Abstract
A mutation in the rat GIMAP5 gene predisposes for autoimmunity, most famously in the BB rat model of autoimmune type 1 diabetes mellitus. This mutation is associated with severe peripheral T lymphopenia, as is mutation of the same gene in mice, but the mechanism by which GIMAP5 normally protects T cells from death is unknown. GIMAP5 is a putative small GTPase, a class of proteins which often fulfil their functions in the vicinity of cellular membranes. The objective of this study was to determine the normal intracellular location of GIMAP5 in lymphoid cells. Combining studies in rat, mouse and human systems, novel monoclonal antibodies (mAbs) were used to examine the localization of GIMAP5 and the closely-related protein, GIMAP1, in lymphoid cells by means of confocal microscopy and sub-cellular fractionation combined with immunoblotting. Additionally, human Jurkat T cells that inducibly express epitope-tagged GIMAP5 were established and used in electron microscopy (EM). Endogenous GIMAP5 was found to be located in a membraneous compartment/s which was also detected by established markers of lysosomes. GIMAP1, by contrast, was found to be located in the Golgi apparatus. EM studies of the inducible Jurkat T cells also found GIMAP5 in lysosomes and, in addition, in multivesicular bodies. This study establishes that the endogenous location of GIMAP5 is in lysosomes and related compartments and provides a clearer context for hypotheses about its mechanism of action.
Key words: GIMAP, GTPase, golgi apparatus, lysosome
Introduction
Intrinsic to the vertebrate immune system is the potential for reactivity against self, a danger that requires the operation of multiple restraining mechanisms. Disruption of these at the molecular or cellular level (in multiple possible ways) can precipitate autoimmunity. This study concerns just one example of this, i.e., T-cell lymphopenia brought about by an autosomal mutation in the rat. On some genetic backgrounds this mutation, known as lymphopenia or lyp, predisposes for autoimmune disease, e.g., type 1 diabetes mellitus in the BioBreeding (BB) diabetes-prone (BBDP) rat1–4 and eosinophilic bowel disease in the PVG-RT1u, lyp/lyp rat.5 Although the peripheral T-cell lymphopenia caused by this mutation is severe and effects all T-cell sub-populations, it may be its detrimental effect on CD4+CD25+ regulatory T-cell function that lies at the heart of its promotion of autoimmunity,6 although additional impacts cannot be ruled out.
In 2002, the probable basis of the lymphopenia trait was located to a cluster of genes encoding a little-studied family of putative GTPases, now known as the GIMAPs.7,8 Bioinformatic analysis has shown that the mammalian GIMAPs are members of the AIG1 class of GTPases and have relatives in higher plants and non-mammalian vertebrates but not in many convenient experimental organisms such as E. coli, C. elegans or D. melanogaster.9 The specific defect identified in the mutant rat was a frame-shift mutation within the Gimap5 gene. Shortly following that, an in vitro screen independently identified human GIMAP5 as an anti-apoptotic protein.10 More recently, targeted and chemically-induced mutations of the GIMAP5 gene in mice were shown to result in lymphopenic phenotypes very similar to that seen in the rat,11,12 and genetic studies in humans have shown associations of a polyadenylation signal polymorphism in GIMAP5 both with systemic lupus erythematosus risk13 and with levels of IA-2 autoantibodies in type 1 diabetic patients.14 Despite these advances, the mechanism by which GIMAP5 protects lymphocytes from death in vivo and how it might interact with signalling pathways known to be important for cell survival is still unresolved. One study, which may point the way to a full understanding of the mechanism, has described physical interaction of GIMAP5 with bcl-2 family members.9
GIMAP5 is a putative ‘tail-anchored’ protein bearing a predicted carboxy-terminal transmembrane domain, indicative of an intracellular membrane location with exposure to the cytosol.15 Previous studies have placed GIMAP5 in different organelles—claims have been made for its localization to mitochondria, the Golgi apparatus, the centrosome and the endoplasmic reticulum.9,10,16–18 The majority of these studies have depended on the in vitro overexpression of tagged GIMAP5 constructs. The diversity of the reported locations for GIMAP5 raised in our minds the concern that overexpression or mislocation artifacts may have been at play in some of the earlier studies. We therefore undertook the generation of new reagents in the form of highly sensitive monoclonal antibodies for use in identifying the location of GIMAP5 in cells in which it is expressed endogenously. Some of these antibodies proved to be of sufficient quality to enable their use in discriminating confocal microscopy of lymphoid cell lines. With supporting evidence coming from biochemical separations of cellular membrane fractions and from electron microscopy, our studies led us to the conclusion that GIMAP5 is a lysosome-associated protein. We also examined the localization of GIMAP1, a sister protein of GIMAP5, and found that the two GTPases were differentially localised.
Results
Immunodetection of rat GIMAP5 using a novel monoclonal antibody.
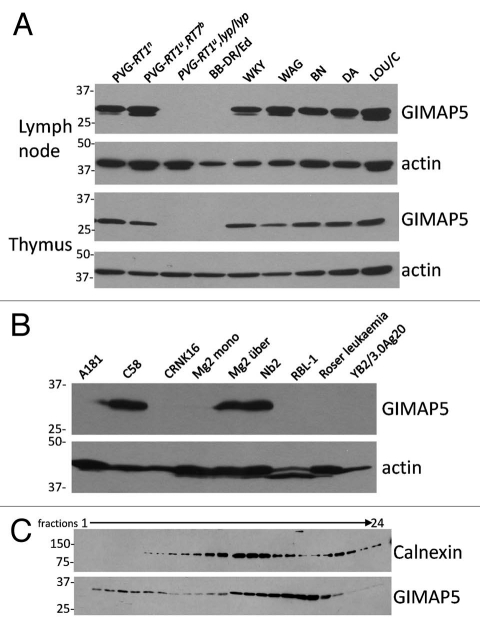
Novel monoclonal antibodies were generated against rat GIMAP5. The reactivity of one of these, MAC427, is illustrated in Figure 1A. A band of the predicted relative molecular mobility of GIMAP5 was seen in lymphoid cell lysates of all rat strains assayed except PVG-RT1u,lyp/lyp and BB-DR/Ed,19 both of which bear the mutant lyp allele of rat GIMAP5. In some samples e.g., WKY, an additional minor slightly smaller immunoreactive species was seen. It is unclear whether this represents an alternative form of GIMAP5 or reflects low levels of proteolysis during isolation. Figure 1B shows, in addition, the endogenous expression of GIMAP5 protein by the rat T-cell lines C58, Nb2 and Mg2über. In the light of a report by Keita and colleagues, who suggested that GIMAP5 occupies an intracellular compartment distinct from the mitochondria or the endoplasmic reticulum (ER),20 we proceeded to prepare homogenates of Nb2 cells and subject post-nuclear supernatant fractions to density separation by centrifugation through self-forming Percoll gradients, as described in Materials and Methods. This was followed by immuno-staining of the fractions obtained. Figure 1C shows that the distribution of rat GIMAP5 immunoreactivity in the Percoll gradient was bimodal and clearly distinct from that of the ER marker calnexin. Despite repeated attempts, we were unable to obtain satisfactory confocal microscopy images of rat GIMAP5 using our antibodies, presumably because of masking of the epitopes during fixation. In addition, we did not find any antibodies which were capable of convincingly detecting rat LAMP proteins on western blots. Because of these limitations we moved to study GIMAP5 in mouse cells.
