells were counted per strain.
Lrs4 and Csm1 localize to kinetochores during anaphase.
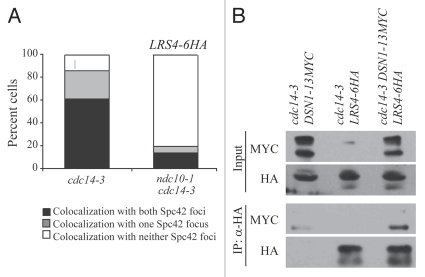
During anaphase, kinetochores are closely associated with spindle pole bodies (SPBs).19 Using light microscopy, it is therefore not possible to distinguish a kinetochore-localized from an SPB-localized protein. To assess whether Lrs4 and Csm1 bind spindle poles or kinetochores, we analyzed Lrs4 localization in relation to Spc42-marked spindle pole bodies in cells that lack NDC10, a gene that encodes a central kinetochore component. Cells carrying a temperature-sensitive ndc10-1 allele progress through mitosis but cannot segregate chromosomes because kinetochore structures are absent.29 To capture cells in anaphase, when Lrs4 is fully released from the nucleolus, we conducted this analysis in cdc14-3 mutants. In ndc10-1 cdc14-3 double mutants, which arrest as single-lobed cells with anaphase-like spindles, Lrs4 no longer co-localized with Spc42 (Fig. 4A). The residual co-localization observed in the ndc10-1 cdc14-3 mutant is likely due to the incomplete penetrance of the ndc10-1 mutant as evidence from 12% of cells that displayed elongated spindles and divided nuclei. We were unable to detect Lrs4-6HA or Csm1-9MYC at kinetochores in Chromatin Immuno-Precipitation (ChIP) experiments. Lrs4 and Csm1 are likely to bind to the periphery of the kinetochore, where the efficiency of crosslinking between proteins and centromeric DNA is low. We note that even during meiosis when localization of the complex to kinetochores is prominent as judged by indirect in situ immunofluorescence, monopolin complex subunits are poorly detected at kinetochores by ChIP.17 However, our data and that of others, strongly suggest that Lrs4 and Csm1 specifically bind kinetochores, rather than spindle pole bodies, during anaphase.
Figure 4.
Lrs4 and Csm1 localize to kinetochores. (A) cdc14-3 (A16802) and ndc10-1 cdc14-3 (A17569) cells carrying an Spc42-GFP fusion were released from a pheromone-induced G1 arrest at 37°C. Chromosome spreads were performed 105 minutes after release and the percentage of cells with Lrs4-6HA co-localized with both or one Spc42-GFP signal was determined. At least 50 cells were counted per strain. (B) cdc14-3 cells carrying fusions of Dsn1-13MYC (A19333), Lrs4-6HA (A14204) or both (A19297) were released from a pheromone-induced G1 arrest at 37°C. Protein extracts were prepared from cells harvested 150 minutes after release. Lrs4-6HA was immunoprecipitated using an anti-HA antibody. Levels of Lrs4-6HA and Dsn1-13MYC were determined in the extracts (B, upper part) and in the precipitates (B, lower part).
Csm1 has has putative interactions with members of two different sub-complexes of the kinetochore. Csm1 has been shown to interact with Ctf19 of the outer-kinetochore COMA complex by two-hybrid screens and with Dsn1 of the MIND complex by in vitro binding studies.30,31 To determine whether these interactions occur in vivo, we performed co-immunoprecipitation experiments in cdc14-3 anaphase-arrested cells (Fig. 4B). Dsn1-13MYC was detected in immunoprecipitates of Lrs4-6HA (Fig. 4B). Our data indicate that the Lrs4-Csm1 complex binds kinetochores perhaps via the MIND complex.
The monopolin complex is required for faithful chromosome segregation during mitosis.
