Abstract
Dynamic phosphorylation of the RNA polymerase II CTD repeats (YS2PTS5PS7) is coupled to transcription and may act as a “code” that controls mRNA synthesis and processing. To examine the "code" in budding yeast, we mapped genome-wide CTD S2, 5 and 7 phosphorylations (PO4) and compared them with the CTD-associated termination factors, Nrd1 and Pcf11. CTD-PO4 dynamics are not scaled to the size of the gene. At 5’ ends, the onset of S2-PO4 is delayed by about 450 bases relative to S5-PO4, regardless of gene length. Phospho-CTD dynamics are gene-specific, with high S5/7-PO4 at the 5' end being characteristic of well-expressed genes with nucleosome-occupied promoters. Furthermore, the CTD kinases Kin28 and Ctk1 profoundly affect pol II distribution along genes in a highly gene-specific way. The "code" is therefore written differently on different genes, probably under the control of promoters. S7-PO4 is enriched on introns and at sites of Nrd1 accumulation suggesting that this modification may function in splicing and Nrd1 recruitment. Nrd1 and Pcf11 frequently co-localized, suggesting functional overlap between these terminators. Surprisingly, Pcf11 is also recruited to centromeres and pol III transcribed genes.
Keywords: RNA pol II, CTD phosphorylation, Nrd1, Pcf11, Kin28, Ctk1, transcription elongation/termination
INTRODUCTION
RNA polymerase II (pol II) is equipped with a unique C-terminal domain (CTD) on its large subunit that comprises heptad repeats (26 in budding yeast, 52 in humans) with a YSPTSPS consensus sequence that is conserved between yeast and mammals. This domain serves as a landing pad that presents a dynamic surface to proteins that interact with the transcription elongation complex (TEC) and carry out co-transcriptional pre-mRNA processing and histone modification 1. Phosphorylation and dephosphorylation of the heptad repeats at S2, 5 and 7 may constitute a CTD “code” that directs the binding and release of TEC-associated factors and imposes order on the transcription cycle of initiation, elongation and termination 2. It is not known whether the CTD "code" is written in a uniform way on all pol II transcribed genes or conversely, whether it can be inscribed in a variable way to establish gene-specific patterns of phosphorylation. To test this idea we mapped S2, S5, and S7 CTD phosphorylation genome-wide in yeast relative to total pol II and the CTD-associated transcription termination factors, Nrd1 and Pcf11.
Seminal ChIP studies of CTD phosphorylation revealed that the S5-PO4: S2-PO4 ratio is high at 5’ ends and low at 3’ ends 3 suggesting a dual gradient model of increasing S2 and decreasing S5 phosphorylation 5’-3’ across genes 1,2,4. It has been suggested that dual gradients of S2 and S5 phosphorylation could function as a metric of polymerase position within a gene 5, but this model has yet to be tested at high resolution genome-wide.
CTD S5 and S7 phosphorylation (S7-PO4) in yeast is catalyzed by the TFIIH-associated cyclin-dependent kinase, Kin28 6–8. S5 phosphorylation has been implicated in recruitment of capping enzymes and Nrd1 5,8,9. The function of S7 phosphorylation remains to be determined. Inhibition of Kin28 in an analogue sensitive (as) mutant revealed no effect on transcription, but this analysis was limited to a few genes 8,10. The major CTD S2 kinase in yeast is Ctk1 11,12, which facilitates recruitment of cleavage/polyadenylation factors 13. Surprisingly, Δctk1 and loss of CTD S2-PO4 has not been found to cause transcription defects in vivo 13(W.L. D.B unpublished).
Two mechanisms that are not mutually exclusive have been proposed for how pol II transcription terminates. In the “allosteric” model, the TEC is destabilized by interacting factors such as Pcf11 and Nrd1 that bind the CTD. In the “torpedo” model, the 5’-3’ RNA exonuclease, Rat1, 14, destabilizes the TEC by degrading the nascent transcript starting at a 5’ monophosphate entry site. Poly (A) sites, Rnt1 cleavage sites, and uncapped 5’ ends can provide such entry sites for Rat1 14–16. The termination factors Nrd1 and Pcf11 bind S5 and S2 phosphorylated heptads, respectively, through conserved CTD interaction domains 9,17. Pcf11 mediates termination at promoter-distal positions downstream of poly (A) sites where S2-PO4 is high. In contrast, Nrd1/Nab3/Sen1 mediated termination operates at promoter-proximal positions where S5-PO4 is high 5,9 particularly on short non-coding (nc) RNA genes including snoRNAs and cryptic unstable transcripts (CUTs) 18–22. Although Pcf11 and Nrd1-mediated termination appear to operate predominantly at 5’ and 3’ positions, respectively, their functions overlap under some circumstances 15,18,23,24. The degree to which Pcf11 is really confined to promoter-distal locations, and Nrd1 to promoter-proximal locations has not been analyzed genome-wide.
In this paper we investigated the “CTD code” in yeast by genome-wide analysis of S2, 5 and 7-PO4 relative to total pol II. These experiments revealed that CTD-PO4 dynamics are not constant but instead differ between genes with different promoter structures and expression levels. Our results also suggest new functions for CTD phosphorylation in control of pol II progression through genes and its association with termination factors Nrd1 and Pcf11.
RESULTS
CTD-PO4 dynamics are gene-specific and not scaled to gene length
We analyzed distributions of total pol II and the S2, S5 and S7 phosphorylated isoforms of the CTD in budding yeast by ChIP-Chip. DNA immunoprecipitated with anti-pan-CTD, -S2-PO4, -S5-PO4 (H14) or S7-PO4 (4E12, see Methods) was hybridized to whole genome arrays of 50mers tiled every 32 bases (Nimblegen). Background signals for these immunoprecipitations at rDNA were low (Supp. Fig. 1A, B). The results for total pol II were highly reproducible and in good agreement with previous ChIP-Chip studies using other antibodies 19,25 (Supp. Fig. 1C, D). The validity of the pol II density profiles is further supported by the fact that they demonstrate the expected slower termination at genes with weak poly (A) sites relative to those with strong sites (Supp. Fig. 1E).