Figure 1.
Rat strain, cell line and subcellular distribution of rat GIMAP5. (A) Lymph node and thymus cell lysates from a part of rat strains were assayed for the expression of rat GIMAP5 by western blotting with the anti rat GIMAP5 monoclonal antibody MAC427. Anti-β-actin served as a loading control. (B) Lysates of the rat cell lines A181 (NK), C58 (thymoma), CRNK16 (NK), Mg2mono (myeloid-thymic adherent), Mg2über (thymoma), Nb2 (T lymphoma), RBL-1 (basophilic leukemia), Roser leukemia (T) and YB2/3.0Ag20 (hybrid plasmacytoma) were assayed for the expression of rat GIMAP5 by western blotting with the anti rat GIMAP5 monoclonal antibody. (C) Analysis of the distribution of GIMAP5 and calnexin in Nb2 subcellular fractions. Post-nuclear supernatants prepared from rat Nb2 cells were fractionated on a self-establishing Percoll gradient as described in the Materials and Methods section. The gradient was split into 24 fractions and an aliquot of each assayed for the presence of GIMAP5 and calnexin by western blotting using mouse monoclonal antibody MAC427 against rat GIMAP5 and rabbit anti-calnexin antiserum. Note that in this part, and in Figures 2B and 6B, fractions 1–12 and 13–24 were resolved on separate gels but were immunoblotted simultaneously. In all three parts of Figure 1 the mobility of co-electrophoresed protein molecular weight markers is indicated (in kDa).
Intracellular localization of mouse GIMAP5.
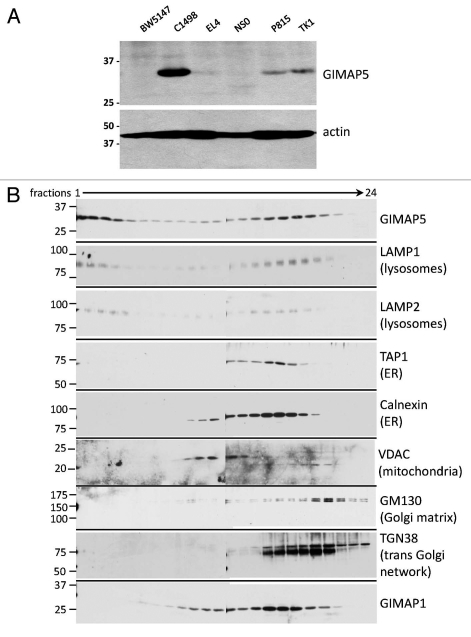
We had previously generated a monoclonal antibody specific for mouse GIMAP5, MAC421.12,21 Among a part of mouse lymphoid cell lines MAC421 was found to react strongly with the NKT cell line C1498,22 (Fig. 2A) at the predicted molecular weight for mGIMAP5. Weak positive reactions were also seen with TK1, P815 and EL4 lysates. On the basis of these results, C1498 cells were chosen for further study. Cultured cells were homogenised and post-nuclear fractions subjected to density separation as described for Nb2 cells. The resulting fractions were immunoblotted for GIMAP5 (using MAC421) and for a number of standard compartmental ‘marker’ proteins. Figure 2B shows that the distribution of GIMAP5 in the Percoll gradient was broad but noticeably similar to the distributions of the lysosomal marker proteins LAMP1 and LAMP2. Characteristically, as seen for the LAMP proteins, some of the mouse GIMAP5 signal was found in the densest fractions towards the bottom of the gradient (fractions 1–4), as had been seen earlier for rat GIMAP5 (Fig. 1C). The distributions of marker proteins for the endoplasmic reticulum (ER-calnexin), the Golgi (GM130 and TGN38) and mitochondria (VDAC) were unlike that of GIMAP5 and did not extend into the densest fractions.
Figure 2.
Cell-line and subcellular distribution of mouse GIMAP5. (A) Lysates of the mouse cell lines BW5147 (thymoma), C1498 (NKT), EL4 (T) NS0 (plasmacytoma), P815 (mastocytoma) and TK1 (T) were tested for expression of GIMAP5 by western blotting using an anti-mouse GIMAP5 monoclonal antibody, MAC421. (B) Analysis of the distribution of GIMAP5 in subcellular fractions from the mouse C1498 cells. Post-nuclear supernatants prepared from mouse C1498 cells were fractionated on a self-establishing Percoll gradient as described in the Materials and Methods section. The gradient was split into 24 fractions and an aliquot of each assayed for the presence of GIMAP5 and subcellular organelle marker proteins as indicated, using rat or rabbit antibodies as indicated in Table S2. In both parts of the figure the mobility of co-electrophoresed protein molecular weight markers is indicated (in kDa).