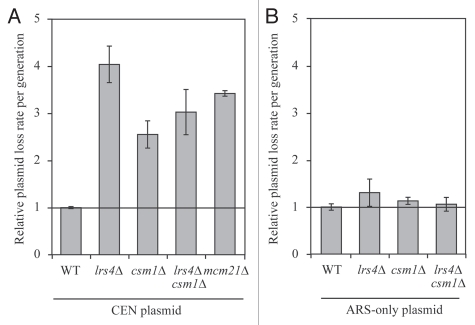
Do Lrs4 and Csm1, which associate with kinetochores during anaphase, play a role in mitotic chromosome segregation? To address this question we measured the loss rates of a centromere-containing plasmid in lrs4Δ and csm1Δ single and double mutants using fluctuation analysis. We observed a 2.5- to 4-fold increase in plasmid loss rates in the two mutants (Fig. 5A). This increase in plasmid loss was comparable to that observed in cells carrying deletions of genes encoding non-essential kinetochore components. For example, cells lacking MCM21, encoding a non-essential member of the kinetochore COMA sub-complex showed plasmid loss rates within this range. Loss rates of plasmids carrying a replication origin (ARS) but not a centromere were not increased in lrs4Δ and csm1Δ single and double mutant (Fig. 5B) indicating that it is kinetochore defects that bring about the CEN plasmid segregation defect observed in the mutants. We conclude that LRS4 and CSM1 are required for faithful chromosome segregation.
Figure 5.
Lrs4 and Csm1 are required for the faithful segregation of CEN plasmids. (A) Fluctuation analysis was performed to determine the rate of plasmid loss per generation (Materials and Methods) of wild-type (A18996), lrs4Δ (A18998), csm1Δ (A19000), lrs4Δ csm1Δ (A19002) or mcm21Δ (A20436) mutants carrying a centromeric plasmid (CEN plasmid). Each experiment consisted of three independent cultures and was repeated at least three times. Error bars represent standard error of the mean. (B) Fluctuation analysis was performed to determine the rate of plasmid loss per generation on wild-type (A19004), lrs4Δ (A19005), csm1Δ (A19006) and lrs4Δ csm1Δ (A19007) cells carrying a plasmid that contains only an autonomous replication sequence (ARS-only). Each experiment consisted of three independent cultures and was repeated at least three times. Error bars represent standard error of the mean.
Spindle integrity is not compromised in cells lacking LRS4 or CSM1.
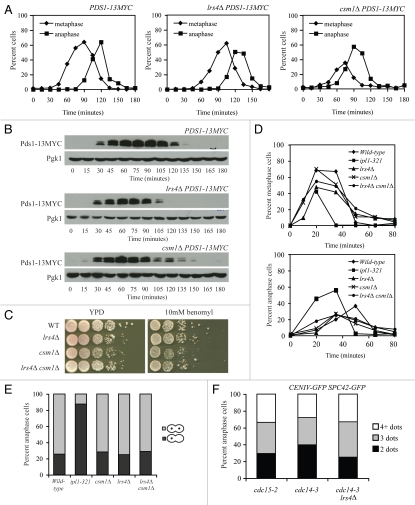
We were not able to determine the role of the Lrs4-Csm1 complex in mitotic chromosome segregation. Cells lacking LRS4 or CSM1 did not exhibit signs of spindle structure or kinetochore attachment defects. Even subtle defects in either process lead to activation of the spindle assembly checkpoint (SAC), which causes a cell cycle delay in metaphase and stabilization of the anaphase inhibitor Pds1 (reviewed in ref. 2). Cells lacking LRS4 or CSM1 did not exhibit a significant delay in Pds1 degradation or anaphase entry (Fig. 6A and B).
Figure 6.
