Abstract
Background
. The National Heart, Lung, and Blood Institute’s Candidate Gene Association Resource (CARe), a planned cross-cohort analysis of genetic variation in cardiovascular, pulmonary, hematological, and sleep-related traits, comprises more than 40,000 participants representing four ethnic groups in nine community-based cohorts. The goals of CARe include the discovery of new variants associated with traits using a candidate gene approach and the discovery of new variants using the genome-wide association mapping approach specifically in African Americans.
Methods and Results
. CARe has assembled DNA samples for more than 40,000 individuals self-identified as European-American, African-American, Hispanic, or Chinese-American, with accompanying data on hundreds of phenotypes that have been standardized and deposited in the CARe Phenotype Database. All participants were genotyped for seven single-nucleotide polymorphisms (SNPs) selected based on prior association evidence. We performed association analyses relating each of these SNPs to lipid traits, stratified by gender and ethnicity and adjusted for age and age2. In at least two of the ethnic groups, SNPs near CETP, LIPC, and LPL strongly replicated for association with high-density lipoprotein cholesterol concentrations, PCSK9 with low-density lipoprotein cholesterol levels, and LPL and APOA5 with serum triglycerides. Notably, some SNPs showed varying effect sizes and significance of association in different ethnic groups.
Conclusions
. The CARe Pilot Study validates the operational framework for phenotype collection, SNP genotyping, and analytical pipeline of the CARe project and validates the planned candidate gene study of ~2,000 biologic candidate loci in all participants and genome-wide association study in ~8,000 African-American participants. CARe will serve as a valuable resource for the scientific community.
Keywords: Genetics, lipids, diabetes, blood pressure, epidemiology
INTRODUCTION
A key goal of biomedical research is to understand how genetic variation contributes to inter-individual differences in risk for disease. Despite many years of effort, the DNA sequence variants and underlying genes that affect complex genetic diseases such as myocardial infarction or type 2 diabetes and risk factors such as blood lipoprotein levels or body weight in humans remain mostly unknown. Critical questions that remain largely unanswered include which biological pathways are altered in patients in a manner that contributes causally to disease and might therefore be the best targets for interventions and therapies.
Genetic association studies, both genome-wide and those based on biologic candidates, have proven to be robust tools to discover genes associated with disease processes, potentially leading to novel therapeutics, personalized medicine and preventive programs. With genotyping efforts and association studies being conducted at an increasing number of institutions, it has become critical to establish collaborations or a centralized database where shared resources for analyses of the association of genotypes with phenotypes relevant to the biomedical community are located. The majority of large-scale genetic studies that have been completed have focused on whites of European ancestry, and have had limited representation of other ethnic groups; specifically, there has been a paucity of large-scale population-based studies that have included African Americans, Hispanics, and Chinese Americans. The application of candidate gene and genome-wide association approaches to a large number of individuals from a variety of ethnic groups promises to comprehensively advance our understanding of the heritability and biology of many diseases and traits.
The Candidate Gene Association Resource (CARe), a unique initiative of the National Heart, Lung, and Blood Institute (NHLBI), seeks to assemble well-characterized phenotypic data from nine NHLBI cohorts, to generate new genotype data across candidate genes and/or the whole genome, and to perform genotype-phenotype association analyzes across more than 100 cardiovascular, pulmonary, hematological, and sleep-related traits available from the different cohorts. The nine cohorts include the Atherosclerosis Risk In Communities (ARIC) study,1 the Coronary Artery Risk Development in Young Adults (CARDIA) study,2 the Cleveland Family Study (CFS),3 the Cardiovascular Health Study (CHS),4 the Cooperative Study of Sickle Cell Disease (CSSCD),5 the Framingham Heart Study (FHS),6–8 the Jackson Heart Study (JHS),9 the Multi-Ethnic Study of Atherosclerosis (MESA),10 and the Sleep Heart Health Study (SHHS)11 (Table 1).
Table 1.
Study design and participant characteristics*
| Study | ARIC | JHS | MESA | CSSCD | CFS | FHS | CHS | CARDIA | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gen. 1 | Gen. 2 | Gen. 3 | All | |||||||||||||||
