Abstract
The cyclin-dependent kinase inhibitor p21cip/CDKN1A is induced to promote growth arrest in response to a variety of stimuli in normal cells and loss of correct regulation of this gene is frequently observed in cancer. In particular, the upregulation of CDKN1A by p53 is considered to be a central mechanism of tumor suppression. Other transcription factors with tumor suppressor activity can also regulate CDKN1A, including the developmentally regulated factor, TFAP2A. Here we identify a novel AP-2 binding site within the proximal promoter of the CDKN1A gene and show this is required for optimal, p53-independent expression of p21cip/CDKN1A. We further describe a non-tumorgenic breast epithelial cell line model to study the role of endogenous TFAP2A and p53 in the control of drug-induced p21cip expression using ChIP. Maximal expression of CDKN1A requires TFAP2A which binds to two regions of the promoter: the proximal region where the AP-2 site lies and upstream near the major p53 binding site. The pattern of binding alters with time post-induction, with the proximal, p53-independent site becoming more important at later stages of p21cip induction. This pattern of promoter interaction by TFAP2A is distinct from that seen for the TFAP2C family member which represses CDKN1A expression.
Key words: AP-2, p21cip, ChIP assay, p53
Introduction
AP-2α-ε proteins represent a family of developmentally regulated transcription factors, each encoded by a separate gene. The region of greatest homology overlaps the C-terminal DNA binding and helix-span-helix dimerisation domains; the N-terminal sequences required for transcriptional activation being less well conserved. These factors have been shown to bind to palindromic GC-rich DNA recognition sequences as either homo- or heterodimers1 and can act as either transcriptional activators or repressors depending on the promoter context (reviewed in ref. 2). The function of the AP-2α, β and γ (TFAP2A-C) family members during embryogenesis has been examined by knockout studies in a variety of model organisms and these genes have also been linked to human birth defects affecting the heart, eyes, limbs and face. TFAP2A in particular has been shown to be critical for many aspects of embryonic patterning in structures that undergo complex changes in morphology, including the skin, neural tube and neural crest (reviewed in ref. 2).
Although, quite widely expressed during development, the expression of this transcription factor family in adult tissues is largely confined to keratinocytes and germ cells. However, there is an extensive literature documenting the deregulated expression of AP-2 factors in common solid tumors, particularly in melanoma and breast cancer. In the majority of these studies, expression of TFAP2A has been associated with a favorable outcome.3,4 Expression profiling studies in breast cancer5 have shown that TFAP2A mRNA expression correlated inversely with tumor grade (p < 0.01). This agreed with an immunohistochemical study which reported reduced nuclear TFAP2A staining associated with more aggressive, high-grade breast cancers6 and a further study which found that TFAP2A levels were highest in normal breast, declined in DCIS cases and were lowest in invasive cancers.7 In addition, there was a positive correlation between TFAP2A staining and expression of the universal cell cycle inhibitor p21cip/CDKN1A and an association with a lower rate of proliferation. These studies have led to the suggestion that TFAP2A may have a tumor suppressive role, consistent with the observation that its expression level is reduced with advancing stages of breast cancer and also with its suggested role in other cancer types (reviewed in ref. 3). Increased methylation over the first exon of the gene has been proposed as the mechanism behind the progressive loss of TFAP2A expression in invasive breast cancer.8 These findings are supported by observations in transgenic mice overexpressing TFAP2A in the mammary gland where transgenic tissues were shown to undergo significantly reduced proliferation compared to controls.9
The in vivo studies have been complemented by parallel in vitro examination of TFAP2A activity on its direct transcriptional targets which, in breast cancer, include ERBB2, oestrogen receptor-α (ESR1) and CDKN1A. In particular, target genes involved in induction of growth arrest and apoptosis have been suggested to mediate the proposed tumor suppressor role of TFAP2A. Initial work by Zeng and colleagues showed that exogenous TFAP2A expression in hepatoblastoma and colon carcinoma cells inhibited their growth and this was accompanied by activation of p21cip/CDKN1A expression. TFAP2A was demonstrated to activate transcription from a CDKN1A reporter construct and this activity depended on a single AP-2 binding site mapped within the proximal promoter region.10 Further support for a direct role for TFAP2A in regulating CDKN1A has come from adenoviral overexpression causing increased expression11 while use of TFAP2A siRNA lead to repression of CDKN1A,12 in a variety of tumor-derived lines.
