Abstract
Although macrophages were originally recognized as major immune effector cells, it is now appreciated that they also play many important roles in the maintenance of tissue homeostasis, and are involved in a variety of pathological conditions including cancer. Several studies have demonstrated the contributions of tumor-associated macrophages (TAMs) to tumor initiation, progression and metastasis. However, the detailed mechanisms underlying how TAMs differ molecularly from their normal counterparts and how the conversion to TAMs occurs have only just begun to be understood. TAMs have been proposed to exhibit phenotypes of ‘alternatively activated’ acrophages, though there has been limited evidence directly linking the phenotypes of TAMs to the alternative activation of macrophages. This review will focus on IL-4, the prototypic cytokine that induces the alternative activation of macrophages, and review current knowledge regarding the contributions of IL-4 to the phenotypes of TAMs and its effects on tumorigenesis.
Key words: interleukin-4, tumor-associated macrophage, tumor microenvironment, cytokines, cathepsin proteases
Diversity and Plasticity of Macrophages
Macrophages are differentiated cells of the mononuclear phagocyte system, which is composed of tissue macrophages, dendritic cells, circulating blood monocytes and the committed myeloid progenitor cells in the bone marrow.1 Both macrophages and their precursor blood monocytes are renowned for their phenotypic and functional heterogeneity, and can change their physiology in response to microenvironmental cues.
Much effort has been made to identify and characterize subsets of blood monocytes. Based on the surface expression of CD14 and CD16, human blood monocytes were categorized into at least two major subsets: CD14highCD16− cells, which are often termed ‘classical’ or ‘inflammatory’ monocytes, and CD14+CD16+ cells, also known as ‘nonclassical’ or ‘resident’ monocytes. Likewise, phenotypic characterization in mice also identified two blood monocyte subsets: Ly6C+CCR2+CX3CR1low and Ly6C-CCR2-CX3CR1high, which recapitulate the human ‘inflammatory’ and ‘resident’ monocytes.2–4 More recently, a third ‘intermediate’ subdivision has been proposed for both human and mouse monocytes.5 These phenotypic subsets have been shown to represent distinct functional classes.2–5
Like blood monocytes, macrophages also exhibit marked heterogeneity. However, there is currently a lack of distinct expression patterns of surface markers that clearly define macrophage subsets. The present definitions of macrophage subpopulations are generally based on their tissue distribution, functional characteristics or differential activation status. Initially, macrophages were classified as ‘classically activated’ and ‘alternatively activated’ macrophages. The ‘classical activation’ of macrophages, which has been delineated since early studies in the 1960s,6 depends on the products of specifically activated T helper 1 (TH1) lymphocytes or natural killer (NK) cells, in particular interferon-gamma (IFNγ), and other activators including tumor necrosis factor-alpha (TNFα) and lipopolysaccharide (LPS). Classical activation results in a population of macrophages with enhanced microbicidal capacity and increased secretion of pro-inflammatory cytokines to further intensify the cell-mediated immunity.4,7 In contrast, the signature cytokines of a TH2-type immune response, namely IL-4 and IL-13, are the major stimuli for the ‘alternative activation’ of macrophages, which induce a different phenotype that is important for the immune response to parasites, tissue remodeling and allergic immune reactions.8,9 The ‘classically activated’ and ‘alternatively activated’ macrophages have been designated as M1 and M2 macrophages respectively by analogy to the TH1 and TH2 nomenclature of helper T cells.10 The M2 designation of macrophages was further subdivided into M2a, macrophages elicited by IL-4 or IL-13; M2b, macrophages triggered by immune complexes in the presence of a Toll-like receptor stimulus; and M2c, which are inactivated by glucocorticoid, IL-10 or TGFβ.11 Although this classification of macrophages provides a useful working scheme, it does not fully represent the complexity of macrophage activation, which is often fine-tuned in response to different microenvironments. Another classification of macrophages has been proposed in which the repertoire of macrophage activation is instead viewed as a continuum spectrum, with the three fundamental functions of macrophages, namely host defense, wound healing and immune regulation, occupying three zones of the spectrum, analogous to the three primary colors in a color wheel.12
Tumor-associated macrophages (TAMs) have emerged as a critical regulatory cell type in the tumor microenvironment, with a potent ability to facilitate tumor initiation, progression and metastasis.13,14 Importantly, TAMs have been shown to acquire the hallmark properties of ‘alternatively activated’ macrophages, such as the ability to tune inflammatory and immune responses, and to promote angiogenesis and tissue remodeling.15 However, until recently there has been limited evidence showing that TAMs and ‘alternatively activated’ macrophages are indeed activated by the same cytokine repertoire. Here we will focus on IL-4, the prototypic TH2 cytokine that elicits the ‘alternative activation’ of macrophages, and review current knowledge concerning its effects on macrophages in different contexts. We will outline the common features shared by TAMs and IL-4-activated macrophages, and further discuss recent studies that identified IL-4 as a major regulator of the phenotypes of TAMs.16,17 Finally we will also discuss the opposing effects of IL-4 on tumorigenesis that act through different cell types in the tumor microenvironment.
IL-4 Receptor Signaling Pathways
IL-4 was initially identified as a T cell-derived factor that stimulates the proliferation of B cells, and was hence first designated as B cell growth factor (BCGF) or B cell stimulating factor (BSF).18 It is now clear that IL-4 plays pivotal roles in the TH2 immune response, serving as both an initiator and effector of TH2 immune reactions.19 It has broad effects on both hematopoietic cells, including cells of the innate and adaptive immune system, and non-hematopoietic cells such as smooth muscle and epithelial cells.
IL-4 is found only in mammals, and it shows a high divergence of amino acid sequence among species.20 Human and murine IL-4 share only ∼50% homology at the amino acid level,21 and do not show ligand-receptor cross-reactivity between these two species at physiological concentrations.22 IL-4 receptors are expressed in a variety of cell types. The numbers of receptors expressed on the cell surface were reported to range from less than 100 to more than 5,000, with the highest expression on helper T cells, endothelial cells, mast cells and macrophages.23
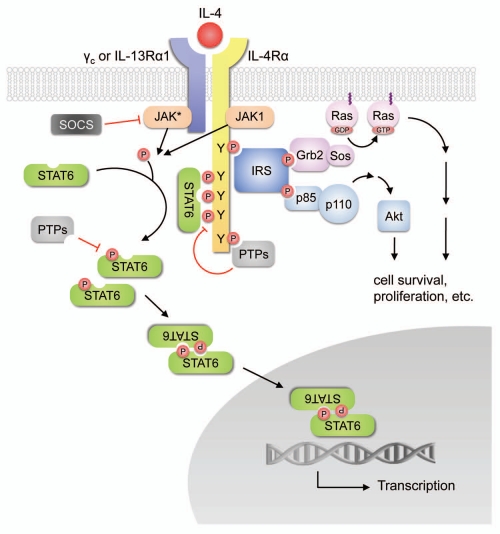
There are two types of receptors that have been identified for IL-4. The type I receptor consists of the IL-4 receptor α-chain (IL-4Rα) and the common gamma chain (γc), while the type II receptor is composed of IL-4Rα and the IL-13 receptor α-chain 1 (IL-13Rα1). The signal transduction pathways of IL-4 receptors have been thoroughly reviewed,24–27 and are summarized in Figure 1. In brief, IL-4 first binds to IL-4Rα with high affinity (Kd at subnanomolar levels), which leads to the recruitment of γc or IL-13Rα1 to form either type I or type II ternary ligand-receptor complexes. Dimerization of the receptor subunits activates the receptor-associated Janus kinases (JAKs), and leads to the phosphorylation of tyrosine residues in the cytoplasmic domain of IL-4Rα, which then serve as docking sites to recruit other signaling molecules including the signal transducer and activator of transcription-6 (STAT6), the primary STAT protein activated in response to IL-4 stimulation. STAT6 can be further tyrosine-phosphorylated by JAKs, then disengages from the receptor and dimerizes through reciprocal interactions between its SH2 domain and the phosphotyrosine residue on a second STAT6 molecule. The dimerized STAT6 complexes translocate to the nucleus, bind to specific DNA motifs within the promoters of responsive target genes, and initiate their transcription. Alternatively, IL-4 can signal through the recruitment of the insulin receptor substrate (IRS) proteins to specific phosphotyrosine residues on IL-4Rα, where IRS can be phosphorylated and recruit other signaling molecules including the p85 subunit of phosphatidylinositol 3-kinase (PI3K) that leads to the activation of the downstream protein serine/threonine kinase Akt pathways. IRS can also interact with Grb2, which is complexed to the guanine nucleotide exchange protein Sos, and leads to the activation of Ras and the downstream MAPK pathways.
Figure 1.
Signaling pathways downstream of IL-4 receptors. IL-4 receptors can signal through the Janus kinase/signal transducer and activator of transcription (JAK/STAT) cascades, or through insulin receptor substrate (IRS)-mediated activation of the phosphatidylinositol 3-kinase (PI3K)-Akt and Ras-MAPK pathways.
The IL-4 signaling pathways are subject to negative regulation by several mechanisms. For example, the SH2-containing tyrosine phosphatases (PTPs) can modulate IL-4 signaling by dephosphorylating JAKs and STAT6,28,29 whereas the suppressor of cytokine signaling (SOCS) family, such as SOCS1 and SOCS3, can inhibit the activity of JAKs by blocking the interaction of the JAK catalytic domain with their STAT protein substrates.30,31 Interestingly, SOCS3 has been shown to play an essential role in the ‘classical activation’ of macrophages, as knockdown of SOCS3 by small-interfering RNA prevents the ‘M1’ activation of bone marrow-derived macrophages by IFNγ and LPS.32
Effects of IL-4 on Macrophages
IL-4-associated gene signatures in macrophages.
Data regarding the gene expression programs in IL-4-stimulated macrophages have accumulated with the advent of genomic technologies in the past decade.9,11,33–37 Tables 1 and 2 summarize the major gene signatures associated with IL-4 in mouse and human studies. In some cases, gene homologs exist in humans and mice and are regulated by IL-4 in both species. For example, mouse Mrc1 is homologous to human MRC1, and both are upregulated by IL-4. In other examples, the regulation of gene expression is confined to one species, although gene homologs exist in both. For instance, the induction of Arg1 expression by IL-4 is only observed in murine macrophages.38 In yet other cases, the IL-4-regulated genes lack homologs in the other species. For example, Chi3l3 and Chi3l4 (also known as Ym1 and Ym2 respectively) are well-characterized markers of IL-4-activated macrophages in mice; however, they do not have direct homologs in humans. These issues of interspecies differences should be carefully taken into consideration when relating information obtained from murine models to the human situations. Among the genes listed in Tables 1 and 2, several of them are also upregulated in TAMs. For example, Arg1, Folr2, Retnla and Trem2 are part of the TAM-associated gene expression signature that Ojalvo and colleagues identified in a mouse model of mammary adenocarcinoma.39
Table 1.
Mouse IL-4-associated macrophage gene signatures
| Gene annotations and functional classifications* | Gene symbols | Other names | Entrez gene ID | Human homologs | Reference |
| Endocytosis and protein transport | |||||
| CD36 antigen | Cd36 | Scarb3 | 12491 | CD36** | 17, 137 |
| Folate receptor 2 (fetal) | Folr2 | 14276 | FOLR2 | 35 | |
| Mannose receptor, C type 1 | Mrc1 | MMR; MR; CD206 | 17533 | MRC1** | 17, 35 |
| Immunity and defense | |||||
| Interferon gamma inducible protein 30 | Ifi30 | 65972 | IFI30 | 34 | |
| Serum amyloid A 3 | Saa3 | 20210 | SAA1 | 33 | |
| Selenoprotein P, plasma, 1 | Sepp1 | Se-P | 20363 | SEPP1** | 35 |
| Chemotaxis | |||||
| Chemokine (C-C motif) ligand 2 | Ccl2 | MCP-1, JE | 20296 | CCL13** | 138 |
| Chemokine (C-C motif) ligand 9 | Ccl9 | Scya9 | 20308 | - | 34 |
| Chemokine (C-C motif) ligand 17 | Ccl17 | Tarc | 20295 | CCL17 | 17, 139 |
| Chemokine (C-C motif) ligand 22 | Ccl22 | MDC | 20299 | CCL22** | 17 |
| Cell adhesion | |||||
| C-type lectin domain family 10, member A | Clec10a | Mgl1 | 17312 | CLEC10A | 35 |
| Cadherin 1 | Cdh1 | Ecad | 12550 | CDH1 | 35 |
| Macrophage galactose N-acetyl-Galactosamine specific lectin 2 | Mgl2 | 216864 | CLEC10A | 35 | |
| Amino acid and protein modification | |||||
| Arginase, liver | Arg1 | 11846 | ARG1 | 17, 34, 35 | |
| Heat shock protein 5 | Hspa5 | 14828 | HSPA5 | 34 | |
| Tissue inhibitor of metalloproteinase 1 | Timp1 | 21857 | TIMP1 | 34 | |
| Carbohydrate metabolism | |||||
| Chitinase 3-like 3 | Chi3l3 | Ym1 | 12655 | - | 34, 35 |
| Chitinase 3-like 4 | Chi3l4 | Ym2 | 104183 | - | 35 |
| Others | |||||
| Resistin like alpha | Retnla | Fizz1; RELMa | 57262 | - | 33, 35 |
| Triggering receptor expressed on myeloid cells 2 | Trem2 | 83433 | TREM2 | 35 |
Genes are categorized based on the Gene Ontology (GO) classes of biological processes in the Database for Annotation, Visualization and Integrated Discovery (DAVID), NIH, and listed alphabetically.
Gene upregulation has been validated in both human and murine macrophages.
Table 2.
Human IL-4-associated macrophage gene signatures
| Gene annotations and functional classifications* | Gene symbols | Other names | Entrez gene ID | Mouse homologs | Reference |
| Endocytosis | |||||
| CD36 molecule | CD36 | SCARB3 | 948 | Cd36** | 37 |
| CD209 molecule | CD209 | DC-SIGN | 30835 | Cd209a | 37 |
| CD302 molecule | CD302 | DCL-1 | 9936 | Cd302 | 37 |
| Macrophage scavenger receptor 1 | MSR1 | SR-A; CD204; SCARA1 | 4481 | Msr1 | 37 |
| Mannose receptor, C type 1 | MRC1 | CD206 | 4360 | Mrc1** | 37 |
| Cell adhesion | |||||
| C-type lectin domain family 4, member F | CLEC4F | CLECSF13 | 165530 | Clec4f | 37 |
| C-type lectin domain family 7, member A | CLEC7A | 64581 | Clec7a | 37 | |
| Fibronectin 1 | FN1 | 2335 | Fn1 | 36, 37 | |
| Chemotaxis | |||||
| Chemokine (C-C motif) ligand 13 | CCL13 | MCP-4 | 6357 | Ccl2** | 37 |
| Chemokine (C-C motif) ligand 14 | CCL14 | 6358 | - | 37 | |
| Chemokine (C-C motif) ligand 17 | CCL17 | TARC | 6361 | Ccl17** | 37 |
| Chemokine (C-C motif) ligand 18 | CCL18 | MIP-4 | 6362 | - | 37 |
| Chemokine (C-C motif) ligand 22 | CCL22 | MDC | 6367 | Ccl22** | 140 |
| Chemokine (C-C motif) ligand 23 | CCL23 | MIP-3 | 6368 | - | 37 |
| Chemokine (C-C motif) ligand 24 | CCL24 | eotaxin-2 | 6369 | Ccl24 | 141 |
| Chemokine (C-X-C motif) receptor 4 | CXCR4 | 7852 | Cxcr4 | 37 | |
| Signal transduction | |||||
| Ceramide kinase | CERK | 64781 | Cerk | 37 | |
| Chimerin (chimaerin) 2 | CHN2 | 1124 | - | 37 | |
| Fibrinogen-like 2 | FGL2 | 10875 | Fgl2 | 37 | |
| Histamine receptor H1 | HRH1 | 3269 | Hrh1 | 37 | |
| Insulin-like growth factor 1 | IGF1 | 3479 | Igf1 | 37 | |
| Lysophosphatidic acid receptor 6 | LPAR6 | P2RY5 | 10161 | Lpar6 | 37 |
| Purinergic receptor P2Y, G-protein coupled, 13 | P2RY13 | GPR86 | 53829 | P2ry13 | 37 |
| Purinergic receptor P2Y, G-protein coupled, 14 | P2RY14 | GPR105 | 9934 | P2ry14 | 37 |
| Transforming growth factor, beta-induced | TGFBI | BIGH3 | 7045 | Tgfbi | 36, 37 |
| Transforming growth factor, beta receptor II | TGFBR2 | 7048 | Tgfbr2 | 37 | |
| Toll-like receptor 5 | TLR5 | 7100 | Tlr5 | 37 | |
| Ion transport | |||||
| Solute carrier family 38, member 6 | SLC38A6 | 145389 | Slc38a6 | 37 | |
| Solute carrier family 4, sodium Bicarbonate cotransporter, member 7 | SLC4A7 | 9497 | Slc4a7 | 37 | |
| Solute carrier organic anion transporter family, member 2B1 | SLCO2B1 | SLC21A9 | 11309 | Slco2b1 | 37 |
| Protein modification | |||||
| Cathepsin C | CTSC | 1075 | Ctsc | 37 | |
| Heparan sulfate (glucosamine) 3-O-sulfotransferase 1 | HS3ST1 | 9957 | Hs3st1 | 37 | |
| Heparan sulfate (glucosamine) 3-O-sulfotransferase 2 | HS3ST2 | 9956 | Hs3st2 | 37 | |
| Tyrosylprotein sulfotransferase 2 | TPST2 | 8459 | Tpst2 | 37 | |
| Lipid, fatty acid and steroid metabolism | |||||
| Arachidonate 15-lipoxygenase | ALOX15 | 246 | Alox15 | 37 | |
| Hexosaminidase B | HEXB | 3074 | Hexb | 37 | |
| Leukotriene A4 hydrolase | LTA4H | 4048 | Lta4h | 37 | |
| Lipase A, lysosomal acid, cholesterol esterase | LIPA | 3988 | Lipa | 37 | |
| Nucleotide metabolism | |||||
| Adenosine kinase | ADK | 132 | Adk | 37 | |
| Early growth response 2 | EGR2 | 1959 | Egr2 | 37 | |
| Others | |||||
| Acyl-malonyl condensing enzyme 1 | AMAC1 | 146861 | Amac1 | 36 | |
| Carbonic anhydrase II | CA2 | 760 | Car2 | 37 | |
| Growth arrest-specific 7 | GAS7 | 8522 | Gas7 | 37 | |
| Histamine N-methyltransferase | HNMT | 3176 | Hnmt | 37 | |
| v-maf musculoaponeurotic fibrosarcoma oncogene homolog | MAF | c-MAF | 4094 | Maf | 37 |
| Membrane-spanning 4-domains, subfamily A, member 4 | MS4A4A | 51338 | Ms4a4a | 37 | |
| Membrane-spanning 4-domains, subfamily A, member 6A | MS4A6A | 64231 | Ms4a6b | 37 | |
| Selenoprotein P, plasma, 1 | SEPP1 | 6414 | Sepp1** | 37 |
Genes are categorized based on the Gene Ontology (GO) classes of biological processes in the Database for Annotation, Visualization and Integrated Discovery (DAVID), NIH, and listed alphabetically.
Gene upregulation has been validated in both human and murine macrophages.
Endocytic activity.
Endocytosis is one of the fundamental biological processes macrophages exhibit, contributing to cell homeostasis, antigen presentation and defense against pathogens among others. It comprises receptor-mediated and fluid phase pinocytosis as well as phagocytosis. IL-4 has been shown to play important roles in regulating different aspects of endocytic activity in macrophages. One of the earliest characterized hallmarks of IL-4-activated macrophages is the induction of macrophage mannose receptor (MMR) expression.40 MMR was first described as a pattern recognition receptor that mediates endocytosis of mannose-rich glycoproteins.41 Subsequent studies showed that it also mediates phagocytosis of mannose and fructose-coated pathogens.42,43 IL-4 enhances both fluid phase pinocytosis and MMR-mediated endocytosis.44
In contrast, the effects of IL-4 on phagocytosis are controversial. It has been shown that IL-4 enhances MMR-mediated phagocytosis of Saccharomyces, but decreases phagocytosis of antibody-opsonized erythrocytes via Fcγ receptors or unopsonized small polystyrene latex microspheres.45–47 A recent study further showed that IL-4 markedly decreases phagocytosis of Neisseria meningitides by macrophages through the inhibition of phagosome formation.48 Thus, it appears the effects of IL-4 on phagocytosis are dependent on the different stimuli and mechanisms of phagocytosis in macrophages.
Chemotaxis.
Chemokines are a superfamily of chemotactic cytokines that direct the movement of circulating leukocytes and play critical roles in inflammatory and immune reactions.49 The chemokine and chemokine receptor repertoire is differentially expressed during macrophage differentiation and activation, and has been thoroughly reviewed previously.50 A number of chemokines are upregulated in macrophages by IL-4, as listed in Tables 1 and 2. Chemokines are also involved in carcinogenesis and play critical roles in directing cellular interactions and tropism in the tumor microenvironment. For example, CCL2 produced by either tumor cells or stromal cells promotes tumor progression in part through the recruitment of TAMs and stimulation of their pro-tumor functions (reviewed in refs. 51 and 52). TAMs not only respond to chemokines, but are in fact one of the major sources of chemokines in the tumor microenvironment. Several of the IL-4-regulated chemokines in macrophages are also found upregulated in TAMs,53,54 again suggesting that IL-4 is an major regulator of TAMs.
Nitric oxide.
IL-4 is also an important regulator of nitric oxide (NO) metabolism in macrophages. NO has a wide range of physiologic and pathophysiologic effects on the immune, nervous, cardiovascular, endocrine and other systems.55 It is a lipidand water-soluble radical gas that can react in water with oxygen and its reactive intermediates to form other radicals which contribute to the cytotoxic activity of macrophages.56 NO is synthesized from L-arginine, oxygen and NADPH by NO synthase (NOS). There are three isoforms of NOS (NOS1, 2 and 3) in mammals. Macrophages primarily express NOS2, and its expression is significantly induced by the typical ‘M1’ activators such as IFNγ, TNFα and LPS.56 In contrast, IL-4 downregulates the expression of NOS2 through a STAT6-dependent mechanism.57 The production of NO by macrophages is also determined by the availability of the enzyme substrate, L-arginine, which can be modulated by another arginine catabolic enzyme, arginase. Arginase functions by degrading arginine to urea and ornithine, which decreases the substrate pool available for NOS, and thus reduces the production of NO.58 The expression of arginase is induced by IL-4 in murine macrophages.59 Like IL-4-stimulated macrophages, TAMs also exhibit defective NO production, which in part accounts for their impaired tumoricidal activity. 60 Taken together, these data suggest IL-4 is likely involved in the attenuation of NO-dependent tumoricidal activity of TAMs by modulating the expression of arginine-catabolizing enzymes.
Macrophage fusion.
Multinucleated giant cells have beenrecognized as a histopathological hallmark of granulomatous conditions such as tuberculosis, schistosomiasis and foreign body reactions. They are also present in normal states and have important physiological functions, for example, as osteoclasts that are responsible for bone resorption.61 These giant cells originate from fusion of cells from the monocyte/macrophage lineage, a process that IL-4 can induce in vitro.62 Depletion of IL-4 by neutralizing anti-IL-4 antibodies decreases the formation of granuloma and multinucleated giant cells in response to Schistosoma mansoni eggs or foreign bodies in mice.63,64
The mechanisms of IL-4-induced macrophage fusion remain poorly understood. Helming and Gordon proposed a multistage model, in which IL-4 stimulation induces the expression of fusogenic molecules on macrophages, which mediate aggregation and membrane adhesion of adjacent macrophages, subsequently leading to cell fusion.65,66 Several cell surface receptor proteins and adhesion molecules that are induced by IL-4 have been implicated in this process, including MMR, the scavenger receptor CD36, dendritic cell-specific transmembrane protein (DC-STAMP) and E-cadherin.47,67–70 It was proposed that these molecules may act as co-receptors for membrane attachment, as illustrated by the example of CD36,69 or as chemokine receptors (e.g., DC-STAMP),71 or they may trigger other yet unknown molecules that initiate the membrane fusion events.
The evidence of macrophage fusion in tumors remains elusive and controversial. It has been suggested that fusion of myeloid cells with malignant cells may confer myeloid traits to the cancer/myeloid cell hybrids and generate aggressive cancer cell clones.72 Several studies have shown that macrophages are able to fuse with tumor cells both in vitro and in vivo, and the resultant hybrid cells are associated with increased metastatic potential.73,74 However, it remains to be determined whether IL-4 plays a role in heterotypic macrophage-tumor cell fusion, and the extent to which this may occur in vivo.
IL-4-Associated TAM Phenotypes
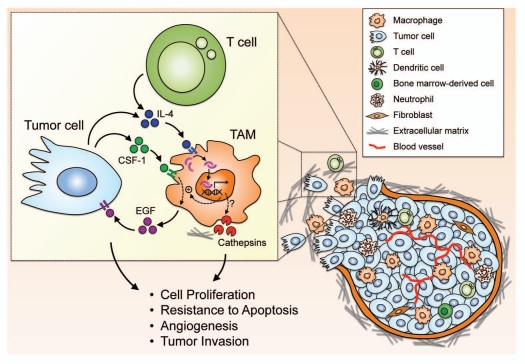
Although TAMs have been shown to display phenotypes of ‘alternatively activated’ macrophages, until recently there had been a lack of direct evidence that links IL-4, the principal inducer of the ‘alternative activation’ of macrophages, to the phenotypes of TAMs. Two studies using different experimental strategies showed that IL-4 can be supplied by different cell types in the tumor microenvironment including T cells and tumor cells, and is a key activator of the tumor-promoting functions of TAMs, acting through different mechanisms (summarized in Fig. 2 and below).16,17
Figure 2.
IL-4 as a major activator of TAM phenotypes in the tumor microenvironment. The tumor microenvironment is comprised of a variety of different cell types and matrix components, and complex cellular interactions are likely involved in the acquisition of tumor-promoting phenotypes by tumor-associated macrophages (TAMs). IL-4 supplied by either T cells or tumor cells can act on TAMs to augment the EGF/CSF-1 paracrine loop between TAMs and tumor cells, and also upregulates cathepsin enzyme activity in TAMs, although the detailed molecular mechanisms remain to be elucidated. These effects of IL-4 collectively prime TAMs with the capability to promote tumor growth and progression through different mechanisms.16,17,76
Intratumoral CD4+ T cell-derived IL-4 stimulates TAM phenotypes.
A recent study by DeNardo and colleagues revealed the essential roles of IL-4 in enhancing the protumor properties of TAMs via CD4+ T cells.16 Using the transgenic MMTV-PyMT mouse model75 of mammary adenocarcinoma development they found that CD4+ T cells can promote pulmonary metastasis of mammary tumors. Elimination of endogenous CD4+ T cells, by generating MMTV-PyMT mice in the Cd4−/− background, dramatically reduced the incidence of lung metastases but did not affect primary tumor development. The authors further demonstrated that the infiltrating CD4+ T cells in the MMTV-PyMT tumors express TH2 cytokines including IL-4, IL-10 and IL-13, and are capable of inducing the ‘M2’ type activation of TAMs. Moreover, they generated MMTV-PyMT mice either harboring a homozygous null mutation of the IL-4 receptor gene (Il4ra) or treated mice with neutralizing anti-IL-4 antibodies, and showed that ablation of IL-4 signaling by these approaches mirrors the phenotypes of Cd4-deficient MMTV-PyMT mice, suggesting that the pro-tumor effects of CD4+ T cells are mediated in part through IL-4-induced TAM phenotypes. Finally, they found that the activation of TAMs by IL-4 significantly augments the induction of epidermal growth factor (EGF) expression in TAMs by CSF-1 derived from the malignant mammary epithelial cells, and this IL-4-enhanced EGF/CSF-1 paracrine loop contributes to the invasive behavior of cancer cells in vitro and metastasis in vivo,16 as shown previously.76 Together, these studies uncovered the tumor-promoting roles of CD4+ T cells, delineated the connections between CD4+ T cells, TAMs and cancer cells, and identified IL-4 as a key inducer of TAM phenotypes.
IL-4 induces cysteine cathepsin activity in TAMs.
A complementary series of experiments linking IL-4 to the phenotypes of TAMs came from our recent studies showing that IL-4 induces the activity of the tumor-promoting cysteine cathepsins in TAMs.17 Cysteine cathepsins are a family of proteases that have emerged as important players in cancer in recent years.77–79 There are 11 human cysteine cathepsin proteases (B, C, H, F, K, L, O, S, L2/V, W, X/Z) that participate in many important physiological and pathological processes.80 Numerous clinical studies have shown that individual cathepsins are frequently highly expressed and correlate with poor patient prognosis in a broad range of human cancers.81 Our group and others have identified critical roles for cathepsins in tumor growth, angiogenesis, invasion and metastasis using both genetic and pharmacological strategies in mouse models of cancer.82–87 For example, during sequential stages of tumor development in the RIP1-Tag2 (RT2) model of pancreatic islet carcinogenesis,88 we found that expression of a subset of cathepsins (B, C, H, L, S, X/Z) progressively increased.82 Moreover, most of these cathepsins, except cathepsin L, were provided predominantly by infiltrating TAMs in different tumor microenvironments.17,89 The importance of TAM-supplied cathepsins in tumor progression was further demonstrated by a series of bone marrow transfer experiments, in which RT2 mice were lethally irradiated prior to neoplastic development, and transplanted with bone marrow cells from mice in which individual Cathepsin genes were deleted (Ctsb, Ctsc, Ctsl or Ctss). Interestingly, removal of BM-derived cathepsin B or S, but not C or L, significantly reduced RT2 tumor growth, angiogenesis and invasion.17
Importantly, the induction of cathepsin activity in TAMs was observed locally within the tumors, indicating that certain tumor microenvironmental factors are involved in triggering this activity switch.17 To identify the factors that upregulate cathepsin activity in macrophages, we developed a cell-based assay in which macrophages were differentiated from bone marrow cells in conditioned media, and cathepsin activity in the bone marrow-derived macrophages (BMDMs) was assessed by flow cytometry using a cathepsin activity-based probe.90 We found that tumor cell-conditioned media significantly induced cathepsin activity in BMDMs, and further identified IL-4 as a tumor-derived factor that triggers this induction. Treatment with IL-4 results in a significant increase in cathepsin activity, along with upregulation of several ‘M2’ marker genes including Ccl17, Ccl22, Arg1, Mrc1 and Cd36, and downregulation of ‘M1’ genes such as Tnf, Il12a and Nos2 in BMDMs. Deletion of Il4 in the RT2 mice also led to a significant reduction in cathepsin-positive TAMs in tumors. Moreover, analyses of samples from the pancreatic islets of RT2 mice revealed that IL-4 expression parallels the increased cathepsin activity during tumor development, and that IL-4 is expressed in both tumor cells and T cells.17
Collectively, our work identified a paracrine network within the tumor microenvironment (Fig. 2) where tumor cells, as well as T cells, produce IL-4, which contributes to the transition of normal macrophages to tumor-promoting TAMs in part by inducing cathepsin activity that facilitates tumor growth, angiogenesis and invasion.17
Opposing Effects of IL-4 on Tumor Growth
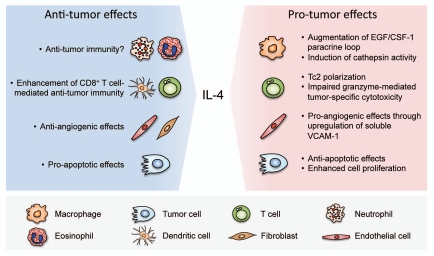
Although this review has mainly focused on the contributions of IL-4 to the tumor-promoting phenotypes of TAMs, it should be noted that both anti- and pro-tumor activities of IL-4 have been reported. It is very likely that the effects of IL-4 are dependent on the types of its source and target cells, the concentration and time of expression, the availability of the signaling components and regulatory pathways in target cells, and other interacting microenvironmental factors; some of these aspects have been previously reviewed.91,92 Here we summarize the current knowledge of the opposing anti- and pro-tumor effects which IL-4 exerts on different target cells (Fig. 3).
Figure 3.
Opposing effects of IL-4 on tumorigenesis through its impact on different cell types in the tumor microenvironment. IL-4 can exert tumorpromoting functions by enhancing the EGF/CSF-1 paracrine loop between TAMs and tumor cells, and through induction of cathepsin enzyme activity in TAMs. It also impedes T cell-mediated immunity against tumor cells through polarization of CD8+ T cells to type 2 cytotoxic T cells (Tc2), or via impairment of granzyme-mediated tumor-specific cytotoxicity of CD8+ T cells. IL-4 has also been shown to induce angiogenesis by stimulating the production of soluble VCAM-1 from endothelial cells. Additionally, IL-4 protects tumor cells from apoptosis through upregulation of anti-apoptotic proteins, and enhances cell proliferation through activation of MAPK signaling pathways. On the other hand, IL-4 also exhibits anti-tumor effects through different mechanisms including recruitment and activation of innate immune cells, such as neutrophils, eosinophils and dendritic cells. CD8+ T cells have been found to mediate the anti-tumor activities of IL-4. In some types of cancers, IL-4 can induce apoptosis of tumor cells. IL-4 has also been reported to inhibit angiogenesis directly through its effects on endothelial cells or indirectly through effects on tumor stromal fibroblasts.
Anti-tumor effects of IL-4.
The first evidence showing that IL-4 exhibits anti-tumor effects was provided by Tepper and colleagues.93,94 In their original studies, they demonstrated that tumor cells engineered to express IL-4 were rejected by the host when inoculated into mice in a syngeneic background, while the parental and the control transfected cells grew rapidly. They further showed that the tumor-inhibiting effects of IL-4 are not cell-autonomous, and are eosinophil-dependent, although this conclusion has been challenged by other reports showing that neutrophils are instead responsible for the growth suppression of IL-4-secreting tumors.95 In addition to eosinophils and neutrophils, dendritic cells can also be recruited to IL-4-expressing tumors,96 and the IL-4-activated tumor-infiltrating dendritic cells were capable of promoting a tumor-specific cytotoxic T cell response97 which was found to be important in the eradication of IL-4-expressing cells during later phases.98,99 The finding that IL-4 expression in tumor cells leads to tumor rejection was subsequently demonstrated in different types of tumors including colon cancer,96 renal cell cancer100 and melanoma.95
Besides the recruitment and activation of immune cells, the anti-tumor effects of IL-4 were also shown to be mediated through inhibition of angiogenesis, as reduced vascularization was observed in the IL-4-expressing tumors.101 IL-4 was found to exhibit anti-angiogenic effects in vitro, in part through downregulation of vascular endothelial growth factor receptor 2 (VEGFR2) expression on endothelial cells, and decreasing their responses to VEGF and other angiogenic factors such as basic fibroblast growth factor (bFGF).101–103 Moreover, the antiangiogenic effects of IL-4 can be mediated indirectly through its effects on tumor stromal fibroblasts.104
In addition to these effects on stromal cells, direct anti-tumor functions of IL-4 on tumor cells have also been reported, such as the induction of apoptosis in several types of cancer cells including breast cancer, renal cell carcinoma and hepatocarcinoma.105–107
Pro-tumor effects of IL-4.
Although these initial studies demonstrated the protective effects of IL-4 against tumor growth, it should be noted that most of these reports employed a similar strategy, by engineering the tumor cells to secrete IL-4. There are conflicting results, however, emerging from more recent studies using different approaches showing instead that IL-4 can lead to tumor-promoting activities in different cell types, such as those we have outlined above for TAMs.16,17
For example, IL-4 can polarize CD8+ T cells to type 2 cytotoxic T cells (Tc2), which have impaired cytolytic activity against tumor cells.108–111 Moreover, Il4 knockout mice exhibit increased resistance to tumor growth, and this tumor resistance was found to be related to an enhanced granzyme-mediated tumor-specific cytotoxicity of CD8+ T cells in the Il4 knockout animals.112,113 Likewise, an increased resistance to tumor growth was also observed in mice deficient in Stat6, in part through an enhanced cytotoxic T cell activity.114–117
In other non-immune stromal cells, IL-4 also induces tumorpromoting activities. For example, IL-4 can stimulate angiogenesis both in vitro and in vivo through the induction of soluble VCAM-1 production from endothelial cells, which acts through autocrine/paracrine mechanisms to activate the VCAM-1/α4 integrin signaling pathways and induce neovascularization.118,119
In addition, IL-4 may promote tumor growth by directly acting on tumor cells. Increased levels of IL-4R have been reported in a variety of human malignancies,120–125 and IL-4 signaling in tumor cells has been shown to protect cells from apoptosis through upregulation of anti-apoptosis proteins including cFLIP, PED, Bcl-xL and Bcl-2.126–129 IL-4 also enhances tumor cell proliferation through activation of MAPK signaling.130 Moreover, both IL-4 and IL-4R were found to be expressed in CD133+ cancer cells, and autocrine IL-4 signaling was shown to be responsible for the marked resistance to chemotherapeutic drugs in these stem-like cancer cells.131
Clinical Significance and Future Directions
While multiple lines of evidence support the importance of the tumor microenvironment in tumorigenesis,132 there are still critical open questions that limit our understanding of cancer and stromal cell interactions: specifically, what are the molecular signals produced by cancer cells that modify the tissue microenvironment, what cellular changes occur in the co-opted stromal cells to promote cancer progression, and can these be targeted therapeutically? From a therapeutic perspective, it is essential to understand the complex interactions between different cell types in the tumor microenvironment to develop approaches that either selectively target these cells, or block the communication between them. Among the complex cellular components in the tumor microenvironment, macrophages have emerged as a major regulatory cell type, with a potent ability to facilitate tumor initiation and progression, although little is known about how TAMs differ at the molecular level from normal macrophages and how they are converted to TAMs. In this review, we have highlighted the common phenotypes that are shared by TAMs and IL-4-polarized macrophages, and summarized current evidence that directly links IL-4 to the activation of TAMs. These data identify IL-4 as a key regulatory cytokine in the tumor microenvironment and provide a rationale for therapeutically targeting IL-4. However, as pointed out above, opposing effects of IL-4 on tumorigenesis have also been reported. In fact, recombinant IL-4 was initially used in clinical trials as an anti-cancer therapy for renal cell carcinoma, non small-cell lung cancer and malignant melanoma, based on earlier reports showing the anti-tumor effects of IL-4. However, these early trials were discontinued due to lack of efficacy.133,134 More recently, an IL-4 cytotoxin was developed by fusing IL-4 to the truncated Pseudomonas exotoxin, which was reported to be highly effective in several pre-clinical tumor models and to show promising results in completed phase I/II clinical trials (reviewed in refs. 135 and 136).
To develop more effective combination therapies against cancer in the future, we need to gain a deeper understanding of the cellular interplay between cancer cells and the tumor microenvironment. For example, it is likely that IL-4 acts in concert with additional factors in the microenvironment to regulate the tumor-promoting functions of TAMs. It will also be important to assess whether multiple cytokine pathways need to be targeted in parallel to achieve optimal inhibition of their tumor-promoting effects. The identification and characterization of these cellular interactions may provide novel strategies to disarm the tumorpromoting functions of TAMs by targeting either the upstream regulators (e.g., IL-4) or their downstream effectors (e.g., cathepsins, EGF signaling), and could have significant potential either as monotherapies or to complement conventional anticancer therapies.
Acknowledgements
We would like to thank members of the Joyce lab for discussion on this topic, and for critical review of the manuscript, in particular Drs. Vasilena Gocheva, Alberto J. Schuhmacher and Lisa Sevenich. Research in the Joyce laboratory is supported by the National Cancer Institute, the Breast Cancer Research Foundation, and the Geoffrey Beene Foundation. H.W.W. was supported by the Frank L. Horsfall Jr. Fellowship and Taiwan Merit Scholarship.
Footnotes
Previously published online: www.landesbioscience.com/journals/cc/article/14322
References
- 1.van Furth R, Cohn ZA, Hirsch JG, Humphrey JH, Spector WG, Langevoort HL. The mononuclear phagocyte system: a new classification of macrophages, monocytes and their precursor cells. Bull World Health Organ. 1972;46:845–852. [PMC free article] [PubMed] [Google Scholar]
- 2.Auffray C, Sieweke MH, Geissmann F. Blood monocytes: development, heterogeneity and relationship with dendritic cells. Annu Rev Immunol. 2009;27:669–692. doi: 10.1146/annurev.immunol.021908.132557. [DOI] [PubMed] [Google Scholar]
- 3.Geissmann F, Jung S, Littman DR. Blood monocytes consist of two principal subsets with distinct migratory properties. Immunity. 2003;19:71–82. doi: 10.1016/s1074-7613(03)00174-2. [DOI] [PubMed] [Google Scholar]
- 4.Gordon S, Taylor PR. Monocyte and macrophage heterogeneity. Nat Rev Immunol. 2005;5:953–964. doi: 10.1038/nri1733. [DOI] [PubMed] [Google Scholar]
- 5.Ziegler-Heitbrock L, Ancuta P, Crowe S, Dalod M, Grau V, Hart DN, et al. Nomenclature of monocytes and dendritic cells in blood. Blood. 2010;116:74–80. doi: 10.1182/blood-2010-02-258558. [DOI] [PubMed] [Google Scholar]
- 6.Mackaness GB. The immunological basis of acquired cellular resistance. J Exp Med. 1964;120:105–120. [PubMed] [Google Scholar]
- 7.Gordon S. The macrophage: Past, present and future. Eur J Immunol. 2007;37:9–17. doi: 10.1002/eji.200737638. [DOI] [PubMed] [Google Scholar]
- 8.Gordon S. Alternative activation of macrophages. Nat Rev Immunol. 2003;3:23–35. doi: 10.1038/nri978. [DOI] [PubMed] [Google Scholar]
- 9.Martinez FO, Helming L, Gordon S. Alternative activation of macrophages: An immunologic functional perspective. Annu Rev Immunol. 2009;27:451–483. doi: 10.1146/annurev.immunol.021908.132532. [DOI] [PubMed] [Google Scholar]
- 10.Mills CD, Kincaid K, Alt JM, Heilman MJ, Hill AM. M-1/M-2 macrophages and the Th1/Th2 paradigm. J Immunol. 2000;164:6166–6173. doi: 10.4049/jimmunol.1701141. [DOI] [PubMed] [Google Scholar]
- 11.Martinez FO, Sica A, Mantovani A, Locati M. Macrophage activation and polarization. Front Biosci. 2008;13:453–461. doi: 10.2741/2692. [DOI] [PubMed] [Google Scholar]
- 12.Mosser DM, Edwards JP. Exploring the full spectrum of macrophage activation. Nat Rev Immunol. 2008;8:958–969. doi: 10.1038/nri2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Condeelis J, Pollard JW. Macrophages: Obligate partners for tumor cell migration, invasion and metastasis. Cell. 2006;124:263–266. doi: 10.1016/j.cell.2006.01.007. [DOI] [PubMed] [Google Scholar]
- 14.Qian BZ, Pollard JW. Macrophage diversity enhances tumor progression and metastasis. Cell. 2010;141:39–51. doi: 10.1016/j.cell.2010.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mantovani A, Sozzani S, Locati M, Allavena P, Sica A. Macrophage polarization: Tumor-associated macrophages as a paradigm for polarized M2 mononuclear phagocytes. Trends Immunol. 2002;23:549–555. doi: 10.1016/s1471-4906(02)02302-5. [DOI] [PubMed] [Google Scholar]
- 16.DeNardo DG, Barreto JB, Andreu P, Vasquez L, Tawfik D, Kolhatkar N, et al. CD4(+) T cells regulate pulmonary metastasis of mammary carcinomas by enhancing protumor properties of macrophages. Cancer Cell. 2009;16:91–102. doi: 10.1016/j.ccr.2009.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gocheva V, Wang HW, Gadea BB, Shree T, Hunter KE, Garfall AL, et al. IL-4 induces cathepsin protease activity in tumor-associated macrophages to promote cancer growth and invasion. Genes Dev. 2010;24:241–255. doi: 10.1101/gad.1874010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Howard M, Farrar J, Hilfiker M, Johnson B, Takatsu K, Hamaoka T, et al. Identification of a T cell-derived b cell growth factor distinct from interleukin 2. J Exp Med. 1982;155:914–923. doi: 10.1084/jem.155.3.914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Paul WE, Zhu J. How are T(H)2-type immune responses initiated and amplified? Nat Rev Immunol. 2010;10:225–235. doi: 10.1038/nri2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mueller TD, Zhang JL, Sebald W, Duschl A. Structure, binding and antagonists in the IL-4/IL-13 receptor system. Biochim Biophys Acta. 2002;1592:237–250. doi: 10.1016/s0167-4889(02)00318-x. [DOI] [PubMed] [Google Scholar]
- 21.Yokota T, Otsuka T, Mosmann T, Banchereau J, DeFrance T, Blanchard D, et al. Isolation and characterization of a human interleukin cDNA clone, homologous to mouse B-cell stimulatory factor 1, that expresses B-cell- and T-cell-stimulating activities. Proc Natl Acad Sci USA. 1986;83:5894–5898. doi: 10.1073/pnas.83.16.5894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mosmann TR, Yokota T, Kastelein R, Zurawski SM, Arai N, Takebe Y. Species-specificity of T cell stimulating activities of IL 2 and BSF-1 (IL 4): comparison of normal and recombinant, mouse and human IL 2 and BSF-1 (IL 4) J Immunol. 1987;138:1813–1816. [PubMed] [Google Scholar]
- 23.Lowenthal JW, Castle BE, Christiansen J, Schreurs J, Rennick D, Arai N, et al. Expression of high affinity receptors for murine interleukin 4 (BSF-1) on hemopoietic and nonhemopoietic cells. J Immunol. 1988;140:456–464. [PubMed] [Google Scholar]
- 24.Hebenstreit D, Wirnsberger G, Horejs-Hoeck J, Duschl A. Signaling mechanisms, interaction partners and target genes of STAT6. Cytokine Growth Factor Rev. 2006;17:173–188. doi: 10.1016/j.cytogfr.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 25.Kelly-Welch AE, Hanson EM, Boothby MR, Keegan AD. Interleukin-4 and interleukin-13 signaling connections maps. Science. 2003;300:1527–1528. doi: 10.1126/science.1085458. [DOI] [PubMed] [Google Scholar]
- 26.Nelms K, Keegan AD, Zamorano J, Ryan JJ, Paul WE. The IL-4 receptor: signaling mechanisms and biologic functions. Annu Rev Immunol. 1999;17:701–738. doi: 10.1146/annurev.immunol.17.1.701. [DOI] [PubMed] [Google Scholar]
- 27.Jiang H, Harris MB, Rothman P. IL-4/IL-13 signaling beyond JAK/STAT. J Allergy Clin Immunol. 2000;105:1063–1070. doi: 10.1067/mai.2000.107604. [DOI] [PubMed] [Google Scholar]
- 28.Myers MP, Andersen JN, Cheng A, Tremblay ML, Horvath CM, Parisien JP, et al. TYK2 and JAK2 are substrates of protein-tyrosine phosphatase 1B. J Biol Chem. 2001;276:47771–47774. doi: 10.1074/jbc.C100583200. [DOI] [PubMed] [Google Scholar]
- 29.Lu X, Malumbres R, Shields B, Jiang X, Sarosiek KA, Natkunam Y, et al. PTP1B is a negative regulator of interleukin 4-induced STAT6 signaling. Blood. 2008;112:4098–4108. doi: 10.1182/blood-2008-03-148726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dickensheets H, Vazquez N, Sheikh F, Gingras S, Murray PJ, Ryan JJ, et al. Suppressor of cytokine signaling-1 is an IL-4-inducible gene in macrophages and feedback inhibits IL-4 signaling. Genes Immun. 2007;8:21–27. doi: 10.1038/sj.gene.6364352. [DOI] [PubMed] [Google Scholar]
- 31.O'Connor JC, Sherry CL, Guest CB, Freund GG. Type 2 diabetes impairs insulin receptor substrate-2-mediated phosphatidylinositol 3-kinase activity in primary macrophages to induce a state of cytokine resistance to IL-4 in association with overexpression of suppressor of cytokine signaling-3. J Immunol. 2007;178:6886–6893. doi: 10.4049/jimmunol.178.11.6886. [DOI] [PubMed] [Google Scholar]
- 32.Liu Y, Stewart KN, Bishop E, Marek CJ, Kluth DC, Rees AJ, et al. Unique expression of suppressor of cytokine signaling 3 is essential for classical macrophage activation in rodents in vitro and in vivo. J Immunol. 2008;180:6270–6278. doi: 10.4049/jimmunol.180.9.6270. [DOI] [PubMed] [Google Scholar]
- 33.Loke P, Nair MG, Parkinson J, Guiliano D, Blaxter M, Allen JE. IL-4 dependent alternatively-activated macrophages have a distinctive in vivo gene expression phenotype. BMC Immunol. 2002;3:7. doi: 10.1186/1471-2172-3-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Welch JS, Escoubet-Lozach L, Sykes DB, Liddiard K, Greaves DR, Glass CK. TH2 cytokines and allergic challenge induce Ym1 expression in macrophages by a STAT6-dependent mechanism. J Biol Chem. 2002;277:42821–42829. doi: 10.1074/jbc.M205873200. [DOI] [PubMed] [Google Scholar]
- 35.Ghassabeh GH, De Baetselie P, Brys L, Noël W, Van Ginderachter JA, Meerschaut S, et al. Identification of a common gene signature for type II cytokine-associated myeloid cells elicited in vivo in different pathologic conditions. Blood. 2006;108:575–583. doi: 10.1182/blood-2005-04-1485. [DOI] [PubMed] [Google Scholar]
- 36.Gratchev A, Guillot P, Hakiy N, Politz O, Orfanos CE, Schledzewski K, et al. Alternatively activated macrophages differentially express fibronectin and its splice variants and the extracellular matrix protein betaIG-H3. Scand J Immunol. 2001;53:386–392. doi: 10.1046/j.1365-3083.2001.00885.x. [DOI] [PubMed] [Google Scholar]
- 37.Martinez FO, Gordon S, Locati M, Mantovani A. Transcriptional profiling of the human monocyteto-macrophage differentiation and polarization: New molecules and patterns of gene expression. J Immunol. 2006;177:7303–7311. doi: 10.4049/jimmunol.177.10.7303. [DOI] [PubMed] [Google Scholar]
- 38.Raes G, Van den Berg R, De Baetselier P, Ghassabeh GH, Scotton C, Locati M, et al. Arginase-1 and Ym1 are markers for murine, but not human, alternatively activated myeloid cells. J Immunol. 2005;174:6561. doi: 10.4049/jimmunol.174.11.6561. [DOI] [PubMed] [Google Scholar]
- 39.Ojalvo LS, King W, Cox D, Pollard JW. High-density gene expression analysis of tumor-associated macrophages from mouse mammary tumors. Am J Pathol. 2009;174:1048–1064. doi: 10.2353/ajpath.2009.080676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stein M, Keshav S, Harris N, Gordon S. Interleukin 4 potently enhances murine macrophage mannose receptor activity: A marker of alternative immunologic macrophage activation. J Exp Med. 1992;176:287–292. doi: 10.1084/jem.176.1.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stahl P, Schlesinger PH, Sigardson E, Rodman JS, Lee YC. Receptor-mediated pinocytosis of mannose glycoconjugates by macrophages: characterization and evidence for receptor recycling. Cell. 1980;19:207–215. doi: 10.1016/0092-8674(80)90402-x. [DOI] [PubMed] [Google Scholar]
- 42.Stahl PD, Ezekowitz RA. The mannose receptor is a pattern recognition receptor involved in host defense. Curr Opin Immunol. 1998;10:50–55. doi: 10.1016/s0952-7915(98)80031-9. [DOI] [PubMed] [Google Scholar]
- 43.Aderem A, Underhill DM. Mechanisms of phagocytosis in macrophages. Annu Rev Immunol. 1999;17:593–623. doi: 10.1146/annurev.immunol.17.1.593. [DOI] [PubMed] [Google Scholar]
- 44.Montaner LJ, da Silva RP, Sun J, Sutterwala S, Hollinshead M, Vaux D, et al. Type 1 and type 2 cytokine regulation of macrophage endocytosis: Differential activation by IL-4/IL-13 as opposed to IFNgamma or IL-10. J Immunol. 1999;162:4606–4613. [PubMed] [Google Scholar]
- 45.Raveh D, Kruskal BA, Farland J, Ezekowitz RA. Th1 and Th2 cytokines cooperate to stimulate mannosereceptor-mediated phagocytosis. J Leukoc Biol. 1998;64:108–113. [PubMed] [Google Scholar]
- 46.Cipriano IM, Mariano M, Freymuller E, Carneiro CR. Murine macrophages cultured with IL-4 acquire a phenotype similar to that of epithelioid cells from granulomatous inflammation. Inflammation. 2003;27:201–211. doi: 10.1023/a:1025084413767. [DOI] [PubMed] [Google Scholar]
- 47.Moreno JL, Mikhailenko I, Tondravi MM, Keegan AD. IL-4 promotes the formation of multinucleated giant cells from macrophage precursors by a STAT6-dependent, homotypic mechanism: contribution of E-cadherin. J Leukoc Biol. 2007;82:1542–1553. doi: 10.1189/jlb.0107058. [DOI] [PubMed] [Google Scholar]
- 48.Varin A, Mukhopadhyay S, Herbein G, Gordon S. Alternative activation of macrophages by IL-4 impairs phagocytosis of pathogens but potentiates microbial-induced signalling and cytokine secretion. Blood. 2010;115:353–362. doi: 10.1182/blood-2009-08-236711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mackay CR. Chemokines: Immunology's high impact factors. Nat Immunol. 2001;2:95–101. doi: 10.1038/84298. [DOI] [PubMed] [Google Scholar]
- 50.Mantovani A, Sica A, Sozzani S, Allavena P, Vecchi A, Locati M. The chemokine system in diverse forms of macrophage activation and polarization. Trends Immunol. 2004;25:677–686. doi: 10.1016/j.it.2004.09.015. [DOI] [PubMed] [Google Scholar]
- 51.Soria G, Ben-Baruch A. The inflammatory chemokines CCL2 and CCL5 in breast cancer. Cancer Lett. 2008;267:271–285. doi: 10.1016/j.canlet.2008.03.018. [DOI] [PubMed] [Google Scholar]
- 52.Zhang J, Lu Y, Pienta KJ. Multiple roles of chemokine (C-C motif ) ligand 2 in promoting prostate cancer growth. J Natl Cancer Inst. 2010;102:522–528. doi: 10.1093/jnci/djq044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hagemann T, Wilson J, Burke F, Kulbe H, Li NF, Plüddemann A, et al. Ovarian cancer cells polarize macrophages toward a tumor-associated phenotype. J Immunol. 2006;176:5023–5032. doi: 10.4049/jimmunol.176.8.5023. [DOI] [PubMed] [Google Scholar]
- 54.Biswas SK. A distinct and unique transcriptional program expressed by tumor-associated macrophages (defective NFκB and enhanced IRF-3/STAT1 activation) Blood. 2006;107:2112–2122. doi: 10.1182/blood-2005-01-0428. [DOI] [PubMed] [Google Scholar]
- 55.Nathan C. Nitric oxide as a secretory product of mammalian cells. FASEB J. 1992;6:3051–3064. [PubMed] [Google Scholar]
- 56.MacMicking J, Xie QW, Nathan C. Nitric oxide and macrophage function. Annu Rev Immunol. 1997;15:323–350. doi: 10.1146/annurev.immunol.15.1.323. [DOI] [PubMed] [Google Scholar]
- 57.Coccia EM, Stellacci E, Marziali G, Weiss G, Battistini A. IFNgamma and IL-4 differently regulate inducible NO synthase gene expression through IRF-1 modulation. Int Immunol. 2000;12:977–985. doi: 10.1093/intimm/12.7.977. [DOI] [PubMed] [Google Scholar]
- 58.Bogdan C. Nitric oxide and the immune response. Nat Immunol. 2001;2:907–916. doi: 10.1038/ni1001-907. [DOI] [PubMed] [Google Scholar]
- 59.Corraliza IM, Soler G, Eichmann K, Modolell M. Arginase induction by suppressors of nitric oxide synthesis (IL-4, IL-10 and PGE2) in murine bone-marrow-derived macrophages. Biochem Biophys Res Commun. 1995;206:667–673. doi: 10.1006/bbrc.1995.1094. [DOI] [PubMed] [Google Scholar]
- 60.Dinapoli MR, Calderon CL, Lopez DM. The altered tumoricidal capacity of macrophages isolated from tumor-bearing mice is related to reduce expression of the inducible nitric oxide synthase gene. J Exp Med. 1996;183:1323–1329. doi: 10.1084/jem.183.4.1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Anderson JM. Multinucleated giant cells. Curr Opin Hematol. 2000;7:40–47. doi: 10.1097/00062752-200001000-00008. [DOI] [PubMed] [Google Scholar]
- 62.McInnes A, Rennick DM. Interleukin 4 induces cultured monocytes/macrophages to form giant multinucleated cells. J Exp Med. 1988;167:598–611. doi: 10.1084/jem.167.2.598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chensue SW, Terebuh PD, Warmington KS, Hershey SD, Evanoff HL, Kunkel SL, et al. Role of IL-4 and IFNgamma in Schistosoma mansoni egg-induced hypersensitivity granuloma formation: Orchestration, relative contribution and relationship to macrophage function. J Immunol. 1992;148:900–906. [PubMed] [Google Scholar]
- 64.Kao WJ, McNally AK, Hiltner A, Anderson JM. Role for interleukin-4 in foreign-body giant cell formation on a poly(etherurethane urea) in vivo. J Biomed Mater Res. 1995;29:1267–1275. doi: 10.1002/jbm.820291014. [DOI] [PubMed] [Google Scholar]
- 65.Helming L, Gordon S. Macrophage fusion induced by IL-4 alternative activation is a multistage process involving multiple target molecules. Eur J Immunol. 2007;37:33–42. doi: 10.1002/eji.200636788. [DOI] [PubMed] [Google Scholar]
- 66.Helming L, Gordon S. The molecular basis of macrophage fusion. Immunobiology. 2007;212:785–793. doi: 10.1016/j.imbio.2007.09.012. [DOI] [PubMed] [Google Scholar]
- 67.McNally AK, DeFife KM, Anderson JM. Interleukin-4-induced macrophage fusion is prevented by inhibitors of mannose receptor activity. Am J Pathol. 1996;149:975–985. [PMC free article] [PubMed] [Google Scholar]
- 68.Yagi M, Miyamoto T, Sawatani Y, Iwamoto K, Hosogane N, Fujita N, et al. DC-STAMP is essential for cell-cell fusion in osteoclasts and foreign body giant cells. J Exp Med. 2005;202:345–351. doi: 10.1084/jem.20050645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Helming L, Winter J, Gordon S. The scavenger receptor CD36 plays a role in cytokine-induced macrophage fusion. J Cell Sci. 2009;122:453–459. doi: 10.1242/jcs.037200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Van den Bossche J, Bogaert P, van Hengel J, Guerin CJ, Berx G, Movahedi K, et al. Alternatively activated macrophages engage in homotypic and heterotypic interactions through IL-4 and polyamine-induced E-cadherin/catenin complexes. Blood. 2009;114:4664–4674. doi: 10.1182/blood-2009-05-221598. [DOI] [PubMed] [Google Scholar]
- 71.Vignery A. Macrophage fusion: The making of osteoclasts and giant cells. J Exp Med. 2005;202:337–340. doi: 10.1084/jem.20051123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pawelek JM. Tumour-cell fusion as a source of myeloid traits in cancer. The Lancet Oncology. 2005;6:988–993. doi: 10.1016/S1470-2045(05)70466-6. [DOI] [PubMed] [Google Scholar]
- 73.Rachkovsky M, Sodi S, Chakraborty A, Avissar Y, Bolognia J, McNiff JM, et al. Melanoma x macrophage hybrids with enhanced metastatic potential. Clin Exp Metastasis. 1998;16:299–312. doi: 10.1023/a:1006557228604. [DOI] [PubMed] [Google Scholar]
- 74.Busund LTR, Killie MK, Bartnes K, Seljelid R. Spontaneously formed tumorigenic hybrids of Meth A sarcoma cells and macrophages in vivo. Int J Cancer. 2003;106:153–159. doi: 10.1002/ijc.11210. [DOI] [PubMed] [Google Scholar]
- 75.Guy CT, Cardiff RD, Muller WJ. Induction of mammary tumors by expression of polyomavirus middle T oncogene: A transgenic mouse model for metastatic disease. Mol Cell Biol. 1992;12:954–961. doi: 10.1128/mcb.12.3.954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wyckoff J, Wang W, Lin EY, Wang Y, Pixley F, Stanley ER, et al. A paracrine loop between tumor cells and macrophages is required for tumor cell migration in mammary tumors. Cancer Res. 2004;64:7022–7029. doi: 10.1158/0008-5472.CAN-04-1449. [DOI] [PubMed] [Google Scholar]
- 77.Mohamed MM, Sloane BF. Cysteine cathepsins: Multifunctional enzymes in cancer. Nat Rev Cancer. 2006;6:764–775. doi: 10.1038/nrc1949. [DOI] [PubMed] [Google Scholar]
- 78.Gocheva V, Joyce JA. Cysteine cathepsins and the cutting edge of cancer invasion. Cell Cycle. 2007;6:60–64. doi: 10.4161/cc.6.1.3669. [DOI] [PubMed] [Google Scholar]
- 79.Palermo C, Joyce JA. Cysteine cathepsin proteases as pharmacological targets in cancer. Trends Pharmacol Sci. 2008;29:22–28. doi: 10.1016/j.tips.2007.10.011. [DOI] [PubMed] [Google Scholar]
- 80.Turk V, Turk B, Turk D. Lysosomal cysteine proteases: Facts and opportunities. EMBO J. 2001;20:4629–4633. doi: 10.1093/emboj/20.17.4629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Jedeszko C, Sloane BF. Cysteine cathepsins in human cancer. Biol Chem. 2004;385:1017–1027. doi: 10.1515/BC.2004.132. [DOI] [PubMed] [Google Scholar]
- 82.Joyce JA, Baruch A, Chehade K, Meyer-Morse N, Giraudo E, Tsai FY, et al. Cathepsin cysteine proteases are effectors of invasive growth and angiogenesis during multistage tumorigenesis. Cancer Cell. 2004;5:443–453. doi: 10.1016/s1535-6108(04)00111-4. [DOI] [PubMed] [Google Scholar]
- 83.Gocheva V, Zeng W, Ke D, Klimstra D, Reinheckel T, Peters C, et al. Distinct roles for cysteine cathepsin genes in multistage tumorigenesis. Genes Dev. 2006;20:543–556. doi: 10.1101/gad.1407406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Vasiljeva O, Papazoglou A, Kruger A, Brodoefel H, Korovin M, Deussing J, et al. Tumor cell-derived and macrophage-derived cathepsin B promotes progression and lung metastasis of mammary cancer. Cancer Res. 2006;66:5242–5250. doi: 10.1158/0008-5472.CAN-05-4463. [DOI] [PubMed] [Google Scholar]
- 85.Wang B, Sun J, Kitamoto S, Yang M, Grubb A, Chapman HA, et al. Cathepsin S controls angiogenesis and tumor growth via matrix-derived angiogenic factors. J Biol Chem. 2006;281:6020–6029. doi: 10.1074/jbc.M509134200. [DOI] [PubMed] [Google Scholar]
- 86.Burden RE, Gormley JA, Jaquin TJ, Small DM, Quinn DJ, Hegarty SM, et al. Antibody-mediated inhibition of cathepsin S blocks colorectal tumor invasion and angiogenesis. Clin Cancer Res. 2009;15:6042–6051. doi: 10.1158/1078-0432.CCR-09-1262. [DOI] [PubMed] [Google Scholar]
- 87.Sevenich L, Schurigt U, Sachse K, Gajda M, Werner F, Müller S, et al. Synergistic antitumor effects of combined cathepsin B and cathepsin Z deficiencies on breast cancer progression and metastasis in mice. Proc Natl Acad Sci USA. 2010;107:2497–2502. doi: 10.1073/pnas.0907240107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hanahan D. Heritable formation of pancreatic betacell tumors in transgenic mice expressing recombinant insulin/simian virus 40 oncogenes. Nature. 1985;315:115–122. doi: 10.1038/315115a0. [DOI] [PubMed] [Google Scholar]
- 89.Gocheva V, Chen X, Peters C, Reinheckel T, Joyce JA. Deletion of cathepsin H perturbs angiogenic switching, vascularization and growth of tumors in a mouse model of pancreatic islet cell cancer. Biol Chem. 2010;391:937–945. doi: 10.1515/BC.2010.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Greenbaum D, Baruch A, Hayrapetian L, Darula Z, Burlingame A, Medzihradszky KF, et al. Chemical approaches for functionally probing the proteome. Mol Cell Proteomics. 2002;1:60–68. doi: 10.1074/mcp.t100003-mcp200. [DOI] [PubMed] [Google Scholar]
- 91.Olver S, Apte S, Baz A, Kienzle N. The duplicitous effects of interleukin 4 on tumour immunity: How can the same cytokine improve or impair control of tumour growth? Tissue Antigens. 2007;69:293–298. doi: 10.1111/j.1399-0039.2007.00831.x. [DOI] [PubMed] [Google Scholar]
- 92.Li Z, Chen L, Qin Z. Paradoxical roles of IL-4 in tumor immunity. Cell Mol Immunol. 2009;6:415–422. doi: 10.1038/cmi.2009.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Tepper RI, Pattengale PK, Leder P. Murine interleukin-4 displays potent anti-tumor activity in vivo. Cell. 1989;57:503–512. doi: 10.1016/0092-8674(89)90925-2. [DOI] [PubMed] [Google Scholar]
- 94.Tepper RI, Coffman RL, Leder P. An eosinophildependent mechanism for the antitumor effect of interleukin-4. Science. 1992;257:548–551. doi: 10.1126/science.1636093. [DOI] [PubMed] [Google Scholar]
- 95.Noffz G, Qin Z, Kopf M, Blankenstein T. Neutrophils but not eosinophils are involved in growth suppression of IL-4-secreting tumors. J Immunol. 1998;160:345–350. [PubMed] [Google Scholar]
- 96.Stoppacciaro A, Paglia P, Lombardi L, Parmiani G, Baroni C, Colombo MP. Genetic modification of a carcinoma with the IL-4 gene increases the influx of dendritic cells relative to other cytokines. Eur J Immunol. 1997;27:2375–2382. doi: 10.1002/eji.1830270936. [DOI] [PubMed] [Google Scholar]
- 97.Eguchi J, Kuwashima N, Hatano M, Nishimura F, Dusak JE, Storkus WJ, et al. IL-4-transfected tumor cell vaccines activate tumor-infiltrating dendritic cells and promote type-1 immunity. J Immunol. 2005;174:7194–7201. doi: 10.4049/jimmunol.174.11.7194. [DOI] [PubMed] [Google Scholar]
- 98.Hock H, Dorsch M, Kunzendorf U, Qin Z, Diamantstein T, Blankenstein T. Mechanisms of rejection induced by tumor cell-targeted gene transfer of interleukin 2, interleukin 4, interleukin 7, tumor necrosis factor or interferon gamma. Proc Natl Acad Sci USA. 1993;90:2774–2778. doi: 10.1073/pnas.90.7.2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Pericle F, Giovarelli M, Colombo MP, Ferrari G, Musiani P, Modesti A, et al. An efficient Th2-type memory follows CD8+ lymphocyte-driven and eosinophil-mediated rejection of a spontaneous mouse mammary adenocarcinoma engineered to release IL-4. J Immunol. 1994;153:5659–5673. [PubMed] [Google Scholar]
- 100.Golumbek PT, Lazenby AJ, Levitsky HI, Jaffee LM, Karasuyama H, Baker M, et al. Treatment of established renal cancer by tumor cells engineered to secrete interleukin-4. Science. 1991;254:713–716. doi: 10.1126/science.1948050. [DOI] [PubMed] [Google Scholar]
- 101.Saleh M, Davis ID, Wilks AF. The paracrine role of tumour-derived mIL-4 on tumour-associated endothelium. Int J Cancer. 1997;72:664–672. doi: 10.1002/(sici)1097-0215(19970807)72:4<664::aid-ijc19>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 102.Lee IY, Kim J, Ko EM, Jeoung EJ, Kwon YG, Choe J. Interleukin-4 inhibits the vascular endothelial growth factor- and basic fibroblast growth factor-induced angiogenesis in vitro. Mol Cells. 2002;14:115–121. [PubMed] [Google Scholar]
- 103.Volpert OV, Fong T, Koch AE, Peterson JD, Waltenbaugh C, Tepper RI, et al. Inhibition of angiogenesis by interleukin 4. J Exp Med. 1998;188:1039–1046. doi: 10.1084/jem.188.6.1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Schüler T, Körnig S, Blankenstein T. Tumor rejection by modulation of tumor stromal fibroblasts. J Exp Med. 2003;198:1487–1493. doi: 10.1084/jem.20030849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Gooch JL, Christy B, Yee D. STAT6 mediates interleukin-4 growth inhibition in human breast cancer cells. Neoplasia. 2002;4:324–331. doi: 10.1038/sj.neo.7900248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Falkensammer C, Jöhrer K, Gander H, Ramoner R, Putz T, Rahm A, et al. IL-4 inhibits the TNFalpha induced proliferation of renal cell carcinoma (RCC) and cooperates with TNFalpha to induce apoptotic and cytokine responses by RCC: implications for antitumor immune responses. Cancer Immunol Immunother. 2006;55:1228–1237. doi: 10.1007/s00262-006-0122-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Aoudjehane L, Podevin P, Scatton O, Jaffray P, Dusanter-Fourt I, Feldmann G, et al. Interleukin-4 induces human hepatocyte apoptosis through a Fas-independent pathway. FASEB J. 2007;21:1433–1444. doi: 10.1096/fj.06-6319com. [DOI] [PubMed] [Google Scholar]
- 108.Kienzle N, Buttigieg K, Groves P, Kawula T, Kelso A. A clonal culture system demonstrates that IL-4 induces a subpopulation of noncytolytic T cells with low CD8, perforin and granzyme expression. J Immunol. 2002;168:1672–1681. doi: 10.4049/jimmunol.168.4.1672. [DOI] [PubMed] [Google Scholar]
- 109.Kienzle N, Olver S, Buttigieg K, Groves P, Janas ML, Baz A, et al. Progressive differentiation and commitment of CD8+ T cells to a poorly cytolytic CD8low phenotype in the presence of IL-4. J Immunol. 2005;174:2021–2029. doi: 10.4049/jimmunol.174.4.2021. [DOI] [PubMed] [Google Scholar]
- 110.Helmich BK, Dutton RW. The role of adoptively transferred CD8 T cells and host cells in the control of the growth of the EG7 thymoma: factors that determine the relative effectiveness and homing properties of Tc1 and Tc2 effectors. J Immunol. 2001;166:6500–6508. doi: 10.4049/jimmunol.166.11.6500. [DOI] [PubMed] [Google Scholar]
- 111.Kemp RA, Ronchese F. Tumor-specific Tc1, but not Tc2, cells deliver protective antitumor immunity. J Immunol. 2001;167:6497–6502. doi: 10.4049/jimmunol.167.11.6497. [DOI] [PubMed] [Google Scholar]
- 112.Stremmel C, Greenfield EA, Howard E, Freeman GJ, Kuchroo VK. B7-2 expressed on EL4 lymphoma suppresses antitumor immunity by an interleukin 4-dependent mechanism. J Exp Med. 1999;189:919–930. doi: 10.1084/jem.189.6.919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Baschuk N, Utermöhlen O, Gugel R, Warnecke G, Karow U, Paulsen D, et al. Interleukin-4 impairs granzyme-mediated cytotoxicity of Simian virus 40 large tumor antigen-specific CTL in BALB/c mice. Cancer Immunol Immunother. 2007;56:1625–1636. doi: 10.1007/s00262-007-0309-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Kacha AK, Fallarino F, Markiewicz MA, Gajewski TF. Cutting edge: Spontaneous rejection of poorly immunogenic P1.HTR tumors by Stat6-deficient mice. J Immunol. 2000;165:6024–6028. doi: 10.4049/jimmunol.165.11.6024. [DOI] [PubMed] [Google Scholar]
- 115.Ostrand-Rosenberg S, Grusby MJ, Clements VK. Cutting edge: STAT6-deficient mice have enhanced tumor immunity to primary and metastatic mammary carcinoma. J Immunol. 2000;165:6015–6019. doi: 10.4049/jimmunol.165.11.6015. [DOI] [PubMed] [Google Scholar]
- 116.Ostrand-Rosenberg S, Clements VK, Terabe M, Park JM, Berzofsky JA, Dissanayake SK. Resistance to metastatic disease in STAT6-deficient mice requires hemopoietic and nonhemopoietic cells and is IFNgamma dependent. J Immunol. 2002;169:5796–5804. doi: 10.4049/jimmunol.169.10.5796. [DOI] [PubMed] [Google Scholar]
- 117.Jensen SM, Meijer SL, Kurt RA, Urba WJ, Hu HM, Fox BA. Regression of a mammary adenocarcinoma in STAT6-/- mice is dependent on the presence of STAT6-reactive T cells. J Immunol. 2003;170:2014–2021. doi: 10.4049/jimmunol.170.4.2014. [DOI] [PubMed] [Google Scholar]
- 118.Fukushi J, Morisaki T, Shono T, Nishie A, Torisu H, Ono M, et al. Novel biological functions of interleukin-4: Formation of tube-like structures by vascular endothelial cells in vitro and angiogenesis in vivo. Biochem Biophys Res Commun. 1998;250:444–448. doi: 10.1006/bbrc.1998.9334. [DOI] [PubMed] [Google Scholar]
- 119.Fukushi J, Ono M, Morikawa W, Iwamoto Y, Kuwano M. The activity of soluble VCAM-1 in angiogenesis stimulated by IL-4 and IL-13. J Immunol. 2000;165:2818–2823. doi: 10.4049/jimmunol.165.5.2818. [DOI] [PubMed] [Google Scholar]
- 120.Puri RK, Leland P, Kreitman RJ, Pastan I. Human neurological cancer cells express interleukin-4 (IL-4) receptors which are targets for the toxic effects of IL4-Pseudomonas exotoxin chimeric protein. Int J Cancer. 1994;58:574–581. doi: 10.1002/ijc.2910580421. [DOI] [PubMed] [Google Scholar]
- 121.Leland P, Taguchi J, Husain SR, Kreitman RJ, Pastan I, Puri RK. Human breast carcinoma cells express type II IL-4 receptors and are sensitive to antitumor activity of a chimeric IL-4-Pseudomonas exotoxin fusion protein in vitro and in vivo. Mol Med. 2000;6:165–178. [PMC free article] [PubMed] [Google Scholar]
- 122.Kawakami M, Kawakami K, Stepensky VA, Maki RA, Robin H, Muller W, et al. Interleukin 4 receptor on human lung cancer: A molecular target for cytotoxin therapy. Clin Cancer Res. 2002;8:3503–3511. [PubMed] [Google Scholar]
- 123.Shimamura T, Royal RE, Kioi M, Nakajima A, Husain SR, Puri RK. Interleukin-4 cytotoxin therapy synergizes with gemcitabine in a mouse model of pancreatic ductal adenocarcinoma. Cancer Res. 2007;67:9903–9912. doi: 10.1158/0008-5472.CAN-06-4558. [DOI] [PubMed] [Google Scholar]
- 124.Ishige K, Shoda J, Kawamoto T, Matsuda S, Ueda T, Hyodo I, et al. Potent in vitro and in vivo antitumor activity of interleukin-4-conjugated Pseudomonas exotoxin against human biliary tract carcinoma. Int J Cancer. 2008;123:2915–2922. doi: 10.1002/ijc.23865. [DOI] [PubMed] [Google Scholar]
- 125.Koller FL, Hwang DG, Dozier EA, Fingleton B. Epithelial interleukin-4 receptor expression promotes colon tumor growth. Carcinogenesis. 2010;31:1010–1017. doi: 10.1093/carcin/bgq044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Conticello C, Pedini F, Zeuner A, Patti M, Zerilli M, Stassi G, et al. IL-4 protects tumor cells from anti-CD95 and chemotherapeutic agents via upregulation of antiapoptotic proteins. J Immunol. 2004;172:5467–5477. doi: 10.4049/jimmunol.172.9.5467. [DOI] [PubMed] [Google Scholar]
- 127.Todaro M, Zerilli M, Ricci-Vitiani L, Bini M, Perez Alea M, Maria Florena A, et al. Autocrine production of interleukin-4 and interleukin-10 is required for survival and growth of thyroid cancer cells. Cancer Res. 2006;66:1491–1499. doi: 10.1158/0008-5472.CAN-05-2514. [DOI] [PubMed] [Google Scholar]
- 128.Todaro M, Lombardo Y, Francipane MG, Alea MP, Cammareri P, Iovino F, et al. Apoptosis resistance in epithelial tumors is mediated by tumor-cell-derived interleukin-4. Cell Death Differ. 2008;15:762–772. doi: 10.1038/sj.cdd.4402305. [DOI] [PubMed] [Google Scholar]
- 129.Li Z, Jiang J, Wang Z, Zhang J, Xiao M, Wang C, et al. Endogenous interleukin-4 promotes tumor development by increasing tumor cell resistance to apoptosis. Cancer Res. 2008;68:8687–8694. doi: 10.1158/0008-5472.CAN-08-0449. [DOI] [PubMed] [Google Scholar]
- 130.Prokopchuk O, Liu Y, Henne-Bruns D, Kornmann M. Interleukin-4 enhance proliferation of human pancreatic cancer cells: evidence for autocrine and paracrine actions. Br J Cancer. 2005;92:921–928. doi: 10.1038/sj.bjc.6602416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Todaro M, Alea MP, Di Stefano AB, Cammareri P, Vermeulen L, Iovino F, et al. Colon cancer stem cells dictate tumor growth and resist cell death by production of interleukin-4. Cell Stem Cell. 2007;1:389–402. doi: 10.1016/j.stem.2007.08.001. [DOI] [PubMed] [Google Scholar]
- 132.Joyce JA, Pollard JW. Microenvironmental regulation of metastasis. Nat Rev Cancer. 2009;9:239–252. doi: 10.1038/nrc2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Whitehead RP, Lew D, Flanigan RC, Weiss GR, Roy V, Glode ML, et al. Phase II trial of recombinant human interleukin-4 in patients with advanced renal cell carcinoma: A Southwest Oncology Group study. J Immunother. 2002;25:352–358. doi: 10.1097/00002371-200207000-00007. [DOI] [PubMed] [Google Scholar]
- 134.Whitehead RP, Unger JM, Goodwin JW, Walker MJ, Thompson JA, Flaherty LE, et al. Phase II trial of recombinant human interleukin-4 in patients with disseminated malignant melanoma: A Southwest Oncology Group study. J Immunother. 1998;21:440–446. doi: 10.1097/00002371-199811000-00006. [DOI] [PubMed] [Google Scholar]
- 135.Kawakami M, Kawakami K, Puri RK. Interleukin-4-Pseudomonas exotoxin chimeric fusion protein for malignant glioma therapy. J Neurooncol. 2003;65:15–25. doi: 10.1023/a:1026294416718. [DOI] [PubMed] [Google Scholar]
- 136.Puri S, Mahapatra AK, Hussain E, Sarkar C, Sinha S, Joshi BH. A review of studies on targeting interleukin 4 receptor for central nervous system malignancy. Curr Mol Med. 2009;9:732–739. doi: 10.2174/156652409788970661. [DOI] [PubMed] [Google Scholar]
- 137.Huang JT, Welch JS, Ricote M, Binder CJ, Willson TM, Kelly C, et al. Interleukin-4-dependent production of PPAR-gamma ligands in macrophages by 12/15-lipoxygenase. Nature. 1999;400:378–382. doi: 10.1038/22572. [DOI] [PubMed] [Google Scholar]
- 138.Kikuchi H, Hanazawa S, Takeshita A, Nakada Y, Yamashita Y, Kitano S. Interleukin-4 acts as a potent stimulator for expression of monocyte chemoattractant JE/MCP-1 in mouse peritoneal macrophages. Biochem Biophys Res Commun. 1994;203:562–569. doi: 10.1006/bbrc.1994.2219. [DOI] [PubMed] [Google Scholar]
- 139.Liddiard K, Welch JS, Lozach J, Heinz S, Glass CK, Greaves DR. Interleukin-4 induction of the CC chemokine TARC (CCL17) in murine macrophages is mediated by multiple STAT6 sites in the TARC gene promoter. BMC Mol Biol. 2006;7:45. doi: 10.1186/1471-2199-7-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Bonecchi R, Sozzani S, Stine JT, Luini W, D'Amico G, Allavena P, et al. Divergent effects of interleukin-4 and interferon-gamma on macrophage-derived chemokine production: An amplification circuit of polarized T helper 2 responses. Blood. 1998;92:2668–2671. [PubMed] [Google Scholar]
- 141.Watanabe K, Jose PJ, Rankin SM. Eotaxin-2 generation is differentially regulated by lipopolysaccharide and IL-4 in monocytes and macrophages. J Immunol. 2002;168:1911–1918. doi: 10.4049/jimmunol.168.4.1911. [DOI] [PubMed] [Google Scholar]