| Ancestry composition | AA | EA | AA | AA | EA | HIS | CHI | AA | AA | EA | EA | EA | EA | EA | AA | EA | AA | EA |
| N (phenotypes) | 4,141 | 11,412 | 2,702 | 1,754 | 2,537 | 1,475 | 778 | 3,205 | 721 | 722 | 2,511 | 3,765 | 4,082 | 10, 358 | 842 | 4,653 | 2,637 | 2,478 |
| Ascertainment scheme | Community-based | Community-based | Population-based | Patient-based | Family-based | Community-based | Community-based | Community-based | Community-based | Community-based | Community-based | |||||||
| Recruitment period | 1987–1998 | 2000–2004 | 2000–2007 | 1979–1988† | 1990–2005 | 1948–1953 | 1971–1975 | 2002–2005 | 1989–1993 | 1985–1986 | ||||||||
| Mean age (y) | 54±6 | 54±6 | 52±13 | 62±10 | 63±10 | 61±10 | 62±10 | 17±20 | 39±20 | 42±20 | 69±8 | 44±10 | 40±9 | 49±15 | 73±6 | 73±6 | 24±4 | 25±3 |
| Female sex (%) | 62 | 53 | 62 | 55 | 52 | 52 | 51 | 51 | 57 | 52 | 60 | 52 | 53 | 54 | 63 | 56 | 56 | 53 |
| Total cholesterol, mg/dl | 215±45 | 215±50 | 197±39 | 190±36 | 196±35 | 198±37 | 192±31 | - | 171±42 | 181±40 | 229±42 | 204±39 | 189±35 | 204±41 | 210±40 | 211±39 | 177±34 | 176±32 |
| LDL cholesterol, mg/dl | 137±43 | 138±40 | 125±36 | 116±33 | 117±30 | 120±33 | 115±29 | - | 96±33 | 101±30 | - | 130±35 | 112±32 | 120±34 | 129±37 | 130±36 | 110±32 | 108±30 |
| HDL cholesterol, mg/dl | 55 ± 18 | 50 ± 17 | 51 ± 15 | 52 ± 15 | 52 ± 16 | 48 ± 13 | 49 ± 12 | - | 48 ± 15 | 46 ± 13 | 50 ± 16 | 48 ± 13 | 54 ± 16 | 51 ± 15 | 60 ± 16 | 53 ± 16 | 54 ± 13 | 51 ± 13 |
| Triglycerides, mg/dl | 114±81 | 138±93 | 109±98 | 105±69 | 133±90 | 157±101 | 143±85 | - | 102±62 | 143±118 | - | - | 116±90 | 112±95 | 116±63 | 144±79 | 67±38 | 79±57 |
| Systolic BP, mmHg | 129±22 | 119±17 | 126±18 | 131 | 124±21 | 127±22 | 125±22 | 106±16 | 123±18 | 121±16 | 137±19 | 122±17 | 117±14 | 124±18 | 142±23 | 136±22 | 111±11 | 109±11 |
| Hypertension(%) | 58 | 33 | 58 | 62 | 44 | 44 | 41 | 4 | 38 | 29 | 56 | 23 | 11 | 26 | 78 | 64 | 5 | 3 |
| Type 2 diabetes mellitus (%)* | 20 | 9 | 18 | 18 | 6 | 18 | 14 | .3 | 14 | 9 | 9 | 3 | 3 | 4 | 25 | 15 | 1 | .5 |
| Body mass index (kg/m2) | 29.6±6 | 27.0±5 | 31.9±7 | 30.0±6 | 27.7±5 | 29.4±5 | 23.9±3 | 18.9±9 | 32.0±10 | 30.1±9 | 26.2±4 | 25.6±4 | 26.9±6 | 26.0±5 | 28.0±6 | 26.3±4 | 25.2±6 | 23.5±4 |
| Lipid lowering therapy (%) | 1 | 3 | 11 | 16 | 18 | 13 | 14 | - | 8 | 8 | 3 | 1 | 7 | 4 | 7 | 5 | - | - |
| Anti-hypertensive therapy (%) | 44 | 26 | 49 | 50 | 33 | 33 | 29 | 1 | 32 | 23 | 32 | 10 | - | 19 | 63 | 45 | 1 | 1 |
| Reference for study design | 1 | 9 | 10 | 5 | 3 | 6–8 | 4 | 2 | ||||||||||
Values with ‘±’ are means ± standard deviation. The body-mass index is the weight in kilograms divided by the square of the height in meters. Hypertension was defined as systolic blood pressure ≥ 140 mmHg, diastolic blood pressure ≥ 90 mmHg, or use of anti-hypertensive therapy. AA = African American; EA = European American; HIS = Hispanic; CHI = Chinese American.
The Sleep Heart Health Study (SHHS)11 is not represented in this table because the subgroup of the SHHS cohort with genotype data comprises individuals originally recruited from ARIC, CHS, and FHS (and who are included in this table under these other cohorts). A number of individuals are participants in both ARIC and JHS; in this table they are included only in ARIC.
1979–1981 >6 months of age; 1979–1988 <6 months of age
The overall goals of CARe include: (1) the discovery of new variants associated with traits using a candidate gene approach; (2) the discovery of new variants using the genome-wide association mapping approach in ~8,000 African Americans across the cohorts; (3) characterization of validated variants across clinical subgroups stratified by ethnicity, gender, or clinical covariates; and (4) exploration of gene-environment interactions. CARe comprises two major components—candidate gene studies in the combined population of more than 40,000 individuals, and genome-wide association studies in ~8,000 African-American participants.
We conducted the CARe Pilot Study to validate the operational framework of CARe with respect to the phenotype collection, SNP genotyping, and construction of an analytical pipeline for genotype-phenotype association studies by (1) studying a pilot set of SNPs originally identified in European-derived cohorts and (2) assessing whether previously reported associations of SNPs with select traits in European Americans would replicate in other ethnic groups, namely African Americans, Hispanics, and Chinese Americans. The latter exercise will set the stage for multi-ethnic genetic association studies (CARe-wide and beyond). In the CARe Pilot Study, we analyzed 7 SNPs with prior evidence of association (Table 2) with respect to plasma levels of high-density lipoprotein cholesterol (HDL-C), low-density lipoprotein cholesterol (LDL-C), or triglycerides.
Table 2.