CDKN1A was originally identified as a transcriptional target of TP53 and a key mediator of p53-dependent cell cycle arrest13 via two TP53 binding sites centerd at −2,250 and −1,344 in the promoter.14 Intriguingly, it has been suggested that the ability of TFAP2A to activate CDKN1A transcription is dependant on a direct interaction with p53 at the more distal binding site.15,16 Partial support for these data has come from an independent study looking at CDKN1A induction following adenovirus mediated overexpression of TFAP2A in derivatives of the HCT116 colon carcinoma line. The degree of p21cip induction upon TFAP2A overexpression in HCT116 p53-/- cells was significantly reduced compared to that seen with wild-type cells. However, p21cip was still induced by TFAP2A in HCT116 p53-/- cells and the extent of cell cycle arrest was equivalent to that observed for the wild-type line.11 These data suggest therefore that TFAP2A can activate p21cip independently of p53, although cooperation with p53 may enhance this activation. Given this controversy, we have re-examined the sites of interaction of TFAP2A at the CDKN1A promoter and investigated using ChIP how this interaction is modulated at the endogenous gene when p53 is induced in breast epithelial cells.
Results and Discussion
Identification of the bona fide AP-2α binding site within the CDKN1A promoter.
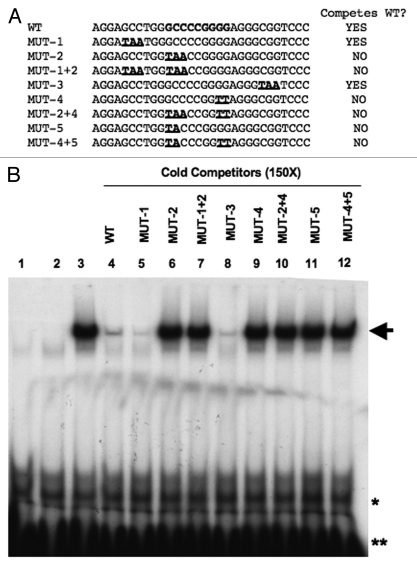
In order to define the AP-2 binding site within the proximal CDKN1A promoter, we performed electromobility shift assays (EMSA). DNA/protein complexes were formed by incubating in vitro translated (IVT) human TFAP2A protein with an oligonucleotide probe containing a well-characterized AP-2 binding site,1 and complex formation was subsequently challenged using a series of cold competitor oligonucleotides representing overlapping 30 bp regions of the CDKN1A promoter sequence from −161 to −31. Surprisingly, only the −121 to −92 sequence (oligonucleotide C) was able to compete for binding, whereas the −101 to −72 oligonucleotide, containing the previously identified AP-2 binding site at −75 to −83,10 did not compete (see Sup. Fig. 1, compare lanes 7 and 8). The reasons behind this discrepancy may be connected with the protein used for the EMSA assays: we have used in vitro translated native AP-2α in contrast to the bacterially expressed Gst-fusion protein employed previously.
The CDKN1A −121/−91 region is highly GC-rich and several potential AP-2 binding sites could be predicted using previously described AP-2 consensus sites.1,17 In order to identify unambiguously the sequence bound by TFAP2A, we performed competition assays using oligonucleotide C as the probe and challenged complex formation with a series of cold competitors carrying mutations in the half-sites of potential palindromic AP-2 binding sites. As illustrated in Figure 1, mutations within half-sites 1 and 3 did not impair competition but mutating half sites 2 or 4, either alone or in combination, completely abrogated competition activity. This defines the AP-2 binding site as the sequence GCC CCG GG at −111/−103 (Chromosome 6, position 36,646,380; UCSC Genome Browser, Feb 2009) within the CDKN1A promoter; the sequence is conserved across primates. Moreover, the site we identify here is closer to the optimal AP-2 consensus sites identified using PCR-based selection and missing phosphate contact analyses which established that GCC N3/4GGG and GCC N3–4GGC are the sequences most avidly bound by TFAP2A.1 Further EMSA assays, using either IVT proteins or nuclear extracts from AP-2 expressing cells, demonstrated that both TFAP2A and TFAP2C can bind equally well to this site (data not shown). This site also conforms with a recent genome-wide analysis of TFAP2C binding using the ChIP-seq technique.18
Figure 1.
TFAP2A specifically binds the GCC CCG GGG site at position −111/−103 in the CDKN1A promoter. (A) Sequence details of oligonucleotide C (−121 to −92) and variants used in the EMSA competition assay. Mutated bases are underlined and shown in bold. The new AP-2 binding site is indicated in bold in the wild-type sequence. Sequence numbering is from the start of transcription at +1 defined by CDKN1A RefSeq NM_078467. (B) Autoradiograph of EMSA assays performed using IVT TFAP2A protein (lanes 3 to 12), IVT luciferase protein (lane 2) or no protein control (lane 1), incubated with radiolabeled ds oligonucleotide C as the probe with or without the indicated “cold” competitors at 150x molar excess over the probe. The arrow indicates the position of the DNA/TFAP2A complex, while * and ** indicate non specific bands and free probe respectively.
TFAP2A activation of CDKN1A in the absence of p53.
