Abstract
We report a novel transgenic mouse model for direct reprogramming of somatic cells by expressing four reprogramming factors from a single genomic locus using a drug-inducible transgene. Multiple somatic cell types explanted from different tissues can generate induced pluripotent stem cells (iPSC) by culture in doxycycline (Dox). Because the reprogramming factors are carried on a single polycistronic construct the transgenic mice can be easily maintained and transferred into another background.
Keywords: 2A peptide, pluripotent, polycistronic, four-factor reprogramming, stem cells
RESULTS
Generation of induced pluripotent stem cells (iPSCs) by primary infection using Moloney based gene delivery generates iPSCs with an efficiency between 0.001–0.1% 1–4. The creation of “secondary” systems that utilize doxycyline (Dox)-inducible lentiviruses to generate iPSCs from murine somatic cells allows the reprogramming of genetically homogenous somatic cell populations and avoids the genetic heterogeneity of primary infections 5–8. However secondary lines harbor multiple (>7) proviruses, each with different capacities for reactivation as indicated by the variegated expression of reprogramming factors after Dox exposure in addition to the inability of factors to reactivate in some somatic tissues 6. Although secondary models have been used to generate transgenic animals carrying all or a subset of factors by germline segregation of the proviruses, it remains difficult to maintain all factors in one mouse because the multiple transgenes segregate in each generation 9
Here we report a transgenic mouse model that overcomes these limitations and minimizes the variegation of factor levels by inducing reprogramming factor expression from a single genomic site. The collagen type I (Col1a1) gene has been used as an expression locus for Oct4 (also called Oct3/4 or Pou5f1) and demonstrated to be capable of transgene induction in multiple somatic cell types including mouse fibroblasts, keratinocytes, adrenal glands, and neural precursor cells 6, 10, 11. We generated “single-gene” transgenic mouse strain(s) that express three or four reprogramming factors from the Col1a1 locus.
Using homologous recombination of embryonic stem cells (ESCs) carrying a drug-inducible reverse tetracycline trans-activator (M2rtTA) in the Rosa26 locus, we inserted four reprogramming factors as a single polycistronic transgene in an expression cassette termed “4F2A,” which harbors four murine reprogramming genes (Oct4, Sox2, Klf4, and c-Myc) separated by three 2A self-cleaving peptides (Fig. 1a,b). This cassette has been shown to reprogram embryonic and adult murine fibroblasts as well as human keratinocytes 12. Transgenic animals carrying one copy of each transgene rtTA(1):4F2A(1) were generated and intercrossed to derive animals carrying various combinations of transgene copies rtTA(2): 4F2A(1), rtTA(1):4F2A(2) and rtTA(2):4F2A(2). Importantly, no tumors or other health problems were detected in any of the transgenic mice for up to 31 weeks of age (Supplementary Table 1).
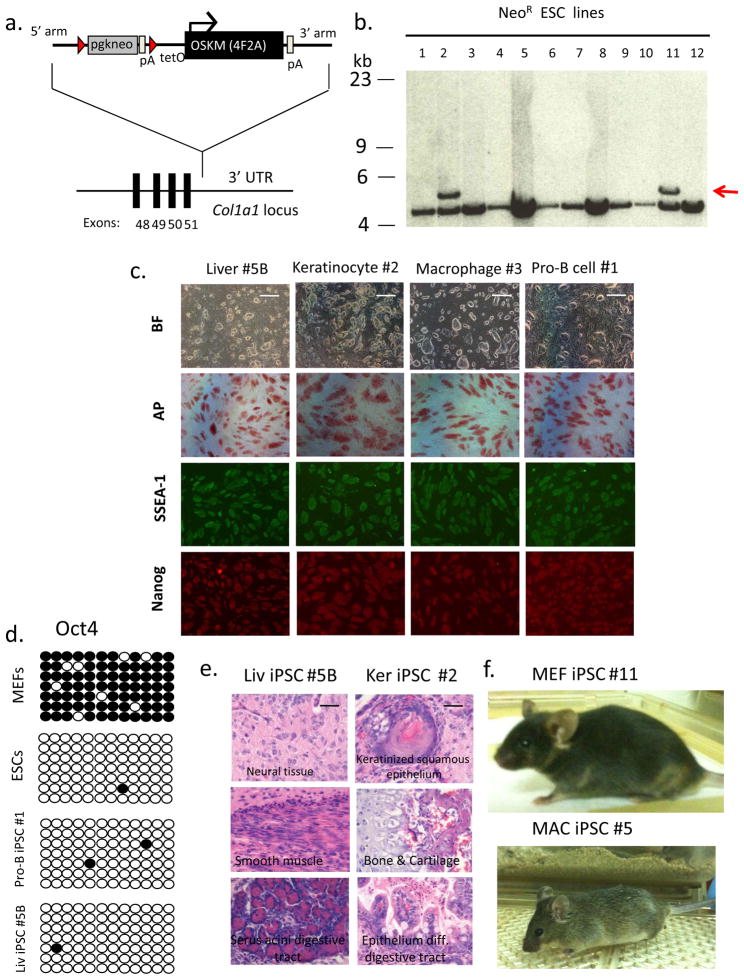
Figure 1. Reprogramming somatic cells from transgenic Col1a1 4F2A mice.