Pilot SNPs and associated phenotypes tested in Design Study
| Gene | Symbol | Polymorphism | Associated phenotype | rs number | CEU MAF | YRI MAF | Reference |
|---|---|---|---|---|---|---|---|
| Apolipoprotein A5 | APOA5 | Ser19Trp | Triglycerides | rs3135506 | 0.06 | 0.05 | 12 |
| Cholesteryl ester transfer protein | CETP | C–1337T | HDL-C | rs17231506 | 0.37 | 0.03 | 13 |
| Cholesteryl ester transfer protein | CETP | G–971A | HDL-C | rs4783961 | 0.46 | 0.42 | 14 |
| Hepatic lipase | LIPC | C–480T | HDL-C | rs1800588 | 0.26 | 0.47 | 15 |
| Lipoprotein lipase | LPL | Ser447X/Ser474X | HDL-C, triglycerides | rs328 | 0.13 | 0.03 | 16 |
| Proprotein convertase subtilisin/kexin type 9 | PCSK9 | Arg46Leu | LDL-C | rs11591147 | 0.00 | 0.00 | 17 |
| Proprotein convertase subtilisin/kexin type 9 | PCSK9 | Glu670Gly | LDL-C | rs505151 | 0.04 | 0.31 | 18 |
CEU = European descent (HapMap); YRI = Yoruban (HapMap); MAF = minor allele frequency in HapMap
DESIGN AND METHODS
CARe Study Conception and Design
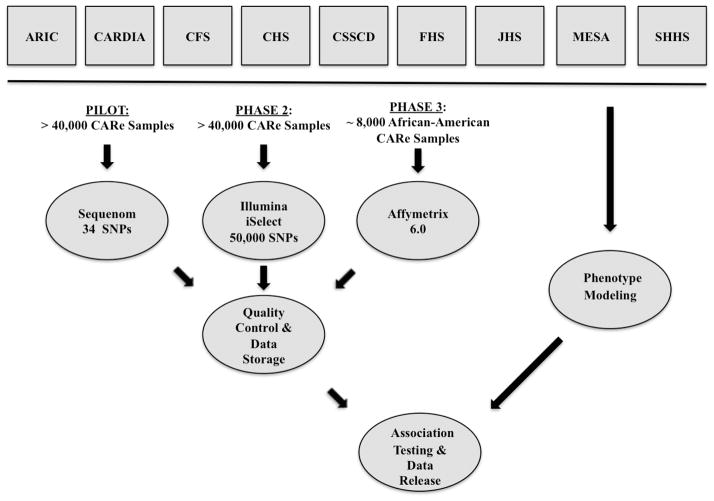
The CARe Study was initiated by the NHLBI in 2006. DNA samples and phenotypic information from the nine NHLBI cohorts—ARIC, CARDIA, CFS, CHS, CSSCD, FHS, JHS, MESA, and SHHS—were sent to the Broad Institute of MIT and Harvard and deposited in the CARe Phenotype Database. The overall CARe procedures include: (1) assembly of phenotypes into a single database; (2) genotyping of assembled DNA samples using three platforms: Sequenom for the Pilot Study, the ITMAT-Broad-CARe (IBC) array for candidate gene studies, and the Affymetrix 6.0 array for genome-wide association studies; (3) quality control procedures on genotype data; (4) establishment of a secure data repository to allow approved researchers to access phenotype and genotype data to pursue hypothesis testing (while meticulously maintaining full confidentiality for the study participants); (5) statistical modeling of the phenotypes of interest; (6) statistical analyses to identify associations between genotypes and phenotypes of interest; and (7) use of existing, public, internet-based resources to disseminate summary data from the GWAS and candidate gene studies to the wider scientific community (Figure 1). The availability of these data to the scientific community will (a) maximize the scientific utility to be gained from the investment in sample collection, genotyping, and phenotyping, (b) facilitate the replication and extension of CARe-derived results in other cohorts, and (c) foster the development of additional analytical and computational methods that can be tested on a large-scale genetic dataset.
Figure 1.
Operational framework of CARe.
Governance
The CARe project is overseen by a Steering Committee comprising representatives from each of the nine NHLBI cohorts as well as the Broad Institute of MIT and Harvard and the University of Pennsylvania; under the Steering Committee are a number of Subcommittees and Working Groups that are responsible for the execution of various aspects of the CARe project (Supplemental Figure 1). NHLBI staff members participate in the meetings and teleconferences of the various committees and groups. An Oversight Committee appointed by the NHLBI monitors the progress of the CARe project.
Phenotype Database Construction
The phenotypes that were assembled for CARe include phenotypes collected at the baseline and follow-up examinations of participants in each of the nine NHLBI cohorts. Each CARe cohort was contacted to contribute the phenotypes, and once each cohort dataset was received, they were deposited in the CARe Phenotype Database. Available phenotypes that have been cataloged in the CARe Phenotype Database range from hundreds to many thousands depending on the cohort. Descriptions of phenotype groupings and examples of phenotypes are available in Table 3 and at the CARe website: http://www.broad.mit.edu/gen_analysis/care/index.php/Main_Page. When they are requested for use by investigators, phenotypes are to be standardized according to the Clinical Data Interchange Standard Consortium Study Data Tabulation Model (CDISC SDTM) available at the website: http://www.cdisc.org/models/sds/v3.1/index.html.
Table 3.
Phenotype categories available in CARe database
| Category | Example phenotypes |
|---|---|
| Aging | Age at examination |
| Age at menopause | |
| Age at birth of first child | |
| Age at birth of last child | |
| Anthropometry | Height |
| Weight | |
| Body-mass index | |
| Waist circumference | |
| Atrial fibrillation/electrocardiography | Atrial fibrillation |
| PR interval | |
| QT interval | |
| Blood biomarkers | C-reactive protein |
| MCP-1 | |
| IL-6 | |
| Fibrinogen | |
| Factor VII | |
| Blood cell counts | |
| Blood pressure/hypertension | Systolic blood pressure |
| Diastolic blood pressure | |
| Pulse pressure | |
| Mean arterial pressure | |
| Hypertension | |
| Hypertension medications | |
| Coronary heart disease | Coronary heart disease |
| Diabetes mellitus/glucose | Type 2 diabetes mellitus |
| Age at diabetes diagnosis | |
| Diabetes medications | |
| Diabetic retinopathy | |
| Hemoglobin A1C | |
| Fasting blood glucose | |
| Fasting insulin | |
| Insulin use | |
| Echocardiography/congestive heart failure | Heart failure |
| Left ventricular mass | |
| Left ventricular ejection fraction | |
| Right ventricular ejection fraction | |
| Kidney disease | Creatinine |
| Estimated glomerular filtration rate | |
| Cystatin C | |
| Urinary albumin:creatinine ratio | |
| Lipids | Total cholesterol |
| HDL cholesterol | |
| LDL cholesterol | |
| Triglycerides | |
| ApoB | |
| Apo A-I | |
| Lipid-lowering therapy | |
| Musculoskeletal | Gout |
| Serum urate | |
| Peripheral arterial disease | Peripheral arterial disease |
| Ankle-brachial index | |
| Pulmonary function | FEV1 |
| FVC | |
| FEF25-75 | |
| Sleep | Sleep apnea |
| Snoring | |
| Epworth Sleepiness Scale | |
| Excessive Daytime Sleepiness | |
| Smoking | Smoking status |
| Pack-year history | |
| Smoking intensity | |
| Stroke | Incident stroke |
| Ischemic stroke | |
| Subclinical atherosclerosis | Coronary artery calcification |
| Carotid intima-media thickness | |
Three of these phenotypes were used for the Pilot Study—HDL-C, LDL-C, and triglycerides. Values of these traits (except LDL-C, which was calculated—see below) determined at baseline visits for each cohort were used. Of note, the subgroup of the SHHS cohort with genotype data comprises individuals originally recruited from ARIC, CHS, and FHS and subsequently evaluated for sleep phenotypes. In this paper, SHHS participants appear under their parent cohorts.