Spindle integrity is not defective in cells lacking LRS4 or CSM1. (A and B) Wild-type (A17064), lrs4Δ (A17061) and csm1Δ (A17063) cells were arrested in G1 using α-factor pheromone (5 µg/ml) and released into medium lacking the pheromone at 25°C. Samples were taken at the indicated times to determine the percent of cells in metaphase (diamonds; A) and anaphase (squares; A) and Pds1-9MYC levels (B). Pgk1 was used as a loading control in (B). (C) Wild-type (A2587), lrs4Δ (A13986), csm1Δ (A8623) and lrs4Δ csm1Δ (A18120) cells were spotted using 10-fold serial dilutions on YPD plates (YEP + 2% glucose) and YPD plates containing 5 µg/ml benomyl. (D and E) Wild-type (A5244; diamonds), ipl1-321 (A16485; squares), lrs4Δ (A15911; triangles), csm1Δ (A24385; exes) and lrs4Δ csm1Δ (A24386; circles) cells all carrying GFP dots at CENIV were arrested in G1 using α-factor pheromone (5 µg/ml) and released into medium containing 15 µg/ml nocodazole. After 80 minutes, cells were washed with YEP D containing 1% DMSO and released into YEP D medium containing 1% DMSO. Samples were taken at the indicated times to determine the percentage of cells in metaphase and anaphase (D) and the percent of anaphase cells in which both GFP dots segregated to the same pole (dark grey bars). (E) 200 cells were counted per time-point in (D) and per strain in (E). (F) cdc15-2 (A23341), cdc14-3 (A16736) and cdc14-3 lrs4Δ (A23340) cells carrying and Spc42-GFP fusion and CENIV GFP dots were arrested in G1 using α-factor (5 µg/ml) and released into medium lacking the pheromone at 37°C. At 240 minutes, the percent of cells with two (indicative of each CENIV region being tightly associated with SPB), three (indicative of only one CENIV region being tightly associated with SPB) or four (indicative of neither CENIV region being tightly associated with SPB) GFP foci was determined in anaphase cells 200 cells were counted per strain.
The SAC appeared to be functional too. Loss of spindle assembly checkpoint function manifests itself as increased sensitivity to the microtubule-depolymerizing drug benomyl.32,33 By this criterion, the SAC is functional in cells deleted for LRS4 or CSM1 (Fig. 6C). Furthermore, unlike cells defective in the sister-kinetochore bi-orientation factor Ipl1, lrs4Δ and csm1Δ single and double mutants reformed mitotic spindles and bi-oriented sister kinetochores normally following transient microtubule depolymerization with nocodazole (Fig. 6D and E).34
Finally, we tested whether LRS4 and CSM1 were required to maintain kinetochores close to the spindle pole body in late anaphase, when chromosome segregation has been completed and cells exit from mitosis.19 In anaphase, the close proximity between centromeres and SPBs can be visualized by marking a centromere (in our case CENIV) and a SPB component (in our case Spc42) with GFP. When the two are closely associated, the GFP dots appear as one. Hence, an anaphase cell contains two GFP dots (Fig. 6F). Straying of the centromere from one of the two GFP dots will lead to 3 or 4 dots being visible in cells. We detected no difference in centromere straying between anaphase-arrested cdc14-3 mutants, where the Lrs4-Csm1 complex is at kinetochores and cdc14-3 lrs4Δ cells or cdc15-2 cells, where the complex is not (Fig. 6F). Our results indicate that LRS4 and CSM1 are required for faithful chromosome segregation during mitosis, but the mechanism whereby the two proteins ensure chromosome segregation fidelity remains to be determined.
Discussion
Lrs4 and Csm1 are essential for rDNA segregation and maintenance.7,8 Our localization studies further raise the possibility that the protein complex has additional functions during anaphase. We considered several possibilities. The first was that Lrs4 and Csm1 control Cdc14 localization. This hypothesis was precipitated by the observation that Lrs4 and Csm1 are released from the nucleolus concomitantly with Cdc14 and that the release of all three proteins depends on the Mitotic Exit Network. Our data argue against this idea. Cdc14 release from the nucleolus is not affected by deletion of LRS4 or CSM1 (Brito IL, unpublished observations) nor are the proteins involved in restraining Cdc14 activity (Suppl. Fig. 1B).
The second possibility we investigated was that Lrs4 and Csm1 function in chromosome segregation because both proteins associate with kinetochores during anaphase. The localization of Lrs4 and Csm1 to kinetochores after their release from the nucleolus may reflect their natural affinity for kinetochore components but lack functional importance. However, we provide evidence to suggest otherwise; lrs4Δ and csm1Δ single and double mutants show elevated rates of centromeric plasmid loss that is comparable to that observed in cells deleted for another non-essential kinetochore component.