(a) A targeting vector to the 3’ UTR was utilized to deliver four murine reprogramming factors (Oct4, Sox2, Klf4, and c-Myc) as a single dox-inducible polycistronic transgene 11,12. Red triangles indicate loxP sites. pA indicates poly-adenylation sequence. TetO is tetracycline operator minimal promoter. (b) Southern analysis of DNA obtained from Neo-resistant V19 ES cell colonies was digested with XbaI and probed for correct targeting of the Col1a1 3’ UTR using a 5’ external Col1a1 probe to the genomic sequence outside the targeting vector homology arms 11. Lines #2 and #11 were correctly targeted as indicated by hybridization to 5.3 kb band (untargeted allele ~ 4.7 kb). (c) Col1a1 4F2A iPS cells express pluripotency markers. Bright field (BF) images and immunostaining for pluripotency markers alkaline phosphatase (AP), SSEA1, and Nanog. Scale bar, 1mm. (d) Oct4 promoter using bisulfite sequencing. Open circles indicate unmethylated and closed circles indicate methylated CpG dinucleotides. Shown are representative sequence analyses from Col1a1 4F2A transgenic MEFs, embryonic stem cells (V6.5), and two Col1a1 4F2A iPSC lines: pro-B-derived #1 and liver-derived line #5B. (e) Hematoxylin and eosin staining of teratomas induced after subcutaneous injection of Col1a1 4F2A Liv iPSC #5B and Ker iPSC #2 into SCID mice indicates Col1a1 4F2A iPSCs contribute to all three germ layers. Scale bar, 100μm (f) Postnatal chimeric mice detected by agouti coat color from Col1a14F2A MEF iPSC #11 and MAC iPSC #5.
We used qRT-PCR analysis to quantify the Dox-induced expression level of the 4F2A transgene (Tg) in mouse embryonic fibroblasts (MEFs) using primers specific to the 4F2A Tg and observed >100-fold induction in MEFs carrying a single copy of both the rtTA and 4F2A (Supplementary Figure 2a). Protein expression for three of the four factors was confirmed by immunoflourescence demonstrating robust Dox-inducible expression (Supplementary Figure 2b). The relative induction as compared to transcript levels in ESCs or MEFs was quantified by qRT-PCR analysis using exon specific primers for each factor demonstrating that the expression of Oct4 and Sox2 was in the same range whereas Klf4 and c-Myc were expressed at a 3- to 5-fold higher than in ES cells (Supplementary Fig. 3a). To test whether factor expression was copy number dependent we quantified transcript levels in adult fibroblasts carrying different copies of the two transgenes. qRT-PCR analysis showed a copy number and Dox-dependent expression of the 4F2A Tg (1:1 < 1:2 < 2:2; Supplementary Fig. 3b).
Mice carrying different copies of each transgene (1:1, 2:1, 1:2, 2:2) were tested for the ability to generate iPSCs from defined tissues (Supplementary Table 2). MEFs isolated from animals carrying single copy rtTA(1):4F2A(1) and cultured in the presence of Dox gave rise to colonies within 2–4 weeks that could be picked and after 2–3 passages generated iPS cell lines that expressed the pluripotency markers alkaline phosphatase (AP), SSEA1, and had reactivated the endogenous Nanog locus as detected by immunostaining (Fig. 1c). However, we failed to obtain iPSCs from any tissue of adult mice carrying single copies of each Tg respectively.
To assess whether higher transgene expression levels would allow the generation of iPSCs from other cell types, tail tip fibroblasts (TTF), epidermis-derived keratinocytes, liver cells, mesenchymal stem cells (MSCs), pro-B cells, macrophages and intestinal epithelial cells were derived from mice carrying different combinations of Tg copies and cultured in the presence of Dox. We were able to isolate iPSCs from adult liver cells as well as keratinocytes from mice carrying 2 copies of rtTA and 1 copy of 4F2A (rtTA(2): 4F2A(1)). Colonies appeared at d12-16 after addition of Dox (Supplementary Figure 4a), were manually picked, expanded without Dox after 2–3 passages and shown to express pluripotency markers AP, SSEA1, and Nanog (Fig. 1c). Other cell types such as CD11b+ macrophages from spleen, CD19+ pro-B cells from bone marrow, intestinal epithelium, adult tail-tip fibroblasts, and mesenchymal stem cells (MSCs) generated iPSCs only in the presence of a second copy of 4F2A transgene. iPSCs from adult fibroblasts, CD11b+ macrophages, and pro-B cells were derived from animals carrying 2 copies of the 4F2A Tg and one copy of the rtTA Tg (rtTA(1):4F2A(2)) whereas intestinal epithelial cells and mesenchymal stem cells (MSCs) required the presence of a second rtTA Tg (rtTA(2):4F2A(2)). The iPSCs grew in the absence of Dox and expressed the pluripotency markers AP, SSEA1, and Nanog (Fig. 1c and Supplementary Figure 4b). Complete demethylation of the endogenous Oct4 promoter was observed in five iPSC lines tested after bisulfite treatment and sequencing (Fig. 1d and Supplementary Figure 4c).