Genotyping and Quality Control
CARe Pilot (Sequenom)
After appropriate Material Transfer Agreements were in place, samples were shipped from each cohort’s central genetics laboratory to the Broad Institute of MIT and Harvard. DNA concentration was determined by the Picogreen assay (Invitrogen, Carlsbad, California) before storage in 2D-barcoded 0.75 mL Matrix tubes at −20°C in the SmaRTStore™ (RTS, Manchester, UK) automated sample handling system. Seven SNPs were selected for the CARe Pilot Study based on previously published evidence of association (see Table 2 for SNPs and references). These SNPs were genotyped using the Sequenom MassArray System platform (Sequenom, San Diego, California). All DNA samples passing initial quality checks were plated at a concentration of 5 ng/μl for processing on the platform. Sequenom SNP genotyping uses bead-less and label-free primer-extension chemistry in a multiplex format to generate allele-specific products with distinct masses that are distinguished by mass spectroscopy. The results were automatically loaded into a database and then scored using SpectroTyper 4.0 Software (Sequenom) and uploaded to the laboratory information management system and data storage system.
Several quality control (QC) procedures were performed on the genotype data, separately for each cohort (Supplemental Table 1). Sample duplicates were identified using sample IDs. For each set of duplicates or monozygotic twins, data from the sample with the highest genotyping success rate was retained. Reported sex and genotype-inferred sex (two independent Sequenom assays for each sample) were compared for concordance. All discordant samples and samples for which no sex information was available were resolved in consultation with the relevant cohort or excluded. SNPs with a missing data rate above 10% and samples with a genotyping success rate below 90% were removed. Only samples with available phenotypic information were used for association studies. In the cohorts with available family information—CFS and FHS—data of descendants from families accounting for the most Mendelian errors in the dataset were excluded. Because several different ethnic groups were represented, with the expectation of differing genotype frequencies and admixture, no filters were applied for minor allele frequency or Hardy-Weinberg P values. All QC analyses were performed in PLINK.19
Candidate Gene Studies (Illumina iSelect–IBC Chip)
The design of the IBC Chip, a custom 50K SNP genotyping array, has been described recently.20 The SNPs (49,320 total) were chosen to densely map about 2,000 candidate gene loci deemed to be relevant to phenotypes available in the CARe Phenotype Database. All DNA samples passing initial quality checks were interrogated with the IBC chip. Analyses with this genotype data are not included in this manuscript, but rather will be the focus of future CARe studies.
GWAS (Affymetrix 6.0 Array Set)
Approximately 8,000 African-American participants from five of the CARe cohorts—ARIC, JHS, CARDIA, CFS, and MESA—were genotyped with the Affymetrix 6.0 (“million-SNP”) Array Set (Affymetrix, Santa Clara, California), typing more than 906,600 SNPs and 946,000 probes for copy-number variation across the genome. The genotype data so obtained will be used for GWAS on phenotypes of interest. Analyses with this genotype data are not included in this manuscript, but rather will be the focus of future CARe studies.
Data Management
A key goal of CARe is to prepare a comprehensive genotype and phenotype dataset that serves as a scientific resource that is broadly accessible to the research community. This was performed taking care to protects the confidentiality and interests of study participants and consistent with the informed consent procedures in each of the cohorts. The Institutional Review Boards (IRBs) of each CARe cohort (i.e., the IRBs for each cohort’s field centers, coordinating center, and laboratory center) have reviewed the cohort’s interaction with CARe. CARe itself has been approved by the Committee on the Use of Humans as Experimental Subjects (COUHES) of the Massachusetts Institute of Technology. Identifiers were removed and codes were assigned to any protected health information (PHI) transmitted to the CARe Data Repository, with a Certificate of Confidentiality issued by the National Institutes of Health. The Data Repository will release limited datasets to qualifying investigators whose projects have been approved by their local IRBs and who have completed a CARe Data Distribution Agreement. Each such dataset will have its own unique, randomly generated set of participant identifiers.
Of note, at the time of study, not all subjects had provided specific consent for data to be made available to non-CARe investigators. Accordingly, upon request public access to the data will be provided to the extent that the informed consent process allows.
Phenotype Modeling
For the CARe Pilot Study, we modeled the pilot phenotypes in the following ways. LDL-C was calculated according to Friedewald’s formula: LDL-C = total cholesterol – HDL-C –(triglycerides ÷ 5). If a triglyceride value was > 400 mg/dL, LDL-C was treated as a missing value. For individuals on lipid-lowering therapy, the LDL-C value was multiplied by 1.42 to model a 30% reduction in LDL-C on therapy. This represents the average expected reduction in LDL-C with a first-generation statin, the most commonly used lipid-lowering medication during the study periods of most of the cohorts.21 Triglyceride values were log(10)-transformed. Sex-specific phenotype residuals were constructed within strata of cohort and ethnicity with adjustment for age and age2 in each individual stratum. Each set of residuals was standardized to a mean of zero and a standard deviation of one. The standardized residual served as the phenotype in genotype-phenotype association analyses.
Generation of residuals was performed with the R statistical package (The R Foundation for Statistical Computing, Vienna, Austria).
