Abstract
The c-myc is a proto-oncogene that manifests aberrant expression at high frequencies in most types of human cancer. C-myc gene amplifications are often observed in various cancers as well. Ample studies have also proved that c-myc has a potent oncogenicity, which can be further enhanced by collaborations with other oncogenes such as Bcl-2 and activated Ras. Studies on the collaborations of c-myc with Ras or other genes in oncogenicity have established several basic concepts and have disclosed their underlying mechanisms of tumor biology, including “immortalization” and “transformation”. In many cases, these collaborations may converge at the cyclin D1-CDK4 complex. In the meantime, however, many results from studies on the c-myc, Ras and cyclin D1-CDK4 also challenge these basic concepts of tumor biology and suggest to us that the immortalized status of cells should be emphasized. Stricter criteria and definitions for a malignantly transformed status and a benign status of cells in culture also need to be established to facilitate our study of the mechanisms for tumor formation and to better link up in vitro data with animal results and eventually with human cancer pathology.
Key words: c-Myc, Cyclin D1, transformation, immortalization, oncogene
C-myc is the first proto-oncogene discovered and is known to participate in many cellular functions,1 including maintenance of stem cell properties.2 Most types of human cancer manifest aberrant expression of c-myc at high frequencies, and gene amplification occurs in many cases of various cancers as well. Ample studies have demonstrated that c-myc has a potent oncogenicity, which can be further enhanced by collaborations with other oncogenes such as a Ras mutant or with many extracellular growth stimuli that activate Ras, such as epidermal growth factor (EGF) or transforming growth factor α (TGFα). Studies on the collaborations of c-myc with Ras and other genes have provided us with mechanistic details behind several basic concepts of cancer biology, including the “two-hit principle”,3 “immortalization” and “transformation”. In the meantime, however, many results from these studies also challenge these basic concepts and thus confuse us. We now discuss the data on the collaborations of c-myc with Ras and other genes and present a perspective that these collaborations may converge at the cyclin D1-CDK4 complex. We also appeal to emphasize the importance of an immortalized status of cells and to establish stricter criteria to better define a transformed and benign statuses, so as to better connect in vitro results with animal data and with human cancer pathology.
Mechanisms for Collaboration of c-myc with Other Oncogenes in Oncogenicity
C-myc is mainly an immortalizer although it also has transforming ability.
Over two decades ago it was proposed that activation of Ras is associated with transformation4 whereas activation of c-myc is linked to immortalization.5,6 In Land et al.'s pioneer work in 1983,4 Ras can transform primary rat embryonic fibroblasts (REFs) by conferring the cells an ability to form colonies in soft agar, but the cells later die of terminal differentiation, whereas co-transfection with c-myc can immortalize the Ras-transformed cells. Many later studies further show that c-myc can also immortalize primary REF, primary mouse embryonic fibroblasts (MEF) and primary human fibroblasts.5,7–12 c-myc, especially its T58A mutant, has a potent immortalizing ability and, for this reason, is used as a routine tool to immortalize neural cells for research.13–16 However, whether c-myc alone can immortalize primary epithelial cells is still less known, although a collaboration with other genes is able to do so, as discussed below. Information available in recent years shows that c-myc can immortalize human primary prostate epithelial cells17 and human breast epithelial cells,18 but it may be a relatively rare event.19 Actually, Wang et al. reported that c-myc only extended the life span of the cells by inducing hTERT,20 thus creating a chance for change(s) in other gene(s), such as inactivation of p16ink4a,21,22 to collaborate with c-myc to immortalize and/or transform the cells. Vaux et al. also reported that c-myc and Bcl-2 together immortalized human B-cells.23,24
After the milestone set by Land et al.4 most studies on the oncogenicity of c-myc have been shifted to the transformation aspect, hardly continuing on the immortalization except for the above described studies. c-myc is shown to be capable of transforming fibroblasts,25–27 although less efficiently than Ras.5 This is probably in part because some c-myc expressing cells spontaneously develop Ras mutations, as seen in the c-myc transgenic mammary tumors28,29 or lymphomas,30 although not in the pancreatic acinar tumors from the Ela-myc transgenic mice.31,32 Kim et al. reported that c-myc rendered immortalized human esophageal squamous cells an ability to form colonies in soft agar, but the cells could not develop tumors in immunodeficient mice.33 Similarly, Zhao et al. have also shown that c-myc can transform immortalized human breast epithelial cells and fibroblasts judged by colony formation in soft agar, but the cells failed to develop tumors in mice;34,35 later Cowling et al. also reported similar results.36 It seems clear that c-myc alone cannot fully transform human epithelial cells if the ability to develop malignant tumor in immunodeficient animal is used as a criterion, unless specific spontaneous mutation occurs as a second hit. Moreover, genes that mediate the transformation by c-myc are still poorly known. As stated by Cowling and Cole et al.36 “the only c-myc-regulated gene demonstrated to be necessary for epithelial cell transformation is CDK4.”37
Oncogenic mutant (but probably not the wild type38) of Ras genes (K-Ras, H-Ras and N-Ras) may be more potent in transformation than c-myc. Ras also has an immortalization ability,5,25–27,39,40 probably in part because it can induce spontaneous c-myc activation,5 amplification41 and protein accumulation.42,43 Actually, transformation by activated Ras depends on the c-myc level.44 Because the oncogenicity of c-myc and Ras involves each other's spontaneous activation, it is actually hard to separate the oncogenic pathways and mechanisms used by these two genes.
The hTERT, c-myc, Ras, p53 and cyclin D1-CDK4 are key oncogenic factors.
In the past 25 years or so, many combinations of genes have been shown to be capable of transforming primary cells of rodent origins. However, the same gene combinations usually fail to transform primary human cells, as reviewed by Zhao et al.35 One of the reasons may be that rodent cells have higher telomerase levels and much longer telomeres, and thus can be more easily immortalized and transformed.35 Kendall et al. have summarized five models of transforming human primary cells reported in the literature, which collectively emphasize the importance of the hTERT, c-myc and activated Ras as well as the inactivation of the p53 tumor suppressor gene.45 Moreover, all five models involve ectopic expression of cyclin D1 (cycD1) and/or CDK4 or involve inactivation of p16ink4a, which suggests a critical role of the cycD1-CDK4-p16ink4a complex in transformation of human cells, presumably mainly via the Rb pathway although Rb-irrelevant mechanisms may also be involved as discussed later.45,46
Sasaki et al. report recently that triple expression of an active CDK4 mutant (CDK4R24C), cycD1 and hTERT, but not just dual expression of CDK4R24C/hTERT or cycD1/hTERT, can immortalize human ovarian surface cells,47 although the evidence is still not convincing enough to prove that the cells are indeed immortalized, not just extended for their life-span. Additional expression of a p53 dominant negative mutant (p53DN) together with a K-Ras mutant can confer the CDK4R24C/cycD1/hTERT cells an ability to form colonies in agar, but the cells still lack the ability to develop tumors in mice.47 Interestingly, the cells would die of apoptosis if only the K-Ras mutant, but not together with the p53DN, is introduced in reference 47. Moreover, further introduction of a c-myc mutant (T58A) into the cells that express all five genes increases the size of colonies but still cannot confer the cells an ability to develop tumors in mice.47 This seems to be the only study showing that a combination of ectopic K-Ras, c-myc and hTERT expression together with inactivation of both Rb (by cycD1-CDK4) and p53 still cannot fully transform a primary human epithelium.
Another system that is still not fully understood is reported by Dajee et al. showing that in the human epidermal keratinocytes transduced with a retroviral laminin 5β3, co-expression of an activated H-Ras and a stable IκBα mutant that represses NFκB activity produces invasive epidermal neoplasm in a xenograft mouse model.48,49 The novelties of this elegant model are that it does not involve inactivation of the p53 or Rb and activation of hTERT or c-myc but it involves inhibition, not activation as one may expect, of NFκB.50 However, the oncogenicity of combined H-Ras and IκBα mutant may only occur in certain special situations wherein the Ras inhibits CDK4 expression, via activating NFκB, to arrest cell proliferation or cause apoptosis,48,50 which may be associated with the reported suppression of cycD1,51 or induction of cycD1 degradatio52 by Ras to cause apoptosis. These situations may be special since in many other circumstances RAS or NFκB induces cell proliferation or increases cell survival in part by activating cycD1 as discussed below.
In the only one of the five models summarized by Kendall et al.45 that does not involve Ras, the OKF6 and OKM1 human oral squamous cells were immortalized by ectopic expression of cycD1 and a p53DN.53,54 Although this system does not involve hTERT or c-myc to induce hTERT, it involves a so-called ALT mechanism to maintain the telomere length in the immortalized cells. Additional expression of the EGF receptor (EGFR) causes transformation as evidenced by colony formation in soft agar.54 Further expression of c-myc in the cycD1/p53DN/EGFR cells, but not in those without EGFR, confers an ability to develop tumors in mice.54 Because EGFR can induces Ras55 and cycD1,56–61 a collaboration of c-myc with Ras or cycD1 may be an underlying mechanism.
Chudnovsky et al. report that human melanocytes can be transformed by ectopic expression of the hTERT, N-RasG12V, p53DN and CDK4R24C; the transformed cells develop invasive melanoma in mice.62 Interestingly, lack of either the p53DN or the CDK4R24C still results in invasive melanoma, suggesting that the p53 pathway and the cycD1-CDK4 pathway (mainly the Rb, see discussion below) are parallel and thus inactivation of either one will do. This may explain why the p53 remains intact in the human dermal keratinocytes transformed by Ras and CDK4,63 and in the Leiden human dermal fibroblasts transformed by c-myc and Ras,64 if these systems target the cycD1-CDK4-Rb pathway instead. Lazarov et al. consider that an intact p53 may be a unique characteristic of dermal cells,63 but this may also be a feature of cancers from other epithelia since the mammary65 and pancreatic31 cancers developed in different c-myc transgenic models also retain a wild type p53 and since knockout of p53 does not facilitate mammary carcinogenesis induced by a c-myc transgene.66,67 However, concomitant inactivation of the p53 and the Rb pathways may have an additive effect because simultaneous cycD1 overexpression and p53 inactivation immortalize human oral squamous cells53,54 as aforementioned.
Collaboration of c-myc with Ras may converge at cyclin D1-CDK4.
CycD1 functions to drive the G1-to-S phase progression of the cell cycle. Once a cell has passed the late stage of the G1 phase and is about to enter the S phase, it can complete the cell cycle without need for any further extracellular stimulus, although an additional stimulus may still further accelerate completion of the cell cycle. Many extracellular mitogenic stimuli induce cycD1 expression and utilize it as a common downstream effector to deliver their signals to the cell cycle machinery.68 These stimuli include EGF, TGFα, various ligands for tyrosine-kinase receptors and steroid/steroid receptors such as estrogen/estrogen receptors.1 Because most of these extracellular stimuli also activate a Ras signaling, induction of cycD1 may actually be a downstream event of Ras activation in many cases (Fig. 1). Ras activates the cycD1 promoter via an AP-1 like sequence69 and achieves predominant cycD1 nuclear localization; as a consequence, the cells may acquire an ability for non-adherent growth70 or anchorage-independent cell cycle progression.71,72 For this reason, cycD1 is regarded as a mitogenic sensor that relays and amplifies extracellular growth signals. Therefore, the observation that cycD1 can transform the NMuMG mouse mammary epithelial cell line to a malignancy73 is inferable if cycD1 is considered a major downstream effector of Ras to exert its transformation activity. Indeed, Ras exerts its oncogenicity via several common downstream pathways that in turn interact at multiple points, including cycD1, as reviewed by Rodriguez-Viciana et al.74
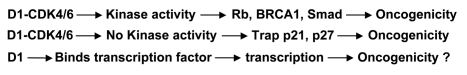
Figure 1.
Ras, which is usually induced by EGF and many other extracellular mitogenic stimuli, activates cycD1 to promote cell replication. However, Ras can also inhibit CDK4 and induce CDK inhibitors (p16, p21 and p27), which sometime causes growth arrest or apoptosis. On the contrary, c-myc can induce CDK4 and inhibit cycD1. Therefore, the c-myc and Ras collaboration may converge at the cycD1-CDK4. c-myc may also directly collaborate with cycD1 when cycD1 is induced by Ras or other growth stimuli. (Arrows and “⊤” indicate stimulation and inhibition, respectively).
Collaboration between c-myc and Ras may simply occur in such a way that each of the two activates separate growth pathway(s) that act additively. For instance, Ras75 and c-myc75–78 have been shown to preferentially induce cycD1 and cycD2, respectively, to drive cell cycle progression and cellular transformation. However, it is also possible that c-myc and Ras correct each other's weakness, as proposed by Dang et al.79 and Larsson et al.80,81 since some of their common downstream effector genes may be suppressed by c-myc but activated by Ras and vice versa. For example, c-myc is known to suppress cycD1,82–85 as reviewed before reference 1 but Ras can prevent this suppression and induce cycD1 expression.63 On the other hand, Ras may inhibit CDK4 and induce several CDK inhibitors to arrest proliferation of dermal keratinocytes and probably some other cell types as well.48–50,63 Ras-transformed cells are also sensitive to apoptosis.86–89 c-myc can reverse these effects of Ras by inducing CDK4,90,91 and inhibiting CDK inhibitors.92–94 Hence, Ras and c-myc collaboration seems to converge at cycD1 and CDK4, leading to a decrease of each other's apoptosis potential (Fig. 1). Moreover, ectopic expression of Ras in pheochromocytoma cells induces differentiation via an upregulation of c-jun but concomitant expression of c-myc inhibits this effect.95 Bazarov et al. have observed that Ras or Raf induces cycD1 to collaborate with c-myc in transformation, and propose that the cooperation occurs in such a way that Ras induces cycD1 while c-myc enhances D1-CDK4 activities,44 likely by induction of CDK4. However, Goga et al. have observed that the Rat1 cells co-expressing an oncogenic Ras mutant and c-myc are about 1.5-fold more sensitive to the apoptosis induced by a CDK1 inhibitory compound, compared with the cells expressing c-myc alone.96 This observation is discrepant to our supposition that Ras collaborates with c-myc by blocking c-myc-induced apoptosis and thus should increase the resistance to a cell death inducer.
Like Ras,4,97,98 TGFα also collaborates with c-myc in the induction of mammary,99,100 liver101–103 and pancreatic carcinogenesis.31,104 Similar to EGF,56–60,105 TGFα protein as another ligand of EGFR can also activate D1 transcription.106 Although c-myc inhibits cycD1,82–84 TGFα seems to override or circumvent this suppression since cycD1 was induced in most cells in the MT-TGFα/MMTV-c-myc dual transgenic mammary tumors, in contrast to a low level of cycD1 in most MMTV-c-myc tumor cells.107 Sandgren et al. also observe that the MT-TGFα transgenic mammary tumors display frequent induction of cycD1, in marked contrast to WAP-c-myc mammary tumors.108 Hence, cycD1 as a major downstream effector of EGFR signaling should also be able to collaborate with c-myc (Fig. 1). Indeed, Roussel et al. have shown earlier that concomitant expression of cycD1 and c-myc in 3T3 cells has additive effects on colony formation,109 although another study fails to show such a cooperation.110 cycD1 has been shown to collaborate with c-myc, N-myc or L-myc to induce lymphomas in transgenic mice.111,112 In human breast cancer, cycD1 and c-myc collaboration may also be elicited via an indirect mechanism that involves co-amplification of the cycD1 gene at 11q13 and the FGFR1 and DDHD2 genes at 8p12.113
c-myc expressing cancer cells may later find ways to restore cycD1 for survival.
c-myc protein mainly acts as a transcription factor to activate or suppress expression of many genes. Considering its potent oncogenicity, c-myc should activate oncogenes but suppress tumor suppressor genes and c-myc indeed does so. However, c-myc also represses some oncogenes such as cycD1,82–84 Neu,114 insulin-like growth factor 2,115 vascular endothelial growth factor116 and several components of NFκB82–85 while inducing the p53 and some other tumor suppressor genes, as reviewed before in reference 1. Collectively, these latter effects of c-myc are considered to be part of the mechanism for c-myc induced apoptosis,1 a unique function of c-myc that will be discussed elsewhere by us (Wang, Lisanti, Liao) in more detail.
Mutations of Ras seem to occur frequently in c-myc-induced mammary tumors.29 Chodosh's laboratory shows that 49% of the mammary tumors developed in an inducible c-myc transgenic mouse model manifest mutations in either K-Ras or N-Ras.28 In the MMTV-c-myc transgenic model, the K-Ras mutations also occur at a frequency of 44%.28 Many c-myc induced mammary tumors regress after the c-myc transgene is turned off,117,118 but K-Ras mutations occur preferentially in those tumors that do not regress after the c-myc transgene is turned off.28 This feature not only suggests that Ras activation renders the tumors independent of c-myc but also explains why certain MMTV-c-myc tumors are independent of a sustained c-myc expression.117,119 Certain c-myc induced tumors in the lung,120 pancreatic islets and skin epidermis121 in transgenic mouse models also fail to regress completely upon c-myc inactivation. It would be intriguing to know whether a Ras mutation is associated with the c-myc independence.
Due to technical constraints, the above-described studies with transgenic mammary tumors are not able to deal with tumor heterogeneity and thus cannot tell us whether the cells with a K-Ras mutation express a higher or a lower level of the c-myc transgene, relative to those that retain the wild type K-Ras. This issue is of importance because gene amplification of both c-myc and cycD1 is highly heterogeneous in human breast cancers, unlike genetic alterations in other genes such as c-erbB2.122 We previously observed that in mammary tumors from MMTV-c-myc mice, the cycD1 level is very low in most tumor areas that express high levels of the c-myc transgene.107 Meanwhile, in the tumors at an advanced stage there are some areas that lose the expression of the c-myc transgene but show a high level of cycD1.107 These specific areas, coined as “tumor-within-a-tumor” foci, basically do not show apoptosis but instead show a very high rate of proliferation, in strong contrast to the surrounding tumor areas that are high in c-myc expression and in apoptotic rate.99,107,123 The appearance of the foci suggests that the MMTV-c-myc transgene may act in a “hit-and-run” manner, disappearing at an advanced stage to allow activation of cycD1 (and probably also other oncogenes inhibited by c-myc) to facilitate tumor progression to a more aggressive phenotype characterized by quicker proliferation and much less apoptosis.99,107 In addition, the reciprocal relationship between the c-myc transgene and the cycD1 in the MMTV-c-myc mammary tumors may be the first in vivo evidence that c-myc inhibits cycD1.82–85
The above-described results from the MMTV-c-myc transgenic mouse model suggest that although c-myc suppresses cycD1,82–84 certain MMTV-c-myc mammary tumor cells may eventually find one or two mechanisms to raise the cycD1 level, i.e., by a spontaneous Ras mutation or by silencing of the c-myc transgene. A third possible mechanism may be a genetic change in the cycD1 per se or in other genes that regulate cycD1, which may be the reason for the high levels of cycD1 mRNA and proteins in some of the Eµ-myc transgenic lymphomas111 but it still awaits studies in MMTV-c-myc mammary models. Interestingly, we used a c-myc-containing retrovirus to transform MCF10A human mammary epithelial cells, judged by efficient colony formation in soft agar, and found that the c-myc level was gradually decreased after the transformed cells are passaged (unpublished observation). A tentative explanation is that the “hit-and-run” feature of c-myc also occurs in vitro and is followed by “clonal evolution.” The runaway of c-myc may allow a rise in the cycD1 level to decrease the apoptotic potential as we and others have shown before.124–126
In the K5-c-myc model, the transgenic epidermis shows a higher cycD1 protein level than the non-myc counterparts, no matter in the CDK4+/+, CDK4+/− or CDK4−/− background.37 The high level of cycD1 in the epidermis may be induced by c-myc via an indirect mechanism or may occur in a chronic situation that allows cycD1 to circumvent the inhibition by c-myc as discussed above, such as a genetic alteration in Ras or other cycD1-regulating genes. A further study on the topographic relationship between the K5-cmyc transgene and the cycD1 to determine whether these two genes have a reciprocal relationship as seen in the aforementioned MMTV-c-myc mammary tumor may be helpful in delineating the mechanism behind the cycD1 induction. cycD1-deficient keratinocytes show reduced susceptibility to chemical tumorigenesis,127,128 suggesting the mediation of cycD1 in this model, whereas Ras can still cause oncogenic transformation of fibroblasts and c-myc can still cause mammary carcinogenesis when cycD1 is deficient.129 A simple explanation for these incongruous data from different systems is that the importance of cycD1 may vary among different situations, so as well the relationship between c-myc and cycD1.
c-myc and cycD1 may be concomitantly expressed as a more aggressive phenotype.
As reviewed before reference 1 c-myc and cycD1 usually show a reciprocal relationship because c-myc inhibits cycD1.82–84 However, ectopic expression of cycD1 in Rat6 embryonic fibroblasts was shown to induce c-myc.130 There are also situations wherein c-myc and cycD1 are co-expressed, as observed in aggressive human blastoid mantle cell lymphoma131 and in those bladder cancers that bear β-catenin mutations.132 Both c-myc and cycD1 are also maintained at high levels in many breast cancers, especially in those aggressive cases.133 Additional expression of c-myc in cycD1-transformed NMuMG cells confers onto the cell aggressivity, including metastatic ability,73 suggesting that these two genes may collaborate in tumor progression. The MMTV-Ron transgenic mice develop mammary tumors that highly express both cycD1 and c-myc, which may be a reason why these tumors are highly aggressive and metastasize to the liver and lung.134
Ras is active throughout the cell cycle, but it is able to induce cycD1 only during the G2 phase.135,136 The level of cycD1 protein in the G2 phase determines whether the cells will continue to enter into another cell cycle or exit the cycle to a G0 phase after completing the M phase of this cell cycle.136 cycD1 protein level needs to be low in the S phase so that DNA can be synthesized.136 Hence, cycD1 serves as a cell cycle regulatory switch in actively proliferating cells.136 Because c-myc is an immediately early growth response gene that drives the G0–G1 transit, when it is co-expressed with cycD1, it may prevent the cell from exiting the cycle as a possible mechanism to collaborate with cycD1 in driving a continuous cell replication.
cycD1 acts via three mechanisms.
Traditionally, cycD1 is considered to associate with its cognate CDK4 or CDK6 to form an active holoenzyme that phosphorylates the Rb protein,137 which is considered the most important substrate of cycD1-CDK4/6 complexes although the BRCA1,138 the NRF-1,139 the MEP50,140 and some Smad members141,142 are later identified as CDK4 substrates as well. However, as reviewed previously in reference 1, increasing evidence suggests that many functions of cycD1 are independent of these CDKs, as exemplified by the complex formation of cycD1 with steroid receptors,143,144 p300,145 or other proteins to regulate transcription of genes, such as the peroxisome proliferator-activated receptor gamma gene shown by us.146 Therefore, cycD1 can regulate gene transcription via CDK-irrelevant mechanisms although cycD1 per se is not a canonical transcription factor (Fig. 2).
Figure 2.
Three mechanisms for the functions of cycD1: (1) cycD1 binds to CDK4 or CDK6 to form a holoenzyme that phosphorylates its substrates like Rb, BRCA1 and Smad. This is the canonical mechanism for cycD1's oncogenic role. (2) cycD1 binds CDK4, CDK6 or their kinase-dead mutants. These cycD1-CDK4/6 complexes cannot phosphorylate their substrates but can still bind CDK inhibitors such as p21 or p27, resulting in a sequestration of these inhibitors. As a consequence, other cyclin-CDK complexes such as cyclin E-CDK2 are less inhibited, i.e., more active. This kinase-irrelevant function of cycD1 may be oncogenic. (3) Instead of binding to CDK4 or CDK6, cycD1 can also form a complex with some transcriptional regulatory proteins to regulate transcription of genes. This CDK-irrelevant function may or may not be oncogenic, depending on the genes that are regulated.
CDK4 knockout (CDK4−/−) mice show a poor development of the mammary gland with impaired ductal branching.147,148 MMTV-Neu induced mammary carcinogenesis is also hindered in CDK4 knockout mice.147,148 However, Wnt-1 can still induce proliferation, preneoplastic lesions and tumors in the mammary gland of CDK4 knockout mice as efficiently as in that of the CDK4 wild type counterparts.147 Similarly, cycD1 is also required for Neu-induced, but not Wnt-induced, mammary carcinogenesis,129 which is in line with the finding that mammary tumors induced by Neu or by Ras share the same genetic markers that are absent in those induced by c-myc.149 Therefore, it may be the cycD1-CDK4 complex, not cycD1 or CDK4 alone, that is required for Neu-induced, but is dispensable for Wnt-induced, mammary carcinogenesis. Moreover, these results also partly explain why cycD1 deficiency does not affect c-myc induced mammary carcinogenesis,129 because c-myc effects via activating the Wnt-pathway,150 not via cycD1, whereas cycD1 may be a downstream effector of Neu (erbB2),151 and thus is required for the oncogenicity of Neu.
Mice with homozygous knock-in of the K112E mutant of cycD1, which can still bind CDK4 and CDK6 but has lost the ability to activate these CDKs,152 show a normal mammary gland development but Neu-induced mammary carcinogenesis was hampered in these mice.153 Therefore, the kinase activity of the cycD1-CDK4 complex seems to be required for Neu-induced carcinogenesis although it is dispensable for a normal development of the mammary gland. In vitro studies also show that concomitant expression of CDK4 inhibitor p16ink4a or p21cip1 inhibits the transformation of REF by cycD1/H-Ras, CDK4/H-Ras or c-myc/H-Ras,154 again showing the need of the CDK4 kinase activity for transformation.
Somewhat opposing the above described data, there are several lines of evidence showing that a third mechanism for function of cycD1 is to bind to CDK4 or CDK6 but it does not require the kinase activity of these CDKs (Fig. 2). For instance, K35M, a CDK4 mutant that has lost the catalytic activity due to its inability to bind ATP, can still collaborate with H-Ras in transforming primary rat fibroblasts as efficiently as the wild type CDK4.154 Conversely, the CDK4R24C mutant, which gains function because it has lost the ability to bind to p16ink4a and thus presumably has a stronger oncogenicity, cannot collaborate with H-Ras in transformation.154 In addition, it has been shown that transformation by cycD1 does not require its binding to the Rb and phosphorylation of the Rb.155 Indeed, cycD1 mutants that are unable to bind the Rb are still oncogenically active.152 S-phase entry due to overexpression of cycD1 and CDK4 can also be independent of the Rb phosphorylation.156 One of the explanations for these phenomena is that the cycD1-CDK4/6 complexes can still bind the p27kip1 or p21cip1, resulting in sequestration of these CDK inhibitors from other cyclin-CDK complexes, such as cyclin E-CDK2, and thus indirectly activating these CDKs.157–161 cycD1 has been shown to locate mainly in the cytoplasm in normal and cancerous cells, irrespective of the time point of the cell cycle.162–164 The cycD1-CDK4 complex is also found in the cytoplasm,158–160,165 probably because cycD1, CDK4 and CDK6 all lack a canonical nuclear localization sequence.166,167 Therefore, binding to p21cip1 or p27kip1 that has such a motif is needed for cycD1-CDK4/6 complexes to internalize into the nucleus to elicit their functions via phosphorylation of the Rb or other substrates or via sequestration of p21cip1 or p27kip1 from binding and inhibiting other CDKs.
Challenges to Basic Concepts of Tumor Biology from Studies of c-myc, Ras and cycD1-CDK4
More convincing evidence for an immortalized status is needed in future studies.
In the above mentioned study by Lazarov et al.63 co-expression of CDK4 and a Ras mutant confers human epidermal keratinocytes an ability to form colonies in soft agar and to develop invasive tumors in mice. However, this co-expression does not support an unlimited growth of the cells in culture.63 An increased hTERT protein level was observed in the xenograft tumor, but not in the cells from culture, suggesting that some factor(s) from the host mouse induce hTERT to support a continuous proliferation, as pointed out by the authors.63 These elegant results of Lazarov et al. together with many data described in section A, raise a serious question as to whether many gene combinations described in section A are really able to immortalize primary human cells as claimed. In many of those studies, the cells have acquired an ability to form colonies in agar and/or tumors in mice, but the evidence is still not convincing enough that the cells have already been immortalized before they were inoculated into a mouse. Probably, in some cases the cells have only acquired a prolonged life span in culture as shown by Lazarov et al. in keratinocytes63 or by Wang et al. in human breast epithelial cells.20 This concern emphasizes a basic concept of pathology that an unlimited cell replication, which is equivalent to “immortalization” in vitro, is a required criterion to define a tumor (Fig. 3). Non-immortalized cells cannot even be considered benign because a benign tumor such as uterine myoma should be able to grow indefinitely, reaching tens of kilograms as shown in some old pathology textbooks. In the studies of Lazarov et al. and probably also of others, the gene combination actually cannot immortalize the cells; it only confers the cells an ability to be immortalized later by factor(s) from the host animal. In other words, factor(s) from the mouse are needed, which is often neglected. Actually, to prove that the cells have been immortalized in vivo, the xenograft or orthotopic tumor needs to be put back in a culture to determine whether the cells can grow indefinitely, which unfortunately is not done in many studies. Future studies need to provide more convincing evidence for an immortalized status, not just an extended life span, of the cells, and to make sure that the malignant transformation is attributed only to the gene combination in vitro without any influence from the host mouse.
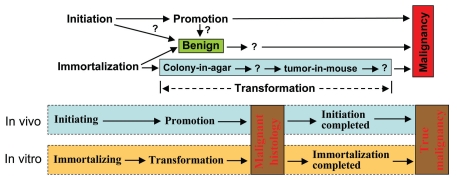
Figure 3.
Coordination of in vitro and in vivo concepts of tumor biology. Up-part: Spontaneous carcinogenesis in animals contains two steps, i.e., initiation and promotion. While immortalization of a cell in culture is equivalent to initiation, transformation may be equivalent to promotion if it includes additional properties (question markers) between colony formation in agar and xenograft tumor in mice and probably also beyond the xenograft tumor. Moreover, It is unclear (question markers) whether a benign tumor results from a clonal expansion of an initiated cell without undergoing promotion or without completion of a promotion stage. A benign status of a cell in culture should have been immortalized and should encompass additional properties towards malignancy that are currently unidentified. Moreover, some, but not all, benign cell lines may encompass a potential to progress to a malignant status, and measurable features of this potential (question marker) should be identified. Low-part: Different from the general situation described in the up-part, in certain animal models of spontaneous carcinogenesis, a cell that may have started but not yet completed an initiation process has already entered and completed the promotion process, thus manifested a malignant histology when the cell propagates to form a tumor. The tumor will regress upon withdrawal of the cancer inducer (sex steroids, chemical carcinogens or oncogenes), but a prolonged presence of the inducer will eventually facilitate the completion of the initiation, manifested as a sustained growth of a truly malignant tumor, even when the inducer is withdrawn. Similarly, in certain in vitro situations, the immortalizing process may not have been completed when the transforming process has been started and completed. The cell may thus develop to a malignant tumor when inoculated into an immunodeficient mouse, and the tumor cells may complete the immortalizing process in the host animal, manifested as a truly malignant xenograft tumor.
Stricter criteria for a transformed status are needed.
Since Freedman and Shin described the anchorage-independent survival and growth of malignant cells in 1974,168 colony formation in agar has been well accepted as a hallmark of malignant transformation by cancer research community. However, many cells can form colonies in agar but cannot form tumor in immunodeficient mice, as described in section A. Moreover, some cell lines such as MCF10AT169,170 can form colonies in agar but form only benign adenoma in mice, not malignant cancer, whereas other cell lines such as NMuMG cannot efficiently form colonies in agar but sometimes can develop benign adenomas in mice [ATCC website (www.atcc.org) and reviewed in ref. 73]. Whether these cells are “transformed” or not is a debatable question. More challenges to the definition of transformation come from the recent finding by Varmus' group that untransformed cells may have already acquired certain metastatic features.171,172 To abide to the basic definition of a cancer described in any pathology textbook, we propose that a malignantly transformed status of a cell in culture needs to meet all four criteria: (1) The cell is immortalized, i.e., can grow indefinitely in culture, (2) the cell can efficiently form colonies in agar, (3) the cell can develop tumor in immunodeficient mice and (4) the xenograft or orthotopic tumor in the mouse shows malignant histology to exclude a pseudo-tumor or a benign tumor. This proposal implies that any cell that meets some but not all of these four criteria cannot be considered “transformed” and thus needs to be redefined. A stricter redefinition or new nomenclature for some of our criteria is a prerequisite for study of the mechanism underlying the corresponding criterion. A system combining cell culture with xenograft or orthotopic animal model can clearly segregate the four criteria and thus has unique merits in dissecting the corresponding mechanism. In contrast, transgenic or carcinogen-treated animals have an intrinsic weakness in this aspect although these animal models mimic a situation of spontaneous carcinogenesis and cannot be superseded by in vitro systems. Therefore, all three systems, i.e., cell culture, immunodeficient animal model and spontaneous tumorigenesis model, are needed for study of tumor biology.
Concepts for in vitro, in vivo and in human studies need to be better connected.
Spontaneous carcinogenesis in animals requires two basic steps, i.e., “initiation” and “promotion,” which can be clearly segregated in many chemical-induced animal models.173–176 The lesions or cells that have completed a promotion step are already malignant, but in many cases they will continue the carcinogenic process to a third step termed “progression” to be more malignant, notably invasive or metastatic. In a cell culture system, immortalization may be roughly equivalent to the “initiation” while transformation, if judged only by anchorage-independent growth in agar, may only be equivalent to part of the “promotion” process because the cells may still lack the ability to form tumor in mice. In other words, “promotion” in a cell culture condition should also have additional biology features beyond colony formation that need to be identified as measurable parameter(s) to reflect that a cell line will certainly develop a malignant, not a benign, xenograft tumor in mice (Fig. 3).
In certain situation, transformation of cultured cells seems to occur before immortalization, i.e., a cell may be transformed but still not be immortalized as has been shown by Land et al. decades ago.4 However, in animal models of spontaneous carcinogenesis, whether promotion can occur before initiation has not been well studied. Nevertheless, it has been well documented even decades ago that tumors induced in rodents by sex steroids still depend on the hormones at their early stages because discontinuation of the inducer causes tumor regression, as has already been thoroughly reviewed decades ago.177–190 This phenomenon is also observed in tumors developed in mouse models of inducible transgenes, although it is more commonly referred to as “tumor dormancy” in recent literature191–194 because the tumor recurs (or probably reoccurs) quickly upon reintroduction of the inducers (steroidal hormones or oncogenes). These histologically malignant but unsustainable tumors, usually at an early stage, cannot be defined as a cancer or even a benign tumor if immortality is a required criterion. Actually, these special cell culture systems and animal models may not really imply “a transformation without immortalization” or “a promotion without initiation.” More likely, the immortalizing or initiating process may actually have been started although not yet completed while the transforming or promoting process is started and completed (Fig. 3). Moreover, progression may occur before promotion, since certain untransformed cells have already acquired some metastatic features.171,172 This partially converse oncogenic process, which still needs to be addressed experimentally, may have its human version because some human cancers can regress spontaneously, although these cases are very rare. For almost sporadic cancers in human, the initiated cells should have been immortalized long before they propagate to an overt tumor, otherwise the tumor will wither before reaching a clinic stage. More studies and discussions are needed to better connect various in vitro data with in vivo results and eventually with human cancer pathology.
In vitro criteria for a benign status need to be established.
Tumorigenesis includes formation of benign tumors, but there are few in vitro studies of a benign status. In an in vivo situation, it is unclear whether a benign tumor simply reflects a clonal expansion of an initiated cell that does not undergo a promotion stage or has not completed a promotion stage. In a cell culture situation, there is also a lack of clear criteria for a benign status. Immortality should be an irrefutable criterion (Fig. 3), because a benign tumor in human can grow indefinitely, but additional criteria are still needed, which in our opinion should include formation of a benign tumor in immunodeficient mice. Other properties of a benign status of cells in culture may reside somewhere between immortality in liquid medium and colony formation in agar (Fig. 3). Moreover, whether those cell lines that can form colonies in agar but not tumors in mice can be considered benign or intermediary, needs a special debate, because this so commonly seen status is not fully malignant whereas tumors are generally diagnosed as benign or malignant although intermediary cases do exist. It also needs to be discussed whether additional parameters are needed to reflect a potential to progress to a malignancy, since in humans some, but not all, types of benign tumors have such a potential. More cell lines like MCF10AT169,170 that can develop a benign tumor in mice need to be developed for identification of these criteria. Establishment of these criteria will facilitate our studies of tumorigenic process in vitro.
Summary and Perspectives
Studies on the oncogenic role of c-myc have provided us with several basic concepts and many underlying mechanisms of cancer biology, such as “immortalization” and “transformation.” Many extracellular mitogenic stimuli, most of which activate Ras, can collaborate with c-myc in oncogenicity, and these collaborations may converge at the cycD1-CDK4 complex. In the meantime, many of the data resulting from studies on the c-myc, Ras and cycD1-CDK4 also confuse us and challenge our criteria used to describe in vitro oncogenicity and in vivo carcinogenesis. Stricter definitions and criteria need to be established to clear up these confusions so that we can better connect in vitro data with animal results and eventually with human cancer pathology.
Acknowledgements
We would like to thank Dr. Fred Bogott at the Medical Center of Austin of Minnesota, for his excellent English editing of the manuscript. This work was supported by a NIH grant (RO1CA100864) to D.J. Liao.
References
- 1.Liao D, Thakur A, Wu J, Biliran H, Sarkar FH. Perspectives on c-Myc, cyclin D1, and their interaction in cancer formation, progression and response to chemotherapy. Crit Rev Oncog. 2007;13:93–158. doi: 10.1615/critrevoncog.v13.i2.10. [DOI] [PubMed] [Google Scholar]
- 2.Laurenti E, Wilson A, Trumpp A. Myc's other life: stem cells and beyond. Curr Opin Cell Biol. 2009;21:844–854. doi: 10.1016/j.ceb.2009.09.006. [DOI] [PubMed] [Google Scholar]
- 3.Knudson AG. Two genetic hits (more or less) to cancer. Nat Rev Cancer. 2001;1:157–162. doi: 10.1038/35101031. [DOI] [PubMed] [Google Scholar]
- 4.Land H, Parada LF, Weinberg RA. Tumorigenic conversion of primary embryo fibroblasts requires at least two cooperating oncogenes. Nature. 1983;304:596–602. doi: 10.1038/304596a0. [DOI] [PubMed] [Google Scholar]
- 5.Kelekar A, Cole MD. Immortalization by c-myc, H-ras and Ela oncogenes induces differential cellular gene expression and growth factor responses. Mol Cell Biol. 1987;7:3899–3907. doi: 10.1128/mcb.7.11.3899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mougneau E, Lemieux L, Rassoulzadegan M, Cuzin F. Biological activities of v-myc and rearranged c-myc oncogenes in rat fibroblast cells in culture. Proc Natl Acad Sci USA. 1984;81:5758–5762. doi: 10.1073/pnas.81.18.5758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Drissi R, Zindy F, Roussel MF, Cleveland JL. c-Myc-mediated regulation of telomerase activity is disabled in immortalized cells. J Biol Chem. 2001;276:29994–30001. doi: 10.1074/jbc.M101899200. [DOI] [PubMed] [Google Scholar]
- 8.Greenberg RA, O'Hagan RC, Deng H, Xiao Q, Hann SR, Adams RR, et al. Telomerase reverse transcriptase gene is a direct target of c-Myc but is not functionally equivalent in cellular transformation. Oncogene. 1999;18:1219–1226. doi: 10.1038/sj.onc.1202669. [DOI] [PubMed] [Google Scholar]
- 9.Kohl NE, Ruley HE. Role of c-myc in the transformation of REF52 cells by viral and cellular oncogenes. Oncogene. 1987;2:41–48. [PubMed] [Google Scholar]
- 10.Simm A, Halle JP, Adam G. Proliferative and metabolic capacity of rat embryo fibroblasts immortalized by c-myc depends on cellular age at oncogenic transfection. Eur J Cell Biol. 1994;65:121–131. [PubMed] [Google Scholar]
- 11.Zoidl G, Brockmann D, Esche H. Deletion of the beta-turn/alpha-helix motif at the exon 2/3 boundary of human c-Myc leads to the loss of its immortalizing function. Gene. 1993;131:269–274. doi: 10.1016/0378-1119(93)90305-m. [DOI] [PubMed] [Google Scholar]
- 12.Benanti JA, Wang ML, Myers HE, Robinson KL, Grandori C, Galloway DA. Epigenetic downregulation of ARF expression is a selection step in immortalization of human fibroblasts by c-Myc. Mol Cancer Res. 2007;5:1181–1189. doi: 10.1158/1541-7786.MCR-06-0372. [DOI] [PubMed] [Google Scholar]
- 13.De Filippis L, Ferrari D, Rota NL, Amati B, Snyder E, Vescovi AL. Immortalization of human neural stem cells with the c-myc mutant T58A. PLoS One. 2008;3:3310. doi: 10.1371/journal.pone.0003310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miljan EA, Hines SJ, Pande P, Corteling RL, Hicks C, Zbarsky V, et al. Implantation of c-mycER TAM immortalized human mesencephalic-derived clonal cell lines ameliorates behavior dysfunction in a rat model of Parkinson's disease. Stem Cells Dev. 2009;18:307–319. doi: 10.1089/scd.2008.0078. [DOI] [PubMed] [Google Scholar]
- 15.De Filippis L, Lamorte G, Snyder EY, Malgaroli A, Vescovi AL. A novel, immortal and multipotent human neural stem cell line generating functional neurons and oligodendrocytes. Stem Cells. 2007;25:2312–2321. doi: 10.1634/stemcells.2007-0040. [DOI] [PubMed] [Google Scholar]
- 16.Kerosuo L, Piltti K, Fox H, Ngers-Loustau A, Hayry V, Eilers M, et al. Myc increases self-renewal in neural progenitor cells through Miz-1. J Cell Sci. 2008;121:3941–3950. doi: 10.1242/jcs.024802. [DOI] [PubMed] [Google Scholar]
- 17.Gil J, Kerai P, Lleonart M, Bernard D, Cigudosa JC, Peters G, et al. Immortalization of primary human prostate epithelial cells by c-Myc. Cancer Res. 2005;65:2179–2185. doi: 10.1158/0008-5472.CAN-03-4030. [DOI] [PubMed] [Google Scholar]
- 18.Thibodeaux CA, Liu X, Disbrow GL, Zhang Y, Rone JD, Haddad BR, et al. Immortalization and transformation of human mammary epithelial cells by a tumor-derived Myc mutant. Breast Cancer Res Treat. 2009;116:281–294. doi: 10.1007/s10549-008-0127-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yaswen P, Stampfer MR. Molecular changes accompanying senescence and immortalization of cultured human mammary epithelial cells. Int J Biochem Cell Biol. 2002;34:1382–1394. doi: 10.1016/s1357-2725(02)00047-x. [DOI] [PubMed] [Google Scholar]
- 20.Wang J, Xie LY, Allan S, Beach D, Hannon GJ. Myc activates telomerase. Genes Dev. 1998;12:1769–1774. doi: 10.1101/gad.12.12.1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bazarov AV, Hines WC, Mukhopadhyay R, Beliveau A, Melodyev S, Zaslavsky Y, et al. Telomerase activation by c-Myc in human mammary epithelial cells requires additional genomic changes. Cell Cycle. 2009;8:3373–3378. doi: 10.4161/cc.8.20.9856. [DOI] [PubMed] [Google Scholar]
- 22.Dimri GP. c-Myc and telomerase activation. Cell Cycle. 2009;8:3075–3076. [PubMed] [Google Scholar]
- 23.Vaux DL, Cory S, Adams JM. Bcl-2 gene promotes haemopoietic cell survival and cooperates with c-myc to immortalize pre-B cells. Nature. 1988;335:440–442. doi: 10.1038/335440a0. [DOI] [PubMed] [Google Scholar]
- 24.Vaux DL. Early work on the function of Bcl-2, an interview with David Vaux. Cell Death Differ. 2004;11:28–32. doi: 10.1038/sj.cdd.4401439. [DOI] [PubMed] [Google Scholar]
- 25.Cole MD. The myc oncogene: its role in transformation and differentiation. Annu Rev Genet. 1986;20:361–384. doi: 10.1146/annurev.ge.20.120186.002045. [DOI] [PubMed] [Google Scholar]
- 26.Keath EJ, Caimi PG, Cole MD. Fibroblast lines expressing activated c-myc oncogenes are tumorigenic in nude mice and syngeneic animals. Cell. 1984;39:339–348. doi: 10.1016/0092-8674(84)90012-6. [DOI] [PubMed] [Google Scholar]
- 27.Kelekar A, Cole MD. Tumorigenicity of fibroblast lines expressing the adenovirus E1a, cellular p53 or normal c-myc genes. Mol Cell Biol. 1986;6:7–14. doi: 10.1128/mcb.6.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.D'Cruz CM, Gunther EJ, Boxer RB, Hartman JL, Sintasath L, Moody SE, et al. c-MYC induces mammary tumorigenesis by means of a preferred pathway involving spontaneous Kras2 mutations. Nat Med. 2001;7:235–239. doi: 10.1038/84691. [DOI] [PubMed] [Google Scholar]
- 29.Jang JW, Boxer RB, Chodosh LA. Isoform-specific ras activation and oncogene dependence during MYC- and Wnt-induced mammary tumorigenesis. Mol Cell Biol. 2006;26:8109–8121. doi: 10.1128/MCB.00404-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Alexander WS, Bernard O, Cory S, Adams JM. Lymphomagenesis in E mu-myc transgenic mice can involve ras mutations. Oncogene. 1989;4:575–581. [PubMed] [Google Scholar]
- 31.Liao DJ, Wang Y, Wu J, Adsay NV, Grignon D, Khanani F. Characterization of pancreatic lesions from MT-tgfalpha, Ela-myc and MT-tgfalpha/Ela-myc single and double transgenic mice. J Carcinog. 2006;5:1–19. doi: 10.1186/1477-3163-5-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schaeffer BK, Terhune PG, Longnecker DS. Pancreatic carcinomas of acinar and mixed acinar/ductal phenotypes in Ela-1-myc transgenic mice do not contain c-K-ras mutations. Am J Pathol. 1994;145:696–701. [PMC free article] [PubMed] [Google Scholar]
- 33.Kim SH, Nakagawa H, Navaraj A, Naomoto Y, Klein-Szanto AJ, Rustgi AK, et al. Tumorigenic conversion of primary human esophageal epithelial cells using oncogene combinations in the absence of exogenous Ras. Cancer Res. 2006;66:10415–10424. doi: 10.1158/0008-5472.CAN-06-2104. [DOI] [PubMed] [Google Scholar]
- 34.Zhao JJ, Gjoerup OV, Subramanian RR, Cheng Y, Chen W, Roberts TM, et al. Human mammary epithelial cell transformation through the activation of phosphatidylinositol-3-kinase. Cancer Cell. 2003;3:483–495. doi: 10.1016/s1535-6108(03)00088-6. [DOI] [PubMed] [Google Scholar]
- 35.Zhao JJ, Roberts TM, Hahn WC. Functional genetics and experimental models of human cancer. Trends Mol Med. 2004;10:344–350. doi: 10.1016/j.molmed.2004.05.005. [DOI] [PubMed] [Google Scholar]
- 36.Cowling VH, D'Cruz CM, Chodosh LA, Cole MD. c-myc Transforms Human Mammary Epithelial Cells through Repression of the Wnt Inhibitors DKK1 and SFRP1. Mol Cell Biol. 2007;27:5135–5146. doi: 10.1128/MCB.02282-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Miliani de Marval PL, Macias E, Rounbehler R, Sicinski P, Kiyokawa H, Johnson DG, et al. Lack of cyclin-dependent kinase 4 inhibits c-myc tumorigenic activities in epithelial tissues. Mol Cell Biol. 2004;24:7538–7547. doi: 10.1128/MCB.24.17.7538-7547.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Singh A, Sowjanya AP, Ramakrishna G. The wild-type Ras: Road ahead. FASEB J. 2005;19:161–169. doi: 10.1096/fj.04-2584hyp. [DOI] [PubMed] [Google Scholar]
- 39.Spandidos DA, Wilkie NM. Malignant transformation of early passage rodent cells by a single mutated human oncogene. Nature. 1984;310:469–475. doi: 10.1038/310469a0. [DOI] [PubMed] [Google Scholar]
- 40.Spandidos DA, Lang JC. Immortalization by truncated myc or ras genes and synergism between myc and ras genes in cell transformation. Anticancer Res. 1989;9:1149–1152. [PubMed] [Google Scholar]
- 41.Elenbaas B, Spirio L, Koerner F, Fleming MD, Zimonjic DB, Donaher JL, et al. Human breast cancer cells generated by oncogenic transformation of primary mammary epithelial cells. Genes Dev. 2001;15:50–65. doi: 10.1101/gad.828901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Prober DA, Edgar BA. Interactions between Ras1, dMyc and dPI3K signaling in the developing Drosophila wing. Genes Dev. 2002;16:2286–2299. doi: 10.1101/gad.991102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sears R, Nuckolls F, Haura E, Taya Y, Tamai K, Nevins JR. Multiple Ras-dependent phosphorylation pathways regulate Myc protein stability. Genes Dev. 2000;14:2501–2514. doi: 10.1101/gad.836800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bazarov AV, Adachi S, Li SF, Mateyak MK, Wei S, Sedivy JM. A modest reduction in c-myc expression has minimal effects on cell growth and apoptosis but dramatically reduces susceptibility to Ras and Raf transformation. Cancer Res. 2001;61:1178–1186. [PubMed] [Google Scholar]
- 45.Kendall SD, Adam SJ, Counter CM. Genetically engineered human cancer models utilizing mammalian transgene expression. Cell Cycle. 2006;5:1074–1079. doi: 10.4161/cc.5.10.2734. [DOI] [PubMed] [Google Scholar]
- 46.Kendall SD, Linardic CM, Adam SJ, Counter CM. A network of genetic events sufficient to convert normal human cells to a tumorigenic state. Cancer Res. 2005;65:9824–9828. doi: 10.1158/0008-5472.CAN-05-1543. [DOI] [PubMed] [Google Scholar]
- 47.Sasaki R, Narisawa-Saito M, Yugawa T, Fujita M, Tashiro H, Katabuchi H, et al. Oncogenic transformation of human ovarian surface epithelial cells with defined cellular oncogenes. Carcinogenesis. 2009;30:423–431. doi: 10.1093/carcin/bgp007. [DOI] [PubMed] [Google Scholar]
- 48.Dajee M, Lazarov M, Zhang JY, Cai T, Green CL, Russell AJ, et al. NFkappaB blockade and oncogenic Ras trigger invasive human epidermal neoplasia. Nature. 2003;421:639–643. doi: 10.1038/nature01283. [DOI] [PubMed] [Google Scholar]
- 49.Bolotin D, Fuchs E. Cancer: More than skin deep. Nature. 2003;421:594–595. doi: 10.1038/421594a. [DOI] [PubMed] [Google Scholar]
- 50.Lazarov M, Green CL, Zhang JY, Kubo Y, Dajee M, Khavari PA. Escaping G1 restraints on neoplasia—Cdk4 regulation by Ras and NFkappaB. Cell Cycle. 2003;2:79–80. [PubMed] [Google Scholar]
- 51.Cheng G, Lewis AE, Meinkoth JL. Ras stimulates aberrant cell cycle progression and apoptosis in rat thyroid cells. Mol Endocrinol. 2002;17:450–459. doi: 10.1210/me.2002-0344. [DOI] [PubMed] [Google Scholar]
- 52.Shao J, Sheng H, DuBois RN, Beauchamp RD. Oncogenic Ras-mediated cell growth arrest and apoptosis are associated with increased ubiquitin-dependent cyclin D1 degradation. J Biol Chem. 2000;275:22916–22924. doi: 10.1074/jbc.M002235200. [DOI] [PubMed] [Google Scholar]
- 53.Opitz OG, Suliman Y, Hahn WC, Harada H, Blum HE, Rustgi AK. Cyclin D1 overexpression and p53 inactivation immortalize primary oral keratinocytes by a telomerase-independent mechanism. J Clin Invest. 2001;108:725–732. doi: 10.1172/JCI11909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Goessel G, Quante M, Hahn WC, Harada H, Heeg S, Suliman Y, et al. Creating oral squamous cancer cells: a cellular model of oral-esophageal carcinogenesis. Proc Natl Acad Sci USA. 2005;102:15599–15604. doi: 10.1073/pnas.0409730102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lo HW, Hsu SC, Hung MC. EGFR signaling pathway in breast cancers: From traditional signal transduction to direct nuclear translocalization. Breast Cancer Res Treat. 2006;95:211–218. doi: 10.1007/s10549-005-9011-0. [DOI] [PubMed] [Google Scholar]
- 56.Kobayashi S, Shimamura T, Monti S, Steidl U, Hetherington CJ, Lowell AM, et al. Transcriptional profiling identifies cyclin D1 as a critical downstream effector of mutant epidermal growth factor receptor signaling. Cancer Res. 2006;66:11389–11398. doi: 10.1158/0008-5472.CAN-06-2318. [DOI] [PubMed] [Google Scholar]
- 57.Petty WJ, Dragnev KH, Dmitrovsky E. Cyclin D1 as a target for chemoprevention. Lung Cancer. 2003;41:155–161. doi: 10.1016/s0169-5002(03)00159-4. [DOI] [PubMed] [Google Scholar]
- 58.Poch B, Gansauge F, Schwarz A, Seufferlein T, Schnelldorfer T, Ramadani M, et al. Epidermal growth factor induces cyclin D1 in human pancreatic carcinoma: evidence for a cyclin D1-dependent cell cycle progression. Pancreas. 2001;23:280–287. doi: 10.1097/00006676-200110000-00009. [DOI] [PubMed] [Google Scholar]
- 59.Ravitz MJ, Yan S, Dolce C, Kinniburgh AJ, Wenner CE. Differential regulation of p27 and cyclin D1 by TGFbeta and EGF in C3H 10T1/2 mouse fibroblasts. J Cell Physiol. 1996;168:510–520. doi: 10.1002/(SICI)1097-4652(199609)168:3<510::AID-JCP3>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 60.Strobl JS, Wonderlin WF, Flynn DC. Mitogenic signal transduction in human breast cancer cells. Gen Pharmacol. 1995;26:1643–1649. doi: 10.1016/0306-3623(95)00062-3. [DOI] [PubMed] [Google Scholar]
- 61.Takuwa N, Takuwa Y. Ras activity late in G1 phase required for p27kip1 downregulation, passage through the restriction point and entry into S phase in growth factor-stimulated NIH 3T3 fibroblasts. Mol Cell Biol. 1997;17:5348–5358. doi: 10.1128/mcb.17.9.5348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chudnovsky Y, Adams AE, Robbins PB, Lin Q, Khavari PA. Use of human tissue to assess the oncogenic activity of melanoma-associated mutations. Nat Genet. 2005;37:745–749. doi: 10.1038/ng1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lazarov M, Kubo Y, Cai T, Dajee M, Tarutani M, Lin Q, et al. CDK4 coexpression with Ras generates malignant human epidermal tumorigenesis. Nat Med. 2002;8:1105–1114. doi: 10.1038/nm779. [DOI] [PubMed] [Google Scholar]
- 64.Drayton S, Rowe J, Jones R, Vatcheva R, Cuthbert-Heavens D, Marshall J, et al. Tumor suppressor p16Ink4a determines sensitivity of human cells to transformation by cooperating cellular oncogenes. Cancer Cell. 2003;4:301–310. doi: 10.1016/s1535-6108(03)00242-3. [DOI] [PubMed] [Google Scholar]
- 65.Nass SJ, Li M, Amundadottir LT, Furth PA, Dickson RB. Role for Bcl-xL in the regulation of apoptosis by EGF and TGFbeta1 in c-myc overexpressing mammary epithelial cells. Biochem Biophys Res Commun. 1996;227:248–256. doi: 10.1006/bbrc.1996.1497. [DOI] [PubMed] [Google Scholar]
- 66.Elson A, Deng C, Campos-Torres J, Donehower LA, Leder P. The MMTV/c-myc transgene and p53 null alleles collaborate to induce T-cell lymphomas, but not mammary carcinomas in transgenic mice. Oncogene. 1995;11:181–190. [PubMed] [Google Scholar]
- 67.McCormack SJ, Weaver Z, Deming S, Natarajan G, Torri J, Johnson MD, et al. Myc/p53 interactions in transgenic mouse mammary development, tumorigenesis and chromosomal instability. Oncogene. 1998;16:2755–2766. doi: 10.1038/sj.onc.1201804. [DOI] [PubMed] [Google Scholar]
- 68.Kim JK, Diehl JA. Nuclear cyclin D1: an oncogenic driver in human cancer. J Cell Physiol. 2009;220:292–296. doi: 10.1002/jcp.21791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Albanese C, Johnson J, Watanabe G, Eklund N, Vu D, Arnold A, et al. Transforming p21ras mutants and c-Ets-2 activate the cyclin D1 promoter through distinguishable regions. J Biol Chem. 1995;270:23589–23597. doi: 10.1074/jbc.270.40.23589. [DOI] [PubMed] [Google Scholar]
- 70.Mori K, Hirao E, Toya Y, Oshima Y, Ishikawa F, Nose K, et al. Competitive nuclear export of cyclin D1 and Hic-5 regulates anchorage dependence of cell growth and survival. Mol Biol Cell. 2009;20:218–232. doi: 10.1091/mbc.E08-04-0428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Resnitzky D. Ectopic expression of cyclin D1 but not cyclin E induces anchorage-independent cell cycle progression. Mol Cell Biol. 1997;17:5640–5647. doi: 10.1128/mcb.17.9.5640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Musgrove EA. Cyclins: Roles in mitogenic signaling and oncogenic transformation. Growth Factors. 2006;24:13–19. doi: 10.1080/08977190500361812. [DOI] [PubMed] [Google Scholar]
- 73.Wang Y, Thakur A, Sun Y, Wu J, Biliran H, Bollig A, et al. Synergistic effect of cyclin D1 and c-Myc leads to more aggressive and invasive mammary tumors in severe combined immunodeficient mice. Cancer Res. 2007;67:3698–3707. doi: 10.1158/0008-5472.CAN-06-4000. [DOI] [PubMed] [Google Scholar]
- 74.Rodriguez-Viciana P, Tetsu O, Oda K, Okada J, Rauen K, McCormick F. Cancer targets in the Ras pathway. Cold Spring Harb Symp Quant Biol. 2005;70:461–467. doi: 10.1101/sqb.2005.70.044. [DOI] [PubMed] [Google Scholar]
- 75.Yu Q, Ciemerych MA, Sicinski P. Ras and Myc can drive oncogenic cell proliferation through individual D-cyclins. Oncogene. 2005;24:7114–7119. doi: 10.1038/sj.onc.1208853. [DOI] [PubMed] [Google Scholar]
- 76.Bouchard C, Thieke K, Maier A, Saffrich R, Hanley-Hyde J, Ansorge W, et al. Direct induction of cyclin D2 by Myc contributes to cell cycle progression and sequestration of p27. EMBO J. 1999;18:5321–5333. doi: 10.1093/emboj/18.19.5321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bouchard C, Dittrich O, Kiermaier A, Dohmann K, Menkel A, Eilers M, et al. Regulation of cyclin D2 gene expression by the Myc/Max/Mad network: Myc-dependent TRRAP recruitment and histone acetylation at the cyclin D2 promoter. Genes Dev. 2001;15:2042–2047. doi: 10.1101/gad.907901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Perez-Roger I, Kim SH, Griffiths B, Sewing A, Land H. Cyclins D1 and D2 mediate myc-induced proliferation via sequestration of p27(Kip1) and p21(Cip1) EMBO J. 1999;18:5310–5320. doi: 10.1093/emboj/18.19.5310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Dang CV, Resar LM, Emison E, Kim S, Li Q, Prescott JE, et al. Function of the c-Myc oncogenic transcription factor. Exp Cell Res. 1999;253:63–77. doi: 10.1006/excr.1999.4686. [DOI] [PubMed] [Google Scholar]
- 80.Hydbring P, Larsson LG. Tipping the balance: Cdk2 enables Myc to suppress senescence. Cancer Res. 2010;70:6687–6691. doi: 10.1158/0008-5472.CAN-10-1383. [DOI] [PubMed] [Google Scholar]
- 81.Hirota K, Ohta K. Transcription of mRNA-type long non-coding RNAs (mlonRNAs) disrupts chromatin array. Commun Integr Biol. 2009;2:25–26. doi: 10.4161/cib.2.1.7378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Jansen-Durr P, Meichle A, Steiner P, Pagano M, Finke K, Botz J, et al. Differential modulation of cyclin gene expression by MYC. Proc Natl Acad Sci USA. 1993;90:3685–3689. doi: 10.1073/pnas.90.8.3685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Marhin WW, Hei YJ, Chen S, Jiang Z, Gallie BL, Phillips RA, et al. Loss of Rb and Myc activation co-operate to suppress cyclin D1 and contribute to transformation. Oncogene. 1996;12:43–52. [PubMed] [Google Scholar]
- 84.Philipp A, Schneider A, Vasrik I, Finke K, Xiong Y, Beach D, et al. Repression of cyclin D1: a novel function of MYC. Mol Cell Biol. 1994;14:4032–4043. doi: 10.1128/mcb.14.6.4032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Cowling VH. Enhanced mRNA cap methylation increases cyclin D1 expression and promotes cell transformation. Oncogene. 2010;29:930–936. doi: 10.1038/onc.2009.368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.di JB, Ulianich L, Racioppi L, D'Armiento F, Feliciello A, Pacifico F, et al. Serum withdrawal induces apoptotic cell death in Ki-ras transformed but not in normal differentiated thyroid cells. Biochem Biophys Res Commun. 1995;214:819–824. doi: 10.1006/bbrc.1995.2360. [DOI] [PubMed] [Google Scholar]
- 87.Shirokawa JM, Elisei R, Knauf JA, Hara T, Wang J, Saavedra HI, et al. Conditional apoptosis induced by oncogenic ras in thyroid cells. Mol Endocrinol. 2000;14:1725–1738. doi: 10.1210/mend.14.11.0559. [DOI] [PubMed] [Google Scholar]
- 88.Cheng G, Meinkoth JL. Enhanced sensitivity to apoptosis in Ras-transformed thyroid cells. Oncogene. 2001;20:7334–7341. doi: 10.1038/sj.onc.1204928. [DOI] [PubMed] [Google Scholar]
- 89.Bond J, Dawson T, Lemoine N, Wynford-Thomas D. Effect of serum growth factors and phorbol ester on growth and survival of human thyroid epithelial cells expressing mutant ras. Mol Carcinog. 1992;5:129–135. doi: 10.1002/mc.2940050208. [DOI] [PubMed] [Google Scholar]
- 90.Hermeking H, Rago C, Schuhmacher M, Li Q, Barrett JF, Obaya AJ, et al. Identification of CDK4 as a target of c-MYC. Proc Natl Acad Sci USA. 2000;97:2229–2234. doi: 10.1073/pnas.050586197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Pawar SA, Szentirmay MN, Hermeking H, Sawadogo M. Evidence for a cancer-specific switch at the CDK4 promoter with loss of control by both USF and c-Myc. Oncogene. 2004;23:6125–6135. doi: 10.1038/sj.onc.1207806. [DOI] [PubMed] [Google Scholar]
- 92.Mateyak MK, Obaya AJ, Sedivy JM. c-Myc regulates cyclin D-Cdk4 and -Cdk6 activity but affects cell cycle progression at multiple independent points. Mol Cell Biol. 1999;19:4672–4683. doi: 10.1128/mcb.19.7.4672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Obaya AJ, Mateyak MK, Sedivy JM. Mysterious liaisons: The relationship between c-Myc and the cell cycle. Oncogene. 1999;18:2934–2941. doi: 10.1038/sj.onc.1202749. [DOI] [PubMed] [Google Scholar]
- 94.Obaya AJ, Kotenko I, Cole MD, Sedivy JM. The proto-oncogene c-myc acts through the cyclin-dependent kinase (Cdk) inhibitor p27(Kip1) to facilitate the activation of Cdk4/6 and early G(1) phase progression. J Biol Chem. 2002;277:31263–31269. doi: 10.1074/jbc.M202528200. [DOI] [PubMed] [Google Scholar]
- 95.Vaque JP, Fernandez-Garcia B, Garcia-Sanz P, Ferrandiz N, Bretones G, Calvo F, et al. c-Myc inhibits Ras-mediated differentiation of pheochromocytoma cells by blocking c-Jun upregulation. Mol Cancer Res. 2008;6:325–339. doi: 10.1158/1541-7786.MCR-07-0180. [DOI] [PubMed] [Google Scholar]
- 96.Goga A, Yang D, Tward AD, Morgan DO, Bishop JM. Inhibition of CDK1 as a potential therapy for tumors overexpressing MYC. Nat Med. 2007;13:820–827. doi: 10.1038/nm1606. [DOI] [PubMed] [Google Scholar]
- 97.Sawey MJ, Hood AT, Burns FJ, Garte SJ. Activation of c-myc and c-K-ras oncogenes in primary rat tumors induced by ionizing radiation. Mol Cell Biol. 1987;7:932–935. doi: 10.1128/mcb.7.2.932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Taya Y, Hosogai K, Hirohashi S, Shimosato Y, Tsuchiya R, Tsuchida N, et al. A novel combination of K-ras and myc amplification accompanied by point mutational activation of K-ras in a human lung cancer. EMBO J. 1984;3:2943–2946. doi: 10.1002/j.1460-2075.1984.tb02236.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Liao DJ, Dickson RB. c-Myc in breast cancer. Endocr Relat Cancer. 2000;7:143–164. doi: 10.1677/erc.0.0070143. [DOI] [PubMed] [Google Scholar]
- 100.Rose-Hellekant TA, Wentworth KM, Nikolai S, Kundel DW, Sandgren EP. Mammary Carcinogenesis Is Preceded by Altered Epithelial Cell Turnover in Transforming Growth Factor-{alpha} and c-myc Transgenic Mice. Am J Pathol. 2006;169:1821–1832. doi: 10.2353/ajpath.2006.050675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Calvisi DF, Thorgeirsson SS. Molecular mechanisms of hepatocarcinogenesis in transgenic mouse models of liver cancer. Toxicol Pathol. 2005;33:181–184. doi: 10.1080/01926230590522095. [DOI] [PubMed] [Google Scholar]
- 102.Thorgeirsson SS, Santoni-Rugiu E. Interaction of c-myc with transforming growth factor alpha and hepatocyte growth factor in hepatocarcinogenesis. Mutat Res. 1997;376:221–234. doi: 10.1016/s0027-5107(97)00047-x. [DOI] [PubMed] [Google Scholar]
- 103.Thorgeirsson SS, Factor VM, Snyderwine EG. Transgenic mouse models in carcinogenesis research and testing. Toxicol Lett. 2000;112:553–555. doi: 10.1016/s0378-4274(99)00224-6. [DOI] [PubMed] [Google Scholar]
- 104.Liao JD, Adsay NV, Khannani F, Grignon D, Thakur A, Sarkar FH. Histological complexities of pancreatic lesions from transgenic mouse models are consistent with biological and morphological heterogeneity of human pancreatic cancer. Histol Histopathol. 2007;22:661–676. doi: 10.14670/hh-22.661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Perry JE, Grossmann ME, Tindall DJ. Epidermal growth factor induces cyclin D1 in a human prostate cancer cell line. Prostate. 1998;35:117–124. doi: 10.1002/(sici)1097-0045(19980501)35:2<117::aid-pros5>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 106.Yan YX, Nakagawa H, Lee MH, Rustgi AK. Transforming growth factor-alpha enhances cyclin D1 transcription through the binding of early growth response protein to a cis-regulatory element in the cyclin D1 promoter. J Biol Chem. 1997;272:33181–33190. doi: 10.1074/jbc.272.52.33181. [DOI] [PubMed] [Google Scholar]
- 107.Liao DJ, Natarajan G, Deming SL, Jamerson MH, Johnson M, Chepko G, et al. Cell cycle basis for the onset and progression of c-Myc-induced, TGFalpha-enhanced mouse mammary gland carcinogenesis. Oncogene. 2000;19:1307–1317. doi: 10.1038/sj.onc.1203430. [DOI] [PubMed] [Google Scholar]
- 108.Sandgren EP, Schroeder JA, Qui TH, Palmiter RD, Brinster RL, Lee DC. Inhibition of mammary gland involution is associated with transforming growth factor alpha but not c-myc-induced tumorigenesis in transgenic mice. Cancer Res. 1995;55:3915–3927. [PubMed] [Google Scholar]
- 109.Roussel MF, Theodoras AM, Pagano M, Sherr CJ. Rescue of defective mitogenic signaling by D-type cyclins. Proc Natl Acad Sci USA. 1995;92:6837–6841. doi: 10.1073/pnas.92.15.6837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Lovec H, Sewing A, Lucibello FC, Muller R, Moroy T. Oncogenic activity of cyclin D1 revealed through cooperation with Ha-ras: link between cell cycle control and malignant transformation. Oncogene. 1994;9:323–326. [PubMed] [Google Scholar]
- 111.Bodrug SE, Warner BJ, Bath ML, Lindeman GJ, Harris AW, Adams JM. Cyclin D1 transgene impedes lymphocyte maturation and collaborates in lymphomagenesis with the myc gene. EMBO J. 1994;13:2124–2130. doi: 10.1002/j.1460-2075.1994.tb06488.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Lovec H, Grzeschiczek A, Kowalski MB, Moroy T. Cyclin D1/bcl-1 cooperates with myc