Abstract
Complementary transcriptional and epigenetic regulatory factors (e.g., histone and chromatin modifying enzymes and non-coding RNAs) regulate genes responsible for mediating neural stem cell maintenance and lineage restriction, neuronal and glial lineage specification and progressive stages of lineage maturation. However, an overall understanding of the mechanisms that sense and integrate developmental signals at the genomic level and control cell type-specific gene network deployment has not emerged. REST and CoREST are central players in the transcriptional and epigenetic regulatory circuitry that is responsible for modulating neural genes, and they have been implicated in establishing cell identity and function, both within the nervous system and beyond it. Herein, we discuss the emerging context-specific roles of REST and CoREST and highlight our recent studies aimed at elucidating their neural developmental cell type- and stage-specific actions. These observations support the conclusion that REST and CoREST act as master regulators of key neural cell fate decisions.
Key words: cell fate, CoREST, gliogenesis, neural stem cell, neurogenesis, neuronal subtype, REST/NRSF
Introduction
In 1995, two laboratories simultaneously reported the identification of the repressor element-1 silencing transcription factor/neuron-restrictive silencer factor (REST/NRSF).1,2 REST was initially implicated in the repression of genes containing the 21 base pair repressor element 1/neuron restrictive silencer element (RE1/NRSE) cis-regulatory DNA sequence. These RE1 motif-associated genes included many neuron-specific genes, such as those encoding ion channels, neurotransmitter receptors, and neurosecretory factors.3 Outside of the nervous system, REST expression was detected ubiquitously in mouse embryos.1 Within the developing nervous system, REST was detected in neural progenitor cells but not in mature neurons.2 These initial observations seemed to suggest that REST acts as a repressor of RE1 motif-associated neuronal gene expression in non-neural, neural stem/progenitor and non-neuronal cells and further that it serves as a master regulator of neurogenesis.
Our knowledge of REST and its actions has expanded significantly since its discovery. We now recognize that the complex biology of REST includes the potential for it to interact directly or indirectly with a broad and increasing array of transcriptional and epigenetic regulatory cofactors (including CoREST4), to be alternatively spliced and differentially transported between the nucleus and cytoplasm,5 to promote gene activation in addition to repression and long-term gene silencing,6,7 to modify the epigenetic status of target gene loci distinct from effects on transcription,8,9 to modulate genes not associated with the canonical RE1 motif,10 and to exhibit high levels of integration with non-coding RNA networks that, among other things, mediate cell identity.11 This remarkable flexibility in all aspects of REST regulation is consistent with our emerging appreciation for the decidedly context-specific effects of REST for establishing and maintaining cell identity and function. Indeed, REST and CoREST seem to play important roles in a growing number of normal and pathological cell types, including embryonic stem cells (ESCs)12–18 and cancer19–28 cells. In this review, we focus on understanding the evolving roles of REST and CoREST in transcriptional and epigenetic regulation of seminal neural cell fate decisions.
Neural Developmental Processes Including Neural Cell Fate Determination
Neural stem cells (NSCs) and their progeny have been the focus of numerous studies aimed at characterizing the molecular mechanisms that orchestrate self-renewal, proliferation, lineage restriction, neuronal and glial lineage specification, progressive maturation and terminal differentiation. Included in these investigations is a quest to determine the regulatory logic for promoting the elaboration of diverse regional neuronal as well as glial cell subtypes and for mediating the complementary processes of progressive oligodendrocyte (OL) lineage maturation and myelination, neuronal-glial interactions and neural connectivity. These studies have begun to define the temporal and spatial profiles of gradient morphogens, cytokines, growth factors, cell-cell signaling cues and combinatorial transcription factor codes that establish neural cell identity and control the elaboration of both neuronal and glial lineages from distinct regional NSC subpopulations contained within the neuraxis.29,30 Gradient morphogens, such as sonic hedgehog (SHH) and bone morphogenetic proteins (BMPs), induce patterning genes in specific areas of the developing neural tube. In ventral domains, SHH establishes a characteristic profile of homeodomain (HD) and basic helix-loop-helix (HLH) transcription factor expression, whereas BMPs mediate a complementary developmental program in the dorsal aspect of the neural tube. Rostrocaudal patterning occurs in a similar manner. Noggin, chordin and follistatin act as inductive signals for forebrain, whereas fibroblast growth factor (FGF) and retinoic acid (RA) represent inductive signals for hindbrain regions. These patterning and lineage specific genes establish discrete progenitor domains through cross-repressive interactions that define the identities of individual neural progenitor species and promote the subsequent elaboration of specific neuronal and glial subtypes. These developmental cues, such as Notch signaling, promote the sequential expression of transcription factor codes that coordinate progressive stages of neuronal and glial lineage elaboration. These combinatorial transcription factor codes include inhibitory HLH transcriptional regulators (Hes/Hey and Id), which interact with other basic HLH and HD genes.31 Dynamic changes in the deployment and redeployment of different combinations and permutations of transcription factors mediate downstream developmental programs, including terminal differentiation and neural network integration.32
More recently, epigenetic mechanisms have emerged as an novel molecular interface for integrating cues from these diverse cell intrinsic, cell-cell associated and environmental signaling pathways and, in turn, for regulating the deployment of specific gene networks that determine neural cell identity. These developmental processes may be mediated through context-dependent gene activation, repression, long-term silencing, graded and coordinate gene expression, establishment of epigenetic memory states and dynamic changes in nuclear architecture. Indeed, an increasing number of studies have begun to uncover how epigenetic processes, such as DNA methylation, histone modifications and chromatin remodeling and short and long non-coding RNAs (ncRNA) regulation, are responsible for promoting neural cell type- and developmental stage-specific gene expression profiles and thus, for mediating seminal neural cell fate decisions.33–35 How transcriptional and epigenetic processes, including those mediated by REST and CoREST, promote key neural developmental events has been the focus of intensive investigations.
REST and CoREST in Developmental Processes
Studies examining the expression, regulation and function of REST suggest a spectrum of developmental roles. In fact, the phenotype exhibited by REST knockout mice clearly establishes that REST is required for embryogenesis, although an integrated understanding of its actions during this process is still emerging. REST knockout animals display malformations in non-neural and neural tissues with lethality at embryonic day 11.5 associated with complex profiles of neural gene deregulation. These wideranging abnormalities are potentially explained by the fact that REST is implicated in a range of development processes, from patterning of ectoderm36 to neuronal maturation (i.e., axon path-finding).37 In addition, REST is modulated by multiple developmental cues, including patterning and specification factors (e.g., RA, BMP and WNT).38,39 These observations imply that REST acts as a regulator of multiple stages of embryonic and neural development through precise regulation of neural developmental genes.
Less is known about the developmental functions of CoREST than those of REST. However, there is evidence to suggest that CoREST has not only cooperative but also distinct roles in mediating neural cell identity. For example, CoREST expression is highly developmentally regulated, found more selectively in the embryonic nervous system compared to REST and downregulated at the time of birth.40–43 Further, CoREST is more evolutionarily conserved than REST.41 These observations suggest that CoREST may have more primordial functions in neural gene regulation.
REST and CoREST as Highly Flexible Transcriptional and Epigenetic Regulatory Factors
REST acts as a modular scaffold for the recruitment of a diverse array of transcriptional and epigenetic factors and macromolecular complexes to specific genomic loci. CoREST binds to the C-terminal domain of REST.4 In turn, CoREST recruits histone deacetylases, HDAC1 and HDAC2; methyl-CpG binding protein 2, MECP2; histone demethylase, LSD1; histone methyltransferases, G9a and Suv39 h1; zinc finger proteins, ZNF198 and ZNF217; the acetyltransferase, Esco2; and components of the SWI/SNF chromatin remodeling complex, BAF57, BRG1 and BAF170. Similarly, mSin3 binds to the N-terminal domain of REST and also recruits histone deacetylases, HDAC1 and HDAC2. Furthermore, it is increasingly being recognized that REST and/or CoREST have the potential to interact with a large array of additional and complementary regulatory factors. These include DNA methyltransferases; additional methyl-CpG binding domain proteins; chromatin remodeling enzymes, such as the histone demethylase, SMCX; the chromodomain Y chromosome family member, CDYL;20 the NADH-binding factor, CtBP;44 the small C-terminal domain phosphatase, Scp1;45 and the transcription factor, Sp3.46 The identification of an increasing number of potential interactors reveals a complex and flexible functional repertoire of transcriptional and epigenetic mechanisms well beyond those initially envisioned for REST and CoREST.