Association Testing
Cohorts were divided into subgroups by ethnicity; association analyses were performed for each subgroup, followed by meta-analysis of the subgroups for each ethnicity. For cohorts in which individuals were largely unrelated or when family information was not available—ARIC, CARDIA, CHS, CSSCD, JHS, and MESA—we used linear regression to test SNP-phenotype associations assuming an additive genetic model. These association analyses were performed in PLINK. For the two cohorts for which there were significant numbers of related individuals, and for which family information was available at the time of the Pilot Study—CFS and FHS—we used a linear mixed effects (LME) model to analyze the traits, with the SNP genotype treated as a fixed effect, and a random effect according to the degree of relatedness within a family.22 Genotype-phenotype associations within each ethnic group were assessed by variance-weighted meta-analyses, and heterogeneity within each ethnic group or between ethnic groups was assessed using Cochran’s Q statistic and/or the I2 inconsistency metric.23
For future GWAS and candidate gene studies, association analyses will include procedures to account for the effects of local and global ancestry, particularly with regard to African-American subjects. Given the small number of SNPs addressed in the Pilot Study, such procedures were not feasible.
The authors had full access to the data and take responsibility for its integrity. All authors have read and agree to the manuscript as written.
RESULTS
Baseline Characteristics of CARe Cohorts
Characteristics of each individual cohort’s study participants with respect to the pilot phenotypes are presented in Table 1. (See also Supplemental Table 1.) At the time of the Pilot Study, the combined sample that had been received and processed at the Broad Institute and undergone successful genotyping and association analyses included 40,324 total individuals, of whom 26,647 were European Americans, 11,550 African Americans, 1,410 Hispanics, and 717 Chinese Americans. The cohorts varied with regard to the mean age, the proportions with T2D, and the BMI of the participants. Mean lipid and SBP values were similar across cohorts. Of note, lipid phenotypes were not available for CSSCD individuals and, thus, these individuals were not included in analyses for HDL-C, LDL-C, and triglycerides.
Evidence for Admixture in African American Cohorts
We compared minor allele frequencies (MAFs) for each of the pilot SNPs in the African American cohorts and European American cohorts (Supplemental Table 2) with the (MAFs) for the SNPs in the Yoruba (YRI) and European-descended (CEU) groups in the International HapMap Project (Table 2).24 These comparisons suggest all of the African American cohorts have a significant degree of admixture of African and European chromosomes. For example, for rs17231506 in the CETP locus, the MAFs range from 10% to 16% in the African American cohorts, whereas the European American cohorts range from 31% to 33%. The European American cohorts’ MAFs are consistent with the CEU MAF of 37%, whereas the African American cohorts’ MAFs are intermediate between those of CEU and YRI (3%). This is generally true for all of the pilot SNPs whose MAFs differ greatly between the CEU and YRI groups.
Pilot Phenotype-Genotype Association Results
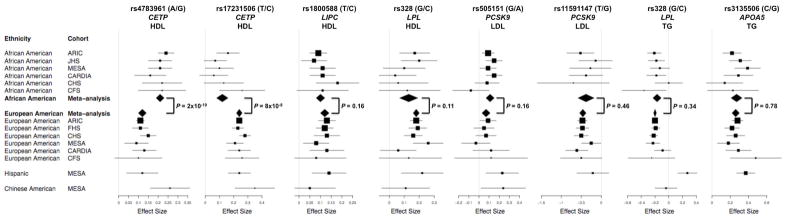
We performed association analyses for each of the seven pilot SNPs against each of the relevant pilot phenotypes (i.e., the phenotypes with which they had previously been shown to have association, Table 2) in each ethnic group within each cohort, assuming additive genetic models. The results of the analyses are presented in Figure 2 and Supplemental Table 2. To account for multiple testing, we considered a P value of 1 × 10−4 to represent the threshold of statistical significance. We performed meta-analyses and heterogeneity analyses for each SNP-phenotype combination (Supplemental Table 2).
Figure 2.
Forest plots for SNP-phenotype effects, with Cochran’s Q P values.
A SNP in the CETP gene, rs17231506, was associated with HDL-C in all four ethnic groups: African Americans, European Americans, Hispanics, and Chinese Americans. Another SNP in CETP, rs4783961, was associated with HDL-C in African Americans, European Americans, and Chinese Americans. Other HDL-C-associated SNPs were rs1800588 in LIPC and rs328 in LPL, both in African Americans and European Americans. Similarly, triglyercide-related SNPs were replicated in both African Americans and European Americans: rs328 in LPL and rs3135506 in APOA5. LDL-C-related SNPs were associated in either African Americans or European Americans but not both: rs505151 in PCSK9 was associated in African Americans but not in European Americans, whereas rs11591147 in PCSK9 was associated with LDL-C in European Americans but not in African Americans.
We found that within each ethnicity, there was low heterogeneity of effect of each SNP on each trait (Supplemental Table 2). For example, for the SNP rs4783961 in CETP and HDL-C, the direction of the association was consistent across all the cohorts, with the G allele representing higher HDL-C levels. The effect sizes (beta coefficients) associated with this allele were remarkably consistent across the African-American cohorts (ranging from 0.17 to 0.24) and across the European American cohorts (0.09 to 0.15). Formal heterogeneity analyses showed very low heterogeneity among the two sets of cohorts, with the I2 inconsistency metric being 0% for each set. Indeed, for all of the statistically significant SNP-phenotype associations, in many cases the I2 metric was 0%, in no case exceeding 50%; similarly, in every case the Cochran’s Q P value for heterogeneity was nonsignificant (P > 0.05).
There were notable differences in effect sizes across ethnic groups for some of the SNPs (Figure 2). For example, at rs4783961 in CETP and HDL-C, the effect size was uniformly larger in African Americans (0.17 to 0.24) than in European Americans (0.09 to 0.15) (P = 2 × 10−10 by Cochran’s Q heterogeneity test). The effect size for this variant in Hispanics (0.12) was more similar to European Americans, and in Chinese Americans (0.26) with African Americans. Unlike rs4783961, the HDL-C-associated SNP rs17231506, also in CETP, had larger effect sizes in European Americans and Hispanics (0.21 to 0.28) compared with African Americans (0.06 to 0.26) (P = 8 × 10−8 between European Americans and African Americans), though smaller than in Chinese Americans (0.35). Thus, even though the same gene locus (CETP) was highly associated with HDL-C across the ethnicities, there were differences between ethnicities in the contributions of individual SNPs at the locus to inter-individual variation in HDL-C levels.