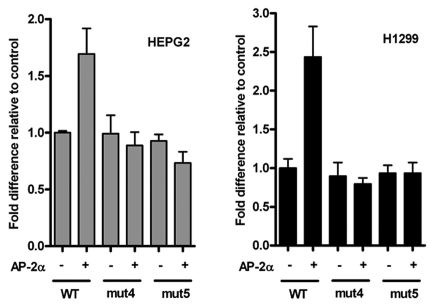
We next examined whether the newly defined AP-2 binding site is essential for TFAP2A regulation of the CDKN1A promoter. Using the HepG2 cell line that lacks endogenous AP-2 factors, we were able to show increased induction of a CDKN1A-luciferase reporter construct in the presence of an escalating dose of TFAP2A expression plasmid in agreement with previous reports10,19 (data not shown). Using the optimal dose of TFAP2A expression construct, we repeated the experiment comparing the activity of CDKN1A reporter constructs containing either the wild type promoter sequence (+8 to −2,250) or versions mutated in one half site of the AP-2 binding site shown to abrogate binding. The induction in reporter activity observed for the wild-type sequence was totally absent when constructs Mut4 or Mut5, containing 2 bp mutations in either half site, were used in the assay, strongly suggesting that the site we have identified at −111/−103 is the only functional AP-2 site in the most proximal 2.25 kb of the CDKN1A promoter (Fig. 2 and left). Moreover, a similar result was observed when the reporter assays were repeated using H1299 cells, which lack both AP-2 and p53 expression, thus demonstrating that the activity of TFAP2A at the proximal AP-2 site is not entirely dependent on the presence of p53 (Fig. 2 and right).
Figure 2.
TFAP2A-mediated activation of the CDKN1A promoter is inhibited by mutation of the −111/−103 AP-2 binding site. Site directed mutagenesis was used to introduce double point mutations into the CDKN1A reporter construct equivalent to oligonucleotides Mut4 or Mut5 (see Fig. 1). HepG2 and H1229 cells were seeded onto 24 well plates and transfected with wild-type or mutant CDKN1A luciferase reporters (150 ng) and pSV-βgalactosidase control vector (300 ng) in the presence of either 500 ng pcDNA3.1 vector or TFAP2A expression construct, as indicated. Cells were assayed 48 hours after transfection. Results (mean ± SD of triplicate wells for three independent experiments) are presented as relative luciferase activity, corrected for βgal activity, with activity in cells transfected with the wt reporter alone set at 1.
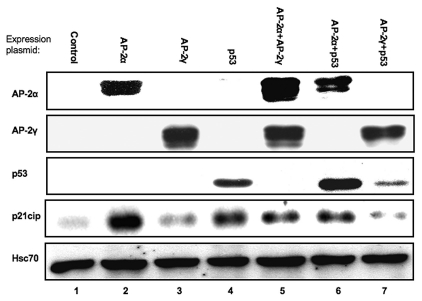
The p53-independent regulation of CDKN1A expression by TFAP2A was further confirmed by examining the induction of endogenous p21cip expression in H1299 cells transfected with expression constructs for AP-2 factors or wild-type p53. Robust induction of p21cip protein was observed 48 hours after exogenous expression of either p53 or TFAP2A (see Fig. 3, lanes 2 and 4) further demonstrating that TFAP2A is able to induce CDKN1A expression even in the absence of p53. Induction of p21cip expression under these conditions appeared to be maximal in the presence of either transcription factor, since expression was not enhanced by co-transfection of both expression plasmids (Fig. 3 and lane 6). In keeping with previous observations,19 expression of TFAP2C failed to induce p21cip and, when co-transfected, tended to reduce the activation potential of both TFAP2A and p53 (Fig. 3 and lanes 3, 5 and 7). Overall therefore, these findings argue against an absolute dependence on p53 by TFAP2A in upregulating p21cip expression15,16 and instead agree with observations in HeLa cells, which are functionally null for p53, where CDKN1A was also shown to be positively regulated by TFAP2A.12
Figure 3.
TFAP2A overexpression induces endogenous CDKN1A expression via a p53 independent pathway. H1229 (p53-/-) cells (which do not express AP-2 factors) were transiently transfected with either pcDNA3.1 (control), or expression vectors for TFAP2A, TFAP2C or TP53 either alone or in combination, as indicated. The total amount of plasmid DNA transfected was held constant using pcDNA3.1. Expression of exogenous (p53, AP-2α and AP-2γ) and endogenous (p21cip, Hsc70) proteins was determined 48 hours later by western blot as indicated; Hsc70 was used as a loading control. Blot shown is a representative example of three independent transfection experiments.
Silencing endogenous TFAP2A expression limits induction of CDKN1A expression.
