Abstract
The Ku70/80 heterodimer is central to non-homologous end joining repair of DNA double-strand breaks and the Ku80 gene appears to be essential for human but not rodent cell survival. The Ku70/80 heterodimer is located at telomeres but its precise function in telomere maintenance is not known. In order to examine the role of Ku80 beyond DNA repair in more detail, we have taken a knockdown approach using a human fibroblast strain. Following targeted Ku80 knockdown, telomere defects are observed and the steady state levels of the TRF2 protein are reduced. Inhibitor studies indicate that this loss of TRF2 is mediated by the proteasome and degradation of TRF2 following Ku depletion appears to involve a decrease in chromatin binding of TRF2, suggesting that the Ku heterodimer enhances TRF2 chromatin association and that non-chromatin bound TRF2 is targeted to the proteasome.
Key words: Ku80, TRF2, chromatin, telomere, fibroblast
Introduction
The Ku 70/80 heterodimer, which is comprised of the Ku70 and 80 subunits, contributes to genomic stability by binding to DNA ends that result from double stranded DNA breaks regardless of structure.1,2 It is believed that this binding represents a key, initiating step in the non-homologous end joining (NHEJ) pathway for DNA repair. In addition, the binding of the Ku heterodimer to a DNA double-strand break serves to both recruit DNA-dependent protein kinase catalytic subunit and activate its kinase activity, initiating DNA damage responses and cell cycle arrest.3–8
In addition to its role in DNA repair, multiple lines of evidence indicate that the Ku heterodimer functions at the telomere. The Ku heterodimer binds to linear telomeric repeat DNA with similar affinity as to unique sequences.9 Ku has been localized to telomeres10 and interacts with telomere repeat-binding factors 1 and 2 (TRF1 and TRF2).11,12 In yeast, mutants in either Ku subunit show a loss of telomere length and telomere silencing, as well as deregulation of the telomere single-strand overhang.13 Other factors, including the Werner syndrome helicase, have been shown to function in regulation of the telomeric overhang.14 Collectively, the yeast data suggests that the Ku heterodimer plays multiple roles at the telomere, regulating access of telomerase, establishing positional effects in subtelomeric regions and establishing peripheral localization of telomeres in the nucleus.15
Evidence from a variety of targeting studies using differing approaches suggests that the function of Ku in telomere maintenance differs in mammalian species. In mice, disruption of the Ku80 gene leads to growth retardation, immunodeficiency and premature aging,16,17 in addition to chromosome abnormalities and telomere dysfunction with telomere elongation.18,19 In mouse cells, Ku depletion leads to telomere elongation,19,20 and Ku seems to mediate the fusion of critically short telomeres, since depletion of Ku prevents fusion in response to uncapping,19,21 but the gene is not essential for growth and survival. In contrast, Ku80 seems to be an essential gene in human cells.22 Despite its central role in NHEJ, reducing the levels of Ku80 in human tumor cell lines leads to telomere shortening, telomere fusion and apoptosis as the primary phenotype,23–25 although certain immortal cell lines may be less sensitive to Ku depletion.26 It has been reported that telomere dysfunction in immortal human cells resulting from complete Ku depletion is accompanied by telomere uncapping and the appearance of telomeric circles.27 These results suggest that the role of Ku in normal telomere maintenance may be one aspect of the critical function that makes Ku essential for the survival of human somatic cells. One important aspect of the cells examined to date in the study of the specific role that Ku plays at the telomere is that these cells have some dysfunction in their cell cycle checkpoint control due to their tumor origin and immortal status in culture. The study of Ku function at the telomere in a “normal” human somatic cell may give additional insight into the specific function of the Ku heterodimer in telomere maintenance.
In order to better understand the functional roles of Ku beyond NHEJ, we have examined the consequences of targeting Ku80 in non-immortal human cells. We find that Ku targeting, in the absence of overt DNA damage, leads to telomere dysfunction involving a loss of TRF2. This TRF2 loss appears to involve a release of the protein from chromatin, suggesting a link between Ku levels and TRF2 stability on telomeres.
Results
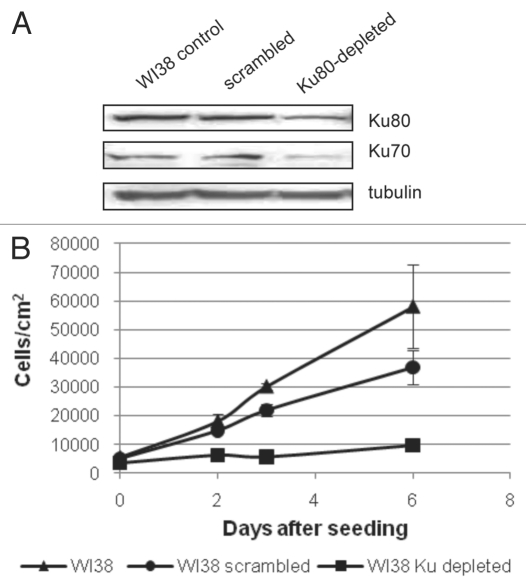
In order to examine the consequences of reducing Ku levels in normal human fibroblasts, an shRNA vector targeting the Ku sequence was tested for its effectiveness in reducing the levels of the Ku80 protein. This targeting vector proved to be effective and, using a lentiviral delivery system that delivers the construct to greater than 90% of the cell population, we were able to achieve a very robust knockdown of Ku80 in WI38 human fibroblasts by 3–4 days (Fig. 1A). Densitometric analysis indicates that the relative reduction in Ku80 levels varied between 40–60% in multiple experiments. Concomitant with the reduction in Ku80, we observed a reduction in Ku70 compared to control cells (Fig. 1A). A similar loss of Ku70 has been reported in mice following disruption of the Ku80 gene.16 We examined growth of Ku80-depleted cells over six days and find that the Ku-depleted cells display a significant growth arrest compared with both control fibroblasts and fibroblasts infected with a scrambled shRNA construct (Fig. 1B). Cell cycle distribution of Ku80-depleted WI38 fibroblasts was analyzed and this analysis indicated that cells accumulated in the G2/M phase of the cell cycle compared with control fibroblasts (Table 1) while the percentage of cells in S phase is reduced following Ku80 depletion, consistent with the dramatic reduction in growth rate that we observed (Table 1 and Fig. 1B).
