Abstract
The protein lysine methyltransferase SET7 regulates DNA methyltransferase-1 (DNMT1) activity in mammalian cells by promoting degradation of DNMT1 and thus allows epigenetic changes via DNA demethylation. Here we reveal an interplay between monomethylation of DNMT1 Lys142 by SET7 and phosphorylation of DNMT1 Ser143 by AKT1 kinase. These two modifications are mutually exclusive, and structural analysis suggests that Ser143 phosphorylation interferes with Lys142 monomethylation. AKT1 kinase colocalizes and directly interacts with DNMT1 and phosphorylates Ser143. Phosphorylated DNMT1 peaks during DNA synthesis, before DNMT1 methylation. Depletion of AKT1 or overexpression of dominant-negative AKT1 increases methylated DNMT1, resulting in a decrease in DNMT1 abundance. In mammalian cells, phosphorylated DNMT1 is more stable than methylated DNMT1. These results reveal cross-talk on DNMT1, through modifications mediated by AKT1 and SET7, that affects cellular DNMT1 levels.
Multiple interdependent post-translational modifications of cellular proteins allow for combinatorial repertoires of interactions. Some of these modifications, including acetylation, phosphorylation, methylation and sumoylation, might participate in cross-talk for dynamic control of cellular signaling under various physiological conditions1. Protein phosphorylation is involved in various cellular processes, including cell growth, development and apoptosis, and regulates essential physiological systems, such as the period of the circadian rhythm within mammalian cells, via cellular signaling pathways2. Protein methylation, especially on lysines, is another important reversible post-translational modification of cellular proteins; for example, histone methylation is involved in the regulation of transcription3–5. Combinations of these modifications on histones (such as histone H3 Lys9 methylation and H3 Ser10 phosphorylation6–8) cooperate to regulate chromatin structure and transcription by allowing or inhibiting binding of specific regulatory proteins9. Similarly, non-histone proteins such as p53 (refs. 10–12) and estrogen receptor13,14 are also regulated by post-translational modifications.
Previously, we have demonstrated that methylation of Lys142 on DNMT1 leads to DNMT1 degradation15. Adjacent to Lys142 is Ser143, a known phosphorylation site16. Therefore, we set out to investigate the interplay between Lys142 monomethylation (Lys142me1) and Ser143 phosphorylation (pSer143), and how this interplay affects DNMT1 stability.
RESULTS
SET7 methylation of DNMT1 blocked by phosphorylation
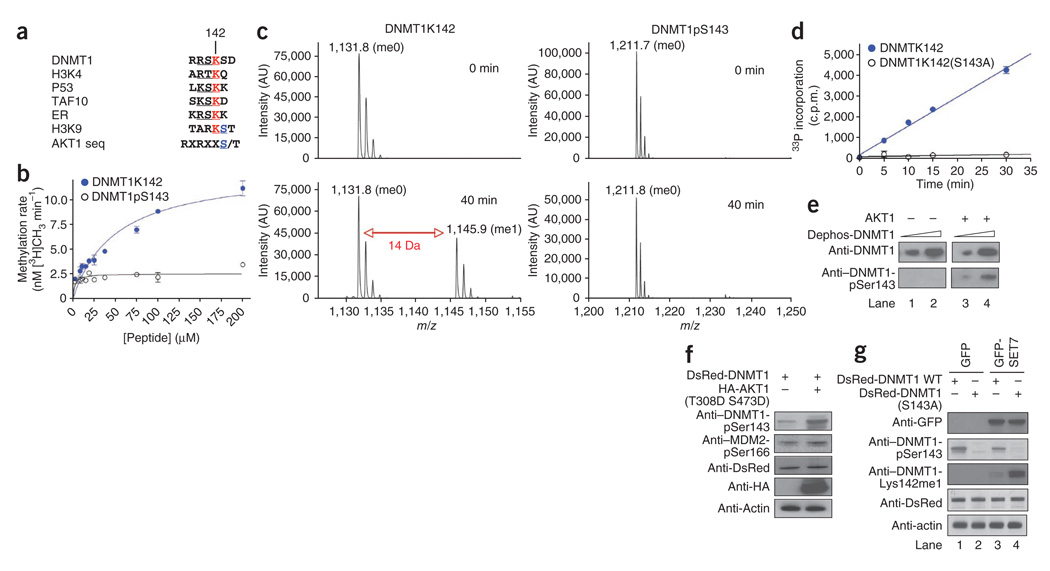
We have previously reported that SET7-mediated methylation of mammalian DNMT1 at Lys142 stimulates its proteasomal degradation15. Conversely, in another study, the absence of lysine-specific demethylase-1 (LSD1) has been found to reduce DNMT1 stability in cells, leading to progressive loss of DNA methylation17. We noticed that the serine residue, Ser143, immediately after DNMT1’s methylation-target lysine, Lys142, is unique among known SET7 substrates (the target-site sequence is RSKS as opposed to the (R/K)(S/T)K(K/Q/D) consensus; Fig. 1a). We speculated that this serine could be subject to phosphorylation, analogous to some histone modification patterns—for example, the ‘methyl-phospho’ cassette of histone H3 Lys9 and Ser10 (ref. 18).
Figure 1.
Lysine methylation and serine phosphorylation on DNMT1. (a) The lysines targeted for methylation by SET7 in various known substrates are shown in red. The serine in the consensus AKT1 kinase sequence is aligned with histone H3 Ser10 and DNMT1 Ser143. (b) SET7 methylation reaction in the presence of various concentrations of DNMT1 peptides representing either native or phosphorylated Ser143. (c) MS of the products of in vitro methylation assays (0 min, before assay; 40 min, after assay) on DNMT1 peptides with either native (left) or phosphorylated Ser143 (right). The mass shift of 14 Da represents addition of one methyl group. (d) AKT1 kinase reaction in the presence of fixed concentrations of wild-type DNMT1 peptides or S143A mutant peptides. (e) Western blot assay showing AKT1 kinase activity on dephosphorylated recombinant full-length DNMT1 on Ser143. (f) Western blot showing Ser143 phosphorylation of full-length DNMT1 in the presence (+) or absence (−) of constitutively active AKT1 kinase. Antibodies used for probe are listed; actin is shown as a loading control. MDM2 pSer166 is a positive control. (g) S143A mutation in full-length DNMT1 facilitates SET7-mediated Lys142 methylation. Western blot demonstrating effect of Ser143 phosphorylation and/or Lys142 methylation in either wild-type (WT) DsRed-DNMT1 or the mutant DsRed-DNMT1(S143A), as indicated by plus and minus signs, when coexpressed with GFP alone (lanes 1 and 2) or GFP-SET7 (lanes 3 and 4).
To examine the possibility of a methyl-phospho dynamic switch on DNMT1, we used a synthetic peptide substrate representing amino acid residues 137–146 of human DNMT1 with either an unmodified Ser143 (DNMT1K142) or phosphorylated Ser143 (DNMT1pS143). We incubated these constructs with recombinant purified SET7 enzyme. SET7 methylated the unmodified substrate but not the phosphorylated peptide (Fig. 1b,c). The SET7-mediated methylation resulted in a mass addition of 14 Da, corresponding to a monomethylation (Fig. 1c).
As the consensus sequence of the AKT1 kinase target site matches the sequence surrounding Ser143 of DNMT1 (Fig. 1a), we tested whether AKT1 is the kinase responsible for DNMT1 Ser143 phosphorylation. In an in vitro kinase assay using recombinant AKT1, the unmodified peptide DNMT1K142 was phosphorylated strongly over time, whereas an otherwise identical peptide substrate with an S143A mutation (DNMT1K142(S143A)) was not, suggesting that Ser143 in this peptide is the target of the AKT1 kinase (Fig. 1d).
To determine whether Ser143 phosphorylation occurs on the full-length human DNMT1 in cells, we raised a specific rabbit polyclonal antibody against DNMT1 with Ser143 phosphorylated (anti–DNMT1-pSer143). We validated this antibody using a panel of peptides representing residues 137–146 in the full-length DNMT1 protein sequence. Various amounts of peptide containing pSer143, Lys142me1, both modifications or no modification were dot-blotted and probed with anti–DNMT1-pSer143. The antibody cross-reacted only with pSer143-modified peptide (Supplementary Fig. 1a).
We also stringently validated the antibody using HeLa cell extract and purified human DNMT1 expressed from baculovirus. The antibody readily detected phosphorylated DNMT1 in the presence of excess unmodified competitor peptide representing the same epitope (Supplementary Fig. 1b,c). By comparison, a catalytically active DNMT1 lacking the N-terminal 580 residues (DNMT1Δ580)19 did not react with anti–phospho AKT1 substrate, suggesting that AKT1-mediated phosphorylation occurs in the N-terminal region of DNMT1 (Supplementary Fig. 1d). Finally, anti–DNMT1-pSer143 did not detect recombinant DNMT1 treated with phosphatase in vitro (Fig. 1e, lanes 1 and 2), but it did cross-react with phosphatase-treated DNMT1 that had been incubated with AKT1 and ATP cofactor (Fig.1e, lanes 3 and 4).
To determine whether AKT1 is the main kinase for DNMT1 Ser143 phosphorylation in mammalian cells, we expressed a constitutively active AKT1 (the double mutant T308D S473D)20 in COS-7 cells along with a construct of DNMT1 fused to the red fluorescent protein DsRed (DsRed-DNMT1), and we monitored Ser143 phosphorylation in the fusion DNMT1. As the endogenous DNMT1 is already phosphorylated, the use of DsRed-DNMT1 allowed us to determine whether AKT1 can phosphorylate newly synthesized DNMT1 fusions. Indeed, a greater amount of phosphorylated DsRed-DNMT1 was observed in the extracts when active AKT1 was overexpressed, as shown by anti–DNMT1-pSer143 (Fig. 1f). In another similar experiment, overexpression of constitutively active AKT1 in HeLa cells resulted in hyperphosphorylated DNMT1. In the presence of hemagglutinin (HA)-tagged AKT1 T308D S473D, endogenous DNMT1 phosphorylation at Ser143 increased ~35% 24 h post-transfection (Supplementary Fig. 1e). Additionally, we stimulated the AKT1 pathway by adding insulin to the cells and observed gradual accumulation of DNMT1-pSer143 (Supplementary Fig. 1f). The DNMT1-pSer143 profile over 24 h looked very similar to ordinary, cell cycle–mediated phosphorylation of Ser143 (see below). Therefore, AKT1-mediated DNMT1 phosphorylation may be dependent on the cell cycle.
To elucidate the functional role of pSer143, we mutated a DsRed-DNMT1 fusion construct (S143A) and compared it side by side with the wild-type DsRed-DNMT1 fusion in the presence or absence of SET7. When the wild-type fusion protein was expressed without SET7, there was two-fold more DNMT1-pSer143 than in cells where SET7 was coexpressed (Fig. 1g, second row, lanes 1 and 3). However, in the mutant DNMT1 fusion (S143A), we detected more Lys142me1 than in the wild-type fusion construct (Fig. 1g, third row, lanes 3 and 4), using a highly specific antibody for DNMT1-Lys142me1 (Supplementary Fig. 1g). These results show that there is a balance between methylated and phosphorylated DNMT1 in cells.
Structure of the SET7–DNMT1 peptide complex
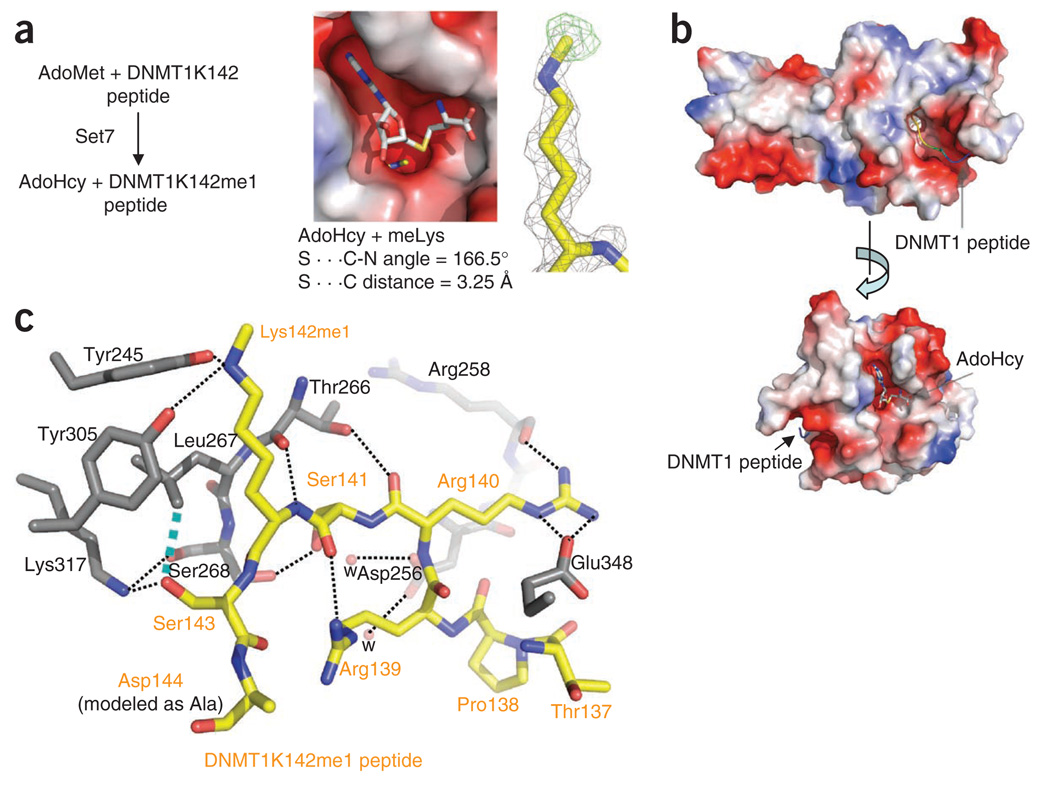
To analyze the molecular mechanism of DNMT1 recognition and methylation by SET7, we solved the structure of a ternary complex of SET7, S-adenosyl-l-methionine (AdoMet) and DNMT1 peptide, to a resolution of 1.69 Å (Table 1). The DNMT1 peptide encompassed residues 137–146 and was cocrystallized in the presence of the methyl donor AdoMet (Fig. 2a). The reaction occurred during the crystal formation of the complex, with the methyl group transferred to the ε-amino of Lys142 and AdoMet converted to AdoHcy and retained in the complex. A single methyl group was transferred, as seen in the methylation reaction in solution with either peptide (Fig. 1c) or DNMT1 N-terminal fragment (residues 1–350; Supplementary Fig. 2a,b). In addition, a point mutation of the target lysine to arginine, K142R, in DNMT1 abolished methylation at this site (Supplementary Fig. 2a) in solution.
Table 1.
Data collection and refinement statistics
| Data collection | Set7–DNMT1–AdoMet |
|---|---|
| Space group | P21212 |
| Cell dimensions | |
| a, b, c (Å) | 101.36, 38.54, 66.54 |
| Resolution (Å) | 31.68–1.69 (1.75–1.69) |
| Rmerge | 0.068 (0.604) |
| I/σI | 21.0 (2.2) |
| Completeness (%) | 99.5 (95.4) |
| Redundancy | 6.2 (3.7) |
| Refinement | |
| Resolution (Å) | 1.69 |
| No. reflections | 28,645 |
| Rwork/Rfree | 0.211/0.238 (0.265/0.309) |
| No. atoms | |
| Protein | 1,896 |
| AdoHcy | 26 |
| DNMT1 peptide | 69 |
| Water | 117 |
| B-factors (Å2) | |
| Protein | 27.6 |
| AdoHcy | 24.9 |
| DNMT1 peptide | 43.4 |
| Water | 36.3 |
| R.m.s. deviations | |
| Bond lengths (Å) | 0.005 |
| Bond angles (°) | 1.4 |
Values in parentheses are for highest-resolution shell.
Figure 2.
Structure of the SET7–DNMT1 complex. (a) The reaction occurred during crystallization. Omit electron densities, Fo − Fc contoured at 4σ above the mean, are shown for the monomethylated Lys142 (black mesh) and the methyl group (green mesh). (b) The substrate peptide and the reaction product AdoHcy occupy binding sites in the opposite ends of a narrow target-lysine channel. (c) Electrostatic interactions, hydrogen bonds and van der Waals interactions define SET7–DNMT1 peptide interactions (dashed lines). The network of interactions includes the following: (i) DNMT1 Arg139 forms a hydrogen bond with the main chain carbonyl oxygen atom of SET7 Gly336 (not shown) and an intramolecular interaction with the main chain carbonyl oxygen of DNMT1 Ser141; (ii) DNMT1 Arg140 forms an electrostatic salt bridge to SET7 Glu348 and a hydrogen bond with the main chain carbonyl oxygen atom of SET7 Arg258; (iii) DNMT1 Ser141 forms a serineserine interaction with SET7 Ser268 and a water-mediated network with DNMT1 His252 (not shown) and SET7 Asp256; (iv) the target nitrogen atom of DNMT1 Lys142me1 forms two hydrogen bonds with the side chain hydroxyl oxygen atoms of SET7 Tyr305 and Tyr245; and (v) DNMT1 Ser143 is involved in a polar interaction with SET7 Lys317 and a van der Waals contact with SET7 Leu267. In addition, two main chain atoms of DNMT1, the carbonyl oxygen of Arg140 and the amide nitrogen of Lys142me1, link through SET7 Thr266.
Like all structurally characterized SET-domain proteins, SET7 binds its ligand, the DNMT1 peptide, in a surface groove (Fig. 2b). Specificity for the DNMT1 peptide is determined primarily through recognition of ordered side chains of DNMT1 (Arg139, Arg140, Ser141 and Ser143) before and after the target Lys142 (Fig. 2c). The complex structure indicates that modification of the side chains of DNMT1 Arg139, Arg140, Ser141 or Ser143 should interfere with DNMT1 Lys142 methylation by SET7. Arg140 and Ser141 match the consensus recognition sequence for SET7-mediated lysine methylation, [R/K][S/T]K21, whereas Arg139 and Ser143 are unique to DNMT1. Ser143 of DNMT1 is involved in a polar interaction with Lys317 and a van der Waals contact with Leu267 of SET7. Adding a phospho-group to the side chain hydroxyl oxygen of Ser143 should result in repulsion from SET7 Leu267. As noted above, such phosphorylation of Ser143 caused complete loss of Lys142 methylation in the context of peptide substrate (Fig. 1b,c).
DNMT1 colocalizes and associates with AKT1
If DNMT1 were an in vivo substrate of AKT1 (whose recognition sequence of RXRXX(S/T), where X is any residue, partially matches the sequence PRRSKS in DNMT1), these two proteins would be predicted to colocalize and physically interact in nuclei. For colocalization studies, COS-7 cells transfected with DsRed-DNMT1 were fixed and immunostained for endogenous AKT1. In a parallel set of experiments, the fixed cells were probed for proliferating cell nuclear antigen (PCNA), a cell cycle marker, and PCNA’s nuclear distribution was used as an indicator for DNA synthesis and replication.
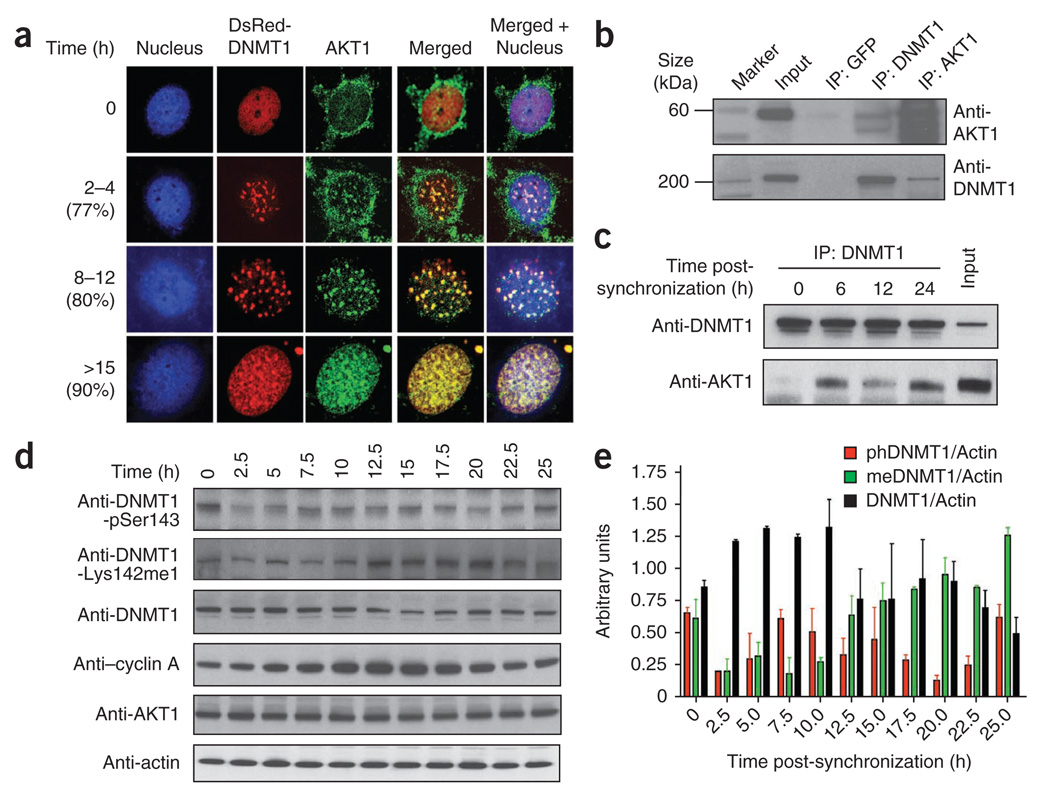
During early S phase, AKT1 appeared in the cytoplasm and nucleus. During mid- and late S phase (8–12 h or >15 h), both DNMT1 and AKT1 were substantially (>75%) colocalized, as shown by a punctate yellow merged pattern (Fig. 3a and Supplementary Fig. 3), similar to what was observed for DNMT1 and SET7 (ref. 15). To validate DNMT1 and AKT1 interactions in other cell systems, HeLa and NIH3T3 were also used for colocalization studies. All three cell lines showed strong DNMT1-AKT1 interaction (Fig. 3a and Supplementary Figs. 3 and 4).
Figure 3.
DNMT1 association with AKT1 kinase. (a) Colocalization of DNMT1 and AKT1 in COS-7 cells, as shown by transiently expressed DsRed-DNMT1 (red) and endogenous AKT1 kinase (green; stained with fluorescent anti-AKT1). Nuclei are Hoechst-stained (blue). Cells were released from G1/S arrest and followed through S and G2 phase for the time indicated at left. Percentages of cells showing similar pattern of colocalization are shown in parentheses. (b) Immunoprecipitation (IP) of AKT1, DNMT1 or GFP (control) from HEK293 nuclear extracts followed by western blot with indicated antibodies. (c) Coimmunoprecipitation of DNMT1–AKT1 complex at different time points after cell synchronization. (d) Expression different post-translationally modified (or total) DNMT1 species during the cell cycle. Cells were released from G1/S arrest and followed for the time shown at top. Extracts were western blotted and probed with the antibodies indicated. Anti-DNMT1 shows total enzyme. Cyclin A was used as the cell cycle marker. A representative western blot for actin (which was used as a loading control) is shown. (e) Quantification of cell cycle–dependent expression of different DNMT1 species, normalized to actin levels. phDNMT1, DNMT1-pSer143; meDNMT1, DNMT1-Lys142me1. Data shown represent two independent western blots as in d. Error bars show s.d.
To further investigate this physical interaction, we immunoprecipitated both endogenous DNMT1 and endogenous AKT1 from HEK293 nuclear extract and probed the western blots with anti-DNMT1 or anti-AKT1. Our results suggest a robust interaction between DNMT1 and AKT1 (Fig. 3b). Similarly, using a series of glutathione S-transferase (GST) fusions of various fragments of DNMT1 and AKT1, we observed the kinase domain of AKT1 interacting with the N terminus (residues 1–446) of DNMT1 (Supplementary Fig. 5). Together, these results indicate that DNMT1 and AKT1 proteins interact directly.
As DNMT1 and AKT1 interaction is cell cycle dependent, we immunoprecipitated DNMT1 and measured the relative amount of AKT1 pulled down from cells 0, 6, 12 and 24 h after synchronization. As expected, at synchronization (0 h), when DNMT1 was predominately nuclear and AKT1 was cytoplasmic, the binding between them was poor. At 6 and 24 h after synchronization, more AKT1 precipitated than at 12 h after synchronization (Fig. 3c). To determine whether the differential binding between DNMT1 and AKT1 is mediated by the cell cycle, we analyzed expression of endogenous DNMT1 and its phosphorylated and methylated forms as the cell cycle progressed (Fig. 3d). Indeed, differential expression of each DNMT1 species was observed during the cell cycle.
To quantify the expression profile, we western blotted each sample with different DNMT1 modification–specific antibodies, or with actin-specific antibody as a loading control, using the ratio between DNMT1 and actin to measure relative expression of DNMT1 protein. The quantitative expression profile of DNMT1, DNMT1-Lys142me1 and DNMT1-pSer143 is shown in Figure 3e. After release from G1 arrest, DNMT1-pSer143 levels increased as cells progressed from early S to G2 phase (typically between 5.0 and 12.5 h), mirroring the expression of the cell cycle–regulated marker cyclin A. In contrast, DNMT1-Lys142me1 levels peaked between 12.5 and 25.0 h. Notably, levels of AKT1 remained approximately constant throughout the cell cycle (Fig. 3d), suggesting that the DNMT1 phosphorylation and methylation switch may be regulated by cell cycle–specific communication between AKT1, SET7 and DNMT1. Furthermore, upon release from cell cycle arrest (0–2.5 h), the amount of both the methylated and phosphorylated DNMT1 species dropped substantially (Fig. 3d,e), implying that differential turnover of both species may be dependent on cell cycle progression.
AKT1 stabilizes DNMT1 and affects genome methylation
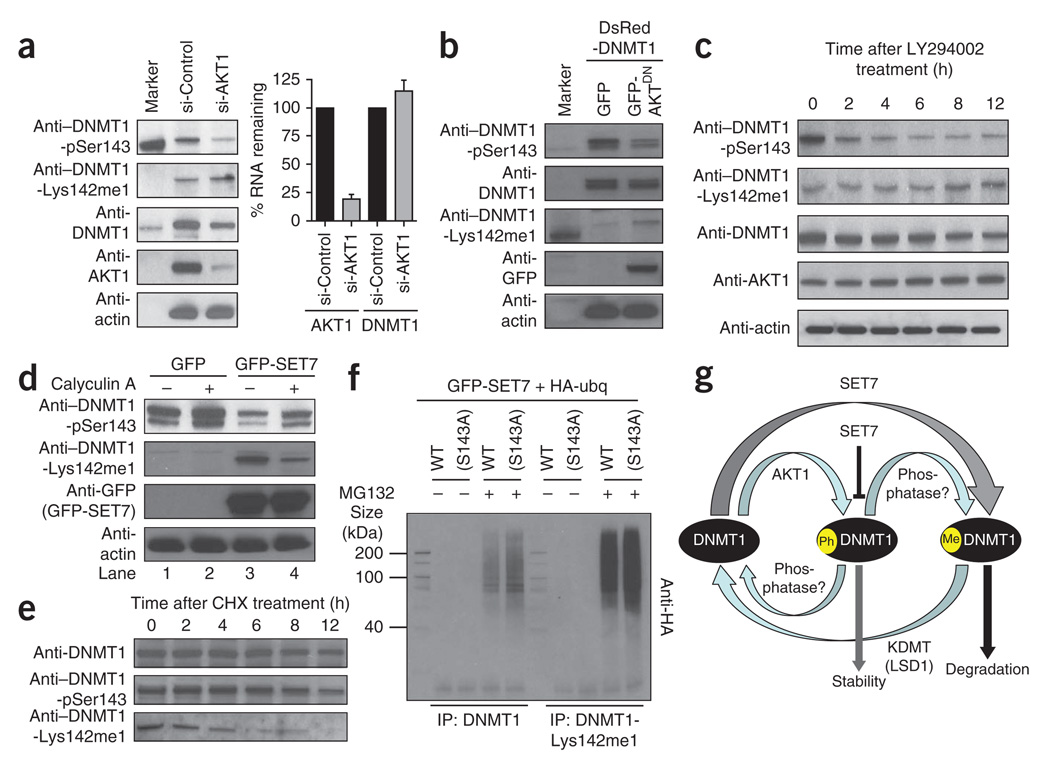
We next investigated the effects of DNMT1 Ser143 phosphorylation in cells by knocking down AKT1 in HeLa cells. Depletion of AKT1 resulted in reduction of total DNMT1 as well as DNMT1-pSer143 species (~70%), along with a moderate increase in endogenous DNMT1-Lys142me1 species (~25%). However, the expression level of DNMT1 mRNA remained the same between AKT1-knockdown cells and control cells, suggesting that AKT1 affects DNMT1 protein stability in cells (Fig. 4a).
Figure 4.
AKT1 kinase–mediated phosphorylation stabilizes DNMT1. (a) Western blotting and RNA expression analysis of extracts from HeLa cells with AKT1 knockdown (si-AKT1) or control knockdown (si-Control). Left, western blot with indicated antibodies. Marker, biotinylated protein ladder showing relative molecular mass of DNMT1 as ~200 kDa. Right, RNA expression measured from quantitative PCR; error bars represent s.d. of three independent experiments. DNMT1-pSer143 is reduced in AKT1-knockdown cells, resulting in moderately higher DNMT1-Lys142me1 levels but no change in DNMT1 RNA levels. (b) Overexpression of the AKT1 dominant-negative mutant (AKT1DN) reduces DNMT1-pSer143 and increases DNMT1-Lys142me1 levels, as shown by western blotting with the indicated antibodies. (c) The AKT kinase inhibitor LY294002 reduces DNMT1-pSer143 levels and increases DNMT1-Lys142me1 levels in a time-dependent manner, as shown by western blotting with the indicated antibodies. (d) Effect of SET7 on DNMT1-Lys142me1 in the absence (−) or presence (+) of calyculin A, a serine/threonine phosphatase inhibitor, as shown by western blotting with indicated antibodies. Cells were co-transfected with constructs expressing GFP alone with DsRed-DNMT1 (lanes 1 and 2) or GFP-SET7 with DsRed-DNMT1 (lanes 3 and 4). (e) Degradation of DNMT1 modified and unmodified species after cells were treated with cycloheximide (CHX). At indicated times, cells were lysed and western blotted with indicated antibodies. (f) Ubiquitinylation of total DNMT1 and DNMT1-Lys142me1. Wild-type DNMT1 or the DNMT1(S143A) mutant was overexpressed in cells, along with HA-ubiquitin and GFP-SET7, in the presence (+) or absence (−) of the proteasome inhibitor MG132. Total DNMT1 or DNMT1-Lys142me1 was immunoprecipitated (IP) from the nuclear extract and western blotted with antibody to HA. (g) A proposed model of cross-talk between DNMT1 (black), SET7 and AKT1 in mammalian cells. The other interactors that can reverse the post-translational modifications are an unknown phosphatase and an unknown lysine-specific demethylase (KDMT). One such KDMT may be LSD1 (ref. 17).
To further examine the role of AKT1 in DNMT1 stabilization, we used a dominant-negative AKT1 mutant (K179M)22. A plasmid expressing green fluorescent protein (GFP)-tagged dominant-negative AKT1 (GFP-AKT1DN) was transfected into COS-7 cells, and extracts were western blotted and probed for DNMT1, DNMT1-Lys142me1 and DNMT1-pSer143 species. Indeed, in the presence of GFP-AKT1DN, both DNMT1-pSer143 and total DNMT1 were less abundant, and DNMT1-Lys142me1 levels increased (Fig. 4b). This seems to be a result of direct regulation of DNMT1 protein stability, as AKT1 knockdown did not change protein synthesis: actin levels remained similar (Fig. 4a,b), as did levels of protein in Ponceau-stained blots (data not shown).
To confirm that DNMT1 stability is AKT1 dependent, we treated cultured HeLa cells that lack a functional p53, and therefore are more amenable to drug treatment, with the AKT1 inhibitor LY294002 and measured DNMT1 protein levels. DNMT1-pSer143 levels decreased substantially within 6 h of the treatment (Fig. 4c, top row), and DNMT1-Lys142me1 increased proportionally (Fig. 4c, second row). Along with the gradual increase of DNMT1-Lys142me1 species, more degradation of the enzyme was observed, resulting in a decrease in total DNMT1 levels (Fig. 4c, third row), although the total level of AKT1 remained similar throughout the treatment (Fig. 4c, fourth row).
As inhibiting AKT1 reduced overall DNMT1 levels, we expected that treating the cells with LY294002 would result in hypomethylation of the genome. To test this hypothesis, we used genomic DNA from LY294002-treated cells as a substrate for M.SssI methyltransferase, which methylates CG dinucleotides. Approximately 20% more radioactive AdoMet was incorporated in the presence of LY294002 than in its absence (Supplementary Fig. 6a). The genomic DNA samples were then hydrolyzed to mononucleosides, and quantitative analysis of the base composition by HPLC indicated that there was about one-fifth less 5-methylcytosine (5mC) in LY294002-treated samples as there was in control samples without the drug (Supplementary Fig. 6b).
We also examined the effect of calyculin A, a general serine/threonine phosphatase inhibitor, on DNMT1 stability. In the presence of calyculin A, strong DNMT1 Ser143 phosphorylation was observed (Fig. 4d, top row, lanes 2 and 4).
Finally, DNMT1 Ser143 phosphorylation decreased substantially (~70%) in the presence of SET7 overexpression (Fig. 4d, top row, compare lane 1 to lane 3 and lane 2 to lane 4), suggesting that methylation of Lys142 impairs phosphorylation of Ser143 in cells. To validate this observation, we also performed the AKT1 kinase assay with the peptide substrates DNMT1K142 (wild type, unmodified) and DNMT1K142me1 (methylated) and observed ~66% less kinase activity in the presence of lysine monomethylation (Supplementary Fig. 6c).
Ser143 phosphorylation stabilizes DNMT1
As Lys142 methylation impairs Ser143 phosphorylation and DNMT1-Lys142me1 is prone to proteasome-mediated protein degradation15, we next sought to determine the half-lives of DNMT1-Lys142me1, DNMT1-pSer143 and unmodified DNMT1. We treated Jurkat cells with the protein synthesis inhibitor cycloheximide so that we could follow DNMT1 degradation patterns by western blot analysis without the complication of additional DNMT1 protein synthesis. In cycloheximide-treated Jurkat cell extracts, total cellular DNMT1 had a half-life of >12 h (Fig. 4e, top row), similar to that of the Ser143-phosphorylated form (Fig. 4e, middle row). In contrast, DNMT1-Lys142me1 species had a half-life of approximately 3.5 h (Fig. 4e, bottom row). Therefore, the phosphorylated DNMT1 species is less likely to be degraded compared to the methylated protein.
To validate this observation, we created a DNMT1 peptide with an S143D substitution (DNMT1K142(S143D)) to mimic constitutive phosphorylation of this site, incubated the peptide with recombinant SET7 and tritiated AdoMet in vitro and compared its methylation with that of DNMT1K142 and DNMT1pS143 peptides. Although the wild-type DNMT1K142 peptide was methylated, neither DNMT1K142(S143D) nor DNMT1pS143 was methylated (Supplementary Fig. 7a). Thus, mimicking constitutive phosphorylation inhibits SET7 activity.
Therefore, we hypothesized that recombinant S143D-mutated DNMT1 would be more stable than wild-type DNMT1. We transfected DsRed-DNMT1(S143A), DsRed-DNMT1(S143D) and wild-type DsRed-DNMT1 into COS-7 cells for robust expression, treated the cells with both cycloheximide and the AKT1 inhibitor LY294002, and monitored DNMT1 levels by western blotting. As expected, DNMT1(S143A) degraded 4 h after treatment, and little or no degradation of DNMT1(S143D) was observed, confirming that Ser143 phosphorylation contributes to DNMT1 stability (Supplementary Fig. 7b).
As proteasome-mediated DNMT1 degradation relies on Lys142 methylation in DNMT1 (ref. 15), and Lys142 methylation was impaired by Ser143 phosphorylation, we next examined whether the S143A mutation facilitated ubiquitin-mediated degradation of DNMT1. Methylated DNMT1 was immunoprecipitated using anti–DNMT1-Lys142me1 from COS-7 cells cotransfected with HA-tagged ubiquitin, GFP-SET7 and either DsRed-DNMT1 or DsRed-DNMT1(S143A). HA antibody was then used to detect ubiquitinylation of the immunoprecipitated proteins. For cells transfected in the presence of proteasome inhibitor MG132, transfection with GFP-SET7 and DsRed-DNMT1 resulted in less HA signal than transfection with GFP-SET7 and DsRed-DNMT1(S143A) (Fig. 4f). This confirms that the S143A mutation facilitates DNMT1 methylation, which is a substrate for ubiquitinylation and protein degradation. A similar observation was made with samples immunoprecipitated with anti-DNMT1 (Fig. 4f). Furthermore, the higher-molecular weight smear of signal indicates that polyubiquitin is more abundant in the presence of DNMT1(S143A).
DISCUSSION
Previous work has shown that covalent post-translational modifications are widely used for regulation of stability15,23,24. Apart from DNMT1, SET7 also mediates methylation of RelA protein, and methylated RelA negatively regulates the transcriptional activation of NF-κB24. Here we show that interplay of Ser143 phosphorylation and Lys142 methylation is a key signal regulating the stability or degradation of DNMT1. The DNMT1 species phosphorylated by AKT1 (DNMT1-pSer143) has a higher half-life than the DNMT1-Lys142me1 species. Therefore, the methyl-phospho switch between these adjacent residues, Lys142 and Ser143, determines the stability of DNMT1.
According to our results, this cross-talk could involve dynamic interaction between SET7, AKT1, a protein lysine demethylase—perhaps LSD1 (ref. 17)—and a yet-to-be-determined protein phosphatase to maintain DNMT1 stability in cells (Fig. 4g). Another noteworthy observation is that endogenous AKT1 stimulation did not result in immediate phosphorylation of Ser143 of DNMT1 (Supplementary Fig. 1f). Clear DNMT1 phosphorylation was visible only after 8 h, suggesting that this phosphorylation is mediated by the cell cycle (Fig. 3d,e). An explanation for the delayed phosphorylation of DNMT1 may be that AKT1 has poor access to the DNMT1 N-terminal region. Indeed, the region surrounding Ser143 of DNMT1 binds to PCNA, Rb, HDACs, G9a and host of other proteins25–28. Therefore, the phosphorylation event on DNMT1 could be regulated by additional, cell cycle–dependent events, as certain binding proteins need to be released before phosphorylation can proceed. Furthermore, misregulation of DNMT1 turnover during the cell cycle may also aid in epigenome stability.
A question emerges from our observations regarding the possible existence of dual modification of Lys142me1 and pSer143 on DNMT1. The results presented here indicate that DNMT1-pSer143 is the predominant stable form of the enzyme and is resistant to Lys142 methylation. However, in our kinase assay on DNMT1K142me1 peptide, a small amount of phosphorylation was observed. Although this result suggests that a small percentage of DNMT1 may have dual modifications, the majority of methylated DNMT1 will be degraded. From all the experiments we have performed, we have little evidence of dually (Lys142me1 and pSer143) modified DNMT1 in cells. However, we cannot rule out its presence absolutely during development or disease stages.
Our observations lead to the hypothesis that if the AKT1-phosphorylated DNMT1 species is more nuclear and is resistant to degradation by the proteasome pathway, then cells with upregulated AKT1 should show aberrant DNA methylation. AKT1 kinase activity has been implicated in the pathophysiology of many different tumors, including multiple myeloma, prostate, breast and liver29,30. For example, promoter hypermethylation of the tumor-suppressor gene cystatin M (CST6) is associated with aberrant AKT1 activation in breast epithelial cells31. Data reported here suggest that the interplay between AKT1-mediated stabilization of DNMT1 and SET7-mediated degradation of DNMT1 creates a delicate balance resulting in epigenetic modulation of gene expression during the cell cycle and development (Fig. 4g). Furthermore, the cell may use such switch mechanism(s) to regulate other key epigenetic modifiers in different cellular processes.
METHODS
Methods and any associated references are available in the online version of the paper at http://www.nature.com/nsmb/.
Supplementary Material
ACKNOWLEDGMENTS
We thank J. Benner for nucleotide analysis of the genomic DNA, S.M. Kinney for critical reading of this manuscript, H.G. Chin (New England Biolabs) for recombinant enzymes, X. Zhang for insightful discussion, and D.G. Comb, R.J. Roberts, J.V. Ellard at New England Biolabs for supporting the basic research. The work in the Cheng laboratory was supported by US National Institutes of Health (NIH) grants GM049245 and GM068680. X.C. is a Georgia Research Alliance Eminent Scholar.
Footnotes
Accession codes. Protein Data Bank: The coordinates and structure factors of the SET7–DNMT1 peptide complex have been deposited with accession code 3OS5.
Note: Supplementary information is available on the Nature Structural & Molecular Biology website.
AUTHOR CONT RIBUTIONS
P.-O.E. performed cell biology and biochemistry experiments. M.S. made constructs, tested kinetics and performed the pull-down assay. G.R.F. performed quantitative PCR. Y.C. performed SET7 purifications, MS-based methylation assays on DNMT1 peptide, mutagenesis of K142R and crystallization of SET7–DNMT1 peptide. A.K.U. purified the DNMT1 N-terminal domain and performed MS-based methylation assays on this fragment. J.R.H. performed crystallographic experiments. X.C. and S.P. organized and analyzed data and wrote the manuscript.
COMPETING FINANCIAL INT ERESTS
The authors declare no competing financial interests.
Reprints and permissions information is available online at http://npg.nature.com/reprintsandpermissions/.
References
- 1.Yang XJ, Seto E. Lysine acetylation: codified crosstalk with other posttranslational modifications. Mol. Cell. 2008;31:449–461. doi: 10.1016/j.molcel.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vanselow K, Kramer A. Role of phosphorylation in the mammalian circadian clock. Cold Spring Harb. Symp. Quant. Biol. 2007;72:167–176. doi: 10.1101/sqb.2007.72.036. [DOI] [PubMed] [Google Scholar]
- 3.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 4.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 5.Aletta JM, Cimato TR, Ettinger MJ. Protein methylation: a signal event in post-translational modification. Trends Biochem. Sci. 1998;23:89–91. doi: 10.1016/s0968-0004(98)01185-2. [DOI] [PubMed] [Google Scholar]
- 6.Chin HG, et al. Sequence specificity and role of proximal amino acids of the histone H3 tail on catalysis of murine G9A lysine 9 histone H3 methyltransferase. Biochemistry. 2005;44:12998–13006. doi: 10.1021/bi0509907. [DOI] [PubMed] [Google Scholar]
- 7.Rea S, et al. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature. 2000;406:593–599. doi: 10.1038/35020506. [DOI] [PubMed] [Google Scholar]
- 8.Jeong YS, Cho S, Park JS, Ko Y, Kang YK. Phosphorylation of serine-10 of histone H3 shields modified lysine-9 selectively during mitosis. Genes Cells. 2010;15:181–192. doi: 10.1111/j.1365-2443.2009.01375.x. [DOI] [PubMed] [Google Scholar]
- 9.Kouzarides T. Chromatin modfications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 10.Chuikov S, et al. Regulation of p53 activity through lysine methylation. Nature. 2004;18:353–360. doi: 10.1038/nature03117. [DOI] [PubMed] [Google Scholar]
- 11.Jansson M, et al. Arginine methylation regulates the p53 response. Nat. Cell Biol. 2008;10:1431–1439. doi: 10.1038/ncb1802. [DOI] [PubMed] [Google Scholar]
- 12.Huang J, et al. P53 is regulated by the lysine demethylase LSD1. Nature. 2007;449:105–108. doi: 10.1038/nature06092. [DOI] [PubMed] [Google Scholar]
- 13.Subramanian K, et al. Regulation of estrogen receptor alpha by the SET7 lysine methyltransferase. Mol. Cell. 2008;30:336–347. doi: 10.1016/j.molcel.2008.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim H, et al. Requirement of histone methyltransferase SMYD3 for estrogen receptor-mediated transcription. J. Biol. Chem. 2009;284:19867–19877. doi: 10.1074/jbc.M109.021485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Estève PO, et al. Regulation of DNMT1 stability through SET7-mediated lysine methylation in mammalian cells. Proc. Natl. Acad. Sci. USA. 2009;106:5076–5081. doi: 10.1073/pnas.0810362106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dephoure N, et al. A quantitative atlas of mitotic phosphorylation. Proc. Natl. Acad. Sci. USA. 2008;105:10762–10767. doi: 10.1073/pnas.0805139105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang J, et al. The lysine demethylase LSD1 (KDM1) is required for maintenance of global DNA methylation. Nat. Genet. 2009;41:125–129. doi: 10.1038/ng.268. [DOI] [PubMed] [Google Scholar]
- 18.Fischle W, Wang Y, Allis CD. Binary switches and modification cassettes in histone biology and beyond. Nature. 2003;425:475–479. doi: 10.1038/nature02017. [DOI] [PubMed] [Google Scholar]
- 19.Pradhan S, Estève PO. Allosteric activator domain of maintenance human DNA (cytosine-5) methyltransferase and its role in methylation spreading. Biochemistry. 2003;42:5321–5332. doi: 10.1021/bi034160+. [DOI] [PubMed] [Google Scholar]
- 20.Alessi DR, et al. Mechanism of activation of protein kinase B by insulin and IGF-1. EMBO J. 1996;15:6541–6551. [PMC free article] [PubMed] [Google Scholar]
- 21.Couture JF, Collazo E, Hauk G, Trievel RC. Structural basis for the methylation site specificity of SET7/9. Nat. Struct. Mol. Biol. 2006;13:140–146. doi: 10.1038/nsmb1045. [DOI] [PubMed] [Google Scholar]
- 22.Franke TF, et al. The protein kinase encoded by the Akt proto-oncogene is a target of the PDGF-activated phosphatidylinositol 3-kinase. Cell. 1995;81:727–736. doi: 10.1016/0092-8674(95)90534-0. [DOI] [PubMed] [Google Scholar]
- 23.Fischle W. Talk is cheap–cross-talk in establishment, maintenance, and readout of chromatin modifications. Genes Dev. 2008;22:3375–3382. doi: 10.1101/gad.1759708. [DOI] [PubMed] [Google Scholar]
- 24.Yang XD, et al. Negative regulation of NF-κB action by Set9-mediated lysine methylation of the RelA subunit. EMBO J. 2009;28:1055–1066. doi: 10.1038/emboj.2009.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chuang LS, et al. Human DNA-(cytosine-5) methyltransferase-PCNA complex as a target for p21WAF1. Science. 1997;277:1996–2000. doi: 10.1126/science.277.5334.1996. [DOI] [PubMed] [Google Scholar]
- 26.Pradhan S, Kim GD. The retinoblastoma gene product interacts with maintenance human DNA (cytosine-5) methyltransferase and modulates its activity. EMBO J. 2002;21:779–788. doi: 10.1093/emboj/21.4.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Robertson KD, et al. DNMT1 forms a complex with Rb, E2F1 and HDAC1 and represses transcription from E2F-responsive promoters. Nat. Genet. 2000;25:338–342. doi: 10.1038/77124. [DOI] [PubMed] [Google Scholar]
- 28.Estève PO, et al. Direct interaction between DNMT1 and G9a coordinates DNA and histone methylation during replication. Genes Dev. 2006;20:3089–3103. doi: 10.1101/gad.1463706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hideshima T, et al. Inhibition of Akt induces significant downregulation of survivin and cytotoxicity in human multiple myeloma cells. Br. J. Haematol. 2007;138:783–791. doi: 10.1111/j.1365-2141.2007.06714.x. [DOI] [PubMed] [Google Scholar]
- 30.Hill KM, et al. The role of PI 3-kinase p110beta in AKT signally, cell survival, and proliferation in human prostate cancer cells. Prostate. 2010;70:755–764. doi: 10.1002/pros.21108. [DOI] [PubMed] [Google Scholar]
- 31.Lin HJ, et al. Breast cancer-associated fibroblasts confer AKT1-mediated epigenetic silencing of Cystatin M in epithelial cells. Cancer Res. 2008;68:10257–10266. doi: 10.1158/0008-5472.CAN-08-0288. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.