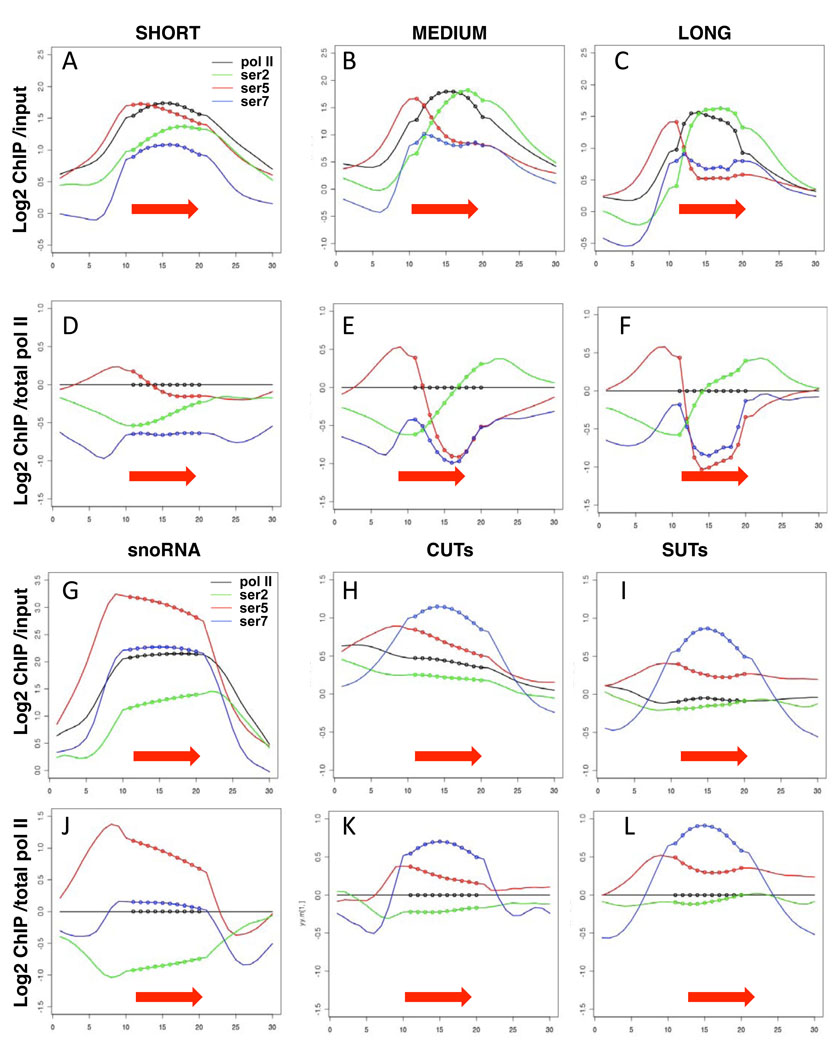
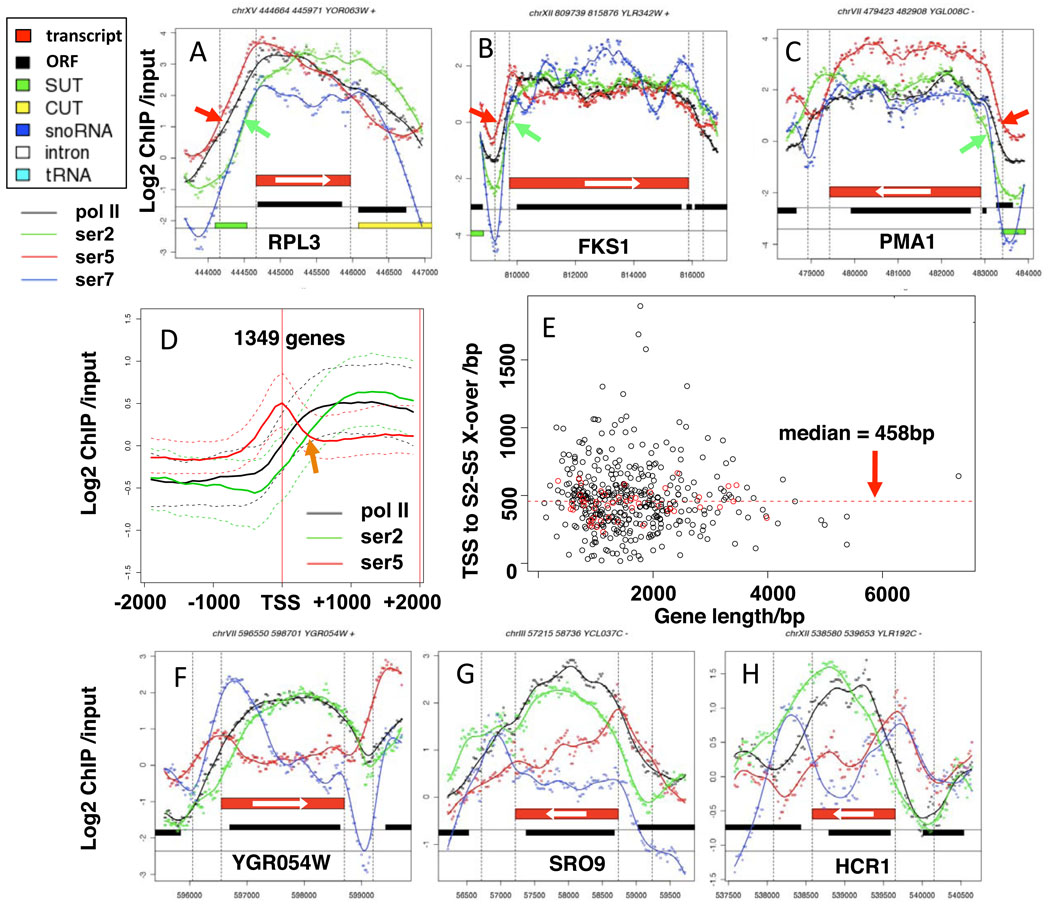
We observed a remarkably constant delay of about 450 bases between the onset of S5 phosphorylation and the onset of S2 phosphorylation near transcription start sites regardless of gene length. Average distributions of CTD S2-, S5-, and S7-PO4 before and after normalization to total pol II are shown for short, medium, and long ORFs in Figure 1A–F. We note however that the widespread overlap of coding and non-coding genes in yeast 22,26 can complicate interpretation of which transcription unit(s) are associated with a particular ChIP signal. On short genes (<800 bp) the level of S2 phosphorylation relative to S5 increases across most of the ORF (Fig. 1A, D) whereas on long genes (>2000 bp) the increase in S2-PO4: S5-PO4 ratio is limited to the 5’ region of the ORF and then remains constant across most of the length of the gene (Fig. 1C, F). This point is illustrated by comparing the CTD phosphorylation profiles on the short RPL3 gene (Fig. 2A) with the long FKS1 and PMA1 genes (Figs. 2B, C). On longer genes the S2-PO4: total pol II ratio reaches higher levels than on short genes (compare Fig. 1D, with E, F) suggesting that S2 phosphorylation might increase with the period of time that polymerase is engaged on the gene. Regardless of gene length, the S2-PO4: total pol II ratio reaches its high point slightly downstream of the 3’ end of the ORF close to the poly (A) site (Fig. 1D–F).
Figure 1. Gene-specific CTD-PO4 dynamics in yeast.
A–C, G–I. Average distributions of total pol II (tot1, black), and phospho-CTD isoforms on highly transcribed short (<800 bases, 248 genes) medium (800–2000 bases, 462 genes) and long (>2000 bases, 128 genes) coding genes, snoRNAs, CUTs and SUTs (red arrows and dotted lines, Supp. Table 2). Transcription units for coding genes are as defined by RNA-seq50. X-axes show transcription units divided into 10 equal intervals (dotted line) with 1 kb of 5’ and 3’ flanking sequence (smooth line). Y-axes represent log2 ChIP signals relative to input DNA.
D–F, J–L. Average distributions of CTD phospho-isoforms normalized to total pol II. Note that the scale for the snoRNA plots differs from that for CUTs and SUTs.
Figure 2. A 450 base interval between S5 and S2 phosphorylation at 5’ ends regardless of gene length.
A–C. Plots of log2 Chip signals relative to input DNA and smoothed curves for total pol II (black) CTD S2-PO4 (green), S5-PO4 (red) and S7-PO4 (blue). Individual points are average ChIP signals at 20 base intervals. Red bars mark transcript boundaries 50. Note the delay between S5- and S2 phosphorylation (red and green arrows) is relatively constant on short (A) and long (B, C) genes. X-axes are nucleotide positions on the respective chromosomes.
D. Median ChIP signals (solid lines) and 50% central range (dashed lines) vs distance from the transcription start site (TSS) for 1349 coding genes (see Supp. Table 2). Arrow marks the cross-over of S2- and S5-PO4 plots
E. Distance from the TSS to the S2–S5 cross-over vs gene length for 438 coding genes (Supp. Table 2). Red dots are ribosomal protein genes. Note the distance to the cross-over is independent of gene length.
F–H. Plots of Chip signals on selected genes showing variable patterns of S7-PO4 distributions across genes.