We do not yet know how Lrs4-Csm1 associates with kinetochores. A recent structural analysis of the complex showed that the proteins form a V-shaped structure with globular domains on each end.31 These could directly bind kinetochore components. Dsn1 could be the Lrs4-Csm1 binding partner at kinetochores. Csm1 has been shown to bind Dsn1 in vitro31 and we detect the two proteins in complexes when precipitated from cell extracts. The function of Lrs4 and Csm1 at kinetochores remains elusive. Dynamic localization of kinetochore components during anaphase has rarely been observed. Fission yeast Scm3 and human hMis18 both localize to kinetochores during anaphase to promote the incorporation of specialized centromeric histone, CENP-A.3–5 Lrs4 and Csm1 are not involved in loading the budding yeast homolog of CENP-A onto DNA (Brito IL, unpublished observations). Nevertheless, their co-regulation with condensin, which provides structural rigidity to centromeres during mitosis in mammals,35 suggests that condensins and monopolins create centromeric rigidity.
Studies of Lrs4-Csm1 in mitosis shed light on the logic of meiosis.
It is generally believed that meiosis is a modulation of the mitotic division with meiosis-specific factors bringing about the meiosis-specific changes to cell cycle progression (reviewed in ref. 16). Our analysis of Lrs4 and Csm1 regulation sheds light on how this is accomplished. Our findings demonstrate that aspects of sister kinetochore co-orientation, the Cdc5-dependent release of Lrs4 and Csm1 from the nucleolus and their association with kinetochores, occur during both mitosis and meiosis but they take place at different times of cell division. In meiosis, Cdc5 promotes the association of the monopolin complex with kinetochores during prophase I;17,36 during mitosis, this does not occur until anaphase.7 It thus appears that establishing meiosis I-specific sister kinetochore co-orientation requires the transposition of Cdc5-dependent mitotic anaphase events, the release of Lrs4-Csm1, to meiotic prophase. Interestingly, changes in chromosome morphogenesis that occur during prophase I also resemble those that take place during late stages of mitosis.37 Furthermore, Cdc5-dependent cohesin removal that is restricted to metaphase and anaphase in mitosis occurs during prophase I in meiosis.38,39 These findings raise the intriguing possibility that some aspects of meiosis I-specific chromosome events are the result of transposing Cdc5-dependent mitotic anaphase events to meiotic prophase I. We speculate that the lynchpin to understanding the meiotic chromosome segregation pattern is to determine the basis for the differential regulation of Cdc5 in the two types of division.
Materials and Methods
Strains and growth conditions.
All strains were derivatives of W303 and are described in Supplemental Table 1. Proteins were tagged using the PCR-based method described in Longtine et al.40 GFP dots were constructed by integrating an array of bacterial TET operator sites 2 kb from the centromere on CENIV.41 Conditions for growth and release are as described in Amon.42 α-factor was re-added to all cultures 90 min after release from the G1 arrest to prevent cells from entering the next cell cycle. Growth conditions for individual experiments are described in the Figure legends.
Localization techniques.
Indirect in situ immunofluorescence was carried out as described in Visintin et al.20 for tubulin, HA- and MYC-tagged proteins. CEN GFP dot visualization was performed as described in Monje-Casas et al.14 Two hundred cells were scored for each time-point. Chromosomes were spread as described in Nairz and Klein.43 HA-tagged proteins were detected with a mouse α-HA.11 antibody (Covance) at a 1:500 dilution. MYC-tagged proteins were detected with a mouse anti-MYC 9E10 antibody (Babco) at a 1:500 dilution. The secondary antibody was an anti-mouse CY3 antibody (Jackson ImmunoResearch) used at a 1:1,000 dilution. Endogenous luminescence was sufficient for visualization of Ndc80-GFP and Spc42-GFP on chromosome spreads. In each experiment, at least fifty cells were counted per strain.
Co-immunoprecipitation.