Figure 1.
Knockdown of Ku proteins in WI38 human fibroblast cells leads to growth arrest. WI38 fibroblast cells were infected with a shRNA vector directed against the Ku80 protein. 24 hours following infection, cultures were placed into puromycin-containing selection medium for 72 hours. (A) Total protein was collected and a representative western blot for Ku80 and Ku70 is shown. Tubulin levels are shown as a loading control. Similar reductions in Ku levels were obtained over multiple trials. (B) WI38 cells were seeded and counted at Days 1, 2, 3 and 6 on a Guava EasyCyte Mini flow cytometer to determine growth kinetics compared to non-infected cells. Each cell type and timepoint was measured in triplicate and standard deviation was measured and is shown.
Table 1.
Cell cycle distribution of WI38 fibroblasts following Ku80 depletion
| 24 hours after infection | Control | Ku-depleted |
| G1 | 61.1% | 61.4% |
| S | 15.1% | 5.7% |
| G2/M | 23.8% | 32.9% |
| 48 hours after infection | Control | Ku-depleted |
| G1 | 64.5% | 58.6% |
| S | 13.7% | 8.5% |
| G2/M | 21.8% | 32.9% |
WI38 cells were infected with Ku80 shRNA for 24 hours then collected for cell cycle analysis either another 24 or 48 hours after infection was complete. Cells were stained with the Guava Cell Cycle reagent and analyzed on the Guava EasyCyte Mini flow cytometer for DNA content. The data are presented as the percentage of cells within either G1, S, or G2/M phases of the cell cycle, with cell debris or apoptotic cells not included.
An examination of the cell population following DAPI staining revealed a large number of cells with double nuclei and incomplete karyokinesis following Ku80 depletion (Fig. 2A). Because telomere fusion events can lead to non-disjunction and Ku is known to play a role in telomere maintenance, we examined metaphase spreads of Ku80-depleted cells for evidence of telomere dysfunction using a fluorescence-labeled telomere probe. Several types of telomere aberrations were observed in the Ku80-depleted cells. We observed apparent sister chromatid fusion events, ring chromosomes and extra telomere signals, presumably resulting from fusion-breakage events (Fig. 2B and C). These abnormalities were present to a much lesser extent in control chromosomes. Because it has been reported that a 50% reduction in Ku levels leads to a decrease in telomere length in immortalized cell lines, we examined the mean telomere length in Ku depleted cells.25 We find that telomere length is not affected by Ku depletion in WI38 normal human fibroblasts (Fig. 2D).
Figure 2.
Loss of Ku80 in human fibroblasts leads to nondisjunction and telomere abnormalities. (A) Nuclear morphology of control and Ku80-depleted WI38 cells. WI38 fibroblasts were infected with Ku80 shRNA or a scrambled control shRNA for 24 hours and selected in puromycin for 72 hours. Seven days after the treatment with shRNA, cells were stained with DAPI. Images were captured at 20X magnification (first 3 parts) or 40X magnification (last part). Non-infected WI38 fibroblasts were included as an additional control. (B and C) Representative chromosomes from telomere FISH analysis of metaphase spreads from control (B) and Ku80-depleted (C) cultures seven days after infection. Images were captured at 100X magnification and enlarged for clarity. Control indicates non-transfected WI38 cells. The percentage of cells containing telomere abnormalities for each treatment is as follows: non-treated controls, 17% (n = 35); scrambled controls, 10% (n = 30); Ku80-depleted, 62.5% (n = 24). Fisher's exact test was performed to determine significance and the differences between Ku80-depleted and both scrambled and untreated controls were significant (p = 0.004 and p = 0.015, respectively). (D) Total genomic DNA from control (1) and Ku80-depleted (2) fibroblasts was subjected to telomere restriction fragment (TRF) analysis seven days after infection as described in Materials and Methods. Representative Southern blot is shown. Molecular weight standards are provided on the side of the blot.
Based on the presence of aberrant telomeres in the metaphase spreads of Ku80-depleted cells and the known interaction between Ku70 and TRF2, an important telomere-binding protein that binds directly to telomere repeats,12 we postulated that Ku may function to regulate the association of proteins with the telomere. In order to gain some insight into the relationship between Ku and telomere-associated proteins, we measured the levels of a subset of known telomere-interacting proteins in cell extracts following Ku80 depletion. Surprisingly, Ku80 depletion leads to a reduction in the level of TRF2 (Fig. 3). However this does not represent a global loss of telomere-binding proteins. In fact, Ku80-depleted cells display an increase in the level of TRF1 compared with control cells (Fig. 3), while POT1 does not seem to vary significantly (data not shown).
Figure 3.