To investigate S2–S5 phosphorylation dynamics in more detail, we plotted their ChIP signals vs absolute distance from the transcription start site (TSS) for 1349 genes of various lengths (Fig, 2D) which revealed that on average the S5-PO4 and S2-PO4 curves cross-over at approximately +450. there was no significant correlation between the position of the S2–S5 cross-over point and gene length for 438 genes with well-defined pol II peaks (Fig. 2E). For ribosomal protein genes (red dots in Fig. 2E) the distance to the cross-over point is quite tightly clustered around the median of 458 bp regardless of gene length. In summary these results demonstrate that the dynamics of S2/5 phosphorylation are not scaled relative to gene length.
To address whether our ChIP resolution is sufficient to detect differences of less than 500 bases at 5' ends, even on short genes, we examined the capping enzyme Abd1, that is predicted to localize at 5' ends with S5-PO4 not S2-PO4 1. The results (Supp. Fig. 1 G–I) clearly resolve a co-localization of Abd1 with S5-PO4 rather than S2-PO4. We conclude that the ~450 base delay between S5- and S2-PO4, even on short genes, does not reflect an artificial limit in ChIP resolution. Consistent with a previous report 6, relative S7-PO4 is higher on snoRNAs than on mRNA genes (Fig. 1 D–F, J) and is even more strongly enriched on CUTs and SUTs (stable untranslated transcripts) (Fig. 1H, I, K, L). Conversely, S5-PO4 is more enriched on snoRNA genes than on CUTs and SUTs (Fig. 1J–L). In summary the results in Figure 1 demonstrate distinct patterns of CTD-PO4 at different classes of pol II transcription unit.
CTD S7-PO4 differs from S5-PO4 and is enriched on introns
We compared the profiles of CTD S7 and S5 phosphorylation that are both added by Kin28. Whereas profiles of S5-PO4 are characterized by a 5’ peak and are relatively invariable between genes, profiles of S7-PO4 were quite variable. At some genes (Figs. 2A–C) S7-PO4 is relatively evenly distributed across the gene. At other genes, discrete peaks of S7-PO4 are found at the 5’ end (Fig. 2F), the 3’ end (Fig. 2G, Supp. Fig. 4G, H) or both 5' and 3' ends (Figs. 2H, 3A, 5E). We conclude that differential S5 and S7 phosphorylation and/or dephosphorylation creates distinct 5’-3’ profiles of these modifications.
Figure 3. Enrichment of S7-PO4 on introns and variation of CTD-PO4 with transcription level and promoter structure.
A, B. Plots of Chip signals on selected genes showing enrichment of S7-PO4 on introns.
C. Average values of CTD S2, S5 and S7 phosphorylation normalized to total pol II on 282 introns (dotted lines) and flanking sequences (smooth lines, 1kb 5' and 3'). Note enrichment of S7-PO4 starting near 5’ splice sites (arrow).
D. Log2 ChIP signals within 123 genuine introns (red) from highly expressed genes and 919 decoy introns were calculated relative to 500 bases of upstream and downstream flanking sequence (Wilcoxon rank sum test P value < 2.2 × 10−16). The decoy introns were extracted from highly expressed intronless genes based on length and positions that match the population of genuine introns.
E, F. Average values of CTD phosphorylation on S2, S5 and S7 normalized to total pol II on ORF’s of similar length (800–1200 bases) in Group 1 (174 genes) and Group 3 (324 genes) with low and high transcription levels respectively 25 (Supp. Table 2) as in Fig. 1. Note that on average pol II on highly expressed genes has higher S5-PO4 and lower S2-PO4 and S7-PO4 than on poorly expressed genes.
G, H. Average values of CTD phosphorylation on S2, S5 and S7 normalized to total pol II on genes with occupied (OPN, 544 genes) vs depleted (DPN, 493 genes) proximal nucleosome promoter structures 27. Note higher S7-PO4 and lower S5-PO4 on DPN genes.
Figure 5. Nrd1 co-localizes with S7-PO4 on ncRNA and mRNA genes.
A–C. Average distributions of Nrd1, S5-PO4 and S7-PO4 across CUTs (230 genes), SUTs (128 genes) and long ORFs (128 genes, Supp. Table 2 as in Fig, 1).
D. Nrd1 localization correlates better with S7-PO4 than with S5-PO4. Absolute values of Pearson’s correlation coefficients for ChIP signals of Nrd1 and S5-PO4 and Nrd1 and S7-PO4 were calculated throughout each gene and 1kb of 5’ and 3’ flanking sequence and plotted relative to one another for 246 highly expressed ORFs 1000–2000 bases long.
E–H. Peaks of Nrd1 and S7-PO4 (*) on an ORF (E), CUT (F), SUT (G) and an intronic snoRNA (H).
I. Apparent premature termination marked by loss of total pol II (arrow) and accumulation of Rat1 (*) downstream of the intronic snR54 in IMD4 (see also Supp. Fig. 5A–C).
J. An exceptionally short transcription unit (blue arrow, left panel) convergent with CLB5 (left panel) that corresponds to a stable unannotated transcript (red arrow, right panel). Right panel is a screenshot from http://steinmetzlab.embl.de/NFRsharing/ with tracks for nucleosome occupancy (green) and hybridization signals (blue heat map) for 5 microarray experiments. (See also Supp. Figs. 5D–G, 6I).
K. Nrd1 enrichment without pol II on ARSs. Average distributions of Nrd1 (black) and total pol II (green) across 209 functional ARS elements 51 (Supp. Table 2).
We noted that peaks of S7-PO4 often coincided with introns (see Fig. 3A, B). Analysis of CTD phosphorylation on all 282 introns showed a specific enrichment for S7-PO4 within introns relative to flanking exons. In particular there is a sharp rise in S7-PO4 relative to total pol II close to 5’ splice sites (Fig. 3C). To test the significance of S7-PO4 enrichment in introns we compared 123 real introns from highly expressed genes with 919 “decoy” intron sequences of similar length and position within highly transcribed genes lacking introns. The results in Figure 3D demonstrate a significant enrichment of S7-PO4 in genuine introns (p < 2.2 × 10−16). Together these results suggest a possible functional relationship between S7 CTD phosphorylation and co-transcriptional spliceosome metabolism.
CTD-PO4 dynamics differ between protein coding genes with different promoter structures
We investigated whether CTD S2, S5 or S7 phosphorylation relative to total pol II varies between genes with low (group 1), medium (group 2) or high (group 3) levels of transcription based on pol II ChIP 25. On genes of equivalent length (800–1200 bases), distinct patterns of CTD phosphorylation distinguish these three groups of genes. At the 5’ ends of well-expressed genes (groups 2 (not shown), 3), S5-PO4 is higher than on poorly expressed group 1 genes (Fig. 3E, F). Unlike S5-PO4, S7-PO4 is higher on poorly expressed group 1 genes than on the highly expressed group 3 genes. Similarly, S2-PO4 is higher at the 5' ends of group 1 genes. The net result is that on average highly transcribed genes have higher S5-PO4: S2-PO4 ratios at their 5’ ends.
To determine whether promoter architecture correlates with CTD phosphorylation, we compared genes with occupied proximal nucleosomes (OPN, 544 genes) with those having depleted proximal nucleosomes (DPN, 493 genes) 27. OPN genes more frequently have TATA containing promoters, and their transcription is noisier and more highly regulated than DPN genes. Our analysis revealed elevated S5-PO4 and reduced S7-PO4 relative to total pol II on OPN genes compared to DPN genes both at the promoter and within the ORF (Fig. 3G, H). Furthermore OPN genes had a greater S5-PO4: S2-PO4 ratio at the transcription start site (p < 2.2 × 10−22) and a greater rate of decline of S5-PO4: S2-PO4 over the first 500 bases (p< 3.78 × 10−6) than DPN genes (see Supplemental Methods). In summary, the results in Figure 3E–H show that different patterns of CTD phosphorylation distinguish genes with different expression levels and promoter structures.