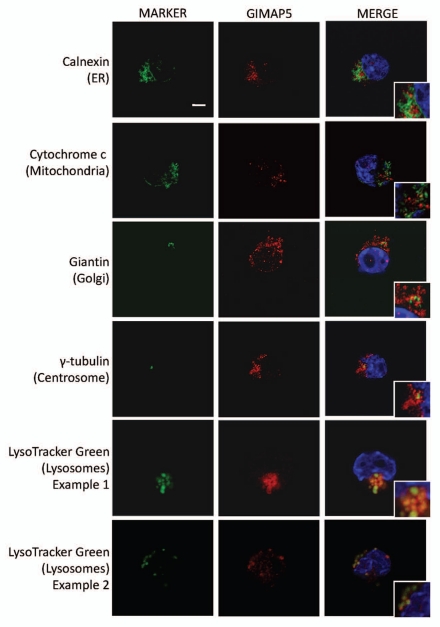
In order to obtain more direct information on the intracellular location(s) of mouse GIMAP5 we performed confocal microscopy using C1498 cells, again comparing anti-GIMAP5 mAb immunostaining with that obtained using marker reagents to identify particular organelles. Lymphoid cells and cell lines possess scanty amounts of cytoplasm. Nevertheless, the analysis gave clear results: GIMAP5 did not co-localise with markers of the ER, mitochondria, Golgi or centrosomes; it did, however, co-localise with a marker of the lysosomal compartment, the reagent Lysotracker Green (Fig. 3).
Figure 3.
Co-localization analysis in C1498 cells of mouse GIMAP5 with markers of different intracellular compartments. Representative images acquired by confocal microscopy of mouse C1498 cells stained as described in the Materials and Methods section. The intracellular compartments detected by the marker reagents (green) (see Table S2) are indicated to the left of the parts. Staining with anti-mGIMAP5 mAb MAC421 is in red. Cropped inserts display the key area of the parent image at ∼2-fold magnification. Nuclei (blue) were stained with DAPI. Scale bar = 5 µm.
Human GIMAP5.
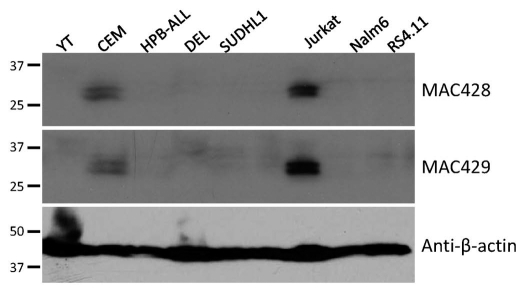
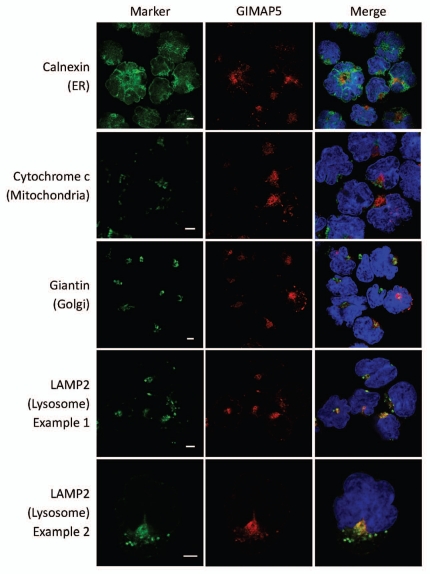
We raised novel mAbs (MAC428 and 429) against human GIMAP5. These gave positive reactions on immunoblots of human peripheral blood lymphocytes and NK cells (data not shown). In a western blot screen of human lymphoid cell lines they gave positive reactions on the human Jurkat and CEM T-ALL cell lines at the predicted relative molecular weight for huGIMAP5 (Fig. 4). Unlike mouse GIMAP5, human GIMAP5 generally resolved on immunoblots as a clear doublet, a feature noted previously by others.23 MAC428 and 429 behaved very similarly to each other and so were used interchangeably in subsequent experiments. Consistent with the findings using mouse C1498 cells, confocal microscopy using CEM cells revealed co-localization of human GIMAP5 immuno-staining with that for the lysosomal proteins LAMP2 but not with markers for the ER, Golgi and mitochondria (Fig. 5). Similar results were seen using Jurkat cells (results not shown).
Figure 4.
Endogenous expression of GIMAP5 by human cells and cell lines. (A) Immunoblotting analysis of a part of human lymphoid cell lines with two monoclonal antibodies, MAC428 and MAC429, against human GIMAP5. CEM, HPB-ALL, SUDHL1, Karpas299 and Jurkat cell lines are all of T lineage origin; Nalm6 and RS4.11 are leukemic pre-B cell lines; YT is an NK-like leukemic cell line and DEL is of myelomonocytic origin. Staining for β-actin was used as a loading control. The mobilities of co-electrophoresed marker proteins are indicated.
Figure 5.
Co-localization analysis in CEM cells of human GIMAP5 with markers of different intracellular compartments. Representative images acquired by confocal microscopy of the human CEM cell-line immunostained as described in the Materials and Methods section. Monoclonal antibody MAC428 was used to detect GIMAP5 (red). The intracellular compartments detected by the marker reagents (green) (see Table S2) are indicated to the left of the parts. Nuclei (blue) were stained with DAPI. Scale bars = 5 µm.
Faithful localization of an epitope-tagged human GIMAP5 in Jurkat T cells.