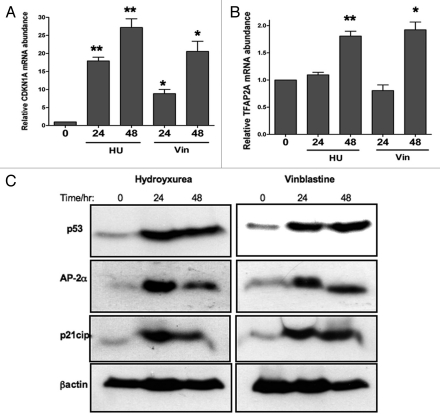
As a model system in which to study the control of expression of the endogenous CDKN1A gene by endogenously expressed transcription factors, we have used the immortalized mammary epithelial line, MCF10A, which expresses TFAP2A, low levels of TFAP2C and wild-type p53. Levels of TFAP2A in these cells can be efficiently knocked down for up to 96 hours at the mRNA and protein level using specific siRNA. Silencing of TFAP2A also led to a significant reduction in p21cip levels, confirming that the CDKN1A gene is regulated by AP-2α in this line (see Sup. Fig. 2). However, in normal cycling MCF10A cells, expression levels of both p53 and p21cip are low, therefore we have induced p53 stabilisation via two distinct pathways by treating cells with either the antimitotic drug, vinblastine or the antimetabolite, hydroxyurea (HU). Drug treatments were optimized to give good p53 induction with minimal cytotoxicity over 24–48 hours. As expected, upregulation of p53 protein was accompanied by robust induction of p21cip expression at both the mRNA and protein levels (see Fig. 4A and C). Of note, the block in CDKN1A transcriptional elongation observed in some colon carcinoma lines upon HU treatment20 was not apparent under our experimental conditions. However, we did also observe a significant induction of TFAP2A mRNA and protein in treated cells (Fig. 4B and C). Since we observe this induction using two drugs that act via separate pathways, it is possible that increased TFAP2A expression is also due to p53 stabilization. In support of this, TFAP2A has been reported to be a p53 target gene, although previously this has only been demonstrated using adenoviral-mediated overexpression of exogenous p53.21 However, comparison of Figures 4B and C suggests that accumulation of TFAP2A mRNA lags behind induction at the protein level, indicating that upregulation of TFAP2A in drug-treated cells occurs via a post-transcriptional pathway. Li et al. also suggested that TFAP2C may be regulated by p53, but we did not observe a consistent alteration in levels of this factor in drug-treated breast epithelial cells (levels declined slightly with vinbastine and rose slightly in HU-treated cells; data not shown). Previously it has been suggested that TFAP2A levels may be regulated in breast epithelial cells via alterations in the efficiency of degradation via the proteasome, particularly between normal and tumor cells.22 Alternatively, prost-transcriptional regulation of TFAP2A may involve miRNAs since targeted inactivation of Dicer in mouse neural crest cells has recently been shown to abolish AP-2α expression in the sympathetic neurons of the mutant mice.23
Figure 4.
Treatment of MCF10A cells with HU or Vin induces p53, CDKN1A and TFAP2A expression. MCF10A cells were treated with 1 mM Hydroxyurea or 60 nM Vinblastine for 24 or 48 hours. Total RNA was extracted, reverse-transcribed and analyzed by quantitative RT-PCR using specific TAQMan primers/probe for CDKN1A (A) and TFAP2A (B). Data for each gene were normalized against GAPDH expression and are represented as fold change (2−ΔΔCT) compared to the control untreated cells. The asterisks denote a significant difference between control untreated and treated cells: **p < 0.001; *p < 0.01; Student's t -test. (C) Whole-cell extracts prepared from control and treated cells were separated by SDS-PAGE and immunoblotted for the indicated proteins, including β-actin to control for protein loading.
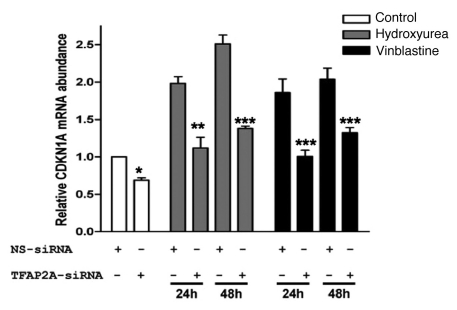
To investigate the role played by TFAP2A in CDKN1A induction in drug-treated MCF10A, cells were transfected with either non-silencing siRNA or the TFAP2A-specifc siRNA and were subsequently treated with either HU or vinblastine, or left untreated as controls. Monitoring mRNA levels revealed that TFAP2A-silencing again significantly reduced CDKN1A expression—both the constitutive, background levels seen in untreated cells but particularly the induced expression in drug-treated cells (see Fig. 5). Therefore, not only is TFAP2A able to induce p21cip expression independently of p53, but maximal induction of CDKN1A requires the presence of endogenous TFAP2A protein. Indeed, the dependence on TFAP2A for optimal CDKN1A expression is likely to be underestimated in this experiment due to the stabilisation of TFAP2A protein noted in drug treat cells (Fig. 4C) resulting in less efficient AP-2α silencing.
