Introduction
Mitochondria are polymorphic structures with fundamental roles in cellular differentiation, aging, and apoptosis. Mitochondria accommodate such diverse functions by establishing varying shapes and distributions in the cell via fusion and fission.1 Thus, the components that mediate fission are likely to be targets for intracellular and extracellular signals that modulate mitochondrial activity in a variety of essential cellular processes.2
Fis1, Mdv1, and Dnm1 are three recently identified proteins essential for the fission of mitochondria in Saccharomyces cerevisiae. Mdv1 and Dnm1 assemble in a Fis1-dependent manner on the mitochondrial outer membrane.2 Dnm1 is a dynamin-related GTPase that presumably oligomerizes to form rings around the outer surface of the mitochondrial tubules at sites of constriction, similar to structures formed during endocytosis by its homologue dynamin.3 Mdv1 associates with Fis1 and Dnm1 and is thought to act late in the fission process.4
The amino acid sequence of Fis1 is conserved from yeast to humans [Supplementary Figure A2], implicating a common mechanism for the control of mitochondrial fission.5 Fis1 is an integral membrane protein anchored in the mitochondrial outer membrane by its C-terminal domain. The cytosolic domain of yeast Fis1 has been shown to bind Mdv1 and has been speculated to bind Dnm1 and to self-associate.4,5 To help identify the mechanism by which Fis1 mediates interactions with itself and other components of the fission machinery, we determined the crystal structure of the cytosolic domain of human Fis1. Here, we present insights into proposed binding interactions of Fis1 from a 2.0 Å resolution structure of an indispensable component of the mitochondrial fission machinery.
Results
Fis1 is an α-helical protein with two divergent TPR motifs
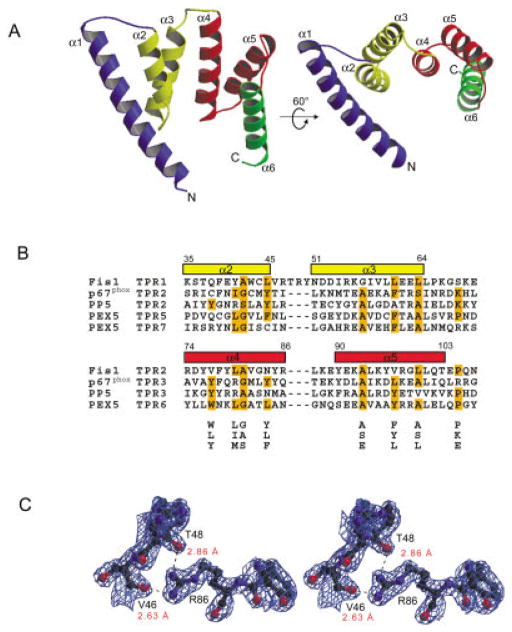
The crystal structure of Fis1 without its C-terminal domain was determined by multiwave-length anomalous dispersion (MAD) phasing (Table I) and reveals an open array of six α-helices [Fig. 1(A) and Supplementary Figure A1]. The 26-residue helix α1 is longer than the other helices and extends away from two three-helix bundles (α2–α4 and α4–α6) that share a common helix (α4). Structural homology searches indicate that Fis1 resembles proteins that possess an α-α super-helix fold, most notably the tetratricopeptide repeat (TPR) motifs. The TPR motif is a well-known protein–protein interaction motif comprised of helix-turn-helix repeats of 34 residues arranged in an antiparallel fashion.6 The Fis1 structure superimposes onto the structures of other TPR proteins reasonably well7–9 (CαRMSD of ~1.5 Å), including the membrane fusion protein Sec17.10 However, the amino acid sequence of Fis1 is only distantly related to TPR proteins; little sequence identity was found from sequence database queries using multiple rounds of PSI-BLAST. Structure-based sequence alignments were required to uncover two divergent TPR motifs in Fis1 [Fig. 1(B)]. TPR1 (residues 35–72) differs from the TPR consensus sequence by a three-residue insertion in the loop between helices α2 and α3. It is interesting that this loop is structurally constrained by hydrogen bonds formed between the backbone carbonyl oxygens of two conserved loop residues, Val46 and Thr48, and the guanidinium group of conserved Arg86 in helix α4 [Fig. 1(C)]. Consequently, the orientation of helices α2 and α4 is fixed, potentially defining a binding site. Loops of other TPR proteins were previously shown to mediate protein–protein interactions.11
TABLE I.