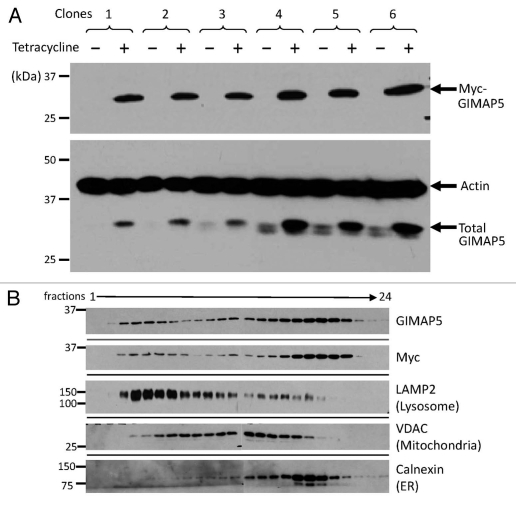
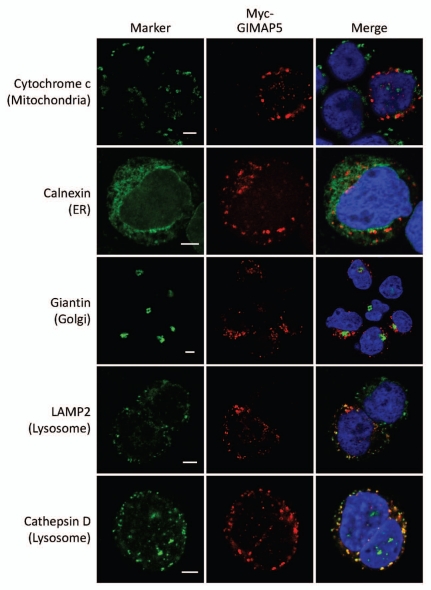
The high nuclear:cytoplasmic ratio of lymphoid cells imposes limitations on the information accessible using light microscopy. In order to facilitate analysis of GIMAP5 localization using electron microscopy (EM) we prepared a construct of human GIMAP5 carrying an amino-terminal myc epitope tag suitable for use in a standard anti-myc detection approach. A tetracycline-inducible form of the construct was transfected into TREx™ Jurkat T cells and stable clones were established. Figure 6A shows the induction of myc-huGIMAP5 by tetracycline in six of the clones. It also shows the parallel blotting with the anti-huGIMAP5 mAb MAC428 which not only reported the induction of expression of myc-huGIMAP5 but also revealed the endogenous levels of huGIMAP5 in the uninduced clones, which was variable. Percoll gradient fractionation of one of these lines showed that part of the myc and the huGI-MAP5 (MAC429) immunoreactivities was located in the heavier membrane fractions towards the bottom of the gradient, where LAMP2 was also detected (Fig. 6B), consistent with localization of some of the GIMAP5 in the lysosomal fraction. In agreement with this, confocal microscopy demonstrated co-localization of the myc immunoreactivity in tetracycline-induced cells with that for the lysosome-related markers cathepsin D and LAMP2 (Figs. 7 and S1).
Figure 6.
Subcellular distribution of human myc-tagged GIMAP5 overexpressed in the TREx™ Jurkat T lymphoid cell line. (A) Immunoblot analysis of human GIMAP5 expression in TREx™ Jurkat T cells carrying a myc-tagged human GIMAP5 cDNA construct under the control of a tetracycline-inducible cassette. Six stable clones (#1–6) of myc-huGIMAP5-TREx™ Jurkat T cells were generated as described in the Materials and Methods section. The upper part shows immunodetection of the myc epitope after 24 hours of culture in medium with (+) or without (™) 1 µg/ml tetracycline. The lower part shows MAC428 detection of total (endogenous + myc-tagged) huGIMAP5 in the same samples, as well as the β-actin loading controls. (B) Immunoblot analysis of fractions from Percoll density gradient sub-cellular fractionation of tetracycline-induced, myc-huGIMAP5-expressing TREx™ Jurkat cells. Aliquots of fractions from the gradient (fractions 1–24) were analysed by western blotting using antibodies/antisera to the indicated markers as described in the Materials and Methods section. Monoclonal antibody MAC429 was used to detect hGIMAP5. Other antibodies used are as detailed in Table S2.
Figure 7.
Co-localization of human GIMAP5 with markers of intracellular compartments in Myc-huGIMAP5-TREx™ Jurkat cells. Representative images of the sub-cellular localization of myc-huGIMAP5 analysed by confocal microscopy are shown. Myc-huGIMAP5-TREx™ Jurkat cells (Fig. 6) were cultured in 1 µg/ml tetracycline for 24 hours before being processed for immunostaining. Localization of the myc-epitope tag (red) was compared with the indicated compartmental markers (green—indicated to the left of the parts) using antibodies as detailed in Table S2. Nuclei are stained blue (DAPI). Note that not all cells responded to tetracycline induction and hence some cells lack ‘red’ staining. Scale bars = 5 µm. An enlarged, 3D version of the image of cells co-stained for myc and cathepsin D is shown in Supplemental Figure 1.
Detection of GIMAP5 by EM in inducible Jurkat T cells.
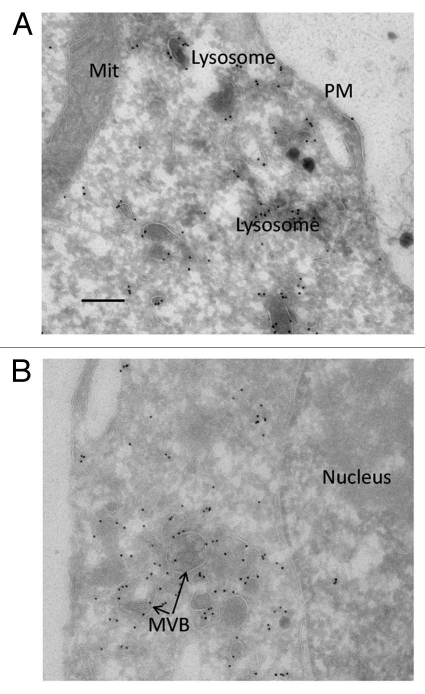
The data described in the previous section suggested that myc-tagged huGI-MAP5 was localising faithfully to the lysosomal compartment in these stable, inducible clones, in a manner consistent with the endogenous expression of the native form. Having established this, studies were initiated using EM and employing gold-labelled staphylococcal protein A as the secondary detection reagent. The results confirmed the association of myc-huGI-MAP5 with lysosomes. Interestingly, label was also evident on multivesicular bodies (MVB) and potentially on related vesicular structures (Fig. 8). However, our analyses did not allow us to determine whether GIMAP5 was localised to a precise membrane within the lysosomes/MVBs or whether the GTPase domain of the protein was localised on the cytoplasmic or luminal face of the vesicles.
Figure 8.
Electron micrographs of anti-myc labelling of myc-hGIMAP5 in TREx™ Jurkat cells. Myc-huGIMAP5-TREx™ Jurkat cells were cultured in 1 g/ml tetracycline for 48 hours and then processed for electron microscopy as described in the Materials and Methods section. (A) Field selected to illustrate gold-labelling of electron-dense membrane vesicles recognisable as lysosomes (or related late endocytic vesicles); this field also shows the absence of label associated with mitochondria (Mit) and the plasma membrane (PM). (B) Field selected to illustrate association of gold label with structures recognisable as multivesicular bodies (MVB); this field also shows the absence of label associated with the nucleus. Scale bar = 200 nm.