Figure 5.
Reduced induction of endogenous CDKN1A expression following TFAP2A silencing. MCF10A cells were transiently transfected with a non-silencing control siRNA (NS-siRNA) or with a TFAP2A targetting siRNA (TFAP2A-siRNA) for 72 hours and then were either left untreated or treated with Hydroxyurea or Vinblastine for 24 or 48 hours, as indicated. Total RNA was extracted, reverse-transcribed and subjected to q-RT-PCR. Data were normalized to GAPDH mRNA and expressed as fold change above untreated control cells transfected with NS-siRNA. The asterisks denote a significant difference between control cells transfected with NS-siRNA and cells transfected with TFAP2A-siRNA: * P<0.05, ** P<0.01, *** P<0.001; Student's t test. Further q-RT-PCR confirmed robust silencing of TFAP2A was maintained throughout the experiment (see Sup. Fig. 2).
TFAP2A binds to both proximal and distal regions of the CDKN1A promoter in induced cells.
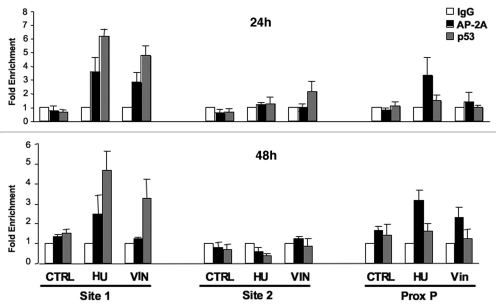
To examine the interaction of both TFAP2A and p53 across the endogenous CDKN1A promoter during expression induction, we performed chromatin immunoprecipitation (ChIP) assays on control and drug-treated cells. Three sets of qPCR primers were designed to amplify individually known regulatory regions across the CDKN1A promoter, namely the well-characterized p53 binding sites at −2,250 (site 1) and −1,344 (site 2)14 and the proximal promoter region including the AP-2 binding site identified above. Figure 6 shows the relative enrichment of each of these regions measured in TFAP2A and p53 ChIP. Association of both factors with the promoter was very low in uninduced cells but, as expected, p53 binding to its high affinity site 1 was enhanced 24 hours after drug induction and this was maintained, although at a reduced level, at 48 hours. We did not detect significant p53 binding to the other regulatory regions. In contrast, TFAP2A was shown to bind both to the proximal promoter and to the distal site 1 p53 binding region. Interaction with both of these regions was induced within 24 hours after drug treatment but at 48 hours, while binding at the proximal promoter was sustained or increased, interaction at the 5′ region had largely returned to background levels, particularly in vinblastine treated cells.
Figure 6.
Endogenous TFAP2A and p53 occupancy across the CDKN1A promoter following drug-induction. Control (Cntrl) or drug-treated (HU or Vin) MCF10A cells were harvested after 24 or 48 hours for ChIP assays. Cross-linked chromatin was immunoprecipitated using control IgG or antibodies specific for TFAP2A or p53. Chromatin samples were analyzed by qPCR using −2,290/−2,185, −935/−864 and −21/+44 CDKN1A promoter-specific primers. Results where normalized to the signal at the SAT2 locus to correct for background variation as described previously in reference 19 and shown as fold enrichment above the control IgG. Data were averaged from three independent experiments, ± standard errors.
Previously, we have shown that TFAP2C, which acts to repress the CDKN1A gene in breast epithelial lines, interacts solely at the proximal promoter.19 However, TFAP2A, which is able to activate CDKN1A, clearly does this in vivo through interactions both at the proximal promoter and at, or near, the predominant p53 binding site. Our ChIP data examining the binding of endogenous factors, therefore, provides some support for previous observations using exogenous protein expression which suggested that TFAP2A needs to interact with p53 site 1 to activate CDKN1A.16 However, in that study the authors omitted to investigate interaction at the proximal promoter and implied that TFAP2A is wholly dependent on p53 to activate CDKN1A. Comparing our ChIP data at 24 and 48 hours, demonstrates that, while the TFAP2A interaction at the −2,250 region was observed at early stages of induction, continued expression of p21cip was associated with increased TFAP2A binding at the proximal promoter, especially in vinblastine treated cells. The increase in TFAP2A binding over time may result from the induction of TFAP2A protein we have observed in drug treated cells at 24 and 48 hours (Fig. 4C).