Summary of Crystallographic Data and Refinement Statistics
| Data collection and phasing | |||
| λ1 | λ2 | λ3 | |
| Wavelength (Å) | 0.9767 | 0.9797 | 0.9645 |
| Resolution (Å) | 20–2.5 | 20–2.5 | 20–2.5 |
| Unique reflectionsa | 7860 | 7890 | 7912 |
| Completeness (%)b | 99.6 (99.6) | 99.7 (99.9) | 99.7 (99.9) |
| Redundancy | 7 | 7 | 7 |
| Rmerge (%)c | 4.2 | 4.3 | 4.4 |
| (〈I〉/〈σ1〉)b | 50.6 (26.8) | 50.6 (26.8) | 50.6 (26.8) |
| Figure of merit | SOLVE 0.65 | RESOLVE 0.79 | |
| Refinement statistics | |||
| Resolution limits (Å) | 20–2.0 | ||
| Rcryst/Rfree (%)e | 23.0/25.5 | ||
| No. of protein atoms | 1010 | ||
| No. of nonprotein atoms | 75 | ||
| RMSD bonds (Å) | 0.006 | ||
| RMSD angles (°) | 1.085 | ||
| Average B (Å2) | 34.1 | ||
Bijvoet pairs were scaled separately for the MAD data sets.
Values in parentheses are for the highest resolution shell.
Σi, h|I(i, h) − 〈I(h)〉|/Σi,h|I(i, h)|
Σ||Fo| − |Fc||/Σ|Fo|
Rcryst for 8% of reflections not used during the refinement.
Fig. 1.
TPR fold of Fis1. A: Two ribbon drawings of Fis1 with labeled helices (α1–α6) and TPR motifs color coded as follows: helix α1 blue, TPR1 (α2–α3) yellow, TPR2 (α4–α5) red and helix α6 green. Electron density was not observed for the first two and last two residues. Figures were prepared with MOLSCRIPT23 and rendered in Raster3d.24 B: Structure-based alignment of TPR motifs. The TPR motifs of p67phox, PP5, and PEX5, structural homologs of Fis1 as found by DALI, VAST, and CE, are aligned with the TPR motifs of Fis1. Residues that conform to the TPR consensus sequence below the alignment are highlighted in orange.25 Fis1 α-helical regions are indicated above the alignment by rectangles color-coded as in Figure 1(A). Helix α6 lacks the 34-residue TPR motif and is, therefore, excluded from the alignment. C: Stereoview of the final σA weighted 2Fo −Fc electron density map displaying interactions between labeled residues on helices α2 and α4 calculated to 2.0 Å resolution and contoured at 1.1α. Hydrogen bonds are indicated by dashes, and distances are labeled in red. The figure was generated using BOBSCRIPT26 and rendered in Raster3d.24
Fis1 forms a dimer in the crystal structure
In Fis1, helices α2, α4, and α6 are arranged to form an amphiphilic, concave surface [Fig. 1(A), right view]. The crystal packing of Fis1 may reveal one manner by which this concave surface mediates binding. In the crystal, a dimer of Fis1 is observed in which the concave surface of a Fis1 molecule makes contacts with helix α1 of a symmetry mate (Fig. 2). The symmetry-related dimer buries ~3000 Å2 of otherwise exposed surface area on monomeric Fis1 (~1500 A2 from helix α1 alone), which supports two-hybrid data that detects yeast Fis1 self-association in vivo.6 However, Fis1 also exists as a monomer in solution as determined for the cytosolic domain of yeast Fis1 by size exclusion chromatography (data not shown). The monomeric Fis1 as seen in the crystal structure suggests that Fis1 may exhibit multiple modes of binding that involve the concave surface and helix α1. Indeed, molecular modeling studies suggest that only a few bond angle rotations about backbone φ, ψ would be required for an intramolecular association of helix α1. Thus, helix α1 may serve as a “switch” for modulating protein–protein interactions by regulating access to the concave binding surface.