To assess pluripotency we tested the ability of Col1a1 4F2A iPSCs to differentiate in vivo. Liver, keratinocyte, and MEF-derived iPSCs were injected subcutaneously into SCID mice, giving rise to teratomas containing cells from all three germ layers (Figure 1e and Supplementary Fig. 4d). Two lines were tested for the ability to contribute to post-natal chimeras. MEF iPSC line #11 contributed to adult chimeras after injection into blastocysts (Fig. 1f). In addition we generated high contribution post-natal chimeras from macrophage-derived iPSC line (#5) that also transmitted the iPSC genome through the germline (Fig. 1f and Supplementary Figure 4e).
Our data demonstrate that iPSCs can be derived from multiple somatic tissues of adult mice carrying the Col1a1 4F2A polycistronic transgene. The observations that some donor cell types were able to generate iPSCs only from animals with two copies of the transgene suggests cell types such as intestinal epithelial cells, MCSs, pro-B cells or TTFs require higher levels of factor expression for reprogramming than other cell types such as MEFs or hepatocytes. Alternatively, differences in the induction level of the Col1a1 locus in certain cell types may require multiple copies of a Col1a1 transgene to reach a threshold of expression capable of achieving an iPS cell state.
We characterized reprogramming efficiencies in two distinct mouse transgene ratios (1:2 and 2:2) using pro-B cells. From each strain CD19+ cells were harvested from bone marrow and plated at similar densities (1×106) per well. Colonies appeared in both genotypes around day 12–14 but with different efficiencies. Alkaline phosphatase (AP) and Nanog immunostaining showed a 100-fold higher efficiency at a 2:2 than at a 1:2 transgene ratio (Supplementary Fig. 5a,b). To quantify the efficiency more precisely we tested the kinetics of reprogramming CD19+ pro-B cells (using ratio 2:2). Approximately 5×105 cells were cultured with Dox and changed to ES media without dox at indicated times. Reprogramming was assessed and quantified as Dox-independent, Nanog-positive iPS cells by immunostaining on day 25. We found Nanog-positive colonies increased with prolonged culture and directly correlated with the number of input donor cells (Supplementary Figure 5c,d).
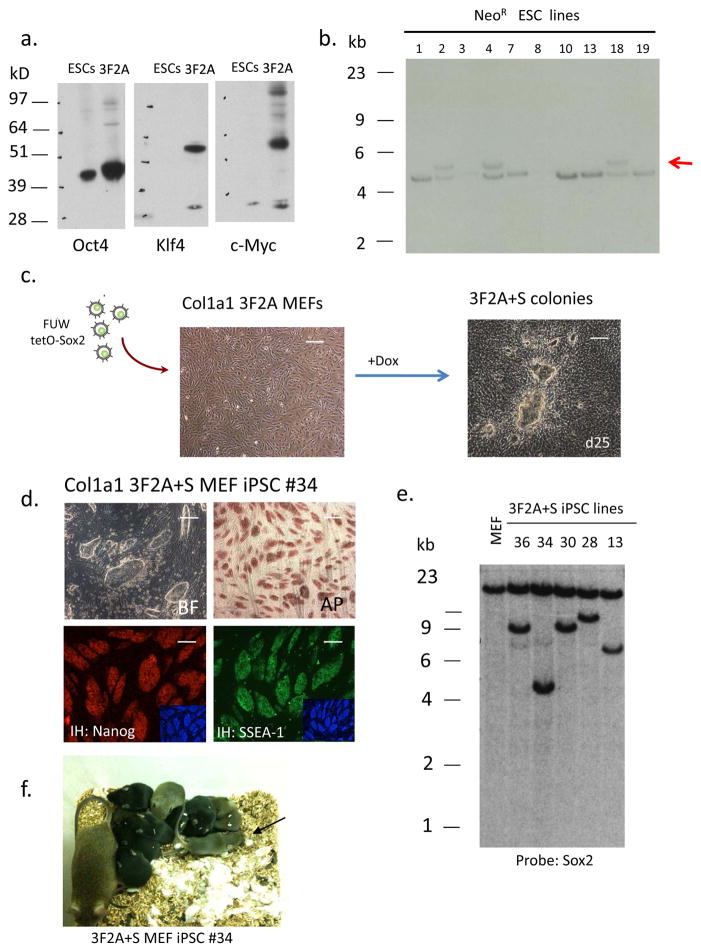
In addition to the Col1a1 4F2A system we established a transgenic model carrying only a subset of reprogramming factors to demonstrate the potential utility for replacement of individual reprogramming factors9. Using a three-factor 2A expression cassette lacking Sox2 (Oct4-T2A-Klf4-E2A-cMyc; abbreviated as “3F2A”) western blot analysis demonstrated that cells transduced with the 3F2A lentivirus were capable of expressing all three reprogramming factors (Fig. 2a). This cDNA was used in a Col1a1 targeting vector to generate Col1a1 3F2A transgenic ESCs that also harbored a drug-inducible M2rtTA at the Rosa 26 locus (Fig. 2b). After injection of 3F2A ESCs into blastocysts we obtained E13.5 chimeric embryos and isolated MEFs. When the cells were cultured in the presence of Dox induction of the transgene was ~ 120-fold as seen by qRT-PCR (Supplementary Fig. 6a). Because the 3F2A-MEFs lacked the essential reprogramming factor Sox2 they were infected with a Sox2 lentivirus (pFUW-tetO Sox2) (Fig 2c). Colonies appeared after 12–16 days and were picked at day 25. iPSCs were established after 2–3 passages staining positive for pluripotency markers AP, SSEA1 and Nanog (Fig. 2d). Four independent iPSC lines (MEF 3F2A+S iPSC) carried a single Sox2 provirus as detected by Southern (Fig. 2e) in addition to the single copy of rtTA and OKM Tg. All MEF 3F2A+S iPSC lines carried the 3F2A Tg as demonstrated by Southern analysis using probes for Oct4, Klf4, and c-Myc which all hybridized to a ~4.8 kb band as expected for the single 3F2A transgene (Supplementary Fig. 6b,c). iPSC line #34 gave rise to high contribution chimeras when injected into blastocysts (Fig 2f and Supplementary Table 3) with two of the four chimeras transmitting the iPSC genome through the germline (Fig. 2f). Somatic cells derived from animals carrying the 3F2A transgene will be useful to screen for small molecules that replace Sox2 for reprogramming.