Cells were arrested in G1 using α-factor (5 µg/ml) and released into fresh medium at 37°C. At 150 minutes, 50 mls of culture (OD of 0.8) were harvested, washed with 10 mM Tris (pH 7.5), frozen in liquid nitrogen and stored at −80°C overnight. Cell pellets were thawed on ice and resuspended in 200 µl of NP40 lysis buffer (1% NP40, 150 mM NaCl, 50 mM Tris (pH 7.5), 1 mM dithiothreitol (DTT), 60 mM β-glycerophosphate, 1 mM NaVO3, 2 µM) Microcystin-LR (EMD Biosciences and complete EDTA-free protease inhibitor cocktail (Roche)). Cells were disrupted with ∼100 µl glass beads in a FastPrep FP120 (Savant) homogenizer for 3 cycles of 45 seconds (6.5 m/s). Equal amounts of extract (5 mgs in 120 µl of NP40 buffer) were used for immunoprecipitation and one-tenth was saved to analyze input protein concentrations. α-HA.11 antibody (Covance) was added to each of the samples (1:50 dilution) and extracts were incubated with rotation for 2 hours at 4°C. 24 µl of protein G-coupled sepharose beads were then added to each sample and incubated with rotation for an additional 2 hrs at 4°C. Samples were washed five times with NP40 buffer, boiled in SDS-based sample buffer and analyzed by western blot analysis.
Western blot analysis.
Immunoblots were all performed as described in Cohen-Fix et al.44 Lrs4-6HA was detected with a mouse α-HA.11 antibody (Covance) at a 1:500 dilution, followed by a horseradish peroxidase (HRP)-conjugated anti-mouse TrueBlot reagent (eBioScience) at 1:1,000 dilution. For cell cycle analysis of Pgk1 and Pds1-13MYC, cells were harvested and incubated in 5% trichloroacetic acid (TCA) and lysed as described in Moll et al.45 The MYC-tagged proteins were detected with a mouse anti-MYC 9E10 antibody (Babco) at a 1:1,000 dilution. Pgk1 was detected with a mouse anti-Pgk1 antibody (Molecular Probes) at a 1:20,000 dilution. The secondary antibody used for the MYC-tagged proteins and the Pgk1 was a goat anti-mouse antibody conjugated to horseradish peroxidase (HRP; Jackson ImmunoResearch) at a 1:2,000 dilution.
Plasmid loss experiments.
Standard fluctuation analysis46 was used to determine the percentage of cells that lose a plasmid per generation. Cells carrying either the YCPlac33 (CEN4, ARS1, URA3) or YRp17 (ARS1, URA3, TRP1) were grown in Ura medium overnight. To begin the experiment, cells were counted using a Coulter Counter and then plated on YPD plates or plates containing 5-FOA to monitor plating efficiency and the percentage of the starting population that contained the plasmid, respectively. After 24 hours in YPD medium, cells were re-counted and plated on YPD plates and plates containing 5-FOA. Viability was similar in all cultures and all cultures were grown for the same number of generations. Plating efficiency and the percentage of cells at the start of the experiment that had lost the plasmid were taken into account. Three or more replicates of each strain were grown on three or more separate occasions.
Acknowledgements
We would like to thank Frank Solomon, Iain Cheeseman and members of the Amon Lab for their comments on the manuscript. This research was supported by National Institutes of Health grant GM62207 to A.A. A.A. is also an Investigator of the Howard Hughes Medical Institute.