Fig. 2.

Fis1 crystallographic dimmer. Ribbon drawing of the Fis1 dimer found in the crystal lattice oriented down the twofold crystallographic symmetry axis. The figure was prepared as in Figure 1(A).
Discussion
Fis1 adopts a TPR fold well-suited for binding interactions necessary for mitochondrial fission. The open structure of Fis1 creates a concave binding surface that can accommodate a helix to potentially promote intermolecular associations (as seen in the crystal packing of the structure) or intramolecular associations (as revealed by modeling). Alternatively, a protein with extended conformation, such as thought for the N-terminal domain of Mdv1, may bind the Fis1 surface in a manner similar to that observed for the extended peptides that bind to HOP, another TPR domain protein that modulates Hsp70/Hsp90 interactions.9
Besides the concave binding surface, Fis1 possesses additional structural features for mediating protein–protein interactions. We propose that helix α1 acts as a molecular switch by promoting either monomeric or dimeric conformations of Fis1 to modulate accessibility of different binding surfaces. Furthermore, the loops of Fis1, especially the loop in TPR1, may be important for interactions with the GTPase Dnm1, as observed in the structure of the TPR domain of p67phox in complex with the GTPase Rac.11 Therefore, Fis1 is structurally equipped to be both free in solution and to associate with itself and with other members of the fission machinery. In addition, the structure of Fis1 is suitable as a scaffold for interactions with proteins involved in other cellular processes such as apoptosis. Several lines of evidence support changes in mitochondrial morphology during caspase-mediated cell death that are consistent with an increase in mitochondrial fission.12–17 Thus, Fis1 might serve as a target for signals that mediate cell death in addition to fission.
Materials and Methods
Protein expression, purification, and crystallization
The Homo sapiens Fis1 gene (CGI-135) without the C-terminal domain (1–123 aa) was subcloned into the plasmid pET15b and overexpressed in E. coli strain pLysS BL21(DE3)codon+ as a His6-tagged fusion protein. The His6-tagged protein was purified by Ni2+ affinity chromatography, cleaved with biotinylated thrombin, incubated with streptavidin agarose beads and further purified by Ni2+ affinity chromatography. The recombinant protein contains three extra vector derived amino acids (Gly-Ser-His) at the N-terminus. Crystals were grown by using hanging drop vapor diffusion at room temperature. Hanging drops consisted of equal volumes of the protein solution (Fis1 at 32 mg mL−1 in 25 mM sodium acetate, 2 mM DTT, pH 5.0) and a reservoir solution (100 mM sodium acetate, 2.0 M ammonium sulfate, 5% glycerol, pH 4.6).
Data collection, structure determination, and refinement
For data collection, crystals were flash-frozen by rapid transfer into mother liquor with 20% (v/v) glycerol followed by direct placement in liquid nitrogen. Multiple three-wavelength MAD data sets were collected at Advanced Photon Source (Argonne, IL) beamline 19-ID on a CCD detector with crystals grown from SeMet-derived protein. All data were processed and scaled with the HKL2000 package.18 Crystals belong to space group P6522 with one molecule in the asymmetric unit (a = b = 48.9 Å, c = 169.1 Å, γ = 120°). SOLVE/RESOLVE was used to locate the positions of the selenium atoms and for initial tracing.19 Reflection intensities were converted to structure factor amplitudes using the CCP4 suite.20 Model building was performed in O21 and followed by refinement with CNS.22
Coordinates
Atomic coordinates have been deposited in the Protein Data Bank (1NZN).
Summary
Fis1 is an integral membrane protein that acts in the fission of mitochondria by controlling the assembly, membrane distribution, and function of the mitochondrial fission machinery. Here we report the 2.0 Å resolution crystal structure of the cytosolic domain of human Fis1. The structure reveals an antiparallel array of α-helices homologous to tetratricopeptide repeat (TPR) proteins. Structure-based sequence alignments of Fis1 uncovered two divergent TPR motifs; the first TPR motif differs from the TPR consensus sequence by a three-residue insertion in a loop that may be important for function. These TPR helices create an amphiphilic, concave surface that can accommodate a helix or, possibly, an extended segment. Indeed, this putative binding surface mediates homodimer formation of Fis1 in the crystal. The structure of Fis1 provides insight into the architecture of the proposed binding interactions that mediate mitochondrial fission.