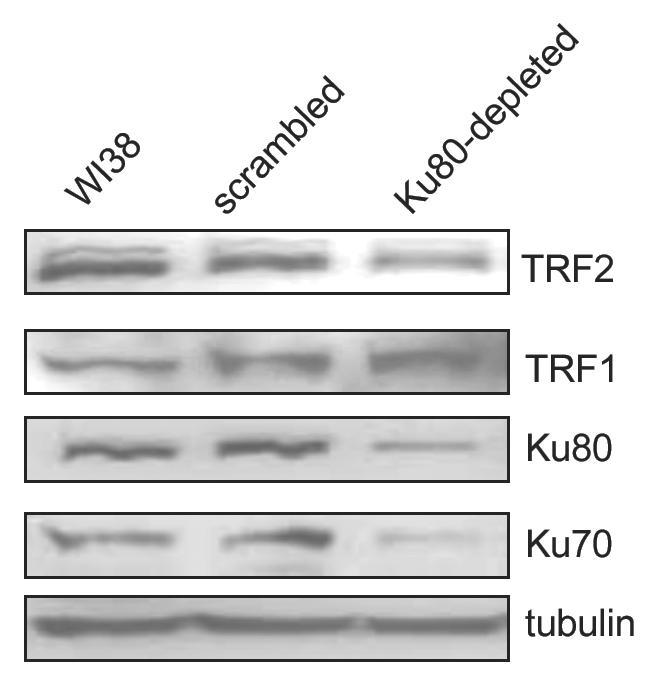
TRF1 and TRF2 levels are altered in Ku depleted cells. WI38 fibroblasts were infected for 24 hours with Ku80 shRNA or a scrambled shRNA construct and selected in puromycin for 72 hours. Non-infected WI38 fibroblasts were included as an additional control. Total protein extracts were then collected, and representative western blots are shown. Protein extracts were examined for TRF2 and TRF1. Results from three independent experiments indicate a 42% decrease in TRF2 and a 1.2-fold increase in TRF1 following Ku80 depletion. Ku80 and Ku70 levels are shown to confirm the knockdown, and actin levels are included as a loading control.
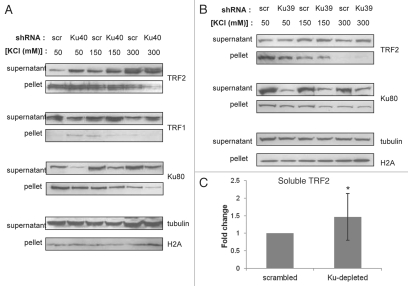
Because TRF2 has been shown to exist in chromatin-bound form,28 one possible explanation for the increased degradation of TRF2 following Ku80 knockdown is increased release from chromatin. In order to test this possibility, we performed chromatin extraction of proteins at increasing salt concentrations. TRF2 showed greater solubility in Ku80-depleted WI38 cells than in scrambled shRNA-treated counterparts at all salt concentrations tested for both Ku80 shRNA constructs (Fig. 4A and B). Because there was variability in the relative degree of TRF2 extraction, quantitation was performed on the relative difference in soluble TRF2 between scrambled and Ku80-depleted extracts over multiple salt concentrations. This analysis indicated a significant difference in soluble TRF2 levels (Fig. 4C). We did not see any difference in chromatin-bound TRF1 between Ku-depleted cells and control cells. These data suggest that depletion of Ku80 from WI38 fibroblasts contributes to the release of TRF2 from chromatin. To confirm our results suggesting that Ku80 reduction leads to an increase in non-chromatin-bound TRF2, we performed chromatin immunoprecipitation (ChIP) analysis. Ku80-depleted WI38 fibroblasts were subjected to ChIP analysis using antibodies targeting TRF1 and TRF2. Consistent with our results suggesting a reduction in chromatin-bound TRF2 following Ku depletion, we found that TRF2 in Ku80-depleted cells pulled down less telomeric DNA than TRF2 in control cells (Fig. 5). Also consistent with our previous results, telomere-bound TRF1 was slightly increased in Ku80-depleted fibroblasts compared with controls (Fig. 5). These data support the interpretation that Ku80 reduction in WI38 fibroblasts leads to a loss of chromatin-bound TRF2 and the ChIP analysis confirmed that the loss of TRF2 from chromatin at least partly represents a loss of telomere-bound TRF2.
Figure 4.
Chromatin-bound TRF2 decreases following Ku80 depletion in WI38 fibroblasts. (A) WI38 cells were infected with Ku80 (shKu40) or scrambled (scr) shRNA constructs, and chromatin extraction of proteins was performed using increasing concentrations of KCl. TRF2, TRF1 and Ku80 levels are shown for both supernatant (whole cell soluble) and pellet (chromatin-bound) fractions. (B) Chromatin extraction results for a second Ku80 shRNA construct (shKu39) are shown. Tubulin and H2A are shown as loading controls for the supernatant and pellet, respectively. Representative western blots are presented. (C) Densitometric analysis of soluble TRF2 in Ku80-depleted extracts compared with scrambled controls. Student's t-test was performed to determine significance, and the difference between Ku80-depleted and scrambled controls was significant (p = 0.03).
Figure 5.
Telomeric association of TRF2 is decreased following Ku80 depletion. Scrambled and Ku80-depleted WI38 fibroblasts (shKu-40) were produced as previously described, and telomere ChIP analysis was performed as described in Materials and Methods. Telomeric DNA bound by TRF1 and TRF2 is indicated, and rabbit IgG is included as a negative control. In addition, 0.5 micrograms of human and mouse genomic DNA was included as a positive control for the efficiency of the telomere detection system.