Kin28 is required for elongation through long genes
We investigated how the major S5/7 kinase Kin28 affects transcription by ChIP-Chip of total pol II in the kin28as point mutant that is inhibited by the ATP analogue NP-PP1 while preserving transcription initiation 8. On short genes, Kin28 inhibition had little effect (Fig. 4A, D Supp. Fig. 2A) but on longer genes it caused a remarkable re-distribution of pol II in favor of 5’ ends that is strongest on genes over 2kb long (Fig. 4 B, C, E–G, Supp. Fig. 2B–E). This relative accumulation of pol II at 5’ ends, strongly suggests either defective transcription elongation or premature termination.
Figure 4. A–G. Kin28 kinase enhances elongation at the 5’ ends of long genes.
A–C. Average distributions of total pol II across well-expressed short, medium and long genes (as in Fig. 1) in WT cells (PMY515, black) and the kin28as mutant (SHY508, red) treated with the inhibitor NP-PP1.
D–G. Plots of ChIP signals on selected genes reveal peaks of pol II accumulation (*) at 5’ ends of long (E–G) but not short genes (D). See also Supp. Fig. 2A–E.
H–L. Ctk1 kinase enhances elongation at the 3’ ends of a subset of genes.
H. Western blot of CTD S2-PO4 at time points after shifting the ctk1 tet-degron mutant (DBY739) to 37° + doxycycline (10 µg/ml) to deplete Ctk1. Pgk1 is a loading control.
I. Average distributions of pol II across 100 highly transcribed genes (Supp. Table 2) with 3' accumulation when Ctk1 is depleted in the ctk1tet-degron (red) relative top WT (black, DBY311). See also Supp. Fig. 2I.
J–L. Peaks of pol II accumulation (*) at 3’ ends after Ctk1 depletion. See also Supp. Fig. 2F.
Ctk1 depletion causes pol II to “pileup” at poly (A) sites
To ask how the S2 kinase, Ctk1, affects transcription we performed anti-pol II ChIP-Chip in a tet-repressible-degron mutant where S2-PO4 is greatly diminished after 2 hours at 37° in doxycycline (Fig. 4H). The advantage of a conditional ctk1 allele is that it is unlikely to acquire suppressor mutations. On many genes including PMA1 and RPL3, there was no effect of Ctk1 depletion on pol II profiles, as previously observed (Supp. Fig. 2G, H)13 however we identified 100 highly expressed genes and 550 genes overall (Supp. Table 2, Fig. 4I–L, Supp Fig. 4F, I) where Ctk1 depletion caused a “pileup” of polymerases close to the poly(A) sites suggesting a strong pause. The subset of genes with strong 3' accumulation of pol II have slightly better than average poly (A) site sequences 28. Downstream of the pause, termination occurred at its normal position. In contrast, on a subset of genes that are well expressed, but have weak poly (A) site consensus sequences, Ctk1 depletion did not induce pausing but did slightly impair termination (Supp. Fig. 2J). In summary, these results show that on a subset of genes with strong consensus poly (A) sites, reduced S2 CTD phosphorylation causes a strong pol II pause just 3’ of the ORF.
CTD S7 phosphorylation and Nrd1 recruitment on coding and non-coding genes
We initially addressed how phosphorylation influences the recruitment and release of factors from the CTD “landing pad” by comparing S2-, S5-, and S7-PO4 with localization of the Nrd1 termination factor. Remarkably, S7-PO4 co-localized closely with Nrd1 on all three classes of pol II transcribed ncRNA genes, CUTS, SUTs and snoRNAs (Fig. 5A, B Supp. Fig. 4A). Nrd1 co-localized with S7-PO4 on intergenic CUTs and SUTs (Supp Fig. 3A, K, L) and those that overlap ORFs (Supp. Fig. 3B–J). Interestingly, Nrd1 was found on PHO84 and GAL10 which are repressed by overlapping antisense transcription (Fig. 5F Supp. Fig. 3I) 29,30. Similarly Nrd1 accumulation and antisense transcription occurred at two meioic genes, NDT80 and IME4 that are repressed in haploid cells 31(Fig. 5G, Supp. Fig. 3J).
High levels of Nrd1 are also recruited to the U1, U2, and U5 snRNA genes (Supp. Fig. 4I, J and data not shown). U4 is an exception with high levels of S5-PO4 and S7-PO4 but little Nrd1 (Supp. Fig. 4K). We also detected specific Nrd1 enrichment without pol II at ARS elements suggesting a chromatin association that is independent of transcription (Fig. 5K). Unexpectedly coincident peaks of CTD S7-PO4 and Nrd1 occur on intronic snoRNAs snR59, snR38 and snR24 in the RPL7B, TEF4 and ASC1 genes (Fig. 5H, Supp. Fig. 4C, D). We observed that pol II occupancy is reduced downstream of the introns that harbor snoRNAs in IMD4, TEF4, ASC1 and RPL7A (Fig. 5I, Supp. Fig. 5A–C and not shown). Furthermore, Rat1 accumulation is evident within all 6 genes harboring intronic Box C/D snoRNAs IMD4, TEF4, ASC1, RPL7A, RPL7B, and EFB1 (Fig. 5I, Supp. Fig. 5A–C and not shown). Together these observations suggest that some level of premature termination involving Rat1 and perhaps Nrd1 is associated with co-transcriptional processing of intronic snoRNAs.
We found that co-transcriptional Nrd1 recruitment is not limited to ncRNA genes but is also a widespread feature of protein coding genes. On these genes, Nrd1 distribution correlates better with S7-PO4 than with S5-PO4 (Fig. 5D). This point is illustrated by the remarkable coincidence of Nrd1 and S7-PO4 across the EXG1, SRO9, HCR1, NAB2, and NPL3 genes (Fig. 5E, Supp. Fig. 4E–H). Note that on each of these genes, peaks of Nrd1 and S7-PO4 co-localize at the 3’ end where S5-PO4 is relatively low. Interestingly, Nrd1 accumulation downstream of NAB2 and NPL3 (Supp. Fig. 4G, H) coincides with where a poly (A) site independent “failsafe” termination mechanism operates 15,32.
We observed very discrete peaks of Nrd1 at the 3’ ends of a few genes where transcript mapping (http://steinmetzlab.embl.de/NFRsharing/) 26 identified small stable unannotated RNAs (≤100 bases) that converge with the 3’ ends of CLB5, UBP14, KAP95, SHE10 and the SUP6 tRNA, which is adjacent to PTR3 (Fig. 5J Supp. Fig. 5D–G). That these loci are genuine pol II transcription units is suggested by co-localization of Nrd1 with phosphorylated pol II, Pcf11 and Abd1 (Fig. 5J, Supp. Fig. 5D–G, 7I and data not shown). These exceptionally short novel short transcription units could influence the expression of adjacent genes. This idea is consistent with the fact that an insertion at SUP6 that separates the short pol II transcription unit from the PTR3 start site (Supp. Fig. 5G) strongly upregulates this gene 33.
Localization of the termination factors Pcf11 and Rat1 relative to Nrd1
We investigated the relationship between CTD S2-PO4 and recruitment of Pcf11, which binds S2-PO4 heptads in vitro. Both S2-PO4 and Pcf11 increase 5’ to 3’ across genes, but the Pcf11 profile is shifted 3’ relative to S2-PO4 (Fig. 6A, Supp. Fig. 6E). S2 phosphorylation therefore appears to precede Pcf11 recruitment. The discrete peak of Pcf11 just 3’ of the ORF suggests that transcription of the poly (A) site may enhance cross-linking of this factor (Fig. 6B–D Supp. Fig. 6A–C). Rat1 closely co-localized with Pcf11 at poly (A) sites consistent with coupling between cleavage/polyadenylation and termination (Fig. 6A, Supp. Fig. 6E).
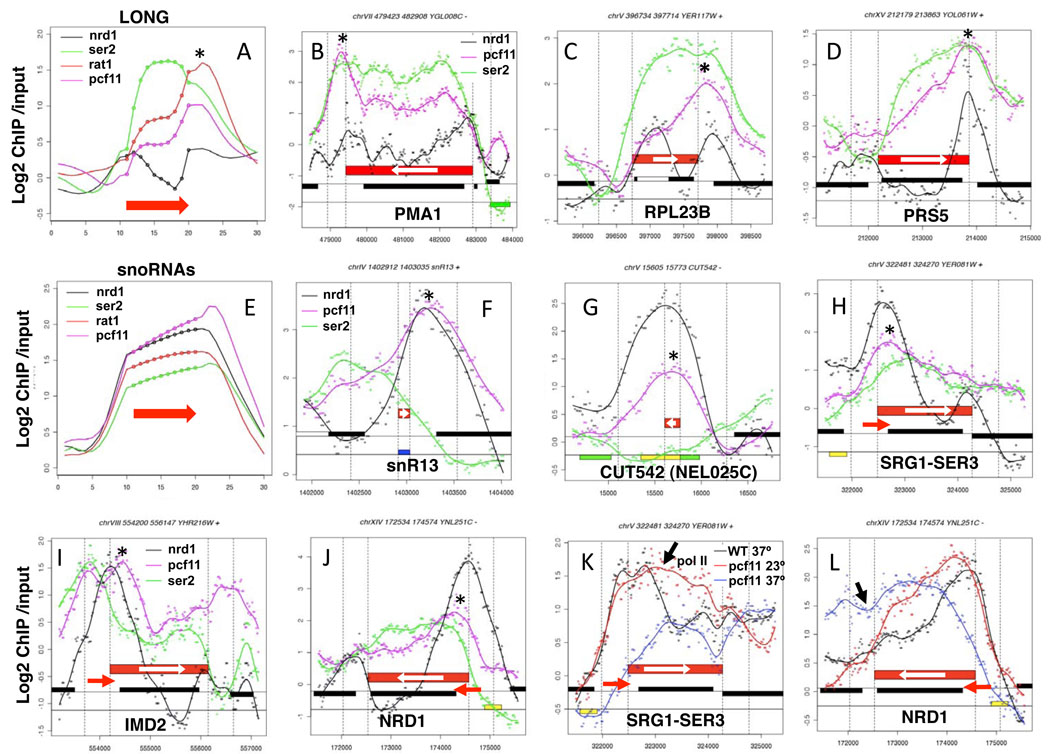
Figure 6. Termination factors Pcf11 and Nrd1 at 3’ ends.
A. Average distributions of Nrd1, S2-PO4, Rat1 and Pcf11 across long ORFs as in Fig. 1. Note coincident peaks of Nrd1, Pcf11 and Rat1 at 3’ ends (*) (see also Supp. Figs. 6E, F).
B–D. Plots of ChIP signals on selected genes showing discrete peaks of Nrd1 and Pcf11 accumulation (*) at poly (A) sites.
E. Average distributions of Nrd1, S2-PO4, Rat1, and Pcf11 across snoRNAs as in A.
F, G. Co-localization of Pcf11 with Nrd1 (*) on a snoRNA and a CUT.
H–J. Recruitment of Pcf11 and Nrd1 (*) to regulatory ncRNAs (red arrows) overlapping the 5’ ends of SER3, IMD2 and NRD1.
K, L. Effects of Pcf11 inactivation on transcription of SER3 and NRD1 with ncRNAs at their 5’ ends. Total pol II in WT (black) and pcf11-9 (PMY518) at 23° (red) and 37° (blue). Apparent readthrough transcription signals in pcf11-9 are marked with black arrows (see also Supp. Fig. 7E, F).
Pcf11 and Rat1 often overlapped with Nrd1 both at sites of poly (A)+ 3' end formation downstream of coding genes (Fig. 6C, D, Supp. Fig.6A–D) and sites of poly (A) − 3' end formation at snoRNAs and CUTs (Fig. 6E–G, Supp. Fig. 6F–H). Notably, Pcf11 is abundantly recruited to snoRNAs (Figs. 6E, F) even though these genes have relatively low levels of S2-PO4 (Fig. 1J). Pcf11 is also strongly enriched, with Nrd1, on regulatory ncRNAs that overlap the 5’ ends of SER3, IMD2, NRD1, URA2, URA8, ADE12, and HRP1 (Fig. 6H–J Supp. Fig. 7A–D) 34. Partial or complete inactivation of Pcf11 in the pcf11-9 ts mutant at 23° or 37° caused apparent defects in termination of the overlapping 5' ncRNAs at SER3, NRD1, URA8, and IMD2 (Fig. 6K, L Supp. Fig. 7E, F) and certain snoRNAs (Supp. Fig. 6K, L) consistent with a previous study 18. However, we cannot exclude the possibility that termination defects in convergent genes when Pcf11 is inactivated may contribute to the elevated pol II signals on some of these genes. Together these observations suggest a substantial functional overlap of Nrd1, Rat1 and Pcf11 in termination at mRNA and ncRNA genes. On the other hand, at ARS elements where pol II levels are low, Nrd1 is present with little or no Pcf11 (Supp. Fig. 8A).
Pcf11 localization at centromeres, pol III genes, and telomeres
Unexpectedly, prominent Pcf11 recruitment occurred at some places where there is little or no pol II transcription. Remarkably discrete peaks of Pcf11 are present at all 16 centromeres, in the absence of associated pol II, Nrd1 or Rat1 (Fig. 7A–C). Supp. Fig. 8B and data not shown). The function of centromeric Pcf11 is unknown. We were surprised to find that tRNA genes, transcribed by pol III, are also major sites of Pcf11 recruitment together with Rat1 (Fig. 7D–G Supp. Fig. 8C, E–H). When pol II density is plotted across all tRNA genes, a clear asymmetry is evident with higher pol II levels downstream of tRNAs than upstream (Fig. 7D). Indeed we observed many cases where a sharp drop in pol II density at the 3’ end of a gene coincided with a convergently transcribed tRNA (Fig. 7E, F, Supp. Fig. 8F–H). Pcf11is also found at the other pol III transcribed genes SCR1, RDN5 and snR52 (Fig. 7H, Supp. Fig. 8G, H) and at rDNA transcribed by pol I (Supp. Fig. 1B). A small amount of Nrd1 is present on tRNAs (Supp. Fig. 8C) but not on other pol III or pol I transcribed genes (Supp. Fig. 8D, Supp. Fig. 1B). Pcf11 is also present at telomeres (Fig. 7I–L) possibly as a result of its association with Rat1 which degrades telomeric TERRA transcripts 35. Consistent with this idea, Rat1 co-localizes with Pcf11 at telomeres whereas Nrd1 does not (Fig. 7I, J).
Figure 7. Pcf11 at centromeres, telomeres and pol III genes.
A–C. Average distributions of Pcf11 and pol II on all 16 centromeres (A, as in Fig. 1) and ChIP profiles of CEN1 and 4 with peaks of Pcf11 marked (*).
D. Average distributions of Pcf11, pol II, Rat1 and Nrd1 on tRNAs (275 genes, Supp. Table 2). Note higher pol II occupancy downstream of tRNAs than upstream (arrow).
E–G ChIP profiles with peaks of Pcf11 (*) over tRNAs and termination of convergent pol II transcription (E, F).
H. Pcf11 localization (*) on the pol III transcribed SCR1 gene for signal recognition particle RNA.
I, J Average distributions of Pcf11, pol II, Rat1, and Nrd1 at the telomeres with and without Y’ elements. Telomere sequences (dotted lines) and 1 kb of subtelomeric sequence (smooth lines) are shown.
K, L. ChIP profiles of Pcf11 and pol II on TEL7L (−Y’) and TEL8L (+Y’) (red bars). Chromosome ends are at left. Peaks of Pcf11 recruitment are marked (*).
DISCUSSION
In this report we analyzed genome-wide localization of RNA pol II CTD phosphorylation and CTD-associated termination factors, Nrd1 and Pcf11. Our results provide strong confirmation that a CTD phosphorylation "code" which "specifies the position of pol II in the transcription cycle" 2 applies generally by showing on a genome-wide scale that polymerases early in the cycle have a distinct pattern of CTD phosphorylation from those that are late in the cycle (Fig. 1A–F). We found that CTD S5 and S2 phosphorylation at 5’ ends is relatively “hard-wired” to produce a delay of about 450 bases between the onset of these two modifications at most mRNA coding genes (Fig. 2D, E). Regardless of gene length, the S2:S5 phosphorylation ratio increases within the first 500 bases of the transcription unit, then remains fairly constant within the body of long genes, and peaks in the 3’ flanking region prior to termination (Fig. 1D–F). As a result, the dual gradient model of increasing S2-PO4 and decreasing S5-PO4 across the entire gene applies well on short genes, but not on long ones. Because S2- and S5-PO4 dynamics are not scaled to compensate for differences in gene length, the ratio of S2-PO4: S5-PO4 is not an unambiguous measure of pol II position within a gene 5.
S5-PO4 and S7-PO4 are regulated quite differently although both are appended by the same kinase, Kin28. Whereas S5-PO4 is remarkably uniform in its profile across genes, high at 5’ ends and low at 3’ ends, S7-PO4 profiles vary in a gene-specific way with discrete peaks at either the 5’ end, the 3’ end or both ends. Intriguingly, S7-PO4 is specifically enriched over introns (Fig. 3A–D) suggesting a potential role in co-transcriptional assembly or disassembly of splicing complexes.
The CTD kinases Kin28 and Ctk1 have quite profound effects on pol II elongation that went unnoticed previously because they are not manifested on all genes. Inhibition of Kin28 impedes pol II progress through long genes resulting in accumulation of pol II at 5’ ends and depletion further downstream (Fig. 4B, C, E–G Supp. Fig. 2B–E). This defect could result from diminished recruitment of the cap methyltransferase Abd1, which can enhance elongation 36, or from premature termination of uncapped transcripts 16. It is also possible that an abnormally high ratio of S2-PO4 to S5/7-PO4 after inactivating Kin28 could impair elongation, particularly on long genes. Consistent with this idea, high S2-PO4: S5-PO4 is characteristic of terminating pol II (Fig. 1). Short genes might be resistant to the effect of Kin28 inhibition because on average they have lower relative levels of S2-PO4 (Fig. 1D–F). In summary, these results reveal a new function for Kin28 in enhancing polymerase progress through long genes, in addition to its previously established role in co-transcriptional capping.
In contrast, Ctk1 depletion caused pol II to pileup at the 3’ ends of over 500 genes with strong consensus poly (A) sites (Fig. 4I–L, Supp. Fig. 2I). At this subset of genes, low S2-PO4 may impair disassembly of 3’ end processing complexes on strong poly (A) sites thereby inducing transcriptional pausing. Previously the Glc7 phosphatase 37, the export factor Mex67 38 and the Tho/Sub2 complex 39 have been implicated in disassembly of 3’ end processing complexes. We speculate that the function of this pause could be to compensate for slower RNP maturation when S2 CTD phosphorylation is low.
Nrd1 is not confined to the 5’ ends of protein coding genes by a strict association with CTD S5-PO4, as previously thought 9. Instead we found a strong correlation between Nrd1 and CTD S7-PO4 profiles on ncRNA and protein-coding genes. We propose that in addition to binding S5-PO4 heptads9 and specific sequences in the nascent RNA 40, S7-PO4 plays an important role in directing recruitment of Nrd1. It is also possible that conversely, Nrd1 binding enhances S7 phosphorylation.
Nrd1 recruitment is a characteristic feature of non-coding CUTs and SUTs including those that overlap protein-coding genes (Fig. 5F, G Supp. Fig. 3B–J). Such overlapping transcription might serve to localize Nrd1 on these genes for the purpose of repressing their expression, possibly through interactions with the nuclear exosome and chromatin silencing factors 41,42. Consistent with this idea, Nrd1 was found at PHO84, GAL1-10, NDT80 and IME4 where antisense transcription is implicated in establishing repressed chromatin states (Fig. 5F, G and Supp. Fig. 3I, J)29–31.
Nrd1 also localized to intronic snoRNAs where we also observed Rat1 recruitment and evidence of premature transcription termination (Fig. 5H, I, Supp. Figs. 4C,D, 5A–C). Co-transcriptional cleavage by Rnt1 43 has been implicated in processing of intronic snoRNAs and might promote premature termination by providing an entry point for the Rat1 exonuclease 15,32. Whether Nrd1 contributes to processing of intronic snoRNAs and/or premature termination remains to be investigated. Together these results suggest that under some conditions, perhaps when splicing is slow, early termination, possibly involving Nrd1 and Rat1, may produce a transcript that serves as a snoRNA precursor, but not as a mRNA precursor.
The cleavage/polyadenylation factor Pcf11 binds S2-PO4 CTD and is enriched at 3’ ends, but its distribution does not precisely mimic that of S2-PO4 (Fig. 6A, Supp. Fig. 6E). Pcf11 is present throughout the length of most genes and often shows a discrete peak at the poly (A) site unlike S2-PO4. In addition to the S2-PO4 mark on the CTD, poly (A) consensus elements in the nascent transcript are therefore likely to enhance Pcf11 binding to the TEC. Frequently, we observed coincident peaks of Pcf11 and Nrd1 at poly (A) sites (Fig. 6D Supp. Fig. 6A–D) and ncRNAs (Fig. 6E–J, Supp. Fig. 6F–H). In addition, Pcf11 inactivation appeared to impair termination downstream of regulatory ncRNAs including those that overlap the 5’ ends of SER3 and NRD1 (Fig. 6 K, L). The widespread overlap of Nrd1 and Pcf11 suggests that these two factors cooperate or back one another up to achieve pol II termination at many yeast genes under normal conditions.
Pcf11 accumulated quite unexpectedly at pol I (Supp. Fig. 1B) and pol III transcribed genes as well as centromeres (Fig. 7, Supp. Fig. 8), presumably by pol II-independent recruitment mechanisms. It is not known whether Pcf11 at these locations is part of the CF1A cleavage/polyadenylation complex or in another form. The function of Pcf11 at centromeres is a mystery. Unlike centromeres, high levels of Rat1 co-localized with Pcf11 at tRNA genes (Fig. 7D). Pol II and associated factors have previously been detected at tRNA genes in yeast and mammalian cells 44,45. There are several possible functions for Pcf11 and Rat1 at tRNAs including termination of convergently transcribed pol II genes (Fig. 7E–G, Supp. Fig. 8F–H), degradation of improperly modified tRNAs 46 and establishment of chromatin boundaries. How Pcf11 functions at telomeres is unclear, but together with Rat1 (Fig. 7A, B), it might influence telomere length by regulating termination and 3' end processing of TERRA transcripts 35. In summary, Pcf11 was previously known only as a cleavage/polyadenylation factor, but our genome-wide localization of this factor at centromeres, and pol I and pol III transcribed genes, shows that this view must be revised.
The most salient result to emerge from genome-wide analysis of CTD-PO4 is that the CTD "code" is inscribed differently on different genes. Distinct patterns of phosphorylation distinguish pol II on the different classes of pol II transcription unit for mRNAs, snoRNAs, CUTs and SUTs (Fig. 1). Furthermore there are significant variations in the CTD "code" among protein-coding genes. Highly expressed protein coding genes and genes with promoters occupied by nucleosomes (OPN) have higher S5-PO4, and lower S7-PO4, relative to total pol II, than poorly expressed genes and those with nucleosome depleted promoters (DPN) (Fig. 3E–H). Although much remains to be learned about exactly what phosphorylation symbols in the CTD “code” mean, the results reported here establish that CTD phosphorylation is differentially regulated between genes. Furthermore they suggest the intriguing hypothesis that promoters can dictate how the CTD "code" will be written.
METHODS
Yeast Strains
Strains used in this study are listed in Supplemental Table 1.
ChIP-Chip
Cells were grown in YPD to O.D. ~0.8 and cross-linked for 15 min or 2 hour at room temperature in 1% formaldehyde. ChIP-Chip and analysis by ChIPViewer is described in Supplemental methods. ChIPViewer profiles will be made available on a searchable database.
Antibodies
Rabbit anti-pan-CTD, -S2-PO4 and -Abd1 antibodies 47,48 4E12 monoclonal anti-S7-PO4 49 have been described. Polyclonal anti-pan-CTD cross-reacts with unphosphorylated, S2, S5 and S7 phosphorylated peptides (Supp. Fig. 1F). Essentially all cross-reactivity of the anti-S2-PO4 polyclonal antibody is abolished by inactivating Ctk1 (Fig. 4H and Zhang et al., 2005). Anti-Ser5-PO4, (H14), anti-HA (12CA5) and anti-myc (9E10) were purchased from Covance, Roche and Santa Cruz.
Supplementary Material
Acknowledgements
Supported by NIH grants GM58613 to D.B., GM066213 to P.M., GM083127 to D.P. and GM072706 to J. G. H. K. was supported by ARRA award 3R01GM063873-06S1. We thank S. Hahn (Fred Hutchinson Cancer Center) and D. Eick (U. Munich) for strains and antibodies, S. Chavez (U. Sevilla), S. Johnson, K. Brannan and S. Kim for valuable discussions, K. Bhatta for help with figures, Anna Lee (Denver School of the Arts) and Amanda Roth (Denver School of Science and Technology) for data analysis and C. Wang and M. Covarrubias (City of Hope Functional Genomics Core) for array hybridization.
REFERENCES
- 1.Phatnani HP, Greenleaf AL. Phosphorylation and functions of the RNA polymerase II CTD. Genes Dev. 2006;20:2922–2936. doi: 10.1101/gad.1477006. [DOI] [PubMed] [Google Scholar]
- 2.Buratowski S. The CTD code. Nat Struct Biol. 2003;10:679–680. doi: 10.1038/nsb0903-679. [DOI] [PubMed] [Google Scholar]
- 3.Komarnitsky P, Cho EJ, Buratowski S. Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes Dev. 2000;14:2452–2460. doi: 10.1101/gad.824700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Egloff S, Murphy S. Cracking the RNA polymerase II CTD code. Trends Genet. 2008;24:280–288. doi: 10.1016/j.tig.2008.03.008. [DOI] [PubMed] [Google Scholar]
- 5.Gudipati RK, Villa T, Boulay J, Libri D. Phosphorylation of the RNA polymerase II C-terminal domain dictates transcription termination choice. Nat Struct Mol Biol. 2008;15:786–794. doi: 10.1038/nsmb.1460. [DOI] [PubMed] [Google Scholar]
- 6.Akhtar M, et al. TFIIH kinase places bivalent marks on the carboxy-terminal domain of RNA polymerase II. Mol. Cell. 2009;34:387–393. doi: 10.1016/j.molcel.2009.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim M, Suh H, Cho EJ, Buratowski S. Phosphorylation of the yeast Rpb1 C-terminal domain at serines 2, 5, and 7. J Biol Chem. 2009;284:26421–26426. doi: 10.1074/jbc.M109.028993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kanin EI, et al. Chemical inhibition of the TFIIH-associated kinase Cdk7/Kin28 does not impair global mRNA synthesis. Proc Natl Acad Sci U S A. 2007;104:5812–5817. doi: 10.1073/pnas.0611505104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vasiljeva L, Kim M, Mutschler H, Buratowski S, Meinhart A. The Nrd1-Nab3-Sen1 termination complex interacts with the Ser5-phosphorylated RNA polymerase II C-terminal domain. Nat Struct Mol Biol. 2008;15:795–804. doi: 10.1038/nsmb.1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hong SW, et al. Phosphorylation of the RNA polymerase II C-terminal domain by TFIIH kinase is not essential for transcription of Saccharomyces cerevisiae genome. Proc Natl Acad Sci USA. 2009;106:14276–14280. doi: 10.1073/pnas.0903642106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jones JC, et al. C-terminal repeat domain kinase I phosphorylates Ser2 and Ser5 of RNA polymerase II C-terminal domain repeats. J Biol Chem. 2004;279:24957–24964. doi: 10.1074/jbc.M402218200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cho E-J, Kobor M, Kim M, Greenblatt J, Buratowski S. Opposing effects of Ctk1 kinase and Fcp1 phosphatase at Ser 2 of the RNA polymerase II C-terminal domain. Genes Dev. 2001;15:3319–3329. doi: 10.1101/gad.935901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ahn SH, Kim M, Buratowski S. Phosphorylation of Serine 2 within the RNA Polymerase II C-Terminal Domain Couples Transcription and 3' End Processing. Mol Cell. 2004;13:67–76. doi: 10.1016/s1097-2765(03)00492-1. [DOI] [PubMed] [Google Scholar]
- 14.Kim M, et al. The yeast Rat1 exonuclease promotes transcription termination by RNA polymerase II. Nature. 2004;432:517–522. doi: 10.1038/nature03041. [DOI] [PubMed] [Google Scholar]
- 15.Rondon AG, Mischo HE, Kawauchi J, Proudfoot NJ. Fail-safe transcriptional termination for protein-coding genes in S. cerevisiae. Mol Cell. 2009;36:88–98. doi: 10.1016/j.molcel.2009.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jimeno-Gonzalez S, Haaning LL, Malagon F, Jensen TH. The yeast 5'-3' exonuclease Rat1p functions during transcription elongation by RNA polymerase II. Mol Cell. 2010;37:580–587. doi: 10.1016/j.molcel.2010.01.019. [DOI] [PubMed] [Google Scholar]
- 17.Meinhart A, Cramer P. Recognition of RNA polymerase II carboxy-terminal domain by 3'-RNA-processing factors. Nature. 2004;430:223–226. doi: 10.1038/nature02679. [DOI] [PubMed] [Google Scholar]
- 18.Kim M, et al. Distinct Pathways for snoRNA and mRNA Termination. Mol Cell. 2006;24:723–734. doi: 10.1016/j.molcel.2006.11.011. [DOI] [PubMed] [Google Scholar]
- 19.Steinmetz EJ, et al. Genome-Wide Distribution of Yeast RNA Polymerase II and Its Control by Sen1 Helicase. Mol Cell. 2006;24:735–746. doi: 10.1016/j.molcel.2006.10.023. [DOI] [PubMed] [Google Scholar]
- 20.Arigo JT, Eyler DE, Carroll KL, Corden JL. Termination of Cryptic Unstable Transcripts Is Directed by Yeast RNA-Binding Proteins Nrd1 and Nab3. Mol Cell. 2006;23:841–851. doi: 10.1016/j.molcel.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 21.Thiebaut M, Kisseleva-Romanova E, Rougemaille M, Boulay J, Libri D. Transcription termination and nuclear degradation of cryptic unstable transcripts: a role for the nrd1-nab3 pathway in genome surveillance. Mol Cell. 2006;23:853–864. doi: 10.1016/j.molcel.2006.07.029. [DOI] [PubMed] [Google Scholar]
- 22.Neil H, et al. Widespread bidirectional promoters are the major source of cryptic transcripts in yeast. Nature. 2009;457:1038–1042. doi: 10.1038/nature07747. [DOI] [PubMed] [Google Scholar]
- 23.Morlando M, et al. Functional analysis of yeast snoRNA and snRNA 3'-end formation mediated by uncoupling of cleavage and polyadenylation. Mol Cell Biol. 2002;22:1379–1389. doi: 10.1128/mcb.22.5.1379-1389.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lykke-Andersen S, Jensen TH. Overlapping pathways dictate termination of RNA polymerase II transcription. Biochimie. 2007;89:1177–1182. doi: 10.1016/j.biochi.2007.05.007. [DOI] [PubMed] [Google Scholar]
- 25.Venters BJ, Pugh BF. A canonical promoter organization of the transcription machinery and its regulators in the Saccharomyces genome. Genome Res. 2009;19:360–371. doi: 10.1101/gr.084970.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu Z, et al. Bidirectional promoters generate pervasive transcription in yeast. Nature. 2009;457:1033–1037. doi: 10.1038/nature07728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tirosh I, Barkai N. Two strategies for gene regulation by promoter nucleosomes. Genome Res. 2008;18:1084–1091. doi: 10.1101/gr.076059.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Graber JH, McAllister GD, Smith TF. Probabilistic prediction of Saccharomyces cerevisiae mRNA 3'-processing sites. Nucleic Acids Res. 2002;30:1851–1858. doi: 10.1093/nar/30.8.1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Camblong J, Iglesias N, Fickentscher C, Dieppois G, Stutz F. Antisense RNA stabilization induces transcriptional gene silencing via histone deacetylation in S. cerevisiae. Cell. 2007;131:706–717. doi: 10.1016/j.cell.2007.09.014. [DOI] [PubMed] [Google Scholar]
- 30.Houseley J, Rubbi L, Grunstein M, Tollervey D, Vogelauer M. A ncRNA modulates histone modification and mRNA induction in the yeast GAL gene cluster. Mol Cell. 2008;32:685–695. doi: 10.1016/j.molcel.2008.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hongay CF, Grisafi PL, Galitski T, Fink GR. Antisense transcription controls cell fate in Saccharomyces cerevisiae. Cell. 2006;127:735–745. doi: 10.1016/j.cell.2006.09.038. [DOI] [PubMed] [Google Scholar]
- 32.Ghazal G, et al. Yeast RNase III triggers polyadenylation-independent transcription termination. Mol Cell. 2009;36:99–109. doi: 10.1016/j.molcel.2009.07.029. [DOI] [PubMed] [Google Scholar]
- 33.Bolton EC, Boeke JD. Transcriptional interactions between yeast tRNA genes, flanking genes and Ty elements: a genomic point of view. Genome Res. 2003;13:254–263. doi: 10.1101/gr.612203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Corden JL. Yeast Pol II start-site selection: the long and the short of it. EMBO Rep. 2008;9:1084–1086. doi: 10.1038/embor.2008.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Luke B, et al. The Rat1p 5' to 3' exonuclease degrades telomeric repeat-containing RNA and promotes telomere elongation in Saccharomyces cerevisiae. Mol Cell. 2008;32:465–477. doi: 10.1016/j.molcel.2008.10.019. [DOI] [PubMed] [Google Scholar]
- 36.Schroeder S, Zorio D, Schwer B, Shuman S, Bentley D. A Function of Yeast mRNA Cap Methyltransferase, Abd1, in transcription by RNA Polymerase II. Mol. Cell. 2004;13:377–387. doi: 10.1016/s1097-2765(04)00007-3. [DOI] [PubMed] [Google Scholar]
- 37.Gilbert W, Guthrie C. The Glc7p nuclear phosphatase promotes mRNA export by facilitating association of Mex67p with mRNA. Mol Cell. 2004;13:201–212. doi: 10.1016/s1097-2765(04)00030-9. [DOI] [PubMed] [Google Scholar]
- 38.Qu X, et al. Assembly of an export-competent mRNP is needed for efficient release of the 3'-end processing complex after polyadenylation. Mol Cell Biol. 2009;29:5327–5338. doi: 10.1128/MCB.00468-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rougemaille M, et al. THO/Sub2p functions to coordinate 3'-end processing with gene-nuclear pore association. Cell. 2008;135:308–321. doi: 10.1016/j.cell.2008.08.005. [DOI] [PubMed] [Google Scholar]
- 40.Carroll KL, Ghirlando R, Ames JM, Corden JL. Interaction of yeast RNA-binding proteins Nrd1 and Nab3 with RNA polymerase II terminator elements. RNA. 2007;13:361–373. doi: 10.1261/rna.338407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vasiljeva L, Buratowski S. Nrd1 interacts with the nuclear exosome for 3' processing of RNA polymerase II transcripts. Mol Cell. 2006;21:239–248. doi: 10.1016/j.molcel.2005.11.028. [DOI] [PubMed] [Google Scholar]
- 42.Vasiljeva L, Kim M, Terzi N, Soares LM, Buratowski S. Transcription termination and RNA degradation contribute to silencing of RNA polymerase II transcription within heterochromatin. Mol Cell. 2008;29:313–323. doi: 10.1016/j.molcel.2008.01.011. [DOI] [PubMed] [Google Scholar]
- 43.Ghazal G, Ge D, Gervais-Bird J, Gagnon J, Abou Elela S. Genome-wide prediction and analysis of yeast RNase III-dependent snoRNA processing signals. Mol Cell Biol. 2005;25:2981–2994. doi: 10.1128/MCB.25.8.2981-2994.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Raha D, et al. Close association of RNA polymerase II and many transcription factors with Pol III genes. Proc Natl Acad Sci U S A. 2010;107:3639–3644. doi: 10.1073/pnas.0911315106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Barski A, et al. Pol II and its associated epigenetic marks are present at Pol III–transcribed noncoding RNA genes. Nat Struct Mol Biol. 2010;17:629–634. doi: 10.1038/nsmb.1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chernyakov I, Whipple JM, Kotelawala L, Grayhack EJ, Phizicky EM. Degradation of several hypomodified mature tRNA species in Saccharomyces cerevisiae is mediated by Met22 and the 5'-3' exonucleases Rat1 and Xrn1. Genes Dev. 2008;22:1369–1380. doi: 10.1101/gad.1654308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schroeder S, Schwer B, Shuman S, Bentley D. Dynamic association of capping enzymes with transcribing RNA polymerase II. Genes Dev. 2000;14:2435–2440. doi: 10.1101/gad.836300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang L, Schroeder S, Fong N, Bentley DL. Altered nucleosome occupancy and histone H3K4 methylation in response to 'transcriptional stress'. EMBO J. 2005;24:2379–2390. doi: 10.1038/sj.emboj.7600711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chapman RD, et al. Transcribing RNA polymerase II is phosphorylated at CTD residue serine-7. Science. 2007;318:1780–1782. doi: 10.1126/science.1145977. [DOI] [PubMed] [Google Scholar]
- 50.Nagalakshmi U, et al. The transcriptional landscape of the yeast genome defined by RNA sequencing. Science. 2008;320:1344–1349. doi: 10.1126/science.1158441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nieduszynski CA, Hiraga S, Ak P, Benham CJ, Donaldson AD. OriDB: a DNA replication origin database. Nucleic Acids Res. 2007;35:D40–D46. doi: 10.1093/nar/gkl758. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.