Supplementary Material
Acknowledgments
We thank Steve Ginnel for help with data collection, Jeremy Berg, Eaton Lattman, Neil Clarke, Dan Leahy, Doug Barrick, and Scott Walsh for reading the manuscript and helpful discussions. RBH thanks Maria Bewley, John Flanagan, and David Jeruzalmi for helpful discussions. Use of the Argonne National Laboratory Structural Biology Center beamlines at the Advanced Photon Source was supported by the U.S. Department of Energy, Office of Biological and Environmental Research, under Contract No. W-31-109-ENG-38. AGG was partially supported by NSF grant MCB9982967. This work was supported by funds to RBH generously provided by the Zanvyl Krieger School of Arts and Sciences at the Johns Hopkins University.
Footnotes
The Supplementary Materials Referred to in this article can be found at http://www.interscience.wiley.com/jpages/0887-3585/suppmat/index.html
References
- 1.Bereiter-Hahn J, Voth M. Dynamics of mitochondria in living cells: shape changes, dislocations, fusion, and fission of mitochondria. Microsc Res Tech. 1994;27(3):198–219. doi: 10.1002/jemt.1070270303. [DOI] [PubMed] [Google Scholar]
- 2.Shaw JM, Nunnari J. Mitochondrial dynamics and division in budding yeast. Trends Cell Biol. 2002;12(4):178–184. doi: 10.1016/s0962-8924(01)02246-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang P, Hinshaw J. Three-dimensional reconstruction of dynamin in the constricted state. Nature Cell Biol. 2001;3:922–926. doi: 10.1038/ncb1001-922. [DOI] [PubMed] [Google Scholar]
- 4.Tieu Q, Okreglak V, Naylor K, Nunnari J. The WD repeat protein, Mdv1p, functions as a molecular adaptor by interacting with Dnm1p and Fis1p during mitochondrial fission. J Cell Biol. 2002 August;158:445–452. doi: 10.1083/jcb.200205031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mozdy AD, McCaffery JM, Shaw JM. Dnm1p GTPase-mediated mitochondrial fission is a multi-step process requiring the novel integral membrane component Fis1p. J Cell Biol. 2000;151(2):367–379. doi: 10.1083/jcb.151.2.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blatch GL, Lassle M. The tetratricopeptide repeat: a structural motif mediating protein-protein interactions. Bioessays. 1999;21(11):932–999. doi: 10.1002/(SICI)1521-1878(199911)21:11<932::AID-BIES5>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 7.Gatto GJ, Geisbrecht BV, Gould SJ, Berg JM. Peroxisomal targeting signal-1 recognition by the TPR domains of human PEX5. Nat Struct Biol. 2000;7:1091–1095. doi: 10.1038/81930. [DOI] [PubMed] [Google Scholar]
- 8.Das AK, Cohen PTW, Barford D. The structure of the tetratricopeptide repeats of protein phosphatase 5: implications for TPR-mediated protein-protein interactions. EMBO J. 1998;17(5):1192–1199. doi: 10.1093/emboj/17.5.1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Scheufler C, Brinker A, Bourenkov G, Pegoraro S, Moroder L, Bartunik H, Hartl F, Moaerefi I. Structure of TPR domain-peptide complexes: critical elements in the assembly of the Hsp70-Hsp90 multichaperone machine. Cell. 2000;101:199–210. doi: 10.1016/S0092-8674(00)80830-2. [DOI] [PubMed] [Google Scholar]
- 10.Rice LM, Brunger AT. Crystal structure of the vesicular transport protein Sec17: implications for SNAP function in SNARE complex disassembly. Mol Cell. 1999 July;4:85–95. doi: 10.1016/s1097-2765(00)80190-2. [DOI] [PubMed] [Google Scholar]
- 11.Lapounge K, Smith SJM, Walker PA, Gamblin SJ, Rittinger K. Structure of the TPR domain of p67phox in complex with RacGTP. Mol Cell. 2000;6:899–907. doi: 10.1016/s1097-2765(05)00091-2. [DOI] [PubMed] [Google Scholar]