This dual association with the CDKN1A promoter by TFAP2A may also reflect a distinct interaction by this factor required for promoter activation, that is in contrast to the binding by TFAP2C during promoter repression which occurred solely via the proximal site.19 Since ChIP can only monitor binding to within a few hundred base pairs, we cannot be certain that TFAP2A interacts with the −2,250 region via an interaction with p53 as has been proposed previously in reference 16. While we cannot rule out that p53 may increase the efficiency of AP-2α activation of this gene, we have established here that robust induction of CDKN1A by TFAP2A clearly can occur in the absence of p53 (Figs. 2 and 3) and, in lines where it is expressed, TFAP2A is required for maximal expression of p21cip (Fig. 4). In terms of tumor biology therefore, the loss of TFAP2A expression observed in invasive breast cancer is likely to contribute to tumorigenesis irrespective of whether the tumor cells express wild-type or mutant p53, since induction of p21cip mediated growth arrest will be impaired.11 This may explain therefore why associations between levels of p21cip and TFAP2A expression in breast tumor studies have been noted while correlations between p53 and TFAP2A have not been observed (reviewed in ref. 3).
Materials and Methods
Cell culture and transient transfection.
MCF10A cells (ATCC) were maintained in DMEM/F12 media supplemented with 5% horse serum, 10 µg/ml insulin, 5 µg/ml hydrocortisone, 100 ng/ml cholera toxin, 20 ng/ml epidermal growth factor in 5% CO2 at 37°C. HepG2 and H1299 cells were grown in DMEM supplemented with 10% foetal bovine serum in 10% CO2 at 37°C. Sub-confluent MCF10A cells where transiently transfected with 20 nM of a non silencing control siRNA (Qiagen, #1022076) or with a validated TFAP2A targeting siRNA (5′-AAC ATC CCA GAT CAA ACT GTA-3′; reviewed in ref. 12) using the Interferin reagent (Polyplus) according to the manufacturer's instructions. HepG2 and H1299 cells were transfected with GeneJuice Transfection Reagent (Merck) according to the instruction manual. For induction of CDKN1A, cells where treated with 1 mM Hydroxyurea or 60 nM Vinblastine (Merck).
Constructs, transfection assays and antibodies.
The pcDNA3-TFAP2A and -TFAP2C expression plasmids24 and CDKN1A luciferase reporter construct25 have been described. Mut4 and Mut5 constructs were generated using the QuickChange Site-Directed Mutagenesis Kit (Stratagene) and mutant oligonucleotide sequences (Fig. 1). The p53 expression vector, pCMV-p53, was the kind gift of Prof Dennis McCance. Reporter assays were controlled by cotransfection with pSV-β-galactosidase Control Vector (Promega). Cells were harvested after 48 hours and processed for western blotting or assayed for luciferase activity (Promega) and β-galactosidase.26 Antibodies for western blotting (from Santa Cruz unless stated otherwise) were: TFAP2A (AP-2a-3B5, #sc12726), p53 (p53-DO-1, #sc-126), TFAP2C (AP-2g 6E4, #sc-53162), p21cip (Cell signaling; #2946), β-actin (C-2, #sc-8432) and HSC70 (#sc7298). Horseradish peroxidase conjugated secondary antibodies were purchased from DakoCytomation.
RNA extraction, cDNA synthesis and q-RT-PCR.
Total RNA was extracted from sub-confluent cells (RNeasy mini kit; Qiagen) and 500 ng was reverse transcribed using the high capacity reverse transcription kit (Applied Biosystem). Ten microlitres of 1:6 diluted cDNA was analyzed by quantitative real time PCR (q-RT-PCR) using Taqman reaction mix and StepOn Real-Time PCR System (Applied Byiosystems) according to the manufacturer's instructions. TaqMan primer/probe sets (Applied Biosystems) were used to analyze the expression of TFAP2A (Hs00231461_m1) and CDKN1A (Hs00355782_m1, spans exons 2 and 3) while GAPDH TaqMan probe (#402869) was used as internal control. Analysis of q-RT-PCR data was carried out using the comparative ΔΔCT method.27 For each sample, the intensity of the amplimer was normalized against that of the GAPDH control and the data were each depicted as fold changes (2−ΔΔCT) with respect to the RNA level observed in the control experiment.
Chromatin immunoprecipitation assay.
Sub-confluent MCF10A cells were cross-linked by addition of formaldehyde directly to the culture medium to a final concentration of 1% for 10 min and subjected to Chromatin Immunoprecipitation (ChIP) assay using the ChIP-IT Express Chromatin Immunoprecipitation Kit (Active Motif) according to the instruction manual. Chromatin was sheared to an average size of 500 bp using a Sonics Vibracell VCX500 sonicator and immunoprecipitated using ChIP-validated antibodies (Active Motif) against TFAP2A (#39001, 5 ml) or p53 (#39334, 2 ml) and 2 mg purified rabbit IgG. Quantification of immunoprecipitated DNA was carried out in triplicate using a StepOne Real-Time PCR System (Applied Biosystems), with SYBR Green I Dye and AmpliTaq Gold DNA polymerase. Primers were chosen at a spacing so as not to amplify overlapping chromatin fragments. Primer sequences and calculation of values as fold enrichment compared with the IgG control versus a control locus (SAT2) were as detailed previously in reference 19.