Footnotes
Previously published online: www.landesbioscience.com/journals/cc/article/12885
Supplementary Material
References
- 1.Kitamura E, Tanaka K, Kitamura Y, Tanaka TU. Kinetochore-microtubule interaction during S phase in Saccharomyces cerevisiae. Genes Dev. 2007;21:3319–3330. doi: 10.1101/gad.449407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Musacchio A, Salmon ED. The spindle-assembly checkpoint in space and time. Nat Rev Mol Cell Biol. 2007;8:379–393. doi: 10.1038/nrm2163. [DOI] [PubMed] [Google Scholar]
- 3.Fujita Y, Hayashi T, Kiyomitsu T, Toyoda Y, Kokubu A, Obuse C, Yanagida M. Priming of centromere for CENP-A recruitment by human hMis18alpha, hMis-18beta and M18BP1. Dev Cell. 2007;12:17–30. doi: 10.1016/j.devcel.2006.11.002. [DOI] [PubMed] [Google Scholar]
- 4.Pidoux AL, Choi ES, Abbott JK, Liu X, Kagansky A, Castillo AG, et al. Fission yeast Scm3: A CENP-A receptor required for integrity of subkinetochore chromatin. Mol Cell. 2009;33:299–311. doi: 10.1016/j.molcel.2009.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Williams JS, Hayashi T, Yanagida M, Russell P. Fission yeast Scm3 mediates stable assembly of Cnp1/CENP-A into centromeric chromatin. Mol Cell. 2009;33:287–298. doi: 10.1016/j.molcel.2009.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.D'Amours D, Stegmeier F, Amon A. Cdc14 and condensin control the dissolution of cohesin-independent chromosome linkages at repeated DNA. Cell. 2004;117:455–469. doi: 10.1016/s0092-8674(04)00413-1. [DOI] [PubMed] [Google Scholar]
- 7.Huang J, Brito IL, Villén J, Gygi SP, Amon A, Moazed D. Inhibition of homologous recombination by a cohesin-associated clamp complex recruited to the rDNA recombination enhancer. Genes Dev. 2006;20:2887–2901. doi: 10.1101/gad.1472706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Waples WG, Chahwan C, Ciechonska M, Lavoie BD. Putting the brake on FEAR: Tof2 promotes the biphasic release of Cdc14 phosphatase during mitotic exit. Mol Biol Cell. 2009;20:245–255. doi: 10.1091/mbc.E08-08-0879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Johzuka K, Horiuchi T. The cis element and factors required for condensin recruitment to chromosomes. Mol Cell. 2009;34:26–35. doi: 10.1016/j.molcel.2009.02.021. [DOI] [PubMed] [Google Scholar]
- 10.Mekhail K, Seebacher J, Gygi SP, Moazed D. Role for pernuclear chromose tethering in maintenance of genome stability. Nature. 2008;456:667–670. doi: 10.1038/nature07460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tóth A, Rabitsch KP, Gαlovα M, Schleiffer A, Buonomo SB, Nasmyth K. Functional genomics identifies monopolin: a kinetochore protein required for segregation of homologs during meiosis I. Cell. 2000;103:1155–1168. doi: 10.1016/s0092-8674(00)00217-8. [DOI] [PubMed] [Google Scholar]
- 12.Rabitsch KP, Petronczki M, Javerzat JP, Genier S, Chwalla B, Schleiffer A, et al. Kinetochore recruitment of two nucleolar proteins is required for homolog segregation in meiosis I. Dev Cell. 2003;4:535–548. doi: 10.1016/s1534-5807(03)00086-8. [DOI] [PubMed] [Google Scholar]
- 13.Petronczki M, Matos J, Mori S, Gregan J, Bogdanova A, Schwickart M, et al. Monopolar attachment of sister kinetochores at meiosis I requires casein kinase 1. Cell. 2006;126:1049–1064. doi: 10.1016/j.cell.2006.07.029. [DOI] [PubMed] [Google Scholar]
- 14.Monje-Casas F, Prabhu VR, Lee BH, Boselli M, Amon A. Kinetochore orientation during meiosis is controlled by Aurora B and the monopolin complex. Cell. 2007;128:477–490. doi: 10.1016/j.cell.2006.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Winey M, Morgan GP, Straight PD, Giddings TH, Mastronarde DN. Three-dimensional ultrastructure of Saccharomyces cerevisiae meiotic spindles. Mol Biol Cell. 2005;16:1178–1188. doi: 10.1091/mbc.E04-09-0765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marston AL, Amon A. Meiosis: cell cycle controls shuffle and deal. Nat Rev Mol Cell Biol. 2004;5:983–997. doi: 10.1038/nrm1526. [DOI] [PubMed] [Google Scholar]