- 12.Chen W, Calvo PA, Malide D, Gibbs J, Schubert U, Bacik I, Basta S, O’Neill R, Schickli J, Palese P, Henklein P, Bennink JR, Yewdell JW. A novel influenza A virus mitochondrial protein that induces cell death. Nat Med. 2001;7(12):1306–1312. doi: 10.1038/nm1201-1306. [DOI] [PubMed] [Google Scholar]
- 13.Frank S, Gaume B, Bergmann-Leitner E, Leitner W, Robert E, Catez F, Smith C, Youle R. The Role of Dynamin-Related Protein 1, a Mediator of Mitochondrial Fission, in Apoptosis. Dev Cell. 2001 October;1:515–525. doi: 10.1016/s1534-5807(01)00055-7. [DOI] [PubMed] [Google Scholar]
- 14.Desagher S, Martinou JC. Mitochondria as the central control point of apoptosis. Trends Cell Biol. 2000;10(9):369–377. doi: 10.1016/s0962-8924(00)01803-1. [DOI] [PubMed] [Google Scholar]
- 15.Mancini M, Anderson BO, Caldwell E, Sedghinasab M, Paty PB, Hockenbery DM. Mitochondrial proliferation and paradoxical membrane depolarization during terminal differentiation and apoptosis in a human colon carcinoma cell line. J Cell Biol. 1997;138(2):449–469. doi: 10.1083/jcb.138.2.449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pinton P, Ferrari D, Rapizzi E, Di Virgilio FD, Pozzan T, Rizzuto R. The Ca2+ concentration of the endoplasmic reticulum is a key determinant of ceramide-induced apoptosis: significance for the molecular mechanism of Bcl-2 action. EMBO J. 2001;20(11):2690–2701. doi: 10.1093/emboj/20.11.2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Karbowski M, Lee YJ, Gaume B, Jeong SY, Frank S, Nechushtan A, Santel A, Fuller M, Smith CL, Youle RJ. Spatial and temporal association of Bax with mitochondrial fission sites, Drp1, and Mfn2 during apoptosis. J Cell Biol. 2002;159(6):931–938. doi: 10.1083/jcb.200209124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Otwinowski Z, Minor W. Processing of X-ray data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 19.Terwilliger TC, Berendzen J. Automated MAD and MIR structure solution. Acta Crystallogr D. 1999;55:849–861. doi: 10.1107/S0907444999000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.CCP4. The CCP4 Suite: programs for protein crystallography. Acta Crystallogr D. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 21.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 22.Brunger A, Adams P, Clore G, DeLano W, Gros P, Grosse-Kunstleve R, Jiang J, Kuszewski J, Nilges M, Pannu N, Read R, Rice L, Simonson T, Warren G. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 23.Kraulis PJ. MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J Appl Crystallogr. 1991;24:946–950. [Google Scholar]
- 24.Merrit EA, Murphy ME. RASTER3D version 2.0. A program for photorealistic molecular graphics. Acta Crystallogr D. 1994;24:946–950. doi: 10.1107/S0907444994006396. [DOI] [PubMed] [Google Scholar]
- 25.Sikorski RS, Boguski MS, Goebl M, Hieter P. A repeating amino acid motif in CDC23 defines a family of proteins and a new relationship among genes required for mitosis and RNA synthesis. Cell. 1990 January;60:307–317. doi: 10.1016/0092-8674(90)90745-z. [DOI] [PubMed] [Google Scholar]
- 26.Esnouf RM. An extensively modified version of MolScript that includes greatly enhanced coloring capabilities. J Mol Graph. 1997;15:132–134. doi: 10.1016/S1093-3263(97)00021-1. [DOI] [PubMed] [Google Scholar]
- 27.DeLano WL. The PyMOL Molecular Graphics System. San Carlos: DeLano Scientific; 2002. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.