We observed inter-ethnic differences with other gene loci vis-à-vis other phenotypes. The rs505151 SNP in the PCSK9 locus had stronger statistical association with LDL-C in African Americans than in European Americans, though this is attributable to the higher minor allele frequency in African Americans (~25%) than in European Americans (~4%).
Other SNPs appear to affect phenotypes to similar degrees across ethnic groups: rs1800588 in LIPC with HDL-C; rs3135506 in APOA5 with triglycerides; and rs328 in LPL with both HDL-C and triglycerides.
DISCUSSION
The CARe Pilot Study was designed to evaluate the operational framework established for the resource, including phenotype collection and integration, routing of a large number of DNA samples for genotyping analyses, quality control procedures on all the collected data, data analysis, and synthesis of the results. Our ability to obtain robust phenotype-genotype associations for SNPs with strong prior evidence in the published literature validates the CARe study framework and sets the stage for the candidate gene and GWAS discovery phases of CARe that are in progress. The Pilot Study also afforded the scientific opportunity to evaluate the effects of DNA variants on clinically important traits in the largest group of African-American individuals with genotype information assembled to date. The critical findings from the pilot phenotype-genotype associations were that: (1) there was low heterogeneity for highly associated SNPs within cohorts of the same ethnicity; (2) for some associated SNPs there were inter-ethnic differences in effect size; and (3) each gene replicated in multiple ethnic groups, although not necessarily through the same SNPs.
The finding of low heterogeneity with the lipid traits suggests that despite the expected variation in phenotypes among the cohorts due to the use of different assays or different disease definitions, at least some of the phenotypes that have been collected in CARe from different cohorts can be standardized and, upon appropriate analysis, yield meaningful scientific results.
We found that for several SNPs in replicated gene loci—most notably in CETP and PCSK9—the associations with phenotype differed between African Americans and European Americans. For example, whereas one CETP SNP (rs4783961) had a larger effect size in African American cohorts, another SNP in the same locus (rs17231506) showed a larger effect size in European American cohorts. One possible explanation for this observation is that African Americans and European Americans share the same causal variant in a gene, but due to ethnicity-specific differences in the major and minor allele frequencies, a SNP may have differing strength of correlation with the causal variant, which manifests as varying effect sizes and the degree of association. In some cases, the inter-ethnic differences are observed despite similar allele frequencies (e.g., rs4783961 in African Americans and European Americans). An alternative explanation is that inter-ethnic differences in the linkage disequilibrium patterns in the gene locus result in SNPs having differing correlations with the casual variants. A third possibility is that different causal variants in the same gene predominate in the ethnic groups, with different SNPs in the locus linked with the variants.
This last possibility appears to be the case for PCSK9, where the SNP that is strongly associated with LDL-C in European Americans (rs11591147) has a weak association in African Americans, and vice versa (rs505151). Both SNPs are coding variants that are likely to be casual for the effect on LDL-C, and the explanation for the strong association in the one ethnic group and the weak association in the other group is that the SNP has a lower MAF in the latter group. For example, rs505151 has a MAF of about 25% in the African American cohorts but only 4% in the European American cohorts. This finding with PCSK9 and LDL-C suggests that for some proportion of lipid-associated loci, the specific SNPs related to lipids will differ among ethnic groups—in contrast with CETP, LIPC, LPL, and APOA5, for which the same SNPs replicated in multiple ethnic groups in this study. Future CARe analyses, particularly large-scale candidate gene studies, will be useful in assessing the prevalence of this phenomenon.
CONCLUSION
CARe represents the successful assembly of DNA samples and phenotype data from over 40,000 participants in nine NHLBI cohort studies into a unique, valuable, publicly available resource to test an array of genes for a variety of phenotypes. The Pilot Study validates CARe’s operational framework and provides an initial evaluation of inter-ethnic differences for selected SNP-phenotype relationships. The ongoing large-scale candidate gene and genome-wide association analyses in CARe will explore the contribution of genetic variation to inter-individual, inter-ethnic, age-related, and cohort-specific differences in cardiovascular, pulmonary, hematological, and sleep-related phenotypes. Thus, CARe should serve as a valuable resource for the scientific community.
Supplementary Material
Acknowledgments
The authors wish to acknowledge the support of the National Heart, Lung, and Blood Institute and the contributions of the research institutions, study investigators, and field staff in creating this resource for biomedical research. Finally, we are most grateful to all of the study participants, without whom this endeavor would not have been possible.
FUNDING SOURCES
The following nine parent studies, funded by the listed National Institutes of Health grants, have contributed parent study data, ancillary study data, and DNA samples through the Broad Institute (N01-HC-65226) to create this genotype/phenotype database for wide dissemination to the biomedical research community: Atherosclerotic Risk in Communities (ARIC): University of North Carolina at Chapel Hill (N01-HC-55015, N01-HC-55018), Baylor Medical College (N01-HC-55016), University of Mississippi Medical Center (N01-HC-55021), University of Minnesota (N01-HC-55019), Johns Hopkins University (N01-HC-55020), University of Texas, Houston (N01-HC-55022); Cardiovascular Health Study (CHS): University of Washington (N01-HC-85079, N01-HC-55222, U01-HL-080295), Wake Forest University (N01-HC-85080), Johns Hopkins University (N01-HC-85081, N01-HC-15103), University of Pittsburgh (N01-HC-85082), University of California, Davis (N01-HC-85083), University of California, Irvine (N01-HC-85084), New England Medical Center (N01-HC-85085), University of Vermont (N01-HC-85086), Georgetown University (N01-HC-35129), University of Wisconsin (N01-HC-75150); Cleveland Family Study (CFS): Case Western Reserve University (R01-HL-46380, M01-RR-00080); Cooperative Study of Sickle Cell Disease (CSSCD): University of Illinois (N01-HB-72982, N01-HB-97062), Howard University (N01-HB-72991, N01-HB-97061), University of Miami (N01-HB-72992, N01-HB-97064), Duke University (N01-HB-72993), George Washington University (N01-HB-72994), University of Tennessee (N01-HB-72995, N01-HB-97070), Yale University (N01-HB-72996, N01-HB-97072), Children’s Hospital-Philadelphia (N01-HB-72997, N01-HB-97056), University of Chicago (N01-HB-72998, N01-HB-97053), Medical College of Georgia (N01-HB-73000, N01-HB-97060), Washington University (N01-HB-73001, N01-HB-97071), Jewish Hospital and Medical Center of Brooklyn (N01-HB-73002), Trustees of Health and Hospitals of the City of Boston, Inc., (N01-HB-73003), Children’s Hospital-Oakland (N01-HB-73004, N01-HB-97054), University of Mississippi (N01-HB-73005), St. Luke’s Hospital-New York (N01-HB-73006), Alta Bates-Herrick Hospital (N01-HB-97051), Columbia University (N01-HB-97058), St. Jude’s Children’s Research Hospital (N01-HB-97066), Research Foundation, State University of New York-Albany (N01-HB-97068, N01-HB-97069), New England Research Institute (N01-HB-97073), Interfaith Medical Center-Brooklyn (N01-HB-97085); Coronary Artery Risk in Young Adults (CARDIA): University of Alabama at Birmingham (N01-HC-48047, N01-HC-95095), University of Minnesota (N01-HC-48048), Northwestern University (N01-HC-48049), Kaiser Foundation Research Institute (N01-HC-48050), Tufts-New England Medical Center (N01-HC-45204), Wake Forest University (N01-HC-45205), Harbor-UCLA Research and Education Institute (N01-HC-05187), University of California, Irvine (N01-HC-45134, N01-HC-95100); Framingham Heart Study (FHS): Boston University (N01-HC-25195, R01-HL-092577, R01-HL-076784, R01-AG-028321); Jackson Heart Study (JHS): Jackson State University (N01-HC-95170), University of Mississippi (N01-HC-95171), Tougaloo College (N01-HC-95172); Multi-Ethnic Study of Atherosclerosis (MESA): University of Washington (N01-HC-95159), University of California, Los Angeles (N01-HC-95160), Columbia University (N01-HC-95161), Johns Hopkins University (N01-HC-95162, N01-HC-95168), University of Minnesota (N01-HC-95163), Northwestern University (N01-HC-95164), Wake Forest University (N01-HC-95165), University of Vermont (N01-HC-95166), New England Medical Center (N01-HC-95167), Harbor-UCLA Research and Education Institute (N01-HC-95169), Cedars-Sinai Medical Center (R01-HL-071205), University of Virginia (subcontract to R01-HL-071205); Sleep Heart Health Study (SHHS): Johns Hopkins University (U01-HL-064360), Case Western University (U01-HL-063463), University of California, Davis (U01-HL-053916), University of Arizona (U01-HL-053938, U01-HL-053934), University of Pittsburgh (U01-HL-077813), Boston University (U01-HL-053941), MedStar Research Institute (U01-HL-063429), Johns Hopkins University (U01-HL-053937).
Footnotes
CONFLICT OF INTEREST DISCLOSURES
None
References
- 1.The Atherosclerosis Risk in Communities (ARIC) Study: design and objectives. The ARIC investigators. Am J Epidemiol. 1989;129:687–702. [PubMed] [Google Scholar]
- 2.Friedman GD, Cutter GR, Donahue RP, Hughes GH, Hulley SB, Jacobs DR, Jr, Liu K, Savage PJ. CARDIA: study design, recruitment, and some characteristics of the examined subjects. J Clin Epidemiol. 1988;41:1105–1116. doi: 10.1016/0895-4356(88)90080-7. [DOI] [PubMed] [Google Scholar]
- 3.Buxbaum SG, Elston RC, Tishler PV, Redline S. Genetics of the apnea hypopnea index in Caucasians and African Americans: I. Segregation analysis. Genet Epidemiol. 2002;22:243–253. doi: 10.1002/gepi.0170. [DOI] [PubMed] [Google Scholar]
- 4.Fried LP, Borhani NO, Enright P, Furberg CD, Gardin JM, Kronmal RA, Kuller LH, Manolio TA, Mittelmark MB, Newman A, O’Leary DH, Psaty B, Rautaharju P, Tracy RP, Weiler PG. The Cardiovascular Health Study: design and rationale. Ann Epidemiol. 1991;1:263–276. doi: 10.1016/1047-2797(91)90005-w. [DOI] [PubMed] [Google Scholar]
- 5.Gaston M, Smith J, Gallagher D, Flournoy-Gill Z, West S, Bellevue R, Farber M, Grover R, Koshy M, Ritchey AK. Recruitment in the Cooperative Study of Sickle Cell Disease (CSSCD) Control Clin Trials. 1987;8:131S–140S. doi: 10.1016/0197-2456(87)90016-x. [DOI] [PubMed] [Google Scholar]
- 6.Dawber TR, Meadors GF, Moore FE., Jr Epidemiological approaches to heart disease: the Framingham Study. Am J Public Health Nations Health. 1951;41:279–281. doi: 10.2105/ajph.41.3.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Feinleib M, Kannel WB, Garrison RJ, McNamara PM, Castelli WP. The Framingham Offspring Study. Design and preliminary data. Prev Med. 1975;4:518–525. doi: 10.1016/0091-7435(75)90037-7. [DOI] [PubMed] [Google Scholar]
- 8.Splansky GL, Corey D, Yang Q, Atwood LD, Cupples LA, Benjamin EJ, D’Agostino RB, Sr, Fox CS, Larson MG, Murabito JM, O’Donnell CJ, Vasan RS, Wolf PA, Levy D. The Third Generation Cohort of the National Heart, Lung, and Blood Institute’s Framingham Heart Study: design, recruitment, and initial examination. Am J Epidemiol. 2007;165:1328–1335. doi: 10.1093/aje/kwm021. [DOI] [PubMed] [Google Scholar]
- 9.Taylor HA., Jr The Jackson Heart Study: an overview. Ethn Dis. 2005;15:S6-1–3. [PubMed] [Google Scholar]
- 10.Bild DE, Bluemke DA, Burke GL, Detrano R, Diez Roux AV, Folsom AR, Greenland P, Jacob DR, Jr, Kronmal R, Liu K, Nelson JC, O’Leary D, Saad MF, Shea S, Szklo M, Tracy RP. Multi-ethnic study of atherosclerosis: objectives and design. Am J Epidemiol. 2002;156:871–881. doi: 10.1093/aje/kwf113. [DOI] [PubMed] [Google Scholar]
- 11.Quan SF, Howard BV, Iber C, Kiley JP, Nieto FJ, O’Connor GT, Rapoport DM, Redline S, Robbins J, Samet JM, Wahl PW. The Sleep Heart Health Study: design, rationale, and methods. Sleep. 1997;20:1077–1085. [PubMed] [Google Scholar]
- 12.Pennacchio LA, Olivier M, Hubacek JA, Krauss RM, Rubin EM, Cohen JC. Two independent apolipoprotein A5 haplotypes influence human plasma triglyceride levels. Hum Mol Genet. 2002;11:3031–3038. doi: 10.1093/hmg/11.24.3031. [DOI] [PubMed] [Google Scholar]
- 13.Frisdal E, Klerkx AH, Le Goff W, Tanck MW, Lagarde JP, Jukema JW, Kastelein JJ, Chapman MJ, Guerin M. Functional interaction between -629C/A, -971G/A and -1337C/T polymorphisms in the CETP gene is a major determinant of promoter activity and plasma CETP concentration in the REGRESS Study. Hum Mol Genet. 2005;14:2607–2618. doi: 10.1093/hmg/ddi291. [DOI] [PubMed] [Google Scholar]
- 14.Le Goff W, Guerin M, Nicaud V, Dachet C, Luc G, Arveiler D, Ruidavets JB, Evans A, Kee F, Morrison C, Chapman MJ, Thillet J. A novel cholesteryl ester transfer protein promoter polymorphism (-971G/A) associated with plasma high-density lipoprotein cholesterol levels. Interaction with the TaqIB and -629C/A polymorphisms. Atherosclerosis. 2002;161:269–279. doi: 10.1016/s0021-9150(01)00641-4. [DOI] [PubMed] [Google Scholar]
- 15.Guerra R, Wang J, Grundy SM, Cohen JC. A hepatic lipase (LIPC) allele associated with high plasma concentrations of high density lipoprotein cholesterol. Proc Natl Acad Sci U S A. 1997;94:4532–4537. doi: 10.1073/pnas.94.9.4532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hata A, Robertson M, Emi M, Lalouel JM. Direct detection and automated sequencing of individual alleles after electrophoretic strand separation: identification of a common nonsense mutation in exon 9 of the human lipoprotein lipase gene. Nucleic Acids Res. 1990;18:5407–5411. doi: 10.1093/nar/18.18.5407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kotowski IK, Pertsemlidis A, Luke A, Cooper RS, Vega GL, Cohen JC, Hobbs HH. A spectrum of PCSK9 alleles contributes to plasma levels of low-density lipoprotein cholesterol. Am J Hum Genet. 2006;78:410–422. doi: 10.1086/500615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen SN, Ballantyne CM, Gotto AM, Jr, Tan Y, Willerson JT, Marian AJ. A common PCSK9 haplotype, encompassing the E670G coding single nucleotide polymorphism, is a novel genetic marker for plasma low-density lipoprotein cholesterol levels and severity of coronary atherosclerosis. J Am Coll Cardiol. 2005;45:1611–1619. doi: 10.1016/j.jacc.2005.01.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Keating BJ, Tischfield S, Murray SS, Bhangale T, Price TS, Glessner JT, Galver L, Barrett JC, Grant SF, Farlow DN, Chandrupatla HR, Hansen M, Ajmal S, Papanicolaou GJ, Guo Y, Li M, Derohannessian S, de Bakker PI, Bailey SD, Montpetit A, Edmondson AC, Taylor K, Gai X, Wang SS, Fornage M, Shaikh T, Groop L, Boehnke M, Hall AS, Hattersley AT, Frackelton E, Patterson N, Chiang CW, Kim CE, Fabsitz RR, Ouwehand W, Price AL, Munroe P, Caulfield M, Drake T, Boerwinkle E, Reich D, Whitehead AS, Cappola TP, Samani NJ, Lusis AJ, Schadt E, Wilson JG, Koenig W, McCarthy MI, Kathiresan S, Gabriel SB, Hakonarson H, Anand SS, Reilly M, Engert JC, Nickerson DA, Rader DJ, Hirschhorn JN, Fitzgerald GA. Concept, design and implementation of a cardiovascular gene-centric 50 k SNP array for large-scale genomic association studies. PLoS ONE. 2008;3:e3583. doi: 10.1371/journal.pone.0003583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kapur NK, Musunuru K. Clinical efficacy and safety of statins in managing cardiovascular risk. Vasc Health Risk Manag. 2008;4:341–353. doi: 10.2147/vhrm.s1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen MH, Yang Q. GWAF: an R package for genome-wide association analyses with family data. Bioinformatics. 2010;26:580–581. doi: 10.1093/bioinformatics/btp710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ. 2003;327:557–560. doi: 10.1136/bmj.327.7414.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.The International HapMap Consortium. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.