EMSA analysis.
In vitro translated (IVT) TFAP2A was prepared from the pcDNA3-TFAP2A construct with the T‘n’T T7 coupled system (Promega). Double-stranded oligonucleotide probes (MTIIA or oligo C) were end-labeled with 32P and purified on autoseq G-50 columns (Amersham). Binding reactions containing probe, IVT protein, poly(dA-dT) (Sigma) non-specific competitor in gel retention buffer (25 mM HEPES pH 7.9, 1 mM EDTA, 5 mM DTT, 150 mM NaCl, 10% Glycerol) and electrophoresis were carried out as described previously in reference 26.
Acknowledgements
This work was supported by a Cancer Research UK program grant (to H.C.H.) number C6775/A6250 and a Breast Cancer campaign grant (to A.G.S.) number 2009MayPR60. K.V.C. was supported by a Cancer Research UK studentship.
Abbreviations
- ATCC
american tissue culture collection
- ChIP
chromatin immunoprecipitation
- DCIS
ductal carcinoma in situ
- EMSA
electromobility shift assay
- HU
hydroxyurea
- IVT
in vitro translated
- MTIIA
metallothionein IIA
- Vin
vinblastine
Footnotes
Previously published online: www.landesbioscience.com/journals/cc/article/13746
Supplementary Material
References
- 1.Mohibullah N, Donner A, Ippolito JA, Williams T. SELEX and missing phosphate contact analyses reveal flexibility within the AP-2[alpha] protein: DNA binding complex. Nucleic Acids Res. 1999;27:2760–2769. doi: 10.1093/nar/27.13.2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Eckert D, Buhl S, Weber S, Jager R, Schorle H. The AP-2 family of transcription factors. Genome Biol. 2005;6:246. doi: 10.1186/gb-2005-6-13-246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pellikainen JM, Kosma VM. Activator protein-2 in carcinogenesis with a special reference to breast cancer—a mini review. Int J Cancer. 2007;120:2061–2067. doi: 10.1002/ijc.22648. [DOI] [PubMed] [Google Scholar]
- 4.Powe DG, Akhtar G, Habashy HO, Abdel-Fatah T, Rakha EA, Green AR, et al. Investigating AP-2 and YY1 protein expression as a cause of high HER2 gene transcription in breast cancers with discordant HER2 gene amplification. Breast Cancer Res. 2009;11:90. doi: 10.1186/bcr2461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sotiriou C, Wirapati P, Loi S, Harris A, Fox S, Smeds J, et al. Gene expression profiling in breast cancer: Understanding the molecular basis of histologic grade to improve prognosis. J Natl Cancer Inst. 2006;98:262–272. doi: 10.1093/jnci/djj052. [DOI] [PubMed] [Google Scholar]
- 6.Pellikainen J, Kataja V, Ropponen K, Kellokoski J, Pietilainen T, Bohm J, et al. Reduced nuclear expression of transcription factor AP-2 associates with aggressive breast cancer. Clin Cancer Res. 2002;8:3487–3495. [PubMed] [Google Scholar]
- 7.Gee JM, Robertson JF, Ellis IO, Nicholson RI, Hurst HC. Immunohistochemical analysis reveals a tumour suppressor-like role for the transcription factor AP-2 in invasive breast cancer. J Pathol. 1999;189:514–520. doi: 10.1002/(SICI)1096-9896(199912)189:4<514::AID-PATH463>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 8.Douglas DB, Akiyama Y, Carraway H, Belinsky SA, Esteller M, Gabrielson E, et al. Hypermethylation of a small CpGuanine-rich region correlates with loss of activator protein-2alpha expression during progression of breast cancer. Cancer Res. 2004;64:1611–1620. doi: 10.1158/0008-5472.can-0318-2. [DOI] [PubMed] [Google Scholar]
- 9.Zhang J, Brewer S, Huang J, Williams T. Overexpression of transcription factor ap-2alpha suppresses mammary gland growth and morphogenesis. Dev Biol. 2003;256:127–145. doi: 10.1016/s0012-1606(02)00119-7. [DOI] [PubMed] [Google Scholar]
- 10.Zeng YX, Somasundaram K, el-Deiry WS. AP2 inhibits cancer cell growth and activates p21WAF1/CIP1 expression. Nat Genet. 1997;15:78–82. doi: 10.1038/ng0197-78. [DOI] [PubMed] [Google Scholar]
- 11.Wajapeyee N, Somasundaram K. Cell cycle arrest and apoptosis induction by activator protein 2alpha (AP-2alpha) and the role of p53 and p21WAF1/CIP1 in AP-2alpha-mediated growth inhibition. J Biol Chem. 2003;278:52093–52101. doi: 10.1074/jbc.M305624200. [DOI] [PubMed] [Google Scholar]