Given the striking reduction in TRF2 levels in Ku80-depleted cells, we sought to further understand the mechanisms involved in TRF2 regulation. We postulated that interaction with the chromatin may serve to stabilize the TRF2 protein and prevent the targeting of TRF2 to the proteasome. In order to test this possibility, Ku-depleted and control cultures were exposed to a proteasome inhibitor and TRF2 levels were examined. The addition of the proteasome inhibitor MG115 either during the first 24 hours or second 24 hours following Ku80 knockdown (0–24 hours or 24–48 hours) prevented the reduction in TRF2 in the Ku80-depleted cells (Fig. 6A). This result suggests that TRF2 is targeted to the proteasome following Ku80 knockdown. Interestingly, Ku70 and Ku80 levels under these same treatment conditions do not increase, suggesting that the majority of Ku is not targeted to the proteasome (Fig. 6B). Interestingly, higher molecular weight forms of Ku80 were visible in the MG115-treated extract, consistent with recent reports that Ku may be ubiquitin-conjugated to facilitate release of the heterodimer during DNA repair.29 Proteasome targeting is accomplished through ubiquitin conjugation and thus ubiquitin-modified TRF2 intermediates should be present in cells following Ku80 depletion. In order to visualize potential ubiquitin-conjugated forms of TRF2 we performed immunoprecipitation of the TRF2 protein and probed for higher molecular weight forms of TRF2 that interacted with ubiquitin. Ku-depleted WI38 cells contained more TRF2-ubiquitin than control cultures and these complexes were further increased in the presence of MG115 (Fig. 6C) suggesting that these ubiquitin-conjugated forms of TRF2 are cleared by the proteasome.
Figure 6.
Proteasome inhibition prevents loss of TRF2 following Ku80 depletion. WI38 cells were infected with either a control shRNA or a shRNA vector against Ku80 for 24 hours, after which they were treated with the proteasome inhibitor MG115 right away for 24 hours (0–24 hours) or after an additional 24 hours for 24 hours (24–48 hours). Protein extracts were then collected and analyzed for TRF2 (A) as well as Ku70 and Ku80 (B). Asterisks (*) indicate higher molecular weight forms of Ku80. (B) Represents extracts from the 24–48 hour time point. (C) MG115 treatment was performed as for A and B, and extracts were immunoprecipitated with anti-TRF2. TRF2-containing complexes were subjected to SDS-PAGE and western blotting using anti-ubiquitin or anti-TRF2 (24–48 hour time point). Actin levels are included as Input. **Heavy and light chains of antibody. EV, empty vector; scr, scrambled shRNA; MW, molecular weight.
Discussion
The role of the Ku heterodimer in telomere maintenance in human cells has been somewhat unclear. The response to heterozygous gene deletions in the Ku80 or Ku70 genes differed in a study comparing human adenocarcinoma and pre-B leukemia cells.30 The adenocarcinoma cells showed a significant growth defect while the pre-B cell lines were minimally affected in terms of growth. Targeting of Ku80 through small interfering RNAs resulted in loss of viability and apoptosis in HeLa cells,23 but despite the difference in growth rates and viability, in all cases a telomere defect was observed. The differences in growth responses and in specifics of the telomere defects in these various tumor cell lines could well be due to the specific genetic alterations acquired during the immortalization process that each of these lines has undergone. Accordingly, an examination of the potential role of the Ku heterodimer at the telomere in a cell that has not undergone the immortalization process should give additional insight because the cells will not continue to cycle but should arrest at the first defect.
As expected, targeting of the Ku80 mRNA in WI38 fibroblasts leads to a significant growth inhibition and the appearance of senescent cells in the population as measured by both morphology and an increase in senescence-associated beta-galactosidase activity (Fig. S1). These results are in agreement with the report that reduced Ku80 enhances senescence induced by high salt.31 Concomitant with growth arrest, we have observed telomere abnormalities and a reduction in steady state levels of the telomere-binding protein TRF2. Additionally, the data suggest that TRF2 is bound less tightly to the chromatin in cells with reduced levels of the Ku heterodimer and that a greater fraction of the TRF2 present in the cell is ubiquitin-conjugated and targeted to the proteasome.
This interpretation is consistent with recent reports that the majority of TRF2 in the cell is bound to chromatin28 and that TRF2 can be degraded by the proteasome.32 Experimental evidence suggests that Ku70 and TRF2 interact directly,12 suggesting the possibility that binding of TRF2 to the Ku heterodimer prevents degradation. Such a relationship would be analogous to that which exists between Ku70 and Bax.33 Ku70 sequesters Bax in the cytoplasm but also prevents ubiquitin conjugation through deubiquitination activity intrinsic to Ku70.34 It is possible that the Ku heterodimer serves a similar function in the nucleus to prevent targeting of TRF2 to the proteasome. A similar relationship has been identified in mouse cells between Gcn5 and TRF1,35 whereby TRF1 is degraded via the proteasome in the absence of Gcn5. It is likely that the interaction of TRF proteins with the telomere is regulated during the cell cycle to allow DNA replication and end processing to proceed, and the differential effects of interference with accessory proteins such as Ku reveals a specific cell cycle-related function at the telomere. For example, it has been suggested that TRF2 acts to inhibit a DNA damage response at the telomere, potentially through the regulation of Chk2,36 as well as stabilizing the Holliday junction structure that is required for T-loop formation during S phase.37 It is also possible that the Ku heterodimer facilitates loading of TRF2 onto telomeric sequences since such a relationship has been described in S. cerevisiae between Ku and Cdc13p.39
Interestingly, in the human WI38 fibroblast cells used in this study, TRF1 levels increase following Ku80 depletion, which suggests either a reciprocal regulation of the telomere repeat factors or that removal of TRF2 from the telomere region allows greater access of TRF1. This increased access may serve to stabilize the protein, since the interaction between TRF1 and Ku has been found to be important for Ku localization to the telomere.11
Given the effect of the proteasome inhibitor in our system, it appears that TRF2 can be targeted to the proteasome under some conditions. This type of regulation is consistent with the recent report that TRF2 appears to be degraded by the proteasome following treatment of tumor cells with a compound that stabilizes G-quadruplex structures.32 This observation would be consistent with the multiple roles of TRF2 as a protein hub for recruitment of proteins to DNA damage sites or telomeres, since unbound TRF2 may be detrimental to cellular function potentially contributing to tumor formation.38 It is interesting to note that ubiquitin conjugation of Ku80 has been described in the release of the Ku heterodimer from double-stranded DNA during the repair process.29 In this case the ubiquitination of Ku80 seems to result from its association with the DNA repair complex. Consistent with this report we observe higher molecular weight forms of Ku80 following treatment of human fibroblasts with a proteasome inhibitor. At the same time, the TRF2 levels are dramatically increased in the presence of the proteasome inhibitor, suggesting that proteasome targeting represents a major route of degradation for TRF2 in this setting. Interestingly, TRF1 has been found to be regulated in a similar manner in that it is rapidly targeted to the proteasome following release from the telomere.39 These results, together with our findings, suggest that the telomere repeat binding factors are subject to rapid degradation when not associated with telomeres.