Figure 2. iPSCs derived from Col1a1 3F2Atransgenic MEFs expressing a subset of reprogramming factors.
(a) Western blot analysis of 293 cells transiently transfected with pFUW-Ubi 3F2A lentivirus. (b) Southern analysis of DNA obtained from Neo-resistant V19 ES cell colonies. DNA was digested with XbaI and probed for correct targeting using a 5’ Col1a1 probe. Three lines (# 2, 4, 18) were correctly targeted as indicated by hybridization to the 5.3kb band (untargeted allele ~ 4.7 kb). (c) 3F2A MEFs were infected with a Dox-inducible lentivirus carrying a Sox2 cDNA (pFUW-tetO-Sox2) and transferred to ES cell medium containing Dox after 48 hours. Typical iPSC colonies appeared around day 12 and were mechanically passaged at day 25. Scale bar, 1mm. (d) Immunostaining for AP, SSEA1 and Nanog of iPSC line #34. DAPI staining is shown in the inset. Scale bar, 1mm. (e) Southern blot analysis of 3F2A+S iPS cell clones. DNA was isolated from each iPSC line and digested with XbaI. The presence of a single Sox2 proviral copy was detected with a Sox2 cDNA probe for hybridization. A similar sized band was detected in lines #36 and 30 suggesting these cells are sibling subclones from the same infected cell. (f) 3F2A+S MEF derived iPS cells give rise to post-natal chimeras. High contribution chimera (as detected by agouti coat color) obtained after injection of 3F2A+S iPSC line #34 into blastocysts. This chimera contained iPSCs that contributed to the germline as shown by agouti pups present after mating to BDF1 females.
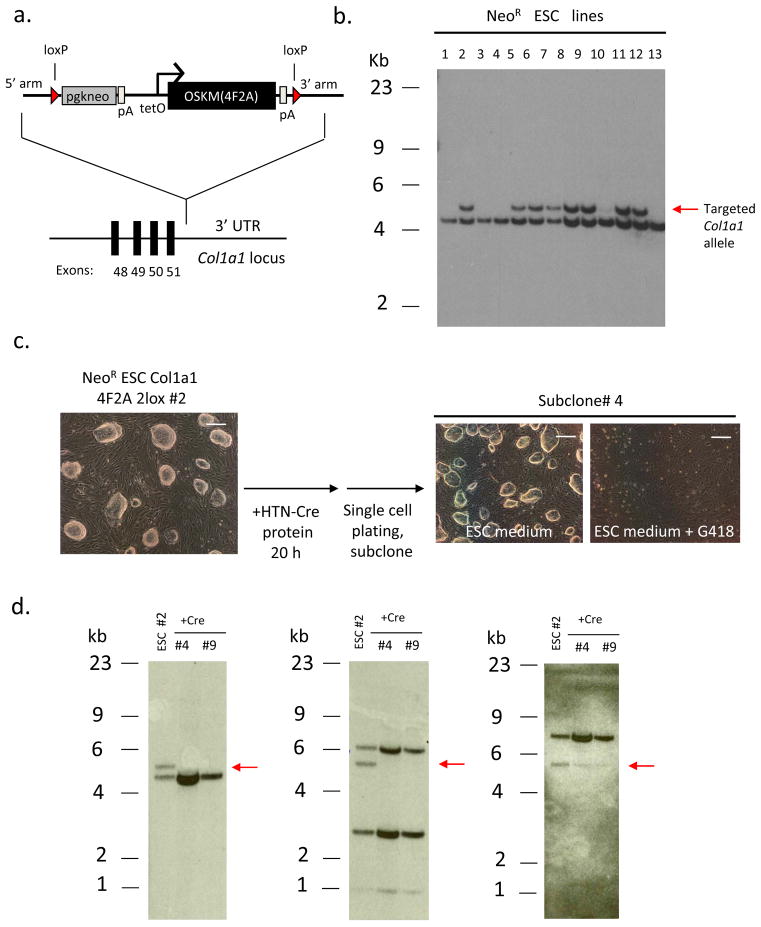
Recently, several strategies have been described that allow the generation of vector-free iPSCs following excision of reprogramming transgenes 13–15. To establish a system that would permit the isolation of vector-free iPSCs from a defined transgenic strain, we created a Col1a1 4F2A targeting vector harboring loxP sites flanking the Tg (Col1a1 2lox 4F2A) (Fig. 3a) leaving a single loxP site in the Col1a1 3’ UTR after Cre-mediated excision. The construct was targeted to the Col1a1 locus in ESCs carrying the M2rtTA transgene at the Rosa 26 locus (Fig 3b). To assess whether the transgene could be excised freshly passaged transgenic ESCs were exposed to a recombinant cell-permeable Cre protein (HTN-Cre) for 20 hours. Deletion of the Col1a1 2lox 4F2A was detected in sibling colonies that were cultured in the presence or absence of G418 giving rise to 2 out of 16 neo sensitive colonies (Fig. 3c) that had deleted the transgene as verified by Southern blot analysis using an external Col1a1 5’ probe as well as probes for Oct4 and c-Myc cDNA (Fig. 3d). Mice derived from this ES cell line will be useful as donors for generating iPSCs that carry no vector sequences.