- 12.Orso F, Penna E, Cimino D, Astanina E, Maione F, Valdembri D, et al. AP-2alpha and AP-2gamma regulate tumor progression via specific genetic programs. FASEB J. 2008;22:2702–2714. doi: 10.1096/fj.08-106492. [DOI] [PubMed] [Google Scholar]
- 13.el-Deiry WS, Harper JW, O'Connor PM, Velculescu VE, Canman CE, Jackman J, et al. WAF1/CIP1 is induced in p53-mediated G1 arrest and apoptosis. Cancer Res. 1994;54:1169–1174. [PubMed] [Google Scholar]
- 14.el-Deiry WS, Tokino T, Waldman T, Oliner JD, Velculescu VE, Burrell M, et al. Topological control of p21WAF1/CIP1 expression in normal and neoplastic tissues. Cancer Res. 1995;55:2910–2919. [PubMed] [Google Scholar]
- 15.McPherson LA, Loktev AV, Weigel RJ. Tumor suppressor activity of AP2alpha mediated through a direct interaction with p53. J Biol Chem. 2002;277:45028–45033. doi: 10.1074/jbc.M208924200. [DOI] [PubMed] [Google Scholar]
- 16.Stabach PR, Thiyagarajan MM, Woodfield GW, Weigel RJ. AP2alpha alters the transcriptional activity and stability of p53. Oncogene. 2006;25:2148–2159. doi: 10.1038/sj.onc.1209250. [DOI] [PubMed] [Google Scholar]
- 17.McPherson LA, Weigel RJ. Ap2alpha and ap2gamma a comparison of binding site specificity and trans-activation of the estrogen receptor promoter and single site promoter constructs. Nucleic Acids Res. 1999;27:4040–4049. doi: 10.1093/nar/27.20.4040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Woodfield GW, Chen Y, Bair TB, Domann FE, Weigel RJ. Identification of primary gene targets of TFAP2C in hormone responsive breast carcinoma cells. Genes Chromosomes Cancer. 2010;49:948–962. doi: 10.1002/gcc.20807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Williams CM, Scibetta AG, Friedrich JK, Canosa M, Berlato C, Moss CH, et al. AP-2gamma promotes proliferation in breast tumour cells by direct repression of the CDKN1A gene. EMBO J. 2009;28:3591–3601. doi: 10.1038/emboj.2009.290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Beckerman R, Donner AJ, Mattia M, Peart MJ, Manley JL, Espinosa JM, et al. A role for Chk1 in blocking transcriptional elongation of p21 RNA during the S-phase checkpoint. Genes Dev. 2009;23:1364–1377. doi: 10.1101/gad.1795709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li H, Watts GS, Oshiro MM, Futscher BW, Domann FE. AP-2alpha and AP-2gamma are transcriptional targets of p53 in human breast carcinoma cells. Oncogene. 2006;25:5405–5415. doi: 10.1038/sj.onc.1209534. [DOI] [PubMed] [Google Scholar]
- 22.Li M, Wang Y, Hung MC, Kannan P. Inefficient proteasomal-degradation pathway stabilizes AP-2alpha and activates HER-2/neu gene in breast cancer. Int J Cancer. 2006;118:802–811. doi: 10.1002/ijc.21426. [DOI] [PubMed] [Google Scholar]
- 23.Huang T, Liu Y, Huang M, Zhao X, Cheng L. Wnt1-cre-mediated conditional loss of Dicer results in malformation of the midbrain and cerebellum and failure of neural crest and dopaminergic differentiation in mice. J Mol Cell Biol. 2010;2:152–163. doi: 10.1093/jmcb/mjq008. [DOI] [PubMed] [Google Scholar]
- 24.Bamforth SD, Braganca J, Eloranta JJ, Murdoch JN, Marques FI, Kranc KR, et al. Cardiac malformations, adrenal agenesis, neural crest defects and exencephaly in mice lacking cited2, a new tfap2 co-activator. Nat Genet. 2001;29:469–474. doi: 10.1038/ng768. [DOI] [PubMed] [Google Scholar]
- 25.el-Deiry WS, Tokino T, Velculescu VE, Levy DB, Parsons R, Trent JM, et al. WAF1, a potential mediator of p53 tumor suppression. Cell. 1993;75:817–825. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- 26.Bosher JM, Williams T, Hurst HC. The developmentally regulated transcription factor AP-2 is involved in c-erbB-2 overexpression in human mammary carcinoma. Proc Natl Acad Sci USA. 1995;92:744–747. doi: 10.1073/pnas.92.3.744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Winer J, Jung CK, Shackel I, Williams PM. Development and validation of real-time quantitative reverse transcriptase-polymerase chain reaction for monitoring gene expression in cardiac myocytes in vitro. Anal Biochem. 1999;270:41–49. doi: 10.1006/abio.1999.4085. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.