The results of Ku depletion in the WI38 fibroblasts differs somewhat from the results of allelic deletion of the Ku80 gene in immortalized HCT116 cells.27 Targeted deletion of both Ku alleles induced telomere loss and cell death in the HCT116 cells while the WI38 fibroblast cells entered a stable senescent arrest. The difference in the response of the two cell types may lie in the fact that some residual Ku protein remains in the WI38 cells while the allelic deletion induces complete lack of Ku80. Additional differences may lie in the fact that the cell cycle checkpoint controls differ between the two cell types, one tumor-derived and one a primary cell strain. Nonetheless, both cell types display alterations in telomere biology and rapid cessation of growth supporting the concept that Ku80 is an essential gene in human cells.
Materials and Methods
Cell culture.
WI38 human fetal lung fibroblasts were obtained from Vincent J. Cristofalo. Cells were maintained in Minimum Essential Medium 1X with Earle's salts and L-glutamine (Mediatech, Manassas VA) containing 10% fetal bovine serum, 1X MEM vitamins, 1X MEM amino acids and penicillin-streptomycin (Mediatech). Fibroblasts were passaged approximately once a week and population doubling level was determined as previously described.40
Chemicals.
All chemicals used were obtained from Sigma Aldrich (St. Louis, MO) unless otherwise noted.
Lentiviral production and infection.
To produce the Ku80 knockdown WI38 cells, we used a pLKO.1 lentiviral vector containing either a Ku80 short hairpin RNA, an empty pLKO.1 vector or a scrambled shRNA sequence. Two Ku80 shRNA constructs were obtained from Sigma Aldrich (MISSION® TRC shRNA TRCN0000039839 and TRCN0000039840) and targeted the 5′ coding region of the human Ku80 messenger RNA. Both constructs provided efficient knockdown of Ku80, however the 39840 construct was slightly more robust and was used for the majority of experiments. Empty pLKO.1 vector was also obtained from Sigma. Scrambled shRNA (Addgene plasmid 1864) had the following hairpin sequence: CCT AAG GTT AAG TCG CCC TCG CTC TAG CGA GGG CGA CTT AAC CTT AGG.41 Ten micrograms of plasmid DNA was transfected into 293T lentiviral packaging cells using LT1 transfection reagent (Mirus Bio LLC, Madison WI) and viral supernatants were collected. WI38 human fibroblasts were infected with Ku80 shRNA, empty vector or scrambled shRNA viral supernatant along with 10 ug/mL polybrene (American Bioanalytical, Natick MA) for 24 hours, followed by a 72 hour selection with 2 ug/mL puromycin (Mediatech), unless otherwise noted.
SDS-PAGE and western blotting.
For SDS-PAGE, 40 micrograms of cell extracts in RIPA buffer (50 mM Tris, 150 mM NaCl, 1% NP-40, 0.1% SDS, 0.5% sodium deoxycholate, PMSF, leupeptin, pepstatin) were loaded onto 8% polyacrylamide gels. Gels were transferred to nitrocellulose membranes and blocked for 1 hour in 5% milk in Tris-buffered saline containing 0.1% Tween-20 (TBST). Membranes were incubated with primary antibodies in 5% milk TBST overnight with shaking. Antibodies used were: anti-Ku80, anti- Ku70 and anti-TRF1 (Abcam Inc., Cambridge MA); anti-TRF2 (Novus Biologicals, Littleton CO); anti-H2A and anti-tubulin (Cell Signaling Technologies, Danvers MA); anti-ubiquitin (Santa Cruz Biotechnologies, Santa Cruz CA); and antiactin (Sigma Aldrich). Membranes were incubated for 1 hour with either goat anti-mouse-HRP or goat anti-rabbit-HRP (Millipore) in 5% milk TBST. Blots were washed in TBST and incubated with West Pico enhanced chemiluminescent substrate (Pierce/Thermo Scientific, Rockford IL) for 1 minute before developing.
Cell cycle analysis.
For cell cycle analysis, cells were harvested in trypsin, washed once in 1X PBS and fixed overnight in 70% ethanol at 4°C. Following fixation, cells were spun at 450x g for 5 minutes to remove ethanol, washed once with 1X PBS and resuspended in Guava Cell Cycle Reagent (Guava Technologies). Cells were incubated in reagent for 30 minutes at room temperature and analyzed for DNA content on a Guava EasyCyte Mini flow cytometer.
Senescence-associated b-galactosidase staining.