Figure 3. Cre-excisable Col1a1 4F2A transgenic ES cells.
(a) Targeting vector containing loxP sites flanking the 4F2A transgene. A similar targeting scheme as described in Figure 1a was used. (b) Targeting of 3F2A to the Col1a1 locus in R26 M2rtTA transgenic ES cells. Southern analysis of DNA obtained from Neo-resistant ES cell colonies digested with XbaI and probed for correct targeting using a 5’ external Col1a1 probe (compare Fig. 1a). Multiple clones were correctly targeted as indicated by hybridization to the 5.3kb band (untargeted allele ~ 4.7kb). Line #2 was chosen for Cre-induced excision. (c) Recombinant cell-permeable HTN-Cre transduced into Col1a1 2lox 4F2AES cells. Cells were grown in ES cell medium containing HTN-Cre protein for 20 hours. 24 hours later the cells were trypsinized, plated at high dilution (1:10,000) and colonies were picked and expanded before being split into ES medium or ES medium + G418. Bright field images depict a subclone that is G418-sensitive. Scale bars, 1mm. (d) Southern blot analysis of HTN-Cre-induced excision of Col1a1 2lox 4F2A transgene in ESCs. DNA obtained from sister clones of Neo-sensitive ES cell colonies was digested with XbaI and probed using a 5’ external Col1a1 probe (compare Fig. 1a). Two subclones (#4 and 9) do not show hybridization to the ~5.3kb band corresponding to the targeted Col1a1 4F2A after exposure to HTN-Cre protein (left). Oct4 and c-Myc cDNA probes do not hybridize to the expected 5.8kb band (also XbaI digested DNA) corresponding to the Col1a1 4F2A Tg (center and right, respectively).
The transgenic models described in this study have several advantages over previous systems: 1) the previous “secondary” iPSCs were derived from primary iPS cells generated by lentivirus-mediated gene transfer of the four reprogramming factors 6, 9. Because the individual proviruses carrying the four factors segregate independently it has been difficult to derive mice homozygous for all factors that could be used as a convenient source for reprogrammable cells. In the present system all reprogramming factors are expressed from one cassette (e.g. 4F2A) inserted into a single locus, facilitating the maintenance of the strain as well as the transfer of the reprogramming factors to mouse strains carrying other genetic determinants of interest. 2) Unlike proviral transgenes that may be silenced by DNA methylation or other epigenetic mechanisms upon germ line transmission, this transgenic system expresses the four reprogramming factors from a specific locus (Col1a1) that is widely expressed in the adult mouse and is capable of reprogramming cells from seven different tissues of the adult. 3) No tumors or other adverse effects have been observed in the transgenic mice suggesting that the Col1a1 4F2A system is both safe and practical for extended breeding. 4) Reprogramming factor stochiometries are fixed within each somatic cell type. This system creates iPSCs from multiple somatic tissues and allows comparison among iPSCs not previously possible. Moreover this system generates iPSCs that can now be compared to genetically identical ESCs. 5) The generation of 3F2A transgenic mice lacking one of the 4 factors will allow screening of somatic cells for small molecules to replace the fourth factor (such as Sox2, Fig. 2). 6) Finally, the generation of mice carrying the loxP flanked 4F2A construct will allow the excision of the transgene and derivation of adult mice created from vector-free iPSCs 13–15.