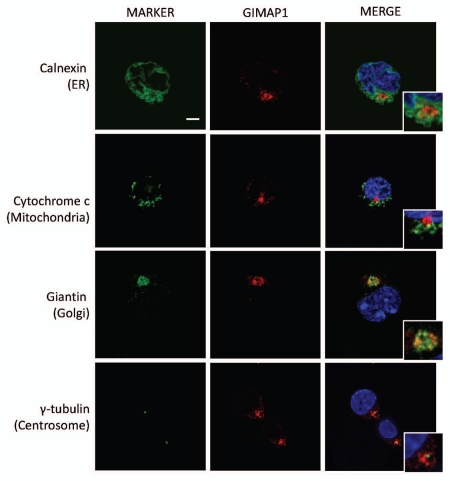
Intracellular localization of mouse GIMAP1.
GIMAP1 is a close relative of GIMAP5 with significant structural similarities, including being a ‘tail-anchored’ protein. In previous work we have shown that this protein, like GIMAP5, is expressed endogenously by C1498 cells.24 Confocal analysis of C1498 cells (Fig. 9) demonstrated, however, that mouse GIMAP1 was located in a site distinct from GIMAP5, namely the Golgi apparatus, defined by the protein giantin. As expected from these findings, GIMAP1 protein distributed differently from GIMAP5 in Percoll gradient analysis (Fig. 2B, bottom part) and behaved similarly to the marker proteins for the ER and the Golgi.
Figure 9.
Co-localization of mouse GIMAP1 with markers of intracellular compartments in C1498 cells. Representative images acquired by confocal microscopy of mouse C1498 cells immunostained as described in the Materials and Methods section. The intracellular compartments detected by the marker reagents (green) (Table S2) are indicated to the left of the parts. Staining with anti-mGIMAP1 monoclonal antibody MAC420 is in red. Cropped inserts display the key area of the parent image at ∼2-fold magnification. Nuclei (blue) were stained with DAPI. Scale bar = 5 µm.
Discussion
As one would expect, where they are known, the functions of ‘tail-anchored’ membrane proteins (of which GIMAP5 is an example) are found to be logically related to their intracellular location. Examples include the localization of Sec61β to the ER membrane where it is a component of the ER translocon25 and the Fzo1/Mfn proteins to mitochondria where they are believed to mediate outer mitochondrial membrane fusion.26 Accordingly, previous proposals for the mechanism of action of GIMAP5 in lymphocytes have been built upon suggestions for its localization in mitochondria or the ER.16,18,27 However, recent work by Keita and colleagues has put in question the claims for either of these sites as the normal intracellular location of GIMAP5.20 These authors used a polyclonal anti-peptide antibody raised against the predicted N-terminal 14 residues of rat GIMAP5 to investigate the behaviour of this protein in analyses involving the sedimentation and density separation of subcellular components of lymphoid cells. Their results showed that GIMAP5 distributed differently from marker proteins present in the ER or in mitochondria. Using a similar strategy, but following different protocols for cell disruption and sub-cellular fractionation and employing highly sensitive mAbs against rat, mouse and human GIMAP5, we obtained evidence in agreement with Keita et al. that GIMAP5 does not distribute like an ER or mitochondrial protein (Figs. 1C and 2B). In addition, we made the important observation that the distribution of mouse GIMAP5 was similar to that of the lysosomal LAMP proteins (Fig. 2B).
Although the correlation of GIMAP5 and LAMP1/2 distribution in Percoll fractionations was highly suggestive, a more direct approach was required in order to determine the intracellular location of GIMAP5. Confocal microscopy of immunostained mouse C1498 NKT cells (Fig. 3) demonstrated the co-localization of GIMAP5 and a marker of lysosomes (Lysotracker Green) and discounted significant co-localization with ER, Golgi and mitochondria. In addition, a lysosomal location for GIMAP5 was indicated in imaging studies of CEM and Jurkat T-ALL cell lines, where co-localization with the lysosomal marker LAMP2 was demonstrated (Fig. 5B).
Having established a lysosomal localization for GIMAP5 in lymphoid cells, we next sought to establish a system for the faithful expression of a tagged form of GIMAP5 in transfected cells. Firstly, to try to avoid mis-localization artifacts associated with the use of inappropriate cell hosts (e.g., of incompatible cell lineage) we chose Jurkat T cells as hosts, encouraged by the fact that this cell line can express GIMAP5 endogenously (Fig. 4). We established stable sublines of Jurkat cells that expressed myc-GIMAP5 when induced by addition of tetracycline to the culture medium (Fig. 6A). Analysis of these by both confocal microscopy (Fig. 7A) and Percoll gradient analysis (Fig. 6B) demonstrated lysosomal localization of the myc-tagged GIMAP5. Subsequent use of these cells for gold-labelling and electron microscopy confirmed the association of gold particles with electron-dense lysosomes (Fig. 8). Furthermore, this analysis also indicated the presence of GIMAP5 on additional vesicles, including the highly recognisable multivesicular bodies, which are also known to be LAMP protein-positive.28 Indeed it was noticeable in all of our Percoll fractionations that the full distributions of both GIMAP5 and the LAMP proteins were broad, including some lower density fractions: it is possible that the latter correspond to the pre-lysosomal structures seen by EM. Interestingly, in some (but not all) density centrifugation analyses, the relative quantitative distributions of GIMAP5 and LAMP were not identical (e.g., Fig. 6B). Without further information this observation is open to different interpretations but it may reflect different relative levels of the respective proteins in sub-compartments of the endo-lysosomal pathway.
What does a lysosomal localization for GIMAP5 suggest? In contrast with the classical view of the lysosomal compartment as the site of catabolic turnover for cellular and extracellular components, there is now substantial evidence for its involvement in other specialised functions. These include downregulation of cell signalling from, for example, receptor tyrosine kinases, exocytic secretion of lysosomal enzymes, provision of a releasable Ca2+ store and the regulated intracellular release of lysosomal hydrolytic enzymes.29–31 Relevant to the involvement of GIMAP5 in cell survival, the last of these has been linked to programmed cell death via a process termed lysosomal membrane permeabilization;30 however, several of the listed functions could plausibly play a role in the complex mechanisms known to underlie T lymphocyte survival.32,33
Significantly, our experiments also demonstrated the differential localization of GIMAP5 and GIMAP1, the latter protein being found predominantly in the Golgi apparatus (Fig. 9). GIMAP1, like GIMAP5, is a tail-anchored membrane protein with a predicted carboxy-terminal transmembrane domain. Of significance to the present discussion, recent work in one of our laboratories has demonstrated that GIMAP1, like GIMAP5, is required for the protection of lymphocytes against cell death.21 Together these data imply that GIMAP proteins are required in different organelles to provide protection against cell death. A mechanistic explanation of these phenomena will therefore need to encompass events in both (endo)-lysosomes and the Golgi apparatus.