Cells were washed twice with 1X PBS, fixed for 5 minutes in 3% formaldehyde and washed again twice in 1X PBS. Cells were stained for 24 hours in X-gal staining solution [5 M NaCl, 1 M MgCl2, 100 mM potassium ferricyanide, 100 mM potassium ferrocyanide, 0.2 M citric acid/sodium phosphate buffer pH 6.0 and 50 mg/mL X-gal (Promega, Madison WI)] in a 37°C incubator. Cells were visualized for blue staining and results are expressed as the percentage of senescent (blue) cells compared to total cell number.
Telomere fluorescence in situ hybridization (FISH).
Cultures were incubated in 10 ug/ml colcimide (Invitrogen, Carlsbad CA) for 4 hours and placed into hypotonic medium (75 mM KCl, 5 mM MgCl) prior to fixation. Metaphase spreads were prepared and hybridized to a telomere specific PNA probe (CCC TAA)4 (DAKO Inc., Carpinteria CA) according to manufacturer's instructions. Control and Ku80-depleted metaphase spreads were analyzed for telomere abnormalities and spreads containing at least one abnormality were scored as positive.
Immunofluorescence.
Cells were seeded onto 8-well chamber slides (Thermo Scientific) or glass coverslips and fixed with 4% paraformaldehyde in PBS. Cells were then permeabilized in 0.2% Triton X-100. Cells were dehydrated in 100% ethanol and mounted using Vectashield containing 4′,6-diamidino-2-phenylindole (DAPI) (Vector Laboratories, Burlingame CA). Nuclei were visualized using an Olympus BX61 microscope and Hamamatsu ORCA-ER camera. Images were analyzed using Slidebook software equipped with deconvolution capabilities.
MG115 treatment.
Fibroblasts were infected with Ku80 or control vector for 24 hours and treated with 10 uM of the proteasome inhibitor MG115 (Biomol, Plymouth Meeting, PA) either right away or after an additional 24 hours following the shRNA treatment. MG115 was prepared in growth medium and added directly to the cells at the appropriate time. Cells were treated with MG115 for 24 hours and total cell extracts were analyzed for Ku and TRF2 levels.
Telomere length assay.
For telomere restriction fragment length (TRF) assays, genomic DNA was digested to completion using Hind III and EcoRI (Promega). Digested DNA was separated by agarose gel electrophoresis and the dried gel was exposed to alkali to denature DNA then hybridized using a 32P end labeled telomere probe (CCC TAA)4.
Immunoprecipitation.
Five hundred micrograms of total cell lysate was pre-cleared with Protein A/G Sepharose Beads (Santa Cruz Biotechnologies) for 20 minutes at 4°C and spun for 5 minutes at 14,000x g. Supernatants were incubated in anti-TRF2 overnight on a shaker at 4°C and complexes were then pulled down with Protein A/G beads for one hour. Complexes were washed three times in ice-cold PBS, resuspended in SDS loading buffer, boiled, loaded onto SDS-polyacrylamide gels as indicated above and blots were probed with anti-ubiquitin.
Chromatin extraction.
Extraction of chromatin-bound proteins was performed using a previously published protocol.42 Cells were lysed in buffer containing 20 mM HEPES pH 7.9, 25% glycerol, 0.1 mM EDTA, 5 mM MgCl2, 0.25% NP-40, 1 mM dithiothreitol, 1 mM sodium orthovanadate, 10 mM sodium fluoride, protease inhibitors, and KCl (either 50, 150 or 300 mM). Briefly, cells were lysed on ice then centrifuged at 14,000x g for 20 minutes at 4°C. Supernatants were removed and used as whole-cell extracts. The remaining pellets were suspended in SDS loading buffer, sonicated using 10 2-second bursts and centrifuged at 14,000x g for 20 minutes at 4°C. The remaining supernatants constituted the chromatin fraction.
Telomere chromatin immunoprecipitation.
For telomere ChIP, the EZ-ChIP (Millipore) and Telo-TAGGG (Roche Applied Science, Indianapolis IN) kits were used with minor modifications, which are described below. All reagents, unless otherwise noted, were provided with the kits. WI38 cells were crosslinked with 3.7% formaldehyde for ten minutes, quenched with glycine, scraped into PBS and pelleted. Chromatin was sheared using a Misonix 3000 microtip at 50% power, with one second on and two seconds off, for a total of 15 seconds of shearing and five total cycles (Qsonica, Newtown CT). Two micrograms of anti-TRF1 (Abcam) or anti-TRF2 (Novus Biologicals) was used for immunoprecipitation of protein/DNA complexes. Rabbit IgG was used as a negatve control for immunoprecipitation. Subsequent washing, elution, crosslink reversal and DNA purification were performed following manufacturer's instructions for the EZ-ChIP kit. DNA was boiled for five minutes and dot-blotted onto Hybond positively-charged nylon membrane. Following the de Lange laboratory protocol, the membrane was denatured in 1.5 M NaCl/0.5 N NaOH for ten minutes and neutralized in 1 M NaCl/0.5 M Tris-HCl pH 7.0 for ten minutes. DNA was crosslinked to the membrane immediately by UV crosslinking, then rinsed in 2X SSC. Telomeric DNA was detected following the manufacturer's instructions included with the Telo-TAG GG telomere length detection kit.
Acknowledgements
We thank Dr. F. Brad Johnson (University of Pennsylvania; Philadelphia, PA) and Dr. Thomas D. Stamato (Lankenau Institute for Medical Research; Wynnewood, PA) for valuable insight and technical assistance; Dr. Claudio Torres for assistance with the MG115 and ubiquitin experiments; and members of our laboratory for helpful advice related to this manuscript. This work was supported by NIH grant AG022443 to C.S. and Drexel University College of Medicine Aging Initiative Fellowship to L.S.F.