MATERIALS AND METHODS
Col1a1 targeting and vectors
(1) Col1a1 4F2A: A ~ 5.8kb EcoRI generated fragment containing Oct4-P2A-Sox2-T2A-Klf4-E2A-c-Myc (also named “4F2A”) cDNA was generated from a previously published plasmid FUW-Ubi-4F2A 12 and ligated into ptet.splicePL3 vector (containing tetO promoter + SV40 intron,pA) similarly digested with EcoRI to obtain ptet.splicePL3-OSKMpA. A 7.3kb NotI generated fragment was obtained from the ptet.splicePL3-OSKMpA and ligated into a NotI digested mCol.loxneo targeting vector that contained both 5’ and 3’ homology arms towards the 3’ UTR of the Col1a1 locus as well as a pgk-driven neo resistance cassette for selection of transgenic cells. The resulting ~18 kb targeting construct (Col1a1 4F2A) was linearized with PvuI restriction enzyme digestion (25μg), precipitated and resuspended in 400μl HEPES, then electroporated into V19 ES cells (V6.5 ES cells containing a reverse tetracycline trans-activator (M2rtTA) driven by the Rosa26 promoter). After 24 hours G418 (350μg per ml) was added to ES cell medium (DMEM supplemented with 10% FBS (Hyclone), leukemia inhibitory factor, β-mercaptoethanol (Sigma-Aldrich), penicillin/streptomycin, L-glutamine and nonessential amino acids (all from Invitrogen). Neo-resistant colonies were picked 10 days later, expanded and DNA isolated for testing of correct targeting by Southen analysis. (2) Col1a1 2lox 4F2A: the 7.3 kb NotI generated fragment from ptet.splicePL3-OSKMpA was blunt-ended and ligated with a EcoRV digested pBS246 plasmid (containing two loxP sites). The resulting pBS246.tetO-OSKMpA was digested with NotI generating a fragment of 7.6 kb that was ligated into the previously mentioned NotI digested mCol.loxneo targeting vector. The resulting vector Col1a1 2lox 4F2A was digested for targeting of V19 ES cells as described above. (3) Col1a1 3F2A: A three-factor cDNA containing Oct4-T2A-Klf4-c-Myc was generated by PCR amplification of Oct4 using 5’ XbaI-EcoRI and SphI containing primers whose product was then TOPO cloned following manufactures instructions (pCR 2.1 TOPO Invitrogen). The TOPO-Oct4 plasmid was then digested with XbaI and SphI and the 1.1 kb Oct4 cDNA was ligated to a similarly digested FUW-Ubi-4F2A ~ 10 kb fragment. The resulting FUW-Ubi-3F2A was digested with EcoRI to generate a ~ 4.0 kb fragment that was cloned into the ptet.splicePL3 vector as described above. A 6.3 kb fragment from ptet.splicePL3-tetO-3F2ApA was generated by NotI digestion and ligated into the mCol.loxneo targeting vector. The resulting 17kb plasmid (Col1a1 3F2A) was linearized with PvuI restriction enzyme (25μg), precipitated, resuspended in 400μl HEPES and electroporated into V19 cells. Selection was the same as described above.
Mouse chimera and teratoma formation
Diploid blastocysts (94–98 h after hCG injection) were placed in a drop of Hepes-CZB medium under mineral oil. A flat tip microinjection pipette with an internal diameter of 16 μm was used for iPS cell injections. Each blastocyst received 8–10 iPS cells or ES cells. After injection, blastocysts were cultured in potassium simplex optimization medium (KSOM) and placed at 37 °C until transferred to recipient females. About 10 injected blastocysts were transferred to each uterine horn of 2.5-day-postcoitum pseudo-pregnant B6D2F1 female. Pups were recovered at day 19.5 and fostered to lactating B6D2F1 mothers when necessary. For summary see Supplementary Table 4. Teratoma formation was performed by depositing 2x10^6 cells under the flanks of recipient SCID mice. Tumors were isolated 3–6 weeks later for histological analysis.
Somatic cell isolation and culture
For MEF isolation, chimeric embryos were isolated at E13.5, and the head and internal organs were removed. The remaining tissues were physically dissociated and incubated in trypsin at 37° C for 20 min after which cells were resuspended in MEF medium. 24 hours later puromycin (2μg/ml) was added and the cells were expanded for two passages before freezing or plating in doxycline containing media (2μg/ml) for reprogramming experiments. Somatic organs were isolated from 4–6 week-old transgenic mice. Epidermal keratinocytes and intestinal epithelium were isolated and cultured as previously described 6. For mesenchymal stem cells (MSCs) and pro-B cells whole marrow was isolated from the femur and tibia after removal of the condyles at the growth plate by flushing with a syringe and 30-guague needle containing DMEM+5% FBS (Hyclone, Thermo Fisher Scientific). CD19+ pro-B cells were isolated by MACS cell separation (Miltenyibiotec Cat# 130-052-201) following manufactures instructions. Purified B cell subsets were resuspended in IMDM with 15% FCS as well as IL-4, IL-7, SCF (10 ng/ml each, Peprotech), doxycyline (2 μg/ml) and plated on OP9 bone marrow stromal cells (ATCC). Three days later the medium was changed to ESC medium plus Dox. Macrophage cells (CD11b+) from freshly isolated spleen were isolated by MACS cell separation and plated by a similar protocol described for CD19+ pro-B cells. Mesenchymal stem cells were selected through differential plating on tissue culture plates (10cm) for 72h in α-MEM supplemented with 15% FBS. Once plates reached a full monolayer cells were split into 6-well dishes and cultured in the presence of Dox. For isolation of liver cells mice were first perfused with 50 ml HBSS buffer (w/o Ca2+ and Mg2+) then 50 ml HBSS (w/o Ca2+ and Mg2+) containing collagenase (type IV) (Sigma Cat# C5138) (100U/ml). Liver was dissected away from surrounding tissues and dissociated in 10ml DAG media (phenol-red free EMEM Gibco-11054-020 and Bovine serum albumin (BSA) 1g/0.5L) and filtered two times through a sterile 100μM cell strainer. Liver cell preparations were centrifuged at 30 g for 3 minutes at 4 °C and the cells were washed two times with DAG media and then plated on γ-irradiated MEFs in ES media + Dox.