- 17.Lee BH, Amon A. Role of Polo-like kinase CDC5 in programming meiosis I chromosome segregation. Science. 2003;300:482–486. doi: 10.1126/science.1081846. [DOI] [PubMed] [Google Scholar]
- 18.Brito IL, Yu HG, Amon A. Condensins promote coorientation of sister chromatids during meiosis I in budding yeast. Genetics. 2010;185:55–64. doi: 10.1534/genetics.110.115139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guacci V, Hogan E, Koshland D. Centromere position in budding yeast: evidence for anaphase A. Mol Biol Cell. 1997;8:957–972. doi: 10.1091/mbc.8.6.957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Visintin R, Hwang ES, Amon A. Cfi1 prevents premature exit from mitosis by anchoring Cdc14 phosphatase in the nucleolus. Nature. 1999;398:818–823. doi: 10.1038/19775. [DOI] [PubMed] [Google Scholar]
- 21.Shou W, Seol JH, Shevchenko A, Baskerville C, Moazed D, Chen ZW, et al. Exit from mitosis is triggered by Tem1-dependent release of the protein phosphatase Cdc14 from nucleolar RENT complex. Cell. 1999;97:233–244. doi: 10.1016/s0092-8674(00)80733-3. [DOI] [PubMed] [Google Scholar]
- 22.Straight AF, Shou W, Dowd GJ, Turck CW, Deshaies RJ, Johnson AD, Moazed D. Net1, a Sir2-associated nucleolar protein required for rDNA silencing and nucleolar integrity. Cell. 1999;97:245–256. doi: 10.1016/s0092-8674(00)80734-5. [DOI] [PubMed] [Google Scholar]
- 23.Bhalla N, Biggins S, Murray AW. Mutation of YCS4, a budding yeast condensin subunit, affects mitotic and nonmitotic chromosome behavior. Mol Biol Cell. 2002;13:632–645. doi: 10.1091/mbc.01-05-0264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Varela E, Shimada K, Laroche T, Leroy D, Gasser SM. Lte1, Cdc14 and MEN-controlled Cdk inactivation in yeast coordinate rDNA decompaction with late telophase progression. EMBO J. 2009;28:1562–1575. doi: 10.1038/emboj.2009.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang B, Yong-Gonzalez V, Strunnikov AV. Cdc14p/FEAR pathway controls segregation of nucleolus in S. cerevisiae by facilitating condensin targeting to rDNA chromatin in anaphase. Cell Cycle. 2004;3:960–967. doi: 10.4161/cc.3.7.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.D'Ambrosio C, Schmidt CK, Katou Y, Kelly G, Itoh T, Shirahige K, Uhlmann F. Identification of cis-acting sites for condensin loading onto budding yeast chromosomes. Genes Dev. 2008;22:2215–2227. doi: 10.1101/gad.1675708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Trautmann S, Rajagopalan S, McCollum D. The S. pombe Cdc14-like phosphatase Clp1p regulates chromosome biorientation and interacts with Aurora kinase. Dev Cell. 2004;7:755–762. doi: 10.1016/j.devcel.2004.10.006. [DOI] [PubMed] [Google Scholar]
- 28.Stegmeier F, Amon A. Closing mitosis: the functions of the Cdc14 phosphatase and its regulation. Annu Rev Genet. 2004;38:203–232. doi: 10.1146/annurev.genet.38.072902.093051. [DOI] [PubMed] [Google Scholar]