The BB rat model of IDDM has long provided an important example of how lymphopenia can perturb the normal controls on autoimmunity. Investigations of the basis of the lymphopenia observed in the BB rat have identified the GIMAP5 GTPase as an important determinant of T lymphocyte survival but have yet to determine the molecular mechanisms involved. In the present study we have generated and used the first monoclonal antibodies against GIMAP5 to re-examine its intracellular location and to correct the assumption that it is a mitochondrial or ER protein.16,18,27 These findings provide a clearer focus for studies aimed at understanding the underpinning mechanisms.
Materials and Methods
Antibody reagents.
Rat and mouse monoclonal antibodies developed in-house against GIMAP proteins are detailed in Supplemental Table 1. Other antibody reagents used for all applications in this study are described in Supplemental Table 2.
Maintenance of cell lines.
Mouse, rat and human lymphoid cell lines were cultured in RPMI 1,640 medium (Invitrogen) with L-glutamine and supplemented with 10% (v/v) fetal calf serum (FCS), 100 U/ml penicillin and 100 µg/ml streptomycin. Parental tetracycline-inducible TREx™ Jurkat cells (Invitrogen) were maintained in RPMI 1,640 medium supplemented as above, with the addition of 100 µg/ml blasticidin S. All cells were cultured in a humidified incubator with 5% (v/v) CO2 atmosphere at 37°C.
Generation of tetracycline-inducible Jurkat cell lines expressing human GIMAP5.
An N-terminally myc-tagged variant of human GIMAP5 was cloned into plasmid pcDNA4/TO (Invitrogen). The resulting plasmid was linearized by digesting with restriction endonuclease Pvu1 and purified from an agarose gel before transfection into parental tetracycline-inducible TREx™ Jurkat cells (Invitrogen) by electroporation (5 µg linearized DNA per 107 cells). Cells were then cultured for 2 days before addition of selective medium containing zeocin. Positive cell lines containing myc-hGIMAP5-pcDNA4/TO were cloned by limiting dilution and maintained in supplemented RPMI 1640-blasticidin S medium as above, with the addition of 200 µg/ml zeocin. To induce expression of myc-hGIMAP5, cells were treated for up to 48 hours with tetracycline to a final concentration of 1 µg/ml.
Sub-cellular membrane fractionations.
These were performed on self-forming Percoll gradients.34 Cells (1–2 × 108) were homogenised in 1 ml Percoll Homogenization Buffer (10 mM acetic acid, 1 mM EDTA, 190 mM sucrose, 10 mM triethanolamine, pH 7.4 with 1:100 Mammalian Protease Inhibitor Cocktail) by 5–10 passes through a ball bearing homogenizer (Isobiotech) with 10 µm clearance. Unbroken cells and nuclei were removed by centrifugation at 1,000 g for 10 min at 4°C. The supernatant was diluted to 2–3 ml and then layered on 21 ml of 27% (v/v) Percoll (Sigma) in Percoll Homogenization Buffer in a 50 ml polycarbonate centrifuge tube (Beckman). A self-forming gradient was established by centrifugation at 48,000 g for 45 min at 4°C using a JA25.50 fixed-angle rotor (Beckman). Fractions of approximately 1 ml were collected from the gradient using a peristaltic pump, starting from the bottom of the gradient. An aliquot of each sample was diluted 1:1 with Laemmli loading buffer containing 50 mM dithiothreitol (DTT) and was analysed by western blotting which was performed as described elsewhere.24
Confocal microscopy.
Cells were suspended in PBS containing 10% FCS at a concentration of 20 × 104 cells/ml. Aliquots of 3 × 104 cells were spun onto glass slides coated with poly-L-lysine (Sigma) in a Shandon Cytospin 2 at 500 rpm (77 g) for 5 min. The cells were washed with PBS, fixed in 4% (w/v) paraformaldehyde in PBS for 15 min at room temperature, washed twice more with PBS and then permeabilized with 0.1% (v/v) Triton X100 in PBS for 15 min at room temperature. After washing with PBS, cells were incubated in DMEM-10% (v/v) FCS for 1 hr at room temperature to block non-specific sites. Primary antibodies were diluted in DMEM-10% (v/v) FCS and then incubated on the slides for 2 h. The cells were washed with PBS three times and secondary, fluorophore-conjugated antibodies were applied in DMEM-10% FCS for 1 h at room temperature. Cells were washed with PBS and were mounted in VectaShield containing 4′,6-diamidino-2-phenylindole (DAPI; Vector Laboratories). Cells were viewed using a Zeiss LSM 510 Meta point scanning confocal microscope and a Plan-Apochromat 63x/1.4 oil DIC objective. Cells to be stained with Lysotracker Green (Invitrogen) were spun onto the microscope slides as detailed and were then incubated with Lysotracker Green diluted 1 in 500 in DMEM-10% FCS for 1 h at 37°C. The cells were then fixed, permeabilized and stained with antibodies as described above.
Immunoelectron microscopy.