Antibodies and Immunostaining
Cells were fixed in 4% paraformaldehyde for 20 minutes at 25 °C, washed 3 times with PBS and blocked for 15 min with 5% FBS in PBS containing 0.1% Triton-X. After incubation with primary antibodies against Oct4 (Santa Cruz H-134), Sox2 (R&D Biosystems), Klf4 (Santa Cruz H-180), Nanog (Bethyl A300-398A), SSEA1 (monoclonal mouse, Developmental Studies Hybridoma Bank) for 1 h in 1% FBS in PBS containing 0.1% Triton-X, cells were washed 3 times with PBS and incubated with fluorophore-labeled appropriate secondary antibodies purchased from Jackson Immunoresearch. Specimens were analyzed on an Olympus Fluorescence microscope and images were acquired with a Zeiss Axiocam camera. Alkaline phosphatase (AP) staining was done following manufactures instructions (Vector Labs).
Bisulfite Sequencing
DNA was isolated from indicated cell lines and conversion of unmethylated CpG dinucleotides was performed using EpiTect bisulfite kit (Qiagen) following manufacturer’s instructions. Following PCR amplification (see Supplementary Table 5) gel purified DNA fragments were subcloned into TOPO TA vector (Invitrogen) and transformed into TOP-10 bacterial cells. 10 clones per cell line were sent for sequencing in both forward and reverse directions (M13F/R).
Southern Blotting
10 μg of restriction enzyme digested genomic DNA was separated on a 0.7% agarose gel, transferred to a nylon membrane (Amersham) and hybridized with 32P random primer (Stratagene) labeled probes for Col1a1 5’ genomic (external) sequences, Col1a1 3’ (internal) located in 3’ arm of Col1a1 targeting vectors (XhoI-PstI fragment of TOPO2.1 5’mCol1a1 & XbaI-PstI 3’mCol1a1 plasmids), Klf4 (full length Klf4 cDNA from pFUW-tetO-Klf4), c-Myc (full length c-Myc cDNA from pFUW-tetO-c-Myc), Oct4 (EcoRI-PstI exon1 fragment of cDNA from pFUW-tetO-Oct4), and Sox2 (full length cDNA from pFUW-tetO-Sox2 plasmid).
Recombinant HTNCre protein delivery to Col1a1 2lox 4F2A transgenic ES cells
Transgenic ES cells were passaged one day prior to Cre protein transduction to obtain a monolayer. ES cell medium containing 10μM HTNCre protein was prepared after appropriate dilution from 200μM stock and incubated with transgenic ES cells for 20 hours. After this time media was changed to ES cell media and cells were incubated for 1 more day until passaging (1:10,000) for single cell colony formation.
Quantitative RT-PCR
Total RNA was isolated using Trizol reagent (Invitrogen). Five micrograms of total RNA was treated with DNase I to remove potential contamination of genomic DNA using a DNA Free RNA kit (Zymo Research). One microgram of DNase I–treated RNA was reverse transcribed using a First Strand Synthesis kit (Invitrogen) and ultimately resuspended in 100 μl of water. Quantitative PCR analysis was performed in triplicate using 1/50 of the reverse transcription reaction in an ABI Prism 7000 (Applied Biosystems) with Platinum SYBR green qPCR SuperMix-UDG with ROX (Invitrogen). Equal loading was achieved by amplifying GAPDH mRNA and all reactions were performed in triplicate. Primers used for amplification are listed in Supplementary Table 5. Data were extracted from the linear range of amplification. All graphs of qRT-PCR data shown represent samples of RNA that were DNase treated, reverse transcribed and amplified in parallel to avoid variation inherent in these procedures. Error bars represent s.d. of the mean of triplicate reactions.
Col 4F2A PCR genotyping
PCR performed using three sequencing primers (Col1a1 frtA F, frtB R, 4F2A R) listed in Supplementary Table 5. PCR reaction: 95C° /1min (1 cycle), 94 C° /30’’, 70 C° /45’’ (2 cycles), 94 C° /30’’, 68 C° /45’’(5 cycles), 94 C° /20’’, 66 C° /1min (29 cycles); 4 C°. Wild-type Col1a1 is ~ 300bp, targeted Col1a1 4F2A product is ~ 550bp.
Supplementary Material
Supplementary Figure 1 Genotyping of Col1a1 4F2A transgenic ES cells and mice
Supplementary Figure 2 Col1a1 dox-inducible expression of 4F2A transgene
Supplementary Figure 3a Expression of the 4F2A transgene in Col1a1 4F2A MEFs
Supplementary Figure 3b Expression of the 4F2A transgene in Col1a1 4F2A TTFs carrying different Tg ratios
Supplementary Figure 4 iPSCs derived from Col1a1 4F2A mice
Supplementary Figure 5 Efficiency of reprogramming pro-B cells
Supplementary Figure 6 Dox-induced transgene expression in 3F2A MEFs and characterization of 3F2A+S iPSCs
Supplementary Table 1 Aging of Col1a1 4F2A mice
Supplementary Table 2 Summary of experiments to derive iPS cells from Col1a1 4F2A mouse strains
Supplementary Table 3 Summary of diploid blastocyst injections of transgenic iPSC and ESC lines
Supplementary Table 4 Summary of iPSC pluripotency tests
Supplementary Table 5 List of primers used in the manuscript
Acknowledgments
B.W.C. would like to thank Chris Lengner and Lorenna Lee-Houghton for advice and assistance on somatic cell isolation techniques. We thank Frank Edenhofer for a kind gift of purified recombinant HTN-Cre protein and Jessie Dousman and Ruth Flannery for mouse husbandry. J.H is supported by a fellowship from the Helen Hay Whitney Foundation. R.J. is supported by grants from the NIH: 5-RO1-HDO45022, 5-R37-CA084198, and 5-RO1-CA087869.