- 29.Goh PY, Kilmartin JV. NDC10: a gene involved in chromosome segregation in Saccharomyces cerevisiae. J Cell Biol. 1993;121:503–512. doi: 10.1083/jcb.121.3.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wong J, Nakajima Y, Westermann S, Shang C, Kang J, Goodner C, et al. A protein interaction map of the mitotic spindle. Mol Biol Cell. 2007;18:3800–3809. doi: 10.1091/mbc.E07-06-0536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Corbett KD, Yip CK, Ee LS, Walz T, Amon A, Harrison SC. The monopolin complex cross-links kinetochore components to regulate chromosome-microtubule attachments. Cell. 2010 doi: 10.1016/j.cell.2010.07.017. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li R, Murray AW. Feedback control of mitosis in budding yeast. Cell. 1991;66:519–531. doi: 10.1016/0092-8674(81)90015-5. [DOI] [PubMed] [Google Scholar]
- 33.Hoyt MA, Totis L, Roberts BT. S. cerevisiae genes required for cell cycle arrest in response to loss of microtubule function. Cell. 1991;66:507–517. doi: 10.1016/0092-8674(81)90014-3. [DOI] [PubMed] [Google Scholar]
- 34.Biggins S, Severin FF, Bhalla N, Sassoon I, Hyman AA, Murray AW. The conserved protein kinase Ipl1 regulates microtubule binding to kinetochores in budding yeast. Genes Dev. 1999;13:532–544. doi: 10.1101/gad.13.5.532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ribeiro SA, Gatlin JC, Dong Y, Joglekar A, Cameron L, Hudson DF, et al. Condensin regulates the stiffness of vertebrate centromeres. Mol Biol Cell. 2009;20:2371–2380. doi: 10.1091/mbc.E08-11-1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Clyne RK, Katis VL, Jessop L, Benjamin KR, Herskowitz I, Lichten M, et al. Polo-like kinase Cdc5 promotes chiasmata formation and cosegregation of sister centromeres at meiosis I. Nat Cell Biol. 2003;5:480–485. doi: 10.1038/ncb977. [DOI] [PubMed] [Google Scholar]
- 37.Kleckner N, Zickler D, Jones GH, Dekker J, Padmore R, Henle J, et al. A mechanical basis for chromosome function. Proc Natl Acad Sci USA. 2004;101:12592–12597. doi: 10.1073/pnas.0402724101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Alexandru G, Uhlmann F, Mechtler K, Poupart MA, Nasmyth K. Phosphorylation of the cohesin subunit Scc1 by Polo/Cdc5 kinase regulates sister chromatid separation in yeast. Cell. 2001;105:459–472. doi: 10.1016/s0092-8674(01)00362-2. [DOI] [PubMed] [Google Scholar]
- 39.Yu HG, Koshland D. Chromosome morphogenesis: condensin-dependent cohesin removal during meiosis. Cell. 2005;123:397–407. doi: 10.1016/j.cell.2005.09.014. [DOI] [PubMed] [Google Scholar]
- 40.Longtine MS, McKenzie A, Demarini DJ, Shah NG, Wach A, Brachat A, et al. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 41.Tanaka T, Fuchs J, Loidl J, Nasmyth K. Cohesin ensures bipolar attachment of microtubules to sister centromeres and resists their precocious separation. Nature Cell Biology. 2000;2:492–499. doi: 10.1038/35019529. [DOI] [PubMed] [Google Scholar]
- 42.Amon A. Synchronization procedures. Meth Enzymol. 2002;351:457–467. doi: 10.1016/s0076-6879(02)51864-4. [DOI] [PubMed] [Google Scholar]
- 43.Nairz K, Klein F. mre11S—a yeast mutation that blocks double-strand-break processing and permits nonhomologous synapsis in meiosis. Genes Dev. 1997;11:2272–2290. doi: 10.1101/gad.11.17.2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cohen-Fix O, Peters JM, Kirschner MW, Koshland D. Anaphase initiation in Saccharomyces cerevisiae is controlled by the APC-dependent degradation of the anaphase inhibitor Pds1p. Genes Dev. 1996;10:3081–3093. doi: 10.1101/gad.10.24.3081. [DOI] [PubMed] [Google Scholar]
- 45.Moll T, Tebb G, Surana U, Robitsch H, Nasmyth K. The role of phosphorylation and the CDC28 protein kinase in cell cycle-regulated nuclear import of the S. cerevisiae transcription factor SWI5. Cell. 1991;66:743–758. doi: 10.1016/0092-8674(91)90118-i. [DOI] [PubMed] [Google Scholar]
- 46.Lea D, Coulson C. The distribution of the numbers of mutants in bacterial populations. J Genetics. 1949;49:264–285. doi: 10.1007/BF02986080. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.