Footnotes
Previously published online: www.landesbioscience.com/journals/cc/article/13129
Supplementary Material
References
- 1.Mimori T, Hardin JA. Mechanism of interaction between Ku protein and DNA. J Biol Chem. 1986;261:10375–10379. [PubMed] [Google Scholar]
- 2.Griffith AJ, Blier PR, Mimori T, Hardin JA. Ku polypeptides synthesized in vitro assemble into complexes which recognize ends of double-stranded DNA. J Biol Chem. 1992;267:331–338. [PubMed] [Google Scholar]
- 3.Walker JR, Corpina RA, Goldberg J. Structure of the Ku heterodimer bound to DNA and its implications for double-strand break repair. Nature. 2001;412:607–614. doi: 10.1038/35088000. [DOI] [PubMed] [Google Scholar]
- 4.Ramsden DA, Gellert M. Ku protein stimulates DNA end joining by mammalian DNA ligases: a direct role for Ku in repair of DNA double-strand breaks. EMBO J. 1998;17:609–614. doi: 10.1093/emboj/17.2.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mahaney BL, Meek K, Lees-Miller SP. Repair of ionizing radiation-induced DNA double-strand breaks by non-homologous end-joining. Biochem J. 2009;417:639–650. doi: 10.1042/BJ20080413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dvir A, Peterson SR, Knuth MW, Lu H, Dynan WS. Ku autoantigen is the regulatory component of a template-associated protein kinase that phosphorylates RNA polymerase II. Proc Natl Acad Sci USA. 1992;89:11920–11924. doi: 10.1073/pnas.89.24.11920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Falck J, Coates J, Jackson SP. Conserved modes of recruitment of ATM, ATR and DNA-PKcs to sites of DNA damage. Nature. 2005;434:605–611. doi: 10.1038/nature03442. [DOI] [PubMed] [Google Scholar]
- 8.McCord RA, Michishita E, Hong T, Berber E, Boxer LD, Kusumoto R, et al. SIRT6 stabilizes DNA-dependent protein kinase at chromatin for DNA double-strand break repair. Aging (Albany NY) 2009;1:109–121. doi: 10.18632/aging.100011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bianchi A, de Lange T. Ku binds telomeric DNA in vitro. J Biol Chem. 1999;274:21223–21227. doi: 10.1074/jbc.274.30.21223. [DOI] [PubMed] [Google Scholar]
- 10.d'Adda di Fagagna F, Hande MP, Tong WM, Lansdorp PM, Wang ZQ, Jackson SP. Effects of DNA nonhomologous end-joining factors on telomere length and chromosomal stability in mammalian cells. Curr Biol. 2001;11:1192–1196. doi: 10.1016/s0960-9822(01)00328-1. [DOI] [PubMed] [Google Scholar]
- 11.Hsu HL, Gilley D, Galande SA, Hande MP, Allen B, Kim SH, et al. Ku acts in a unique way at the mammalian telomere to prevent end joining. Genes Dev. 2000;14:2807–2812. doi: 10.1101/gad.844000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Song K, Jung D, Jung Y, Lee SG, Lee I. Interaction of human Ku70 with TRF2. FEBS Lett. 2000;481:81–85. doi: 10.1016/s0014-5793(00)01958-x. [DOI] [PubMed] [Google Scholar]
- 13.Boulton SJ, Jackson SP. Components of the Ku-dependent non-homologous end-joining pathway are involved in telomeric length maintenance and telomeric silencing. EMBO J. 1998;17:1819–1828. doi: 10.1093/emboj/17.6.1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li B, Reddy S, Comai L. Sequence-specific processing of telomeric 3′ overhangs by the Werner syndrome protein exonuclease activity. Aging (Albany NY) 2009;1:289–302. doi: 10.18632/aging.100032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fisher TS, Zakian VA. Ku: a multifunctional protein involved in telomere maintenance. DNA Repair (Amst) 2005;4:1215–1226. doi: 10.1016/j.dnarep.2005.04.021. [DOI] [PubMed] [Google Scholar]
- 16.Nussenzweig A, Sokol K, Burgman P, Li L, Li GC. Hypersensitivity of Ku80-deficient cell lines and mice to DNA damage: the effects of ionizing radiation on growth, survival and development. Proc Natl Acad Sci USA. 1997;94:13588–13593. doi: 10.1073/pnas.94.25.13588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vogel H, Lim DS, Karsenty G, Finegold M, Hasty P. Deletion of Ku86 causes early onset of senescence in mice. Proc Natl Acad Sci USA. 1999;96:10770–10775. doi: 10.1073/pnas.96.19.10770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Samper E, Goytisolo FA, Slijepcevic P, van Buul PP, Blasco MA. Mammalian Ku86 protein prevents telomeric fusions independently of the length of TTA GGG repeats and the G-strand overhang. EMBO Rep. 2000;1:244–252. doi: 10.1093/embo-reports/kvd051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Espejel S, Franco S, Rodriguez-Perales S, Bouffler SD, Cigudosa JC, Blasco MA. Mammalian Ku86 mediates chromosomal fusions and apoptosis caused by critically short telomeres. EMBO J. 2002;21:2207–2219. doi: 10.1093/emboj/21.9.2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Riha K, Watson JM, Parkey J, Shippen DE. Telomere length deregulation and enhanced sensitivity to genotoxic stress in Arabidopsis mutants deficient in Ku70. EMBO J. 2002;21:2819–2826. doi: 10.1093/emboj/21.11.2819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Celli GB, Denchi EL, de Lange T. Ku70 stimulates fusion of dysfunctional telomeres yet protects chromosome ends from homologous recombination. Nat Cell Biol. 2006;8:885–890. doi: 10.1038/ncb1444. [DOI] [PubMed] [Google Scholar]
- 22.Li G, Nelsen C, Hendrickson EA. Ku86 is essential in human somatic cells. Proc Natl Acad Sci USA. 2002;99:832–837. doi: 10.1073/pnas.022649699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jaco I, Munoz P, Blasco MA. Role of human Ku86 in telomere length maintenance and telomere capping. Cancer Res. 2004;64:7271–7278. doi: 10.1158/0008-5472.CAN-04-1381. [DOI] [PubMed] [Google Scholar]
- 24.Ghosh G, Li G, Myung K, Hendrickson EA. The lethality of Ku86 (XRCC5) loss-of-function mutations in human cells is independent of p53 (TP53) Radiat Res. 2007;167:66–79. doi: 10.1667/RR0692.1. [DOI] [PubMed] [Google Scholar]
- 25.Myung K, Ghosh G, Fattah FJ, Li G, Kim H, Dutia A, et al. Regulation of telomere length and suppression of genomic instability in human somatic cells by Ku86. Mol Cell Biol. 2004;24:5050–5059. doi: 10.1128/MCB.24.11.5050-5059.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Uegaki K, Adachi N, So S, Iiizumi S, Koyama H. Heterozygous inactivation of human Ku70/Ku86 heterodimer does not affect cell growth, double-strand break repair or genome integrity. DNA Repair (Amst) 2006;5:303–311. doi: 10.1016/j.dnarep.2005.10.008. [DOI] [PubMed] [Google Scholar]
- 27.Wang Y, Ghosh G, Hendrickson EA. Ku86 represses lethal telomere deletion events in human somatic cells. Proc Natl Acad Sci USA. 2009;106:430–435. doi: 10.1073/pnas.0903362106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Takai KK, Hooper S, Blackwood S, Gandhi R, de Lange T. In vivo stoichiometry of shelterin components. J Biol Chem. 2010;285:1457–1467. doi: 10.1074/jbc.M109.038026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Postow L, Ghenoiu C, Woo EM, Krutchinsky AN, Chait BT, Funabiki H. Ku80 removal from DNA through double strand break-induced ubiquitylation. J Cell Biol. 2008;182:467–479. doi: 10.1083/jcb.200802146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fattah KR, Ruis BL, Hendrickson EA. Mutations to Ku reveal differences in human somatic cell lines. DNA Repair (Amst) 2008;7:762–774. doi: 10.1016/j.dnarep.2008.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dmitrieva NI, Chen HT, Nussenzweig A, Burg MB. Knockout of Ku86 accelerates cellular senescence induced by high NaCl. Aging (Albany NY) 2009;1:245–253. doi: 10.18632/aging.100022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou WJ, Deng R, Zhang XY, Feng GK, Gu LQ, Zhu XF. G-quadruplex ligand SYUIQ-5 induces autophagy by telomere damage and TRF2 delocalization in cancer cells. Mol Cancer Ther. 2009;8:3203–3213. doi: 10.1158/1535-7163.MCT-09-0244. [DOI] [PubMed] [Google Scholar]
- 33.Cohen HY, Lavu S, Bitterman KJ, Hekking B, Imahiyerobo TA, Miller C, et al. Acetylation of the C terminus of Ku70 by CBP and PCAF controls Bax-mediated apoptosis. Mol Cell. 2004;13:627–638. doi: 10.1016/s1097-2765(04)00094-2. [DOI] [PubMed] [Google Scholar]
- 34.Amsel AD, Rathaus M, Kronman N, Cohen HY. Regulation of the proapoptotic factor Bax by Ku70-dependent deubiquitylation. Proc Natl Acad Sci USA. 2008;105:5117–5122. doi: 10.1073/pnas.0706700105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Atanassov BS, Evrard YA, Multani AS, Zhang Z, Tora L, Devys D, et al. Gcn5 and SAGA regulate shelterin protein turnover and telomere maintenance. Mol Cell. 2009;35:352–364. doi: 10.1016/j.molcel.2009.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Buscemi G, Zannini L, Fontanella E, Lecis D, Lisanti S, Delia D. The shelterin protein TRF2 inhibits Chk2 activity at telomeres in the absence of DNA damage. Curr Biol. 2009;19:874–879. doi: 10.1016/j.cub.2009.03.064. [DOI] [PubMed] [Google Scholar]
- 37.Poulet A, Buisson R, Faivre-Moskalenko C, Koelblen M, Amiard S, Montel F, et al. TRF2 promotes, remodels and protects telomeric Holliday junctions. EMBO J. 2009;28:641–651. doi: 10.1038/emboj.2009.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Begemann S, Galimi F, Karlseder J. Moderate expression of TRF2 in the hematopoietic system increases development of large cell blastic T-cell lymphomas. Aging (Albany NY) 2009;1:122–130. doi: 10.18632/aging.100015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chang W, Dynek JN, Smith S. TRF1 is degraded by ubiquitin-mediated proteolysis after release from telomeres. Genes Dev. 2003;17:1328–1333. doi: 10.1101/gad.1077103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cristofalo VJ, Charpentier R. A standard procedure for cultivating human diploid fibroblastlike cells to study cellular aging. J Tissue Cult Methods. 1980;6:117–121. [Google Scholar]
- 41.Sarbassov DD, Guertin DA, Ali SM, Sabatini DM. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science. 2005;307:1098–1101. doi: 10.1126/science.1106148. [DOI] [PubMed] [Google Scholar]
- 42.Tanaka H, Mendonca MS, Bradshaw PS, Hoelz DJ, Malkas LH, Meyn MS, et al. DNA damage-induced phosphorylation of the human telomere-associated protein TRF2. Proc Natl Acad Sci USA. 2005;102:15539–15544. doi: 10.1073/pnas.0507915102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.