Footnotes
COMPETING INTERESTS: RJ is an advisor to Stemgent and a co-founder of Fate Therapeutics.
AUTHOR CONTRIBUTIONS: B.W.C designed and performed experiments, analyzed data and wrote the paper. S.M. performed all blastocyst injections. J.H. assisted in experiments. C.B. assisted in design of Col1a1 targeting vectors. R.J. participated in experimental design, interpreted data and assisted in writing the paper.
REFERENCES CITED
- 1.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 2.Okita K, Ichisaka T, Yamanaka S. Generation of germline-competent induced pluripotent stem cells. Nature. 2007;448:313–317. doi: 10.1038/nature05934. [DOI] [PubMed] [Google Scholar]
- 3.Wernig M, et al. In vitro reprogramming of fibroblasts into a pluripotent ES-cell-like state. Nature. 2007;448:318–324. doi: 10.1038/nature05944. [DOI] [PubMed] [Google Scholar]
- 4.Maherali N, et al. Directly reprogrammed fibroblasts show global epigenetic remodeling and widespread tissue contribution. Cell Stem Cell. 2007;1:55–70. doi: 10.1016/j.stem.2007.05.014. [DOI] [PubMed] [Google Scholar]
- 5.Hanna J, et al. Direct reprogramming of terminally differentiated mature B lymphocytes to pluripotency. Cell. 2008;133:250–264. doi: 10.1016/j.cell.2008.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wernig M, et al. A drug-inducible transgenic system for direct reprogramming of multiple somatic cell types. Nat Biotechnol. 2008;26:916–924. doi: 10.1038/nbt1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maherali N, et al. A high-efficiency system for the generation and study of human induced pluripotent stem cells. Cell Stem Cell. 2008;3:340–345. doi: 10.1016/j.stem.2008.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hockemeyer D, et al. A drug-inducible system for direct reprogramming of human somatic cells to pluripotency. Cell Stem Cell. 2008;3:346–353. doi: 10.1016/j.stem.2008.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Markoulaki S, et al. Transgenic mice with defined combinations of drug-inducible reprogramming factors. Nat Biotechnol. 2009;27:169–171. doi: 10.1038/nbt.1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hochedlinger K, Yamada Y, Beard C, Jaenisch R. Ectopic expression of Oct-4 blocks progenitor-cell differentiation and causes dysplasia in epithelial tissues. Cell. 2005;121:465–477. doi: 10.1016/j.cell.2005.02.018. [DOI] [PubMed] [Google Scholar]
- 11.Beard C, Hochedlinger K, Plath K, Wutz A, Jaenisch R. Efficient method to generate single-copy transgenic mice by site-specific integration in embryonic stem cells. Genesis. 2006;44:23–28. doi: 10.1002/gene.20180. [DOI] [PubMed] [Google Scholar]
- 12.Carey BW, et al. Reprogramming of murine and human somatic cells using a single polycistronic vector. Proc Natl Acad Sci U S A. 2009;106:157–162. doi: 10.1073/pnas.0811426106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Soldner F, et al. Parkinson's disease patient-derived induced pluripotent stem cells free of viral reprogramming factors. Cell. 2009;136:964–977. doi: 10.1016/j.cell.2009.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kaji K, et al. Virus-free induction of pluripotency and subsequent excision of reprogramming factors. Nature. 2009;458:771–775. doi: 10.1038/nature07864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Woltjen K, et al. piggyBac transposition reprograms fibroblasts to induced pluripotent stem cells. Nature. 2009;458:766–770. doi: 10.1038/nature07863. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1 Genotyping of Col1a1 4F2A transgenic ES cells and mice
Supplementary Figure 2 Col1a1 dox-inducible expression of 4F2A transgene
Supplementary Figure 3a Expression of the 4F2A transgene in Col1a1 4F2A MEFs
Supplementary Figure 3b Expression of the 4F2A transgene in Col1a1 4F2A TTFs carrying different Tg ratios
Supplementary Figure 4 iPSCs derived from Col1a1 4F2A mice
Supplementary Figure 5 Efficiency of reprogramming pro-B cells
Supplementary Figure 6 Dox-induced transgene expression in 3F2A MEFs and characterization of 3F2A+S iPSCs
Supplementary Table 1 Aging of Col1a1 4F2A mice
Supplementary Table 2 Summary of experiments to derive iPS cells from Col1a1 4F2A mouse strains
Supplementary Table 3 Summary of diploid blastocyst injections of transgenic iPSC and ESC lines
Supplementary Table 4 Summary of iPSC pluripotency tests
Supplementary Table 5 List of primers used in the manuscript