REST also associates with Mediator, a large evolutionarily conserved complex that is essential for mediating RNA polymerase II activity.47,48 The Mediator complex is comprised of multiple (∼26) subunits with general and specific roles in gene regulation, including activation and repression. The mechanisms by which Mediator affects transcription are still being characterized but include control of both pre- and post-initiation events. Interestingly, recent studies have shown that Mediator promotes DNA looping between gene enhancers and promoters, in concert with cohesin.49 The profiles of Mediator and cohesin occupancy can vary by cell type, suggesting context-specific developmental roles. The core Mediator subunits, Med19 and Med26, interact with REST directly and synergistically, promoting the recruitment of the entire Mediator complex to genomic RE1 sites occupied by REST.47 In turn, the Mediator complex facilitates the recruitment of G9a to these sites, through interactions between its Med12 subunit and G9a.50 Notably, Med12 expression is enriched in the nervous system, where it plays a role in regulating neuronal development. In zebrafish, Med12 mutants exhibit deficits of monoaminergic neurons and cranial sensory ganglia through deregulation of key neural developmental genes.51 In mouse, mosaic expression of Med12 leads to all known neural tube defects.52 In humans, Med12 missense mutations cause the X-linked mental retardation disorders, FG syndrome and Lujan syndrome, by disturbing its functional interactions with REST.50
REST and CoREST complexes can independently or coordinately regulate certain gene loci. For example, some authors have designated neuronal genes regulated by these factors as Class I or Class II, depending on whether CoREST binds independently to the gene promoter.53 Only the REST complex binds to Class I gene promoters. In contrast, distinct REST and CoREST complexes bind to Class II gene promoters but at different sites. Thus, levels of Class I gene expression are highest when REST is absent. However, levels of activity-dependent Class II gene expression are only sub-maximal when REST is absent because of repressive effects from a distinct CoREST complex bound to the promoters of these genes at different sites. Although these functional classifications are important, our understanding of genome-wide REST and CoREST binding profiles and their site-specific and potentially more long range effects on transcriptional and epigenetic regulation have evolved since the initial description of these two classes.
The REST gene can be spliced into at least five distinct isoforms that may be differentially expressed, regulated, degraded and functionally deployed.5,54–57 Each of these isoforms may have distinct affinities for RE1 motifs. They may have individual profiles of nuclear and cytoplasmic localization, distinct modular partners and differential effects on gene regulation. Further, the stoichiometry of different REST isoforms may impact its regulatory activities. For example, when REST4, a C-terminus truncated isoform, is co-expressed with other REST isoforms, it acts in a dominant-negative fashion, competitively inhibiting REST-induced gene silencing and promoting gene activation.54
The REST complex can function as a transcriptional repressor or activator. A small modulatory ncRNA has been identified that interacts directly with the REST complex and alters its behavior. This double-stranded ncRNA encodes the RE1 sequence (dsNRSE) and is responsible for promoting RE1-associated gene transcription.6,7 Furthermore, the presence of the dsNRSE in certain populations of NSCs leads to their differentiation into neuronal cells.6,7
REST and CoREST can also induce local and more long-range chromatin remodeling, distinct from their effects on transcription. For example, we recently showed that the binding of REST to RE1 sites results in dramatic changes in nucleosome phasing and in histone modifications.8 These included, but were not limited to, significant reductions in profiles of histone acetylation and context-specific increases and decreases in profiles of histone methylation. Interestingly, these REST-mediated chromatin-remodeling events were not restricted to gene promoter regions. This finding may be consistent with previous studies suggesting that REST and CoREST promote chromatin remodeling across extended chromosomal intervals.9
Recent studies have extended our understanding of the REST regulome by utilizing complementary genome-wide approaches to better characterize REST binding sites in the genome.10,15,58 These analyses have identified genomic binding sites for REST associated with canonical RE1 (cRE1) motifs and non-canonical RE1 (ncRE1) motifs. The cRE1 motif is comprised of two sequences that are each ten base pairs (bps) long separated by a single non-conserved nucleotide. By contrast, ncRE1 motifs are of variable length. These ncRE1 motifs are comprised of two segments ten bps long separated by a variable insertion that can be up to nine bps.10,59 These ncRE1 motif-associated genes include many genes that are neuronal but also those that are not. Intriguingly, it has been shown that the interaction between REST and cRE1 motifs is strong, whereas its interaction with other motifs is less robust, suggesting cell- or tissue-specific functions for REST in regulating these genes.60 The differential functional outcomes of REST binding to genes associated with these different binding motifs have yet to be elucidated fully.
The functions of REST and CoREST are intimately linked with those of at least two classes of ncRNAs—microRNAs (miRNAs) and long ncRNAs (lncRNAs).11 miRNAs are relatively well characterized ∼22 nucleotide ncRNA transcripts that participate in post-transcriptional regulation of genes and gene networks, through different degrees of sequence-specific interactions with cognate mRNAs resulting in translational repression or sequestration of these transcripts. By contrast, lncRNAs are an emerging class of ncRNAs defined as transcripts longer than 200 nucleotides. These diverse lncRNAs have a broad range of functions including the ability to modulate transcription, post-transcriptional RNA processing, translation, DNA methylation and chromatin architecture through local and long-distance effects.61 Not surprisingly, both of these classes of ncRNAs are implicated in mediating neural cell identity and function.34,35
A number of miRNAs enriched in the nervous system are associated with RE1 motifs and are regulated by REST.62 Moreover, REST may regulate genes that play roles in miRNA biogenesis including Dicer1, which is associated with an RE1 motif. These observations imply that REST can modulate the expression of miRNAs at multiple levels in their life cycles. These regulatory relationships may be bidirectional. In fact, there are double negative feedback loops between members of the REST complex and the miRNAs they regulate, such as miR-124, miR-9 and miR-132.62,63 Interestingly, miR-9/miR-9* is a bifunctional miRNA that targets REST (miR-9) and CoREST (miR-9*).63 Furthermore, many REST-regulated miRNAs are also subject to modulation by the cAMP response element-binding protein (CREB), another factor with key roles in mediating brain development and adult neurogenesis.62 Indeed, an increasing number of studies has highlighted the complex circuitry involving REST, CoREST, CREB and miRNAs that controls NSC self-renewal, maintenance and differentiation.19,64,65 In addition, a computational analysis comparing the distributions of RE1 motifs and of annotated lncRNA genes in mouse and human genomes suggested that REST also has the potential to regulate 23% of annotated lncRNAs.66 In vitro studies performed utilizing mouse and human tissues validated this hypothesis by showing that REST influenced the expression of various developmentally expressed and nervous system restricted lncRNAs. For example, AK090153 is a REST-regulated lncRNA with enriched expression in the adult mouse pallidum (www.brain-map.org/search.do?queryText=geneid = 143712, Allen Brain Atlas—Allen Institute for Brain Science, Seattle). Intriguingly, one of the critical functions of lncRNAs is to recruit REST and CoREST complexes to specific genomic sites.67,68 We previously reported utilizing in situ hybridization data from the Allen Brain Atlas to identify region-, cell type- or subcellular compartment-specific expression profiles for hundreds of lncRNAs in the adult mouse brain.69 Other groups have identified complementary profiles of lncRNA expression during brain development.70 Together, these observations highlight the complex patterns of neural lncRNA expression present during development and adult life and support the interesting possibility that REST and CoREST act more globally as regulators of lncRNA expression. In turn, REST and CoREST transcriptional and epigenetic regulatory functions may be orchestrated by these lncRNAs.
Evolving Roles of REST and CoREST in Seminal Neural Cell Fate Decisions
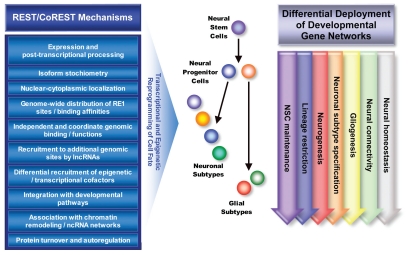
Previous efforts have focused on understanding the role played by REST in regulating cell fate decisions. These have included analyses of REST genome-wide binding profiles in various cell lines.10,15,58 By contrast, we recently examined neural cell type- and developmental stage-specific profiles for REST and CoREST promoter occupancy employing paradigms to generate neuronal and glial subtypes and progressive stages of OL lineage maturation including myelination from mouse forebrain-derived neural stem and progenitor species.71–73 These studies are the first to report such comprehensive and comparative genome-wide binding profiles for REST and for CoREST in primary neural cell types, ranging from immature to mature neural lineage species. We found thousands of occupied genomic loci including those associated with cRE1 and ncRE motifs as well as non-RE1 sites not previously identified as targets for REST and CoREST regulation. We identified both unique and overlapping profiles for gene promoters occupied by REST or CoREST across these cell types. Moreover, we performed complementary gene expression analyses within this neural developmental paradigm. Our data is the first to correlate dynamic profiles of REST and CoREST promoter occupancy with gene expression changes during neural lineage elaboration. We found that REST or CoREST promoter binding was not simply associated with gene downregulation. Rather, we observed that transcriptional activation and repression and the absence of significant shifts in profiles of transcription were all possible outcomes of changes in REST and CoREST occupancy, highlighting the complex, dynamic and flexible actions of REST and CoREST (see above) in neural development functions. Our overall observations suggest that REST and CoREST have the potential to independently and coordinately regulate a very broad range of genes and functional gene networks in an exquisitely context-specific manner, through a spectrum of transcriptional and epigenetic mechanisms (Fig. 1).
Figure 1.
This schematic illustrates the mechanisms by which REST and CoREST are responsible for the transcriptional and epigenetic regulation of developmental gene networks underpinning neural cell fate decisions.
Examining the functions of these genes leads to the conclusion that REST and CoREST modulate diverse signaling pathways, transcription factor codes, epigenetic networks and associated developmental programs, all of which are at the nexus of mediating neural cell identity and function.71–73 For example, we observed that REST and CoREST target members of the SHH gradient morphogen neurodevelopmental signaling pathway. These results are consistent with recent experimental work in zebrafish, which demonstrated that REST modulates SHH signaling.74 This study found that knocking down REST both promotes and inhibits SHH signaling, depending on the developmental context.74 This observation once again highlights the exquisite versatility of REST developmental functions.
We found that REST and CoREST target a large number epigenetic factors in a cell type- and developmental stage-specific manner, including but not limited to those that mediate DNA methylation, histone modification, nucleosome repositioning and higher order chromatin remodeling.71–73 These observations are consistent with and significantly expand on previous findings that have implicated REST in regulating chromatin modulatory factors.10 Many of the epigenetic factors targeted by REST or CoREST individually or in concert help to shape cell fate and produce cellular diversity (see below). For example, REST and CoREST target components of the cohesin complex, suggesting roles in integrating neural enhancer and promoter functions and more precisely regulating the differential deployment of developmental genes (see above).49 Intriguingly, many of these factors are directly recruited by REST and CoREST complexes or indirectly participate in their functions. These observations suggest that REST and CoREST determine cell fate through multiple distinct mechanisms, which include modulation of epigenetic factors at the transcriptional level and operationally through recruitment to their macromolecular complexes.
We also found REST and CoREST targets that are involved in the regulation of REST and CoREST transcription, post-translational modifications and profiles of cellular degradation.71–73 For example, both REST and CoREST bind to the RE1-containing REST promoter in NSCs. In addition, NCoR2 and RARγ regulate transcription of REST, whereas the ubiquitin-proteosome pathway degrades REST and sumoylation inactivates CoREST. Autoregulation utilizing these flexible feed-forward and negative feedback loops may confer significant sensitivity and responsivity to REST and CoREST regulatory control systems, which seem to be differentially exploited throughout NSC-mediated lineage elaboration.
In addition, we found that genes targeted in a developmental stage- and cell type-specific manner encode factors that have roles in cell surface identity and connectivity, homeostasis, stress responses, as well as many additional cellular functions.71–73 These observations suggest that these genes may have selective roles in promoting a particular neural cell identity and/or repressing alternate lineage fates.
Embryonic and neural stem cell maintenance and maturation.
REST is implicated in mediating cell identity and function within a variety of cell types, including ESCs and NSCs. It has been reported that REST is highly expressed in blastocysts and ESCs and that REST promotes the early stages of ESC differentiation by suppressing the expression of pluripotency associated genes, such as Nanog.18 Moreover, it has been suggested that REST is even required for ESC pluripotency, however, this role for REST is controversial.75 Nonetheless, REST transcriptional circuitry is highly integrated with that of pluripotency gene networks.17,76,77 For example, REST is a target of the core pluripotency genes, and it may also regulate them. One of the mechanisms by which REST is implicated in regulating pluripotency gene networks is through effects on miRNAs (i.e., miR-21).17 Indeed, both miRNAs78 and lncRNAs79,80 have roles in ESC pluripotency and differentiation. However, the potential interrelationships between these factors and REST in ESCs have yet to be defined fully.
The role of REST has been examined in the developmental transition from ESCs into NSCs. REST is thought, at least in part, to selectively silence a cohort of neuronal genes in ESCs.14 REST expression decreases with neural lineage commitment.53 Two complementary studies have shown differential REST occupancy profiles in ESCs and NSCs and these findings suggested the presence of distinct and overlapping developmental REST regulons.15,81 For example, one of these studies found 810 and 679 RE1 sites occupied by REST, respectively, in ESCs and NSCs.15 These included only 32 unique sites in NSCs. This study also suggested that REST promotes ESC-mediated neural lineage restriction by repressing a subset of neuronal differentiation genes (i.e., Scg3, Stmn3, Celsr3, Syp, Cplx1 and Syt4) but not those with roles in neural lineage specification (i.e., Mash1 and Math1), implying preferential REST regulation of neuronal terminal differentiation as compared with earlier stages of lineage specification.14,81 A different study found that REST depletion to 1% of wild type levels prevented the formation of NSCs and neurogenesis.82 Further, these deficits could be rescued, in part, by the extracellular matrix component, laminin, which is a target of REST regulation. These observations suggest that REST orchestrates neural cell fate decisions at various cellular stages through multiple interrelated molecular and cellular mechanisms.
Intriguingly, in our NSC studies, we found that REST binds to 322 promoter sites, including those containing RE1 and non-RE1 sequences.71 Differences in the number of REST targets compared with previous studies may be explained by the utilization of distinct cell types—more biological representative primary NSCs as compared to NSC cell lines15 and by a wide variety of methodological considerations related to their isolation, propagation and interrogation. We unexpectedly found that CoREST binds to a significantly greater number of genomic sites (1,820) compared to REST and encompassing a larger proportion of unique target genes in NSCs.71 These included many loci encoding factors that are part of pluripotency networks83 and individual and composite Oct4/Sox2/Nanog transcriptional networks;84 those with critical roles in NSC maintenance, lineage restriction and neurogenesis; those involved in epigenetic regulatory networks, including genes targeted by brain-related miRNAs (e.g., miR-124a, miR-9 and miR-132); and those linked to specific disease states associated with deregulation of NSC functions (i.e., cancer stem cells), including glioblastoma multiforme. By contrast, we found that both REST and CoREST target genes included factors implicated in Huntington's disease, a disorder linked to deregulation of REST subcellular localization and function.85 These observations suggest that CoREST may preferentially promote NSC maintenance and fate restriction compared to REST and that specific core pluripotency genes may have distinct and previously unanticipated roles in early neural development.86
Further, we performed REST and CoREST knockdown experiments utilizing NSC clonal expansion and differentiation assays to examine their roles in mediating neural cell fate decisions. These studies revealed that REST ablation impedes neurogenesis, consistent with previous studies,53,82 whereas CoREST ablation significantly impedes NSC maintenance and altered neural fate restriction, consistent with distinct roles for CoREST in multi-lineage potential and early neural fate decisions.71 Moreover, our studies revealed that both REST and CoREST participate in the process of self-renewal, possibly in a cooperative manner, as revealed by our clonal gene ablation studies and corresponding analysis of downstream REST and CoREST gene targets.71 Further supporting this model, we found that the ncRE1-associated Nr2e1 (Tlx) gene is targeted by CoREST but not REST in NSCs. Nr2e1 interacts directly with the histone demethylase, LSD1, a member of the CoREST complex.87 It promotes NSC maintenance, including self-renewal.88 Nr2e1 also participates in a dual negative transcriptional feedback loop with the microRNA, miR-9,89 which also has bidirectional transcriptional regulatory relationships with REST and CoREST.62,63
REST and CoREST may also interact with other key factors involved in NSC fate decisions, such as the Polycomb group (PcG) proteins, Bmi1 and Ezh2, which are both implicated in regulating NSC maintenance and self-renewal.90,91 It has been hypothesized that PcG complexes that contain these factors (i.e., PRC1 and PRC2) may be recruited to their genomic sites of action by REST.8,92,93 However, an alternative explanation may be that these PcG proteins and REST can both be co-recruited to these genomic sites by lncRNAs.67
Neuronal subtype specification and maturation.
We performed the first study of REST and CoREST occupancy profiles across multiple neuronal subtypes and uncovered large networks of neuronal subtype-specific target genes.72 Intriguingly, only a minority of these gene targets is RE1 motif-associated. Further, REST and CoREST targeted groups of genes that are largely non-overlapping and also distinct within each neuronal subtype. These findings strongly suggest that REST and CoREST do not execute “generic” neuronal transcriptional and epigenetic regulatory programs but rather are involved in sculpting profiles of genes expression involved in terminal neuronal differentiation and regional neuronal subtype identity, maintenance and function, with important implications for our understanding of the pathogenesis of neurodegenerative diseases.85,94
In fact, REST and CoREST each targeted only one site across all four neuronal subtypes that we examined. REST only targeted Ehmt1, a H3K9 methyltransferase closely related to G9a (Ehmt2), that is thought to play a role in regulating p53,95 and lineage-specific genes in neurons.96 Ehmt1 is linked to various cognitive and behavioral processes in animal models96 and associated with the development of intellectual and developmental disabilities97,98 and medulloblastoma99 in humans. CoREST only targeted Defb42, a G9a-regulated member of the ß-defensin family of pleiotropic factors.100 REST recruits G9a to its macromolecular complex, suggesting that the association of both of these factors with G9a is not coincidental but rather that it has a functional and regulatory impact on neurogenesis.
More generally within these neuronal subtypes, we found that REST and CoREST targeted genes encoding members of key neural developmental signaling pathways (e.g., FGF, RA, EGF, Notch, BMP, SHH and WNT) and factors with roles in promoting neuronal subtype identity. For example, we identified many target genes that are protocadherins. These factors are thought to promote neuronal functional diversity and connectivity. We observed that REST and CoREST targets included Pcdh15, Pcdh18, Pcdhb4, Pcdhb5, Pcdhb7, Pcdhb10, Pcdhb14, Pcdhb16, Pcdhb17, Pcdhb18, Pcdhb19, Pcdhb20, Pcdhb21, Pcdhb22, Pcdhga9, Pcdhgb1 and Pcdhgb2. Our observations are consistent with a recent functional analysis showing that REST regulates protocadherin genes, including genes that are not associated with an RE1 motif.101
Our distinct REST and CoREST target gene profiles in neuronal subtypes also included factors not previously associated with the establishment of neuronal subtype identity, such as particular epigenetic factors (e.g., Mbd6, Smcx, Jarid1d, Lsd1 and Smarce1), cell cycle regulators (e.g., Anapc10, Cdkal1, Cdc2l1, Cdkl4 and Mad2l1) and homeostatic modulators (e.g., Arih1, Mib1, Ubb, Ube2j1 and Ubtd1). Furthermore, we found that REST and CoREST also targeted genes associated with neurodegenerative diseases characterized by neuronal subtype-specific profiles of cell loss. These include genes linked to Alzheimer's and Parkinson's diseases, spinocerebellar ataxias, frontotemporal dementia, myoclonic and dopa-responsive dystonias, pantothenate kinase associated neurodegeneration and hemochromatosis, suggesting additional roles of REST and CoREST in mediating the integrity of neuronal subtypes.
In a complementary analysis of neuronal subtype specification, we found differential expression of lncRNAs that may also be targets of REST regulation.102 One of these, AK044422, is a transcript previously implicated in the development of retinal cell types.103 This transcript overlaps a REST regulated miRNA, miR-124a, but is polyadenylated, spliced and likely to also function independently. It is therefore interesting to speculate that REST modulates the expression not only of miR-124a but also of AK044422.64 Mechanistically, miR-124 induces brain-specific alternative splicing by reducing levels of the Ptbp1 splicing regulatory factor and thereby promotes neuronal differentiation.104 An ncRE1 motif is associated with Ptbp1 and with other splicing factors (e.g., Nova2) implying that REST and CoREST may be involved in regulating neurogenesis through effects on alternative splicing. Ptbp1 and AK044422 exhibit complementary expression profiles, suggesting that they may represent components of an integrated AK044422/miR-124/Ptbp1 REST/CoREST-associated developmental gene regulatory network. These observations highlight the complex and integrated nature of REST, CoREST and ncRNA circuitry responsible for the establishment of neuronal cell identity and developmental functions.11,105
Glial subtype specification and progressive stages of oligodendrocyte lineage maturation.
We performed the first study of REST and CoREST in developmental glial cell types and uncovered a large network of glial-selective target genes.73 As in neuronal subtypes, only a minority of these gene targets is RE1 motif-associated. Intriguingly, we identified a much larger cohort of OL developmental stage-selective CoREST as compared to REST target genes in OL precursor cells, which further implies that CoREST plays an important, preferential and previously unappreciated role in uncommitted immature neural cell types. We also found that progressive stages of OL lineage maturation are characterized by increasing numbers of target genes that are common to REST and CoREST and shared between these OL developmental cellular species. These observations imply that, unlike neuronal subtype specification, which seems to employ largely non-overlapping REST and CoREST regulatory modules, OL lineage progression seems to utilize coordinated REST and CoREST regulatory modules. Moreover, profiles of REST and CoREST target genes in NSCs, neuronal subtypes and glial lineage species were largely non-overlapping, highlighting the highly developmental context-dependent nature of REST and CoREST genomic binding profiles. These observations imply that largely independent REST and CoREST gene regulatory networks dynamically sculpt diverse neural fate decisions by integrating multiple layers of cell autonomous, cell-cell associated and environmental cues.
In fact, we found that REST and CoREST targeted genes encoding factors involved in mediating glial cell identity and function. These included members of key developmental pathways that regulate OL specification and progressive maturation (e.g., PDGF, SHH, MAPK and FGF) and transcription factors responsible for governing OL lineage specification, proliferation and terminal differentiation including myelination, such as members of the HLH (e.g., E2A, HEB, Mash1, Olig2 and Id4), Hox (e.g., Hoxa2), POU (e.g., Brn2), Sox (e.g., Sox4, Sox8 and Sox11), Nkx (e.g., Nkx6.1) and zinc finger (e.g., Myt1 and Zfp488) transcription factor families.
Intriguingly, in astroglial cells, we identified a larger number of distinct REST target genes compared to CoREST target genes, suggesting a preferential role for REST in promoting astroglial cell identity. Our observations are consistent with a subsequent study demonstrating that BMP signaling upregulates REST expression which, in turn, promotes astroglial lineage specification and maintenance.39
We also identified other REST and CoREST target genes responsible for mediating a broad array of biological processes that may be involved in determining glial cell identity and function. These included numerous epigenetic (e.g., Hist1 h4a, Mbd6, Scmh1, Smarca2 and Smc4l1) and cell cycle regulatory (e.g., Ccna2, Ccnd1, Cdc34, Cdk5r1 and Cdkn2c) genes as well as many classes of cell surface identity (e.g., protocadherin, olfactory receptors, vomeronasal receptors and cell adhesion and additional transmembrane proteins) factors. Furthermore, REST and CoREST also targeted genes associated with oligodendrogliopathies, including multiple sclerosis and multisystem atrophy, suggesting previously unanticipated roles for REST and CoREST in the pathogenesis of glial disease states.
Intriguingly, in a complementary study, we also identified a number of developmentally regulated lncRNAs transcribed from genomic loci encompassing protein-coding genes important for OL lineage elaboration, in antisense, bidirectional or intronic configurations relative to these genes.102 For example, Sox8OT is a bidirectional lncRNA differentially expressed during OL lineage maturation that is associated with Sox8, a RE1-associated gene that plays key roles in OL differentiation. We found that REST and CoREST targeted the Sox8 promoter in post-mitotic and myelinating OLs, raising the intriguing possibility that REST and CoREST may coordinately regulate the bidirectional transcripts, Sox8 and Sox8OT. In addition, REST may also regulate miRNAs that are important for OL lineage elaboration. For example, the miR-9 is downregulated during OL differentiation.106 In addition, miR-9/9* has a bidirectional regulatory relationship with REST and CoREST,63 highlighting the complex gene regulatory relationships that may underpin OL lineage elaboration. These overall observations highlight the potential importance of REST and CoREST in mediating the dynamic transcriptional and epigenetic modulation of neural fate decisions through the actions of short and long ncRNAs.
Conclusion
Individual transcriptional and epigenetic states define cell identity and function in the nervous system.94,107 Progressive changes in profiles of gene activation and repression, long-term gene silencing and hierarchies of epigenetic modifications (including alterations at single genomic loci as well as those that are genome-wide and associated with nuclear reorganization108) that occur in response to discreet cell autonomous, cell-cell and environmental signals are therefore responsible for mediating neural lineage elaboration. Individual and cooperative REST and CoREST complexes with distinct macromolecular assemblies are likely deployed in a genome-wide and cell type-specific manner; are key nodes in the regulatory circuitry mediating diverse transcriptional, post-transcriptional, as well as local- and long-distance epigenetic processes; and are highly integrated with neural developmental signaling and effector pathways. Thus, REST and CoREST appear to define the overall transcriptional and epigenetic status of individual neural cell types and modulate the robustness, plasticity and responsivity of these states in the context of neural cell fate decisions (Fig. 1). Better characterizing how REST and CoREST promote transcriptional and epigenetic programming of neural cell identity and function has important implications for understanding neural development and disease states associated with CoREST or REST (e.g., neurodevelopmental50,93,97,98,109 and neurodegenerative63,85,110–112 disorders, epilepsy,44,113 stroke114,115 and cancer19–28) and for promoting more effective neural regeneration.
Acknowledgements
We regret that space constraints have prevented the citation of many relevant and important references. M.F.M. is supported by grants from the National Institutes of Health (NS38902, MH66290, NS071571), as well as by the Roslyn and Leslie Goldstein, the Mildred and Bernard H. Kayden, the F.M. Kirby and the Alpern Family Foundations.
Abbreviations
- bps
base pairs
- BMP
bone morphogenic protein
- CREB
cAMP response element-binding protein
- cRE1
canonical RE1 motif
- CoREST
corepressor for element-1-silencing transcription factor
- dsNRSE
double-stranded neuron-restrictive silencing element
- ESC
embryonic stem cell
- FGF
fibroblast growth factor
- HLH
helix-loop-helix
- HD
homeodomain
- lncRNA
long non-coding RNA
- miRNA
microRNA
- ncRE1
non-canonical RE1 motif
- NRSF
neuron-restrictive silencer factor
- NSC
neural stem cell
- OL
oligodendrocyte
- PcG
polycomb group
- RA
retinoic acid
- RE1
repressor element-1
- REST
RE1 silencing transcription factor
- SHH
sonic hedgehog
Footnotes
Previously published online: www.landesbioscience.com/journals/cc/article/13973
References
- 1.Chong JA, Tapia-Ramirez J, Kim S, Toledo-Aral JJ, Zheng Y, Boutros MC, et al. REST: A mammalian silencer protein that restricts sodium channel gene expression to neurons. Cell. 1995;80:949–957. doi: 10.1016/0092-8674(95)90298-8. [DOI] [PubMed] [Google Scholar]
- 2.Schoenherr CJ, Anderson DJ. The neuron-restrictive silencer factor (NRSF): A coordinate repressor of multiple neuron-specific gene. Science. 1995;267:1360–1363. doi: 10.1126/science.7871435. [DOI] [PubMed] [Google Scholar]
- 3.Schoenherr CJ, Paquette AJ, Anderson DJ. Identification of potential target genes for the neuronrestrictive silencer factor. Proc Natl Acad Sci USA. 1996;93:9881–9886. doi: 10.1073/pnas.93.18.9881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Andres ME, Burger C, Peral-Rubio MJ, Battaglioli E, Anderson ME, Grimes J, et al. CoREST: A functional corepressor required for regulation of neural-specific gene expression. Proc Natl Acad Sci USA. 1999;96:9873–9878. doi: 10.1073/pnas.96.17.9873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shimojo M. Characterization of the nuclear targeting signal of REST/NRSF. Neurosci Lett. 2006;398:161–166. doi: 10.1016/j.neulet.2005.12.080. [DOI] [PubMed] [Google Scholar]
- 6.Kuwabara T, Hsieh J, Nakashima K, Taira K, Gage FH. A small modulatory dsRNA specifies the fate of adult neural stem cells. Cell. 2004;116:779–793. doi: 10.1016/s0092-8674(04)00248-x. [DOI] [PubMed] [Google Scholar]
- 7.Kuwabara T, Hsieh J, Nakashima K, Warashina M, Taira K, Gage FH. The NRSE smRNA specifies the fate of adult hippocampal neural stem cells. Nucleic Acids Symp Ser (Oxf) 2005:87–8. doi: 10.1093/nass/49.1.87. [DOI] [PubMed] [Google Scholar]
- 8.Zheng D, Zhao K, Mehler MF. Profiling RE1/REST-mediated histone modifications in the human genome. Genome Biol. 2009;10:9. doi: 10.1186/gb-2009-10-1-r9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lunyak VV, Burgess R, Prefontaine GG, Nelson C, Sze SH, Chenoweth J. Corepressor-dependent silencing of chromosomal regions encoding neuronal genes. Science. 2002;298:1747–1752. doi: 10.1126/science.1076469. [DOI] [PubMed] [Google Scholar]
- 10.Otto SJ, McCorkle SR, Hover J, Conaco C, Han JJ, Impey S, et al. A new binding motif for the transcriptional repressor REST uncovers large gene networks devoted to neuronal functions. J Neurosci. 2007;27:6729–6739. doi: 10.1523/JNEUROSCI.0091-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Qureshi IA, Mehler MF. Regulation of non-coding RNA networks in the nervous system—what's the REST of the story? Neurosci Lett. 2009;466:73–80. doi: 10.1016/j.neulet.2009.07.093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dovey OM, Foster CT, Cowley SM. Histone deacetylase 1 (HDAC1), but not HDAC2, controls embryonic stem cell differentiation. Proc Natl Acad Sci USA. 2010;107:8242–8247. doi: 10.1073/pnas.1000478107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Foster CT, Dovey OM, Lezina L, Luo JL, Gant TW, Barlev N, et al. Lysine-Specific Demethylase 1 Regulates the Embryonic Transcriptome and CoREST Stability. Mol Cell Biol. 2010;30:4851–4863. doi: 10.1128/MCB.00521-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jorgensen HF, Terry A, Beretta C, Pereira CF, Leleu M, Chen ZF, et al. REST selectively represses a subset of RE1-containing neuronal genes in mouse embryonic stem cells. Development. 2009;136:715–721. doi: 10.1242/dev.028548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johnson R, Teh CH, Kunarso G, Wong KY, Srinivasan G, Cooper ML, et al. REST regulates distinct transcriptional networks in embryonic and neural stem cells. PLoS Biol. 2008;6:256. doi: 10.1371/journal.pbio.0060256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Canzonetta C, Mulligan C, Deutsch S, Ruf S, O'Doherty A, Lyle R, et al. DYRK1A-dosage imbalance perturbs NRSF/REST levels, deregulating pluripotency and embryonic stem cell fate in Down syndrome. Am J Hum Genet. 2008;83:388–400. doi: 10.1016/j.ajhg.2008.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Singh SK, Kagalwala MN, Parker-Thornburg J, Adams H, Majumder S. REST maintains self-renewal and pluripotency of embryonic stem cells. Nature. 2008;453:223–227. doi: 10.1038/nature06863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yamada Y, Aoki H, Kunisada T, Hara A. Rest promotes the early differentiation of mouse ESCs but is not required for their maintenance. Cell Stem Cell. 2010;6:10–15. doi: 10.1016/j.stem.2009.12.003. [DOI] [PubMed] [Google Scholar]
- 19.Westbrook TF, Hu G, Ang XL, Mulligan P, Pavlova NN, Liang A, et al. SCFbeta-TRCP controls oncogenic transformation and neural differentiation through REST degradation. Nature. 2008;452:370–374. doi: 10.1038/nature06780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mulligan P, Westbrook TF, Ottinger M, Pavlova N, Chang B, Macia E, et al. CDYL bridges REST and histone methyltransferases for gene repression and suppression of cellular transformation. Mol Cell. 2008;32:718–726. doi: 10.1016/j.molcel.2008.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guardavaccaro D, Frescas D, Dorrello NV, Peschiaroli A, Multani AS, Cardozo T, et al. Control of chromosome stability by the beta-TrCP-REST-Mad2 axis. Nature. 2008;452:365–369. doi: 10.1038/nature06641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Su X, Gopalakrishnan V, Stearns D, Aldape K, Lang FF, Fuller G, et al. Abnormal expression of REST/NRSF and Myc in neural stem/progenitor cells causes cerebellar tumors by blocking neuronal differentiation. Mol Cell Biol. 2006;26:1666–1678. doi: 10.1128/MCB.26.5.1666-1678.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Majumder S. REST in good times and bad: roles in tumor suppressor and oncogenic activities. Cell Cycle. 2006;5:1929–1935. doi: 10.4161/cc.5.17.2982. [DOI] [PubMed] [Google Scholar]
- 24.Blom T, Tynninen O, Puputti M, Halonen M, Paetau A, Haapasalo H, et al. Molecular genetic analysis of the REST/NRSF gene in nervous system tumors. Acta Neuropathol. 2006;112:483–490. doi: 10.1007/s00401-006-0102-8. [DOI] [PubMed] [Google Scholar]
- 25.Westbrook TF, Martin ES, Schlabach MR, Leng Y, Liang AC, Feng B, et al. A genetic screen for candidate tumor suppressors identifies REST. Cell. 2005;121:837–848. doi: 10.1016/j.cell.2005.03.033. [DOI] [PubMed] [Google Scholar]
- 26.Fuller GN, Su X, Price RE, Cohen ZR, Lang FF, Sawaya R, et al. Many human medulloblastoma tumors overexpress repressor element-1 silencing transcription (REST)/neuron-restrictive silencer factor, which can be functionally countered by REST-VP16. Mol Cancer Ther. 2005;4:343–349. doi: 10.1158/1535-7163.MCT-04-0228. [DOI] [PubMed] [Google Scholar]
- 27.Coulson JM. Transcriptional regulation: cancer, neurons and the REST. Curr Biol. 2005;15:665–668. doi: 10.1016/j.cub.2005.08.032. [DOI] [PubMed] [Google Scholar]
- 28.Lawinger P, Venugopal R, Guo ZS, Immaneni A, Sengupta D, Lu W, et al. The neuronal repressor REST/NRSF is an essential regulator in medulloblastoma cells. Nat Med. 2000;6:826–831. doi: 10.1038/77565. [DOI] [PubMed] [Google Scholar]
- 29.Guillemot F. Cell fate specification in the mammalian telencephalon. Prog Neurobiol. 2007;83:37–52. doi: 10.1016/j.pneurobio.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 30.Dessaud E, McMahon AP, Briscoe J. Pattern formation in the vertebrate neural tube: a sonic hedgehog morphogen-regulated transcriptional network. Development. 2008;135:2489, 2503. doi: 10.1242/dev.009324. [DOI] [PubMed] [Google Scholar]
- 31.Mehler MF. Mechanisms regulating lineage diversity during mammalian cerebral cortical neurogenesis and gliogenesis. Results Probl Cell Differ. 2002;39:27–52. doi: 10.1007/978-3-540-46006-0_2. [DOI] [PubMed] [Google Scholar]
- 32.Gokhan S, Marin-Husstege M, Yung SY, Fontanez D, Casaccia-Bonnefil P, Mehler MF. Combinatorial profiles of oligodendrocyte-selective classes of transcriptional regulators differentially modulate myelin basic protein gene expression. J Neurosci. 2005;25:8311–8321. doi: 10.1523/JNEUROSCI.1850-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hirabayashi Y, Gotoh Y. Epigenetic control of neural precursor cell fate during development. Nat Rev Neurosci. 2010;11:377–388. doi: 10.1038/nrn2810. [DOI] [PubMed] [Google Scholar]
- 34.Li X, Jin P. Roles of small regulatory RNAs in determining neuronal identity. Nat Rev Neurosci. 2010;11:329–338. doi: 10.1038/nrn2739. [DOI] [PubMed] [Google Scholar]
- 35.Qureshi IA, Mattick JS, Mehler MF. Long non-coding RNAs in nervous system function and disease. Brain Res. 2010;1338:20–35. doi: 10.1016/j.brainres.2010.03.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Olguin P, Oteiza P, Gamboa E, Gomez-Skarmeta JL, Kukuljan M. RE-1 silencer of transcription/neural restrictive silencer factor modulates ectodermal patterning during Xenopus development. J Neurosci. 2006;26:2820–2829. doi: 10.1523/JNEUROSCI.5037-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Paquette AJ, Perez SE, Anderson DJ. Constitutive expression of the neuron-restrictive silencer factor (NRSF)/REST in differentiating neurons disrupts neuronal gene expression and causes axon pathfinding errors in vivo. Proc Natl Acad Sci USA. 2000;97:12318–12323. doi: 10.1073/pnas.97.22.12318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nishihara S, Tsuda L, Ogura T. The canonical Wnt pathway directly regulates NRSF/REST expression in chick spinal cord. Biochem Biophys Res Commun. 2003;311:55–63. doi: 10.1016/j.bbrc.2003.09.158. [DOI] [PubMed] [Google Scholar]
- 39.Kohyama J, Sanosaka T, Tokunaga A, Takatsuka E, Tsujimura K, Okano H, et al. BMP-induced REST regulates the establishment and maintenance of astrocytic identity. J Cell Biol. 2010;189:159–170. doi: 10.1083/jcb.200908048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Grimes JA, Nielsen SJ, Battaglioli E, Miska EA, Speh JC, Berry DL, et al. The co-repressor mSin3A is a functional component of the REST-CoREST repressor complex. J Biol Chem. 2000;275:9461–9467. doi: 10.1074/jbc.275.13.9461. [DOI] [PubMed] [Google Scholar]
- 41.Lakowski B, Roelens I, Jacob S. CoREST-like complexes regulate chromatin modification and neuronal gene expression. J Mol Neurosci. 2006;29:227–239. doi: 10.1385/JMN:29:3:227. [DOI] [PubMed] [Google Scholar]
- 42.de la Calle-Mustienes E, Modolell J, Gomez-Skarmeta JL. The Xiro-repressed gene CoREST is expressed in Xenopus neural territories. Mech Dev. 2002;110:209–211. doi: 10.1016/s0925-4773(01)00565-2. [DOI] [PubMed] [Google Scholar]
- 43.Tontsch S, Zach O, Bauer HC. Identification and localization of M-CoREST (1A13), a mouse homologue of the human transcriptional co-repressor CoREST, in the developing mouse CNS. Mech Dev. 2001;108:165–169. doi: 10.1016/s0925-4773(01)00477-4. [DOI] [PubMed] [Google Scholar]
- 44.Garriga-Canut M, Schoenike B, Qazi R, Bergendahl K, Daley TJ, Pfender RM, et al. 2-Deoxy-D-glucose reduces epilepsy progression by NRSF-CtBP-dependent metabolic regulation of chromatin structure. Nat Neurosci. 2006;9:1382–1387. doi: 10.1038/nn1791. [DOI] [PubMed] [Google Scholar]
- 45.Visvanathan J, Lee S, Lee B, Lee JW, Lee SK. The microRNA miR-124 antagonizes the anti-neural REST/SCP1 pathway during embryonic CNS development. Genes Dev. 2007;21:744–749. doi: 10.1101/gad.1519107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kim CS, Choi HS, Hwang CK, Song KY, Lee BK, Law PY, et al. Evidence of the neuron-restrictive silencer factor (NRSF) interaction with Sp3 and its synergic repression to the mu opioid receptor (MOR) gene. Nucleic Acids Res. 2006;34:6392–6403. doi: 10.1093/nar/gkl724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ding N, Tomomori-Sato C, Sato S, Conaway RC, Conaway JW, Boyer TG. MED19 and MED26 are synergistic functional targets of the RE1 silencing transcription factor in epigenetic silencing of neuronal gene expression. J Biol Chem. 2009;284:2648–2656. doi: 10.1074/jbc.M806514200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Taatjes DJ. The human Mediator complex: A versatile, genome-wide regulator of transcription. Trends Biochem Sci. 2010;35:315–322. doi: 10.1016/j.tibs.2010.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kagey MH, Newman JJ, Bilodeau S, Zhan Y, Orlando DA, van Berkum NL, et al. Mediator and cohesin connect gene expression and chromatin architecture. Nature. 2010;467:430–435. doi: 10.1038/nature09380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ding N, Zhou H, Esteve PO, Chin HG, Kim S, Xu X, et al. Mediator links epigenetic silencing of neuronal gene expression with x-linked mental retardation. Mol Cell. 2008;31:347–359. doi: 10.1016/j.molcel.2008.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang X, Yang N, Uno E, Roeder RG, Guo S. A subunit of the mediator complex regulates vertebrate neuronal development. Proc Natl Acad Sci USA. 2006;103:17284–17289. doi: 10.1073/pnas.0605414103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rocha PP, Bleiss W, Schrewe H. Mosaic expression of Med12 in female mice leads to exencephaly, spina bifida and craniorachischisis. Birt Defects Res A Clin Mol Teratol. 2010;88:626–632. doi: 10.1002/bdra.20693. [DOI] [PubMed] [Google Scholar]
- 53.Ballas N, Grunseich C, Lu DD, Speh JC, Mandel G. REST and its corepressors mediate plasticity of neuronal gene chromatin throughout neurogenesis. Cell. 2005;121:645–657. doi: 10.1016/j.cell.2005.03.013. [DOI] [PubMed] [Google Scholar]
- 54.Tabuchi A, Yamada T, Sasagawa S, Naruse Y, Mori N, Tsuda M. REST4-mediated modulation of REST/NRSF-silencing function during BDNF gene promoter activation. Biochem Biophys Res Commun. 2002;290:415–420. doi: 10.1006/bbrc.2001.6194. [DOI] [PubMed] [Google Scholar]
- 55.Lee JH, Shimojo M, Chai YG, Hersh LB. Studies on the interaction of REST4 with the cholinergic repressor element-1/neuron restrictive silencer element. Brain Res Mol Brain Res. 2000;80:88–98. doi: 10.1016/s0169-328x(00)00129-7. [DOI] [PubMed] [Google Scholar]
- 56.Lee JH, Chai YG, Hersh LB. Expression patterns of mouse repressor element-1 silencing transcription factor 4 (REST4) and its possible function in neuroblastoma. J Mol Neurosci. 2000;15:205–214. doi: 10.1385/JMN:15:3:205. [DOI] [PubMed] [Google Scholar]
- 57.Shimojo M, Paquette AJ, Anderson DJ, Hersh LB. Protein kinase A regulates cholinergic gene expression in PC12 cells: REST4 silences the silencing activity of neuron-restrictive silencer factor/REST. Mol Cell Biol. 1999;19:6788–6795. doi: 10.1128/mcb.19.10.6788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Johnson DS, Mortazavi A, Myers RM, Wold B. Genome-wide mapping of in vivo protein-DNA interactions. Science. 2007;316:1497–1502. doi: 10.1126/science.1141319. [DOI] [PubMed] [Google Scholar]
- 59.Johnson R, Gamblin RJ, Ooi L, Bruce AW, Donaldson IJ, Westhead DR, et al. Identification of the REST regulon reveals extensive transposable element-mediated binding site duplication. Nucleic Acids Res. 2006;34:3862–3877. doi: 10.1093/nar/gkl525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bruce AW, Lopez-Contreras AJ, Flicek P, Down TA, Dhami P, Dillon SC, et al. Functional diversity for REST (NRSF) is defined by in vivo binding affinity hierarchies at the DNA sequence level. Genome Res. 2009;19:994–1005. doi: 10.1101/gr.089086.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mattick JS, Amaral PP, Dinger ME, Mercer TR, Mehler MF. RNA regulation of epigenetic processes. Bioessays. 2009;31:51–59. doi: 10.1002/bies.080099. [DOI] [PubMed] [Google Scholar]
- 62.Wu J, Xie X. Comparative sequence analysis reveals an intricate network among REST, CREB and miRNA in mediating neuronal gene expression. Genome Biol. 2006;7:85. doi: 10.1186/gb-2006-7-9-r85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Packer AN, Xing Y, Harper SQ, Jones L, Davidson BL. The bifunctional microRNA miR-9/miR-9* regulates REST and CoREST and is downregulated in Huntington' disease. J Neurosci. 2008;28:14341–14346. doi: 10.1523/JNEUROSCI.2390-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Conaco C, Otto S, Han JJ, Mandel G. Reciprocal actions of REST and a microRNA promote neuronal identity. Proc Natl Acad Sci USA. 2006;103:2422–2427. doi: 10.1073/pnas.0511041103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Laneve P, Gioia U, Andriotto A, Moretti F, Bozzoni I, Caffarelli E. A minicircuitry involving REST and CREB controls miR-9-2 expression during human neuronal differentiation. Nucleic Acids Res. 2010 doi: 10.1093/nar/gkq604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Johnson R, Teh CH, Jia H, Vanisri RR, Pandey T, Lu ZH, et al. Regulation of neural macroRNAs by the transcriptional repressor REST. RNA. 2009;15:85–96. doi: 10.1261/rna.1127009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, et al. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689–693. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci USA. 2009;106:11667–11672. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mercer TR, Dinger ME, Sunkin SM, Mehler MF, Mattick JS. Specific expression of long noncoding RNAs in the mouse brain. Proc Natl Acad Sci USA. 2008;105:716–721. doi: 10.1073/pnas.0706729105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ponjavic J, Oliver PL, Lunter G, Ponting CP. Genomic and transcriptional co-localization of protein-coding and long non-coding RNA pairs in the developing brain. PLoS Genet. 2009;5:1000617. doi: 10.1371/journal.pgen.1000617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Abrajano JJ, Qureshi IA, Gokhan S, Molero AE, Zheng D, Bergman A, et al. Corepressor for element-1-silencing transcription factor preferentially mediates gene networks underlying neural stem cell fate decisions. Proc Natl Acad Sci USA. 2010;107:16685–16690. doi: 10.1073/pnas.0906917107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Abrajano JJ, Qureshi IA, Gokhan S, Zheng D, Bergman A, Mehler MF. REST and CoREST modulate neuronal subtype specification, maturation and maintenance. PLoS ONE. 2009;4:7936. doi: 10.1371/journal.pone.0007936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Abrajano JJ, Qureshi IA, Gokhan S, Zheng D, Bergman A, Mehler MF. Differential deployment of REST and CoREST promotes glial subtype specification and oligodendrocyte lineage maturation. PLoS ONE. 2009;4:7665. doi: 10.1371/journal.pone.0007665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gates KP, Mentzer L, Karlstrom RO, Sirotkin HI. The transcriptional repressor REST/NRSF modulates hedgehog signaling. Dev Biol. 2010;340:293–305. doi: 10.1016/j.ydbio.2010.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Jorgensen HF, Fisher AG. Can controversies be put to REST? Nature. 2010;467:3–4. doi: 10.1038/nature09305. [DOI] [PubMed] [Google Scholar]
- 76.Jorgensen HF, Chen ZF, Merkenschlager M, Fisher AG. Is REST required for ESC pluripotency? Nature. 2009;457:4–5. doi: 10.1038/nature07783. [DOI] [PubMed] [Google Scholar]
- 77.Buckley NJ, Johnson R, Sun YM, Stanton LW. Is REST a regulator of pluripotency? Nature. 2009;457:5–6. doi: 10.1038/nature07784. [DOI] [PubMed] [Google Scholar]
- 78.Martinez NJ, Gregory RI. MicroRNA gene regulatory pathways in the establishment and maintenance of ESC identity. Cell Stem Cell. 2010;7:31–35. doi: 10.1016/j.stem.2010.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Dinger ME, Amaral PP, Mercer TR, Pang KC, Bruce SJ, Gardiner BB, et al. Long noncoding RNAs in mouse embryonic stem cell pluripotency and differentiation. Genome Res. 2008;18:1433–1445. doi: 10.1101/gr.078378.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sheik Mohamed J, Gaughwin PM, Lim B, Robson P, Lipovich L. Conserved long noncoding RNAs transcriptionally regulated by Oct4 and Nanog modulate pluripotency in mouse embryonic stem cells. RNA. 2010;16:324–337. doi: 10.1261/rna.1441510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sun YM, Greenway DJ, Johnson R, Street M, Belyaev ND, Deuchars J, et al. Distinct profiles of REST interactions with its target genes at different stages of neuronal development. Mol Biol Cell. 2005;16:5630–5638. doi: 10.1091/mbc.E05-07-0687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sun YM, Cooper M, Finch S, Lin HH, Chen ZF, Williams BP, et al. Rest-mediated regulation of extracellular matrix is crucial for neural development. PLoS ONE. 2008;3:3656. doi: 10.1371/journal.pone.0003656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Muller FJ, Laurent LC, Kostka D, Ulitsky I, Williams R, Lu C, et al. Regulatory networks define phenotypic classes of human stem cell lines. Nature. 2008;455:401–405. doi: 10.1038/nature07213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Buckley NJ, Johnson R, Zuccato C, Bithell A, Cattaneo E. The role of REST in transcriptional and epigenetic dysregulation in Huntington's disease. Neurobiol Dis. 2010;39:28–39. doi: 10.1016/j.nbd.2010.02.003. [DOI] [PubMed] [Google Scholar]
- 86.Molero AE, Gokhan S, Gonzalez S, Feig JL, Alexandre LC, Mehler MF. Impairment of developmental stem cell-mediated striatal neurogenesis and pluripotency genes in a knock-in model of Huntington's disease. Proc Natl Acad Sci USA. 2009;106:21900–21905. doi: 10.1073/pnas.0912171106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yokoyama A, Takezawa S, Schule R, Kitagawa H, Kato S. Transrepressive function of TLX requires the histone demethylase LSD1. Mol Cell Biol. 2008;28:3995–4003. doi: 10.1128/MCB.02030-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Qu Q, Sun G, Li W, Yang S, Ye P, Zhao C, et al. Orphan nuclear receptor TLX activates Wnt/betacatenin signalling to stimulate neural stem cell proliferation and self-renewal. Nat Cell Biol. 2010;12:31–40. doi: 10.1038/ncb2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zhao C, Sun G, Li S, Shi Y. A feedback regulatory loop involving microRNA-9 and nuclear receptor TLX in neural stem cell fate determination. Nat Struct Mol Biol. 2009;16:365–371. doi: 10.1038/nsmb.1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wang Y, Guan Y, Wang F, Huang A, Wang S, Zhang YA. Bmi-1 regulates self-renewal, proliferation and senescence of human fetal neural stem cells in vitro. Neurosci Lett. 2010;476:74–78. doi: 10.1016/j.neulet.2010.04.006. [DOI] [PubMed] [Google Scholar]
- 91.Pereira JD, Sansom SN, Smith J, Dobenecker MW, Tarakhovsky A, Livesey FJ. Ezh2, the histone methyltransferase of PRC2, regulates the balance between self-renewal and differentiation in the cerebral cortex. Proc Natl Acad Sci USA. 2010;107:15957–15962. doi: 10.1073/pnas.1002530107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ku M, Koche RP, Rheinbay E, Mendenhall EM, Endoh M, Mikkelsen TS, et al. Genomewide analysis of PRC1 and PRC2 occupancy identifies two classes of bivalent domains. PLoS Genet. 2008;4:1000242. doi: 10.1371/journal.pgen.1000242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Tahiliani M, Mei P, Fang R, Leonor T, Rutenberg M, Shimizu F, et al. The histone H3K4 demethylase SMCX links REST target genes to X-linked mental retardation. Nature. 2007;447:601–605. doi: 10.1038/nature05823. [DOI] [PubMed] [Google Scholar]
- 94.Hobert O, Carrera I, Stefanakis N. The molecular and gene regulatory signature of a neuron. Trends Neurosci. 2010;33:435–445. doi: 10.1016/j.tins.2010.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Chen L, Li Z, Zwolinska AK, Smith MA, Cross B, Koomen J, et al. MDM2 recruitment of lysine methyltransferases regulates p53 transcriptional output. EMBO J. 2010;29:2538–2552. doi: 10.1038/emboj.2010.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Schaefer A, Sampath SC, Intrator A, Min A, Gertler TS, Surmeier DJ, et al. Control of cognition and adaptive behavior by the GLP/G9a epigenetic suppressor complex. Neuron. 2009;64:678–691. doi: 10.1016/j.neuron.2009.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kleefstra T, van Zelst-Stams WA, Nillesen WM, Cormier-Daire V, Houge G, Foulds N, et al. Further clinical and molecular delineation of the 9q subtelomeric deletion syndrome supports a major contribution of EHMT1 haploinsufficiency to the core phenotype. J Med Genet. 2009;46:598–606. doi: 10.1136/jmg.2008.062950. [DOI] [PubMed] [Google Scholar]
- 98.Kleefstra T, Brunner HG, Amiel J, Oudakker AR, Nillesen WM, Magee A, et al. Loss-of-function mutations in euchromatin histone methyl transferase 1 (EHMT1) cause the 9q34 subtelomeric deletion syndrome. Am J Hum Genet. 2006;79:370–377. doi: 10.1086/505693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Northcott PA, Nakahara Y, Wu X, Feuk L, Ellison DW, Croul S, et al. Multiple recurrent genetic events converge on control of histone lysine methylation in medulloblastoma. Nat Genet. 2009;41:465–472. doi: 10.1038/ng.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Tachibana M, Nozaki M, Takeda N, Shinkai Y. Functional dynamics of H3K9 methylation during meiotic prophase progression. EMBO J. 2007;26:3346–3359. doi: 10.1038/sj.emboj.7601767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Tan YP, Li S, Jiang XJ, Loh W, Foo YK, Loh CB, et al. Regulation of protocadherin gene expression by multiple neuron-restrictive silencer elements scattered in the gene cluster. Nucleic Acids Res. 2010;38:4985–4997. doi: 10.1093/nar/gkq246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Mercer TR, Qureshi IA, Gokhan S, Dinger ME, Li G, Mattick JS, et al. Long noncoding RNAs in neuronalglial fate specification and oligodendrocyte lineage maturation. BMC Neurosci. 2010;11:14. doi: 10.1186/1471-2202-11-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Blackshaw S, Harpavat S, Trimarchi J, Cai L, Huang H, Kuo WP, et al. Genomic analysis of mouse retinal development. PLoS Biol. 2004;2:247. doi: 10.1371/journal.pbio.0020247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Makeyev EV, Zhang J, Carrasco MA, Maniatis T. The MicroRNA miR-124 promotes neuronal differentiation by triggering brain-specific alternative pre-mRNA splicing. Mol Cell. 2007;27:435–448. doi: 10.1016/j.molcel.2007.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Valadkhan S, Nilsen TW. Reprogramming of the noncoding transcriptome during brain development. J Biol. 2010;9:5. doi: 10.1186/jbiol197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Lau P, Verrier JD, Nielsen JA, Johnson KR, Notterpek L, Hudson LD. Identification of dynamically regulated microRNA and mRNA networks in developing oligodendrocytes. J Neurosci. 2008;28:11720–11730. doi: 10.1523/JNEUROSCI.1932-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Lunyak VV, Rosenfeld MG. Epigenetic regulation of stem cell fate. Hum Mol Genet. 2008;17:28–36. doi: 10.1093/hmg/ddn149. [DOI] [PubMed] [Google Scholar]
- 108.Qureshi IA, Mehler MF. Impact of nuclear organization and dynamics on epigenetic regulation in the central nervous system: implications for neurological disease states. Ann NY Acad Sci. 2010;1204:20–37. doi: 10.1111/j.1749-6632.2010.05718.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Lepagnol-Bestel AM, Zvara A, Maussion G, Quignon F, Ngimbous B, Ramoz N, et al. DYRK1A interacts with the REST/NRSF-SWI/SNF chromatin remodelling complex to deregulate gene clusters involved in the neuronal phenotypic traits of Down syndrome. Hum Mol Genet. 2009;18:1405–1414. doi: 10.1093/hmg/ddp047. [DOI] [PubMed] [Google Scholar]
- 110.Johnson R, Zuccato C, Belyaev ND, Guest DJ, Cattaneo E, Buckley NJ. A microRNA-based gene dysregulation pathway in Huntington's disease. Neurobiol Dis. 2008;29:438–445. doi: 10.1016/j.nbd.2007.11.001. [DOI] [PubMed] [Google Scholar]
- 111.Zuccato C, Belyaev N, Conforti P, Ooi L, Tartari M, Papadimou E, et al. Widespread disruption of repressor element-1 silencing transcription factor/neuronrestrictive silencer factor occupancy at its target genes in Huntington's disease. J Neurosci. 2007;27:6972–6983. doi: 10.1523/JNEUROSCI.4278-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Saijo K, Winner B, Carson CT, Collier JG, Boyer L, Rosenfeld MG, et al. A Nurr1/CoREST pathway in microglia and astrocytes protects dopaminergic neurons from inflammation-induced death. Cell. 2009;137:47–59. doi: 10.1016/j.cell.2009.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Bassuk AG, Wallace RH, Buhr A, Buller AR, Afawi Z, Shimojo M, et al. A homozygous mutation in human PRICKLE1 causes an autosomal-recessive progressive myoclonus epilepsy-ataxia syndrome. Am J Hum Genet. 2008;83:572–581. doi: 10.1016/j.ajhg.2008.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Formisano L, Noh KM, Miyawaki T, Mashiko T, Bennett MV, Zukin RS. Ischemic insults promote epigenetic reprogramming of mu opioid receptor expression in hippocampal neurons. Proc Natl Acad Sci USA. 2007;104:4170–4175. doi: 10.1073/pnas.0611704104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Calderone A, Jover T, Noh KM, Tanaka H, Yokota H, Lin Y, et al. Ischemic insults derepress the gene silencer REST in neurons destined to die. J Neurosci. 2003;23:2112–2121. doi: 10.1523/JNEUROSCI.23-06-02112.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]