TRex™Jurkat T cells, carrying a plasmid directing tetracycline-inducible myc-hGIMAP5 expression, were induced to express the protein by treatment with 1 µg/ml tetracycline for 48 h. The cells were washed with PBS, fixed with 4% paraformaldehyde/0.1% glutaraldehyde in 0.1 M sodium cacodylate buffer and pelleted in an eppendorf tube (15,000 g for 5 min). The fixative was aspirated and the cell pellet was re-suspended in warm 10% gelatin. The cells were then pelleted (15,000 g for 5 min) and the gelatin-enrobed pellet was set on ice. The pellet was trimmed into 1 mm3 blocks and infused with 1.7 M sucrose/15% polyvinylpyrrolidone for 24 h at 4°C. The blocks were subsequently mounted on cryostubs and snap-frozen in liquid nitrogen. Frozen ultrathin sections were cut using a diamond knife in a cryochamber attachment (Leica, Milton Keynes, UK), collected from the knife-edge with 50:50 2% methyl cellulose: 2.3 M sucrose35 and mounted on formvar-carbon coated EM grids. Immunolabelling was performed using the protein A-gold technique at room temperature.36 Sections were incubated with 50 mM NH4Cl in PBS for 10 min to quench unreacted aldehydes, transferred to 2% gelatin in PBS for 10 min and then 1% BSA in PBS for 10 min. Sections were incubated for 30 min with 5 µl of mouse anti-myc mAb (clone 4A6, Upstate, Temecula, USA) diluted 1:50 in PBS containing 5% FCS and 0.1% BSA, washed with PBS/0.1% BSA (6 × 3 min) and incubated for 30 min with rabbit anti-mouse IgG (Z0412, DAKO, Ely, UK) diluted 1:50 in PBS containing 5% FCS and 0.1% BSA. The sections were then washed with PBS/0.1% BSA (6 × 3 min) and incubated for 30 min with PBS/0.1% BSA containing protein A conjugated to 15 nm colloidal gold (purchased from the Department of Cell Biology, University of Utrecht, The Netherlands). The sections were washed with PBS/0.1% BSA (2 × 5 min), PBS (4 × 5 min), distilled water (5 × 3 min) and contrasted by embedding in 1.8% methyl cellulose/0.3% uranyl acetate. Sections were allowed to air dry prior to observation in a Philips CM100 transmission electron microscope at an operating voltage of 80 kV.
Acknowledgements
This work was supported by funding to the Babraham Institute from BBSRC UK and Ph.D. studentship funding from BBSRC UK (to A.S.) and MRC UK (to V.W.). N.A.B. was supported by the MRC. We thank Gillian Griffiths, Jane Stinchcombe, Paul Luzio, Simon Cook, Suzanne Turner, Nick Holmes and Hugh Reyburn for advice and/or the provision of antibodies or cell lines. We thank Margaret Graham, Martyn Cooke and Fiona Kemp for technical assistance.
Footnotes
Previously published online: www.landesbioscience.com/journals/selfnonself/article/12819
Supplementary Material
References
- 1.Jackson R, Rassi N, Crump T, Haynes B, Eisenbarth GS. The BB diabetic rat. Profound T-cell lymphocytopenia. Diabetes. 1981;30:887–889. doi: 10.2337/diab.30.10.887. [DOI] [PubMed] [Google Scholar]
- 2.Elder ME, Maclaren NK. Identification of profound peripheral T lymphocyte immunodeficiencies in the spontaneously diabetic BB rat. J Immunol. 1983;130:1723–1731. [PubMed] [Google Scholar]
- 3.Guttmann RD, Colle E, Michel F, Seemayer T. Spontaneous diabetes mellitus syndrome in the rat. II. T lymphopenia and its association with clinical disease and pancreatic lymphocytic infiltration. J Immunol. 1983;130:1732–1735. [PubMed] [Google Scholar]
- 4.Ramanathan S, Poussier P. BB rat lyp mutation and Type 1 diabetes. Immunol Rev. 2001;184:161–171. doi: 10.1034/j.1600-065x.2001.1840115.x. [DOI] [PubMed] [Google Scholar]
- 5.Cousins L, Graham M, Tooze R, Carter C, Miller JR, Powrie FM, et al. Eosinophilic bowel disease controlled by the BB rat-derived lymphopenia/Gimap5 gene. Gastroenterology. 2006;131:1475–1485. doi: 10.1053/j.gastro.2006.09.023. [DOI] [PubMed] [Google Scholar]
- 6.Poussier P, Ning T, Murphy T, Dabrowski D, Ramanathan S. Impaired post-thymic development of regulatory CD4+ 25+ T cells contributes to diabetes pathogenesis in BB rats. J Immunol. 2005;174:4081–4089. doi: 10.4049/jimmunol.174.7.4081. [DOI] [PubMed] [Google Scholar]
- 7.Hornum L, Romer J, Markholst H. The diabetes-prone BB rat carries a frameshift mutation in Ian4, a positional candidate of Iddm1. Diabetes. 2002;51:1972–1979. doi: 10.2337/diabetes.51.6.1972. [DOI] [PubMed] [Google Scholar]
- 8.MacMurray AJ, Moralejo DH, Kwitek AE, Rutledge EA, Van*Yserloo B, Gohlke P, et al. Lymphopenia in the BB rat model of type 1 diabetes is due to a mutation in a novel immune-associated nucleotide (Ian)-related gene. Genome Res. 2002;12:1029–1039. doi: 10.1101/gr.412702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nitta T, Nasreen M, Seike T, Goji A, Ohigashi I, Miyazaki T, et al. IAN family critically regulates survival and development of T lymphocytes. PLoS Biol. 2006;4:103. doi: 10.1371/journal.pbio.0040103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sandal T, Aumo L, Hedin L, Gjertsen BT, Doskeland SO. Irod/Ian5: an inhibitor of gamma-radiation- and okadaic acid-induced apoptosis. Mol Biol Cell. 2003;14:3292–3304. doi: 10.1091/mbc.E02-10-0700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schulteis RD, Chu H, Dai X, Chen Y, Edwards B, Haribhai D, et al. Impaired survival of peripheral T cells, disrupted NK/NKT cell development and liver failure in mice lacking gimap5. Blood. 2008 doi: 10.1182/blood-2008-03-146555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barnes MJ, Aksoylar H, Krebs P, Bourdeau T, Arnold CN, Xia Y, et al. Loss of T cell and B cell quiescence precedes the onset of microbial flora-dependent wasting disease and intestinal inflammation in Gimap5-deficient mice. J Immunol. 2010;184:3743–3754. doi: 10.4049/jimmunol.0903164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hellquist A, Zucchelli M, Kivinen K, Saarialho-Kere U, Koskenmies S, Widen E, et al. The human GIMAP5 gene has a common polyadenylation polymorphism increasing risk to systemic lupus erythematosus. J Med Genet. 2007;44:314–321. doi: 10.1136/jmg.2006.046185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shin JH, Janer M, McNeney B, Blay S, Deutsch K, Sanjeevi CB, et al. IA-2 autoantibodies in incident type I diabetes patients are associated with a polyadenylation signal polymorphism in GIMAP5. Genes Immun. 2007;8:503–512. doi: 10.1038/sj.gene.6364413. [DOI] [PubMed] [Google Scholar]
- 15.Borgese N, Colombo S, Pedrazzini E. The tale of tail-anchored proteins: coming from the cytosol and looking for a membrane. J Cell Biol. 2003;161:1013–1019. doi: 10.1083/jcb.200303069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pandarpurkar M, Wilson-Fritch L, Corvera S, Markholst H, Hornum L, Greiner DL, et al. Ian4 is required for mitochondrial integrity and T cell survival. Proc Natl Acad Sci USA. 2003;100:10382–10387. doi: 10.1073/pnas.1832170100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zenz T, Roessner A, Thomas A, Frohling S, Dohner H, Calabretta B, et al. hIan5: the human ortholog to the rat Ian4/Iddm1/lyp is a new member of the Ian family that is overexpressed in B-cell lymphoid malignancies. Genes Immun. 2004;5:109–116. doi: 10.1038/sj.gene.6364044. [DOI] [PubMed] [Google Scholar]
- 18.Dalberg U, Markholst H, Hornum L. Both Gimap5 and the diabetogenic BBDP allele of Gimap5 induce apoptosis in T cells. Int Immunol. 2007;19:447–453. doi: 10.1093/intimm/dxm009. [DOI] [PubMed] [Google Scholar]
- 19.Joseph S, Diamond AG, Smith W, Baird JD, Butcher GW. BB-DR/Edinburgh: a lymphopenic, non-diabetic subline of BB rats. Immunology. 1993;78:318–328. [PMC free article] [PubMed] [Google Scholar]
- 20.Keita M, Leblanc C, Andrews D, Ramanathan S. GIMAP5 regulates mitochondrial integrity from a distinct subcellular compartment. Biochem Biophys Res Commun. 2007;361:481–486. doi: 10.1016/j.bbrc.2007.07.048. [DOI] [PubMed] [Google Scholar]
- 21.Saunders A, Webb LM, Janas ML, Hutchings A, Pascall J, Carter C, et al. Putative GTPase GIMAP1 is critical for the development of mature B and T lymphocytes. Blood. 2010;115:3249–3257. doi: 10.1182/blood-2009-08-237586. [DOI] [PubMed] [Google Scholar]
- 22.LaBelle JL, Truitt RL. Characterization of a murine NKT cell tumor previously described as an acute myelogenous leukemia. Leuk Lymphoma. 2002;43:1637–1644. doi: 10.1080/1042819021000002974. [DOI] [PubMed] [Google Scholar]
- 23.Chadwick N, Zeef L, Portillo V, Fennessy C, Warrander F, Hoyle S, et al. Identification of novel Notch target genes in T cell leukaemia. Mol Cancer. 2009;8:35. doi: 10.1186/1476-4598-8-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Saunders A, Lamb T, Pascall J, Hutchings A, Dion C, Carter C, et al. Expression of GIMAP1, a GTPase of the immunity-associated protein family, is not upregulated in malaria. Malaria Journal. 2009;8:53. doi: 10.1186/1475-2875-8-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hartmann E, Sommer T, Prehn S, Gorlich D, Jentsch S, Rapoport TA. Evolutionary conservation of components of the protein translocation complex. Nature. 1994;367:654–657. doi: 10.1038/367654a0. [DOI] [PubMed] [Google Scholar]
- 26.Hoppins S, Lackner L, Nunnari J. The machines that divide and fuse mitochondria. Annu Rev Biochem. 2007;76:751–780. doi: 10.1146/annurev.biochem.76.071905.090048. [DOI] [PubMed] [Google Scholar]
- 27.Pino SC, O'Sullivan-Murphy B, Lidstone EA, Yang C, Lipson KL, Jurczyk A, et al. CHOP mediates endoplasmic reticulum stress-induced apoptosis in Gimap5-deficient T cells. PLoS ONE. 2009;4:5468. doi: 10.1371/journal.pone.0005468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Futter CE, Pearse A, Hewlett LJ, Hopkins CR. Multivesicular endosomes containing internalized EGF-EGF receptor complexes mature and then fuse directly with lysosomes. J Cell Biol. 1996;132:1011–1023. doi: 10.1083/jcb.132.6.1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Luzio JP, Pryor PR, Bright NA. Lysosomes: fusion and function. Nat Rev Mol Cell Biol. 2007;8:622–632. doi: 10.1038/nrm2217. [DOI] [PubMed] [Google Scholar]
- 30.Kirkegaard T, Jaattela M. Lysosomal involvement in cell death and cancer. Biochim Biophys Acta. 2009;1793:746–754. doi: 10.1016/j.bbamcr.2008.09.008. [DOI] [PubMed] [Google Scholar]
- 31.Calcraft PJ, Ruas M, Pan Z, Cheng X, Arredouani A, Hao X, et al. NAADP mobilizes calcium from acidic organelles through two-pore channels. Nature. 2009;459:596–600. doi: 10.1038/nature08030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marrack P, Kappler J. Control of T cell viability. Annu Rev Immunol. 2004;22:765–787. doi: 10.1146/annurev.immunol.22.012703.104554. [DOI] [PubMed] [Google Scholar]
- 33.Guimond M, Veenstra RG, Grindler DJ, Zhang H, Cui Y, Murphy RD, et al. Interleukin 7 signaling in dendritic cells regulates the homeostatic proliferation and niche size of CD4+ T cells. Nat Immunol. 2009;10:149–157. doi: 10.1038/ni.1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Casey TM, Meade JL, Hewitt EW. Organelle proteomics: identification of the exocytic machinery associated with the natural killer cell secretory lysosome. Mol Cell Proteomics. 2007;6:767–780. doi: 10.1074/mcp.M600365-MCP200. [DOI] [PubMed] [Google Scholar]
- 35.Liou W, Geuze HJ, Slot JW. Improving structural integrity of cryosections for immunogold labeling. Histochem Cell Biol. 1996;106:41–58. doi: 10.1007/BF02473201. [DOI] [PubMed] [Google Scholar]
- 36.Griffiths GE. Fine Structure Immunocytochemistry. Berlin: Springer-Verlag; 1993. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.