Abstract
A canonical biogenesis pathway involving sequential cleavage by the Drosha and Dicer RNAse III enzymes governs the maturation of most animal microRNAs. However, there exist a variety of alternative miRNA biogenesis pathways, most of which bypass Drosha processing. Recently, three groups described for the first time a vertebrate microRNA pathway that bypasses Dicer cleavage. This mechanism was characterized with respect to the highly conserved vertebrate gene mir-451, for which Drosha processing yields a short (42 nucleotide) hairpin that is directly loaded into Ago2, the sole vertebrate “Slicer” Argonaute. Ago2-mediated cleavage of this hairpin yields a 30 nucleotide intermediate, whose 3′ end is resected to generate the dominantly cloned ∼23 nucleotide mature miR-451. Knowledge of this pathway provides an unprecedented tool with which to express microRNAs and small interfering RNAs in Dicer mutant cells. More generally, the mir-451 backbone constitutes a new platform for gene silencing that complements existing shRNA technology.
Key words: mir-451, Ago2, Slicer, Dicer-independent, erythropoiesis
The “Dogma” of MicroRNA Biogenesis
MicroRNAs (miRNAs) are ∼22-nucleotide (nt) RNAs that are expressed in temporal- and spatial-specific manners to regulate diverse developmental and physiological processes in higher eukaryotes.1,2 Animal miRNAs generally modulate gene expression via complementary target sequences in mRNAs, usually in 3′ untranslated region (3′ UTRs), to effect translational repression or degradation.3 Target recognition frequently involves consecutive Watson-Crick pairing of positions 2–8 of the mature miRNA, also known as the “seed region.”4,5 This modest amount of complementarity has permitted many animal miRNAs to accumulate hundreds of targets, and their aggregate target networks may encompass a majority of animal transcripts.6,7
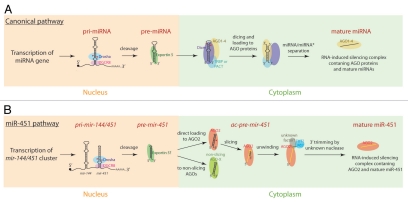
The best-studied “canonical pathway” for miRNA biogenesis is broadly conserved amongst vertebrates and invertebrates (Fig. 1A).8,9 Primary miRNAs (pri-miRNAs) bearing hairpin structures are first recognized and cleaved by the nuclear “Microprocessor” complex composed of the RNase III enzyme Drosha and its dsRNA binding partner DGCR8 (also known as Pasha in invertebrates). The resulting pre-miRNA hairpins are transported to the cytoplasm, where they are cleaved on their terminal loop ends by the RNase III enzyme Dicer to yield miRNA/miRNA* duplexes. One of the strands is typically preferentially loaded into an Argonaute (Ago) protein-containing effector complex to form the RNA-induced silencing complex (RISC), which is guided to target transcripts according to the miRNA sequence.
Figure 1.
(A) Canonical miRNA biogenesis. Primary miRNA (pri-miRNA) transcripts are cleaved in the nucleus by a complex of the Drosha RNAse III enzyme and its partner DGCR8 to yield a ∼55–70 nt pre-miRNA hairpin. Following its translocation to the cytoplasm via Exportin-5, the pre-miRNA hairpin is cleaved on its loop end by a complex of the Dicer RNAse III enzyme and its partner TRBP/PACT. The resultant ∼22 nt miRNA/miRNA* duplex is loaded into any of the 4 vertebrate Argonaute (AGO) proteins and one strand is released leaving behind the mature miRNA. (B) miR-451 biogenesis. mir-451 resides on a primary transcript operon with mir-144, which matures via the canonical miRNA pathway. Drosha/DGCR8 cleavage generates a 42 nt pre-mir-451 hairpin, which resembles an Exportin-5 substrate (although this has not been directly demonstrated, thus the “?”). The pre-mir-451 hairpin is directly loaded into AGO proteins, but loading into non-slicing AGO proteins is abortive and these cannot mature mir-451 further. Loading into AGO2, the sole vertebrate “Slicer” capable of cleaving target strands, generates a 30 nt “Ago2-cleaved (ac)-pre-mir-451 hairpin.” This is then subject to a trimming reaction, which may occur in conjunction with a tailing reaction, to yield the dominant 23 nt mature miR-451.
Expanding the miRNA Reservoir via Non-Canonical Biogenesis Pathways
Studies from the past few years have provided ample evidence for the existence of alternative pathways that generate functional miRNAs.10,11 A steady stream of surprises has emerged, as researchers have focused their attention on unconventional substrates that deviate from canonical miRNAs in terms of their genomic locations, predicted secondary structures and conservation patterns.
The first major non-canonical pathway came with the recognition of mirtrons.12,13 Like canonical miRNA loci, mirtrons generate cloned ∼22 nt RNAs that adopt miRNA/miRNA* duplexes with 3′ overhangs; however, their hallmark is that the genomic termini of miRNA/miRNA* read pairs are defined precisely by splice donor and acceptor sites. Such a configuration suggested that the splicing machinery might generate hairpin ends, thereby bypassing Drosha cleavage. Detailed studies in the Drosophila system indeed showed this to be the case, and further indicated that subsequent linearization by lariat debranching enzyme was required for these introns to adopt hairpin structures that serve as pre-miRNA mimics. On the basis of these characteristic read mappings to short hairpin introns, mirtrons have been further annotated in C. elegans13 and diverse vertebrate genomes.14–16
Mirtrons, in turn, have fostered a diversity of apparent “add-on” pathways, in which splicing is coupled to other exonucleolytic (or potentially endonucleolytic) pathways. For example, in the Drosophila 3′ tailed mirtron pathway, only the 5′ end of the mirtron is coincident with a splice site; a 3′ extension ensues to the 3′ splice acceptor. This type of spliced substrate was shown to be processed by both lariat debranchase and the nuclear RNA exosome complex, which removes the 3′ “tail” to generate the pre-miRNA intermediate.17 The existence of 5′ tailed mirtrons in avians16 and mammals15 is suggestive of yet another pathway for miRNA biogenesis, since the RNA exosome is specific for 3′-5′ processing. However, the biochemical details of their maturation remain to be worked out.
In addition to these sundry mirtron pathways, a dazzling array of other mechanisms can produce miRNAs by Drosha/DGCR8-independent strategies. For example, some small nucleolar RNAs (snoRNAs) utilize Dicer to generate both precursor and mature miRNA-like species;18–21 transfer RNA (tRNA)-derived small RNAs (tsRNAs) are produced via precursor tRNAs cleavage by Dicer.22,23 RNA polymerase III can generate defined transcripts that are not further modified by polyadenylation, and this process may produce endogenous short hairpin RNAs (endo-shRNAs) with 2-nt 3′ overhangs that mimic pre-miRNAs and directly serve as Dicer substrates.15 In yet another variation, the murine γ-herpesvirus MHV68 miRNA, which is fused to a tRNA in the primary transcript, is produced by tRNase Z cleavage 3′ to the tRNA moiety to liberate a pre-miRNA hairpin that undergoes a second cleavage by Dicer.24 Collectively, the miRNA reservoir is expanding through the usage of different Drosha/DGCR8-independent, Dicer-dependent pathways.
Many Clues Regarding the Potentially Unusual Biogenesis of miR-451
These myriad alternative miRNA pathways all converge upon the Dicer enzyme. Moreover, Dicer is the focal point for production of small interfering RNAs (siRNAs), either from endogenous substrates or from exogenously applied substrates.25 Therefore, a Dicer enzyme was widely considered to be essential for in vivo production of all miRNAs and siRNAs. However, there eventually seems to be an exception to every rule, and miRNA biogenesis is indeed no exception.
Many animal miRNAs genes are clustered in the genome, and such clustering provided an additional measure of confidence in the likelihood of computationally-derived hairpin candidates as bona fide miRNA genes. One such candidate was mir-451, a highly conserved hairpin located just downstream of the extant (and also highly conserved) locus mir-144, and shown to generate short RNA reads.26 Subsequent sequencing of small RNAs from red blood cells showed that miR-451 is abundant in both total RNAs27 and Ago2 immunoprecipitates (IP) in this cell type,28 providing further evidence of it as a genuine miRNA. Nevertheless, it was clear from these early experiments that miR-451 was no typical miRNA: its dominant 23-nt reads extended across the terminal loop,28 which never happens in other known miRNAs. Moreover, the stem of predicted pre-mir-451 structure is only 17 basepairs (bp) in length. This is seemingly too short to serve as substrate for Dicer, which requires >19 bp stem in addition to a 2-nt 3′ overhang for efficient hairpin cleavage,29 and most pre-miRNAs have longer stem than this minimum.30
Another clue as to the non-canonical processing of mir-451 came from the details of its evolution. Virtually all miRNA genes that are reasonably well-conserved (e.g., amongst Drosophilids or amongst vertebrates) have a distinctive evolutionarily profile, in which the divergence of the terminal loop is far greater than either of the hairpin arms.31,32 In contrast, most structural RNAs are characterized by consistent or compensatory mutations within duplex regions, signifying that the structure but not primary sequence is under selective pressure.33 This characteristic serves as an effective classifier to distinguish genuine miRNA loci from other conserved genomic hairpins that do not generate ∼22 nt short RNAs.34 As it happens, mir-451 is a rare miRNA locus that defies the signature evolutionary profile of conserved miRNA genes: in addition to its mature sequence, its terminal loop is also invariant from human to fish, while certain positions on the complementary hairpin arm are subject to divergence.35
A final consideration is the fact that the base pairing in pre-mir-451 is unusually perfect, whereas most other miRNA hairpins contain multiple unpaired nucleotides within the stem. Such hairpin imperfections are believed to promote unwinding of the miRNA/miRNA* duplex.36 Taken together with the strict conservation of the mir-451 loop, these details hinted that this locus might not in fact mature via a typical Dicer-generated miRNA/miRNA* duplex. The underlying mechanism that generates this peculiar miRNA was finally unveiled by recent papers from the Hannon, Giraldez and Lai labs.35,37,38 A combination of genetic and biochemical evidence yielded an astonishing conclusion: miR-451 is generated by a Dicer-independent, Ago2-dependent mechanism (Fig. 1B).
miR-451 is Generated by a Dicer-Independent, Ago2-Mediated Pathway
The lower stem of pri-mir-451 between the stem-ssRNA junction and the 5′ terminus of pre-mir-451 is ∼11 bp, implying that pri-mir-451 is a canonical substrate for Drosha.39 Indeed, knockdown35 or conditional knockout37 of Microprocessor components prevented miR-451 processing, and in vitro processing assays demonstrated that Drosha could directly cleave pri-mir-451 to generate a ∼42 nt hairpin.37 However, this short hairpin does not mature via Dicer, since miR-451 accumulated effectively in Dicer conditional knockout ES cells,37 MEFs stably deleted for Dicer35 and in MZdicer mutant zebrafish.38 All other miRNAs were obviously depleted under these conditions; thus miR-451 represented the first Dicer-independent miRNA known.
The perfect pre-mir-451 hairpin structure suggested it as a possible substrate for mammalian Ago2, which can cleave the passenger strand of an extensively paired pre-miRNA at positions 10–11 across from hairpin 5′ end.40 Consistent with this, miR-451 northern blotting revealed a series of hybridized bands larger than typical mature miRNAs, extending to ∼30 nt.35,37,38 Indeed, small RNA sequencing revealed cloned intermediates extending to 30 nt, which corresponds precisely to the position in the hairpin that would be the site of putative Ago2 slicing.35,37,38
By microarray analyses, the three groups and Rasmussen et al. found that miR-451 was the most downregulated miRNA in Ago2 mutant mouse fetal liver,37 MZago2 mutant zebrafish embryos,38 and Ago2-/- mouse bone marrow35 and erythroblasts,41 indicating that Ago2 is genetically required for miR-451 maturation. The links were strengthened by the findings that Ago2,37,42 and mir-451,41,43–47 both play important roles during erythroid differentiation. Perhaps most impressively, the Hannon group showed that knockin of a catalytically dead form of Ago2 exhibited hematopoietic defects and specific loss of miR-451, implying that slicing activity is uniquely required for the generation of this miRNA.37
Since loss of mir-451 also depletes erythrocytes,41,45 the main location of miR-451 expression, it was possible that the decreased level of miR-451 partly represented an indirect effect on altered cell distribution in Ago2 mutants. A direct and essential role of Ago2 cleavage in miR-451 biogenesis was demonstrated by several methods. Tests of mutant mir-451 hairpins that were unpaired at the putative Ago2 cleavage site, and tests of catalytically-dead Ago2 mutant in reconstituted Ago2 knockout MEFs or in vitro processing assays, all directly showed that Ago2 Slicer activity was essential for production of mature miR-451.35,37,38 While Ago2-catalytic mutant and catalytic-incompetent Ago1 could also incorporate pre-mir-451, no intermediate or mature species were detected in their immunoprecipitate (IP) as could be seen in Ago2-IP.35,37 Finally, purified wildtype, but not catalytically dead, Ago2 complex could generate the 30 nt species from synthetic pre-mir-451 hairpin in vitro.37 Taken together, the unconventional structure of mir-451 provides a platform for Ago2 Slicer-dependent, Dicer-independent non-canonical miRNA biogenesis.
Application of the mir-451 Backbone for Gene Silencing
The broad and flexible usage of canonical miRNAs is reflected in the wide diversity of miRNA hairpin sequences and structures. In keeping with this, miRNA hairpins are easily reprogrammed with arbitrary sequences to silence target transcripts of choice. Indeed, sh-miRNA constructs that mimic endogenous miRNA substrates may circumvent host responses against foreign nucleic acids.48 However, as there is apparently only one vertebrate miRNA known to date that utilizes the Dicer-independent pathway, it was less clear that mir-451 could necessarily be reprogrammed.
The Hannon group used synthetic mir-451-like RNA hairpins reprogrammed with let-7c or p53-targeting sequence, and showed that these could repress a GFP reporter containing let-7c target sites as well as the protein level of p53, respectively.37 However, it was not explicitly shown that these hairpins are processed by the miR-451 pathway, and other studies showed that Ago2 is capable of directly incorporating pre-miRNAs and other long RNA species and using these as guide strands for target cleavage.49 The Giraldez group used an in vivo genetic assay to test the biological function of reprogrammed mir-451 hairpins.38 They previously showed that as miRNAs in the miR-430 family are the first and most highly expressed miRNAs during zebrafish embryogenesis, many aspects of the MZDicer phenotype are specifically due to loss of this miRNA activity.50,51 They found that miR-430 expressed from a synthetic pre-mir-451 RNA backbone, but not from a pre-mir-430 backbone, could rescue MZDicer mutant embryonic phenotypes.38 By in vitro processing assay, they further demonstrated the processing of remodeled miR-430 was dependent on Ago2 but not Dicer. Finally, the Lai group performed extensive reprogramming experiments using plasmid vectors in which mir-451 backbone was reprogrammed to express a variety of other canonical miRNA sequences.35 These were shown to be highly active at suppressing sensor constructs in Dicer-/- MEFs and to generate mature miRNA products in this context. In addition, suppression of seed-bearing sensors by reprogrammed mir-451 constructs was shown. This indicated that 5′ ends of reprogrammed small RNAs were generated accurately when they were processed from primary transcripts, as opposed to synthetic pre-mir-451 hairpins bearing pre-defined 5′ ends.
These encouraging reprogramming experiments indicate that the mir-451 backbone is an exciting tool for in vivo expression of short regulatory RNAs. For example, ∼50 publications have described the consequences of systemic or conditional ablation of Dicer, with the general conclusion that every place and time assayed in the mouse probably requires Dicer function. However, the prospects of assigning specific miRNAs to these phenotypes have been largely limited to ex vivo settings using derived cell lines that could be transfected with short RNAs. Although the miRNA repertoire of animal genomes is broad, a relatively small pool of miRNAs often accounts for the bulk of miRNA expression in particular cell types. So it is conceivable that individual miRNAs could contribute disproportionately to Dicer mutant phenotypes. For example, the similarities between Dicer conditional knockout52 and a specific deletion of the mir-17-92 cluster53 during B lymphocyte development suggested that this miRNA cluster plays a major role in this setting. Now, using the mir-451 system, one can envision sophisticated experiments to express individual miRNAs in various Dicer knockout tissues using retroviral/lentiviral vectors, or even genomically encoded transgenes. Evidence from genetic rescues could go a long way towards pinpointing miRNAs of particular phenotypic importance in particular developmental or physiological settings.
Experiments using the p53:mir-451 mimic37 provided evidence for the utility of the mir-451 backbone for designer gene silencing, constituting a complementary system to the widely used sh-miRNA technology for siRNA expression.54 An ongoing issue with gene silencing is the off-target effects, which can occur with unintended miRNA* strand activity or by unintended silencing of seed-bearing targets.55–57 In principle, mir-451-based strategies may offer some advantages since its maturation does not involve a star strand, and its functional species accumulate exclusively in Slicer Argonaute. It remains to be seen if these unique aspects of miR-451 biogenesis correlate with any improved functional characteristics. Finally, it is worthwhile considering that mechanistic studies which demonstrate that loss of Dicer potentiates tumorigenesis in animal models,58,59 and loss of Dicer is correlated with a growing number of human diseases and cancers, and to have prognostic value in predicting their severity and/or aggressivity.60–64 If there are key target genes or pathways that become deregulated in these settings, they may potentially be alleviated by silencing appropriate genes. mir-451 backbones should have an advantage over sh-miRNA vectors in Dicer compromised cells, and would be the only possibility for siRNA expression in Dicer-deficient cells.
Open Questions Regarding mir-451 Biogenesis
Several interesting questions remain unsolved regarding the biogenesis of miR-451. First, how is the 30-nt, Ago2-cleaved pre-mir-451 (ac-pre-mir-451) trimmed back to yield the mature product? Using in vitro processing assay with synthetic 42-nt pre-mir-451 and recombinant Ago2, the Giraldez group showed that the processing of mir-451 stopped at ac-pre-mir-451, but that addition of RNAse I promoted the generation of shorter products.38 This is consistent with the view that a cellular nuclease(s) is required in the trimming step. It has been reported that pre-let-7 stability is regulated post-transcriptionally by terminal uridylyl transferase 4 (TUT4),65,66 which adds multiple uridine residues to the 3′ end of a pre-miRNA (tailing) that possibly recruit a nuclease to trigger miRNA degradation (trimming). Recently, mature miRNAs that encounter perfectly complementary targets were also found to be subject to a tailing and trimming mechanism resulting in their downregulation.67 Untemplated nucleotide additions were detected at the 3′ end of ac-pre-mir-451 intermediates,37,38 suggesting that a possible tailing/trimming mechanism might be involved in maturation of miR-451.
Second, if the mir-451 backbone is going be applied effectively in the future, is it possible to increase the miRNA expression level and knockdown efficiency by manipulating the mir-451 hairpin to make it a better substrate for Ago2 and/or the resection pathway? Structure-function studies should be able to address what parameters of mir-451 sequence, hairpin length, and basepairing type are most critical for optimal maturation and activity. Such knowledge will permit the rational design of reprogrammed mir-451 hairpins towards arbitrary targets of choice.
Third, and perhaps most interestingly, we do not understand the underlying biology to the existence of the mir-451 pathway. Why hasn't this locus evolved as a conventional miRNA? Despite the facile capacity of reprogrammed mir-451 backbones to produce diverse functional miRNAs, the fact is that mir-451 currently stands as structurally unique amongst endogenous miRNA loci. No doubt, the discovery of the direct Ago2-loading of short hairpins has stimulated searches for additional loci of this class using the vast published catalogs of short RNA sequences. However, it should also be noted that mir-451 was itself annotated many years ago,26 prior to recognition of its exotic biogenesis mechanism. Many computational studies have focused on the recovery of conserved hairpins, and by this criterion one might have expected additional candidates to have emerged already.
One wonders then, what is the special utility of the mir-451 pathway that has not only led to its strict conservation amongst all vertebrates, but has apparently been coupled to a suppression of similar hairpins in vertebrate genomes. At present, the only possible clue comes from the cell specificity of miR-451, which is expressed at extraordinary levels in erythrocytes. Three studies of mir-451 mouse knockouts (some in combination with mir-144) provide evidence for the endogenous influence of miR-451 on erythrocyte maturation.41,46,47 Rasmussen et al. showed that the phenotype of mir-451-deficient mice recapitulated that of the mir-144/451-deficient mice, which had a late erythroblast maturation defect.41 More dramatically, these mutant mice were strongly sensitive to oxidative stress induced by phenylhydrazine, which induces hemolytic anemia. Wildtype mice can recover their red blood cells in this situation, but half of the miRNA mutant mice expired within 6 days.41 Similarly, Patrick et al.46 and Yu et al.45 observed that mir-451 knockout and mir-144/451 knockout mice, respectively, had erythroid differentiation defects in both embryonic stage and adulthood, and were unable to sustain robust erythropoiesis following phenylhydrazine challenge. However, unlike knockin of catalytically dead Ago2, which is lethal,37 these three studies showed that mice deleted for miR-451 are viable and only have a mild anemia that was exacerbated when encountering oxidative stress. Thus, the Slicer activity of Ago2 may play other additional roles than miR-451 processing in erythroid differentiation.
At the stage of erythroblast differentiation, the expression level of hemoglobin starts to accumulate, which sensitizes the cells to oxidative damage. miR-451 may be important at this stage to protect erythroblast from oxidative stress by downregulating the level of 14-3-3ζ, a negative regulator of the transcriptional factor FoxO3 that mediates the expression of many anti-oxidant-encoding genes.45,46 It should also not escape notice that red blood cells are notable as the sole cell type that exists lacking its nucleus in mammals. Perhaps some aspect of this exceptional lifestyle is linked to the simplification of its miRNA profile to consist largely of miR-451, and perhaps this is in turn linked to the disposal of Dicer-dependent pathways. The level of Dicer in “elderly” red blood cells, which can circulate for 3–4 months, has not been specifically examined, nor have the properties of old erythrocytes that lack miR-451. The genetic tools exist to study this with clean knockouts, so their analysis may prove revealing.
In summary, the study of vertebrate miR-451 biogenesis has revealed a Dicer-independent, Ago2-mediated strategy for miRNA biogenesis, and provided a new platform for gene suppression. It is interesting to note that archaebacterial genomes encode Argonaute proteins,68–70 but not apparently Drosha or Dicer enzymes. It has been reported that Dicer-independent “primal” short RNAs are directly incorporated to Ago1 in Schizosaccharomyces pombe, and serve to initiate Dicer-dependent siRNA amplification during heterochromatin formation.71 It has also been previously reported that the miRNA-like RNA-2 (milR-2) in Neurospora can be produced by direct loading of the pre-miRNA-like hairpin to Argonaute protein QDE-2, whose Slicer activity is responsible for the processing of pre-milR-2.72 Such studies may suggest that the direct loading and processing of small RNA by Argonaute proteins is an ancient strategy, and conceivably may have preceded the incorporation of RNAse III enzymes into small RNA pathways.
Acknowledgements
Work in E.C.L.'s group was supported by the Burroughs Wellcome Fund, the Alfred Bressler Scholars Fund, the Starr Cancer Consortium (#I3-A139) and the NIH (R01-GM083300).
Footnotes
Previously published online: www.landesbioscience.com/journals/cc/article/13958
References
- 1.Flynt AS, Lai EC. Biological principles of microRNA-mediated regulation: shared themes amid diversity. Nat Rev Genet. 2008;9:831–842. doi: 10.1038/nrg2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Herranz H, Cohen SM. MicroRNAs and gene regulatory networks: Managing the impact of noise in biological systems. Genes Dev. 2010;24:1339–1344. doi: 10.1101/gad.1937010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 4.Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 5.Lai EC. microRNAs are complementary to 3′ UTR sequence motifs that mediate negative post-transcriptional regulation. Nat Genet. 2002;30:363–364. doi: 10.1038/ng865. [DOI] [PubMed] [Google Scholar]
- 6.Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bartel DP. MicroRNAs: Target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol. 2009;10:126–139. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- 9.Winter J, Jung S, Keller S, Gregory RI, Diederichs S. Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat Cell Biol. 2009;11:228–234. doi: 10.1038/ncb0309-228. [DOI] [PubMed] [Google Scholar]
- 10.Miyoshi K, Miyoshi T, Siomi H. Many ways to generate microRNA-like small RNAs: Non-canonical pathways for microRNA production. Mol Genet Genomics. 2010;284:95–103. doi: 10.1007/s00438-010-0556-1. [DOI] [PubMed] [Google Scholar]
- 11.Okamura K, Lai EC. Diversity and complexity of dicer dependent small RNA networks in animals. Seikagaku. 2009;81:904–909. [PMC free article] [PubMed] [Google Scholar]
- 12.Okamura K, Hagen JW, Duan H, Tyler DM, Lai EC. The mirtron pathway generates microRNA-class regulatory RNAs in Drosophila. Cell. 2007;130:89–100. doi: 10.1016/j.cell.2007.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ruby JG, Jan CH, Bartel DP. Intronic microRNA precursors that bypass Drosha processing. Nature. 2007;448:83–86. doi: 10.1038/nature05983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Berezikov E, Chung WJ, Willis J, Cuppen E, Lai EC. Mammalian mirtron genes. Mol Cell. 2007;28:328–336. doi: 10.1016/j.molcel.2007.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Babiarz JE, Ruby JG, Wang Y, Bartel DP, Blelloch R. Mouse ES cells express endogenous shRNAs, siRNAs, and other Microprocessor-independent, Dicer-dependent small RNAs. Genes Dev. 2008;22:2773–2785. doi: 10.1101/gad.1705308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Glazov EA, Cottee PA, Barris WC, Moore RJ, Dalrymple BP, Tizard ML. A microRNA catalog of the developing chicken embryo identified by a deep sequencing approach. Genome Res. 2008;18:957–964. doi: 10.1101/gr.074740.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Flynt AS, Chung WJ, Greimann JC, Lima CD, Lai EC. microRNA biogenesis via splicing and exosome-mediated trimming in Drosophila. Mol Cell. 2010;38:900–907. doi: 10.1016/j.molcel.2010.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ender C, Krek A, Friedlander MR, Beitzinger M, Weinmann L, Chen W, et al. A human snoRNA with microRNA-like functions. Mol Cell. 2008;32:519–528. doi: 10.1016/j.molcel.2008.10.017. [DOI] [PubMed] [Google Scholar]
- 19.Glazov EA, Kongsuwan K, Assavalapsakul W, Horwood PF, Mitter N, Mahony TJ. Repertoire of bovine miRNA and miRNA-like small regulatory RNAs expressed upon viral infection. PLoS ONE. 2009;4:6349. doi: 10.1371/journal.pone.0006349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Taft RJ, Glazov EA, Lassmann T, Hayashizaki Y, Carninci P, Mattick JS. Small RNAs derived from snoRNAs. RNA. 2009;15:1233–1240. doi: 10.1261/rna.1528909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brameier M, Herwig A, Reinhardt R, Walter L, Gruber J. Human box C/D snoRNAs with miRNA like functions: Expanding the range of regulatory RNAs. Nucleic Acids Res. 2010 doi: 10.1093/nar/gkq776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cole C, Sobala A, Lu C, Thatcher SR, Bowman A, Brown JW, et al. Filtering of deep sequencing data reveals the existence of abundant Dicer-dependent small RNAs derived from tRNAs. RNA. 2009;15:2147–2160. doi: 10.1261/rna.1738409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Haussecker D, Huang Y, Lau A, Parameswaran P, Fire AZ, Kay MA. Human tRNA-derived small RNAs in the global regulation of RNA silencing. RNA. 2010;16:673–695. doi: 10.1261/rna.2000810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bogerd HP, Karnowski HW, Cai X, Shin J, Pohlers M, Cullen BR. A mammalian herpesvirus uses noncanonical expression and processing mechanisms to generate viral MicroRNAs. Mol Cell. 2010;37:135–142. doi: 10.1016/j.molcel.2009.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Okamura K, Lai EC. Endogenous small interfering RNAs in animals. Nat Rev Mol Cell Biol. 2008;9:673–678. doi: 10.1038/nrm2479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Altuvia Y, Landgraf P, Lithwick G, Elefant N, Pfeffer S, Aravin A, et al. Clustering and conservation patterns of human microRNAs. Nucleic Acids Res. 2005;33:2697–2706. doi: 10.1093/nar/gki567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rathjen T, Nicol C, McConkey G, Dalmay T. Analysis of short RNAs in the malaria parasite and its red blood cell host. FEBS Lett. 2006;580:5185–5188. doi: 10.1016/j.febslet.2006.08.063. [DOI] [PubMed] [Google Scholar]
- 28.Nelson PT, De Planell-Saguer M, Lamprinaki S, Kiriakidou M, Zhang P, O'Doherty U, et al. A novel monoclonal antibody against human Argonaute proteins reveals unexpected characteristics of miRNAs in human blood cells. RNA. 2007;13:1787–1792. doi: 10.1261/rna.646007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Siolas D, Lerner C, Burchard J, Ge W, Linsley PS, Paddison PJ, et al. Synthetic shRNAs as potent RNAi triggers. Nat Biotechnol. 2005;23:227–231. doi: 10.1038/nbt1052. [DOI] [PubMed] [Google Scholar]
- 30.Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: Tools for microRNA genomics. Nucleic Acids Res. 2008;36:154–158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lai EC, Tomancak P, Williams RW, Rubin GM. Computational identification of Drosophila microRNA genes. Genome Biol. 2003;4:1–20. doi: 10.1186/gb-2003-4-7-r42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Berezikov E, Guryev V, van de Belt J, Wienholds E, Plasterk RH, Cuppen E. Phylogenetic shadowing and computational identification of human microRNA genes. Cell. 2005;120:21–24. doi: 10.1016/j.cell.2004.12.031. [DOI] [PubMed] [Google Scholar]
- 33.Stark A, Lin MF, Kheradpour P, Pedersen JS, Parts L, Carlson JW, et al. Discovery of functional elements in 12 Drosophila genomes using evolutionary signatures. Nature. 2007;450:219–232. doi: 10.1038/nature06340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lai EC. microRNAs: Runts of the genome assert themselves. Curr Biol. 2003;13:925–936. doi: 10.1016/j.cub.2003.11.017. [DOI] [PubMed] [Google Scholar]
- 35.Yang JS, Maurin T, Robine N, Rasmussen KD, Jeffrey KL, Chandwani R, et al. Conserved vertebrate mir-451 provides a platform for dicer-independent, Ago2-mediated microRNA biogenesis. Proc Natl Acad Sci USA. 2010;107:15163–15168. doi: 10.1073/pnas.1006432107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kawamata T, Seitz H, Tomari Y. Structural determinants of miRNAs for RISC loading and slicer-independent unwinding. Nat Struct Mol Biol. 2009;16:953–960. doi: 10.1038/nsmb.1630. [DOI] [PubMed] [Google Scholar]
- 37.Cheloufi S, Dos Santos CO, Chong MM, Hannon GJ. A dicer-independent miRNA biogenesis pathway that requires Ago catalysis. Nature. 2010;465:584–589. doi: 10.1038/nature09092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cifuentes D, Xue H, Taylor DW, Patnode H, Mishima Y, Cheloufi S, et al. A novel miRNA processing pathway independent of dicer requires argonaute2 catalytic activity. Science. 2010;328:1694–1698. doi: 10.1126/science.1190809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Han J, Lee Y, Yeom KH, Nam JW, Heo I, Rhee JK, et al. Molecular basis for the recognition of primary microRNAs by the Drosha-DGCR8 complex. Cell. 2006;125:887–901. doi: 10.1016/j.cell.2006.03.043. [DOI] [PubMed] [Google Scholar]
- 40.Diederichs S, Haber DA. Dual role for argonautes in microRNA processing and posttranscriptional regulation of microRNA expression. Cell. 2007;131:1097–1108. doi: 10.1016/j.cell.2007.10.032. [DOI] [PubMed] [Google Scholar]
- 41.Rasmussen KD, Simmini S, Abreu-Goodger C, Bartonicek N, Di Giacomo M, Bilbao-Cortes D, et al. The miR-144/451 locus is required for erythroid homeostasis. J Exp Med. 2010;207:1351–1358. doi: 10.1084/jem.20100458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.O'Carroll D, Mecklenbrauker I, Das PP, Santana A, Koenig U, Enright AJ, et al. A Slicer-independent role for Argonaute 2 in hematopoiesis and the microRNA pathway. Genes Dev. 2007;21:1999–2004. doi: 10.1101/gad.1565607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dore LC, Amigo JD, Dos Santos CO, Zhang Z, Gai X, Tobias JW, et al. A GATA-1-regulated microRNA locus essential for erythropoiesis. Proc Natl Acad Sci USA. 2008;105:3333–3338. doi: 10.1073/pnas.0712312105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pase L, Layton JE, Kloosterman WP, Carradice D, Waterhouse PM, Lieschke GJ. miR-451 regulates zebrafish erythroid maturation in vivo via its target gata2. Blood. 2009;113:1794–1804. doi: 10.1182/blood-2008-05-155812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Papapetrou EP, Korkola JE, Sadelain M. A genetic strategy for single and combinatorial analysis of mirna function in mammalian hematopoietic stem cells. Stem Cells. 2010;28:287–296. doi: 10.1002/stem.257. [DOI] [PubMed] [Google Scholar]
- 46.Yu D, dos Santos CO, Zhao G, Jiang J, Amigo JD, Khandros E, et al. miR-451 protects against erythroid oxidant stress by repressing 14-3-3zeta. Genes Dev. 24:1620–1633. doi: 10.1101/gad.1942110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Patrick DM, Zhang CC, Tao Y, Yao H, Qi X, Schwartz RJ, et al. Defective erythroid differentiation in miR-451 mutant mice mediated by 14-3-3zeta. Genes Dev. 24:1614–1619. doi: 10.1101/gad.1942810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chang K, Elledge SJ, Hannon GJ. Lessons from Nature: microRNA-based shRNA libraries. Nat Methods. 2006;3:707–714. doi: 10.1038/nmeth923. [DOI] [PubMed] [Google Scholar]
- 49.Tan GS, Garchow BG, Liu X, Yeung J, Morris JPt, Cuellar TL, et al. Expanded RNA-binding activities of mammalian Argonaute 2. Nucleic Acids Res. 2009;37:7533–7545. doi: 10.1093/nar/gkp812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Giraldez AJ, Mishima Y, Rihel J, Grocock RJ, Van Dongen S, Inoue K, et al. Zebrafish MiR-430 promotes deadenylation and clearance of maternal mRNAs. Science. 2006;312:75–79. doi: 10.1126/science.1122689. [DOI] [PubMed] [Google Scholar]
- 51.Giraldez AJ, Cinalli RM, Glasner ME, Enright AJ, Thomson JM, Baskerville S, et al. MicroRNAs regulate brain morphogenesis in zebrafish. Science. 2005;308:833–838. doi: 10.1126/science.1109020. [DOI] [PubMed] [Google Scholar]
- 52.Koralov SB, Muljo SA, Galler GR, Krek A, Chakraborty T, Kanellopoulou C, et al. Dicer ablation affects antibody diversity and cell survival in the B lymphocyte lineage. Cell. 2008;132:860–874. doi: 10.1016/j.cell.2008.02.020. [DOI] [PubMed] [Google Scholar]
- 53.Ventura A, Young AG, Winslow MM, Lintault L, Meissner A, Erkeland SJ, et al. Targeted deletion reveals essential and overlapping functions of the miR-17 through 92 family of miRNA clusters. Cell. 2008;132:875–886. doi: 10.1016/j.cell.2008.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Silva JM, Li MZ, Chang K, Ge W, Golding MC, Rickles RJ, et al. Second-generation shRNA libraries covering the mouse and human genomes. Nat Genet. 2005;37:1281–1288. doi: 10.1038/ng1650. [DOI] [PubMed] [Google Scholar]
- 55.Jackson AL, Burchard J, Schelter J, Chau BN, Cleary M, Lim L, et al. Widespread siRNA “off-target” transcript silencing mediated by seed region sequence complementarity. Rna. 2006;12:1179–1187. doi: 10.1261/rna.25706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schwarz DS, Hutvagner G, Du T, Xu Z, Aronin N, Zamore PD. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115:199–208. doi: 10.1016/s0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- 57.Khvorova A, Reynolds A, Jayasena SD. Functional siRNAs and miRNAs exhibit strand bias. Cell. 2003;115:209–216. doi: 10.1016/s0092-8674(03)00801-8. [DOI] [PubMed] [Google Scholar]
- 58.Kumar MS, Lu J, Mercer KL, Golub TR, Jacks T. Impaired microRNA processing enhances cellular transformation and tumorigenesis. Nat Genet. 2007;39:673–677. doi: 10.1038/ng2003. [DOI] [PubMed] [Google Scholar]
- 59.Kumar MS, Pester RE, Chen CY, Lane K, Chin C, Lu J, et al. Dicer1 functions as a haploinsufficient tumor suppressor. Genes Dev. 2009;23:2700–2704. doi: 10.1101/gad.1848209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lin RJ, Lin YC, Chen J, Kuo HH, Chen YY, Diccianni MB, et al. microRNA signature and expression of Dicer and Drosha can predict prognosis and delineate risk groups in neuroblastoma. Cancer Res. 2010;70:7841–7850. doi: 10.1158/0008-5472.CAN-10-0970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pampalakis G, Diamandis EP, Katsaros D, Sotiropoulou G. Downregulation of dicer expression in ovarian cancer tissues. Clin Biochem. 43:324–327. doi: 10.1016/j.clinbiochem.2009.09.014. [DOI] [PubMed] [Google Scholar]
- 62.Karube Y, Tanaka H, Osada H, Tomida S, Tatematsu Y, Yanagisawa K, et al. Reduced expression of Dicer associated with poor prognosis in lung cancer patients. Cancer Sci. 2005;96:111–115. doi: 10.1111/j.1349-7006.2005.00015.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Merritt WM, Lin YG, Han LY, Kamat AA, Spannuth WA, Schmandt R, et al. Dicer, Drosha and outcomes in patients with ovarian cancer. N Engl J Med. 2008;359:2641–2650. doi: 10.1056/NEJMoa0803785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hill DA, Ivanovich J, Priest JR, Gurnett CA, Dehner LP, Desruisseau D, et al. DICER1 mutations in familial pleuropulmonary blastoma. Science. 2009;325:965. doi: 10.1126/science.1174334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Heo I, Joo C, Kim YK, Ha M, Yoon MJ, Cho J, et al. TUT4 in concert with Lin28 suppresses microRNA biogenesis through pre-microRNA uridylation. Cell. 2009;138:696–708. doi: 10.1016/j.cell.2009.08.002. [DOI] [PubMed] [Google Scholar]
- 66.Hagan JP, Piskounova E, Gregory RI. Lin28 recruits the TUTase Zcchc11 to inhibit let-7 maturation in mouse embryonic stem cells. Nat Struct Mol Biol. 2009;16:1021–1025. doi: 10.1038/nsmb.1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ameres SL, Horwich MD, Hung JH, Xu J, Ghildiyal M, Weng Z, et al. Target RNA-directed trimming and tailing of small silencing RNAs. Science. 2010;328:1534–1539. doi: 10.1126/science.1187058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Song JJ, Smith SK, Hannon GJ, Joshua-Tor L. Crystal structure of Argonaute and its implications for RISC slicer activity. Science. 2004;305:1434–1437. doi: 10.1126/science.1102514. [DOI] [PubMed] [Google Scholar]
- 69.Parker JS, Roe SM, Barford D. Crystal structure of a PIWI protein suggests mechanisms for siRNA recognition and slicer activity. EMBO J. 2004;23:4727–4737. doi: 10.1038/sj.emboj.7600488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yuan YR, Pei Y, Ma JB, Kuryavyi V, Zhadina M, Meister G, et al. Crystal structure of A. aeolicus argonaute, a site-specific DNA-guided endoribonuclease, provides insights into RISC-mediated mRNA cleavage. Mol Cell. 2005;19:405–419. doi: 10.1016/j.molcel.2005.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Halic M, Moazed D. Dicer-independent primal RNAs trigger RNAi and heterochromatin formation. Cell. 2010;140:504–516. doi: 10.1016/j.cell.2010.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lee HC, Li L, Gu W, Xue Z, Crosthwaite SK, Pertsemlidis A, et al. Diverse pathways generate microRNA-like RNAs and Dicer-independent small interfering RNAs in fungi. Mol Cell. 38:803–814. doi: 10.1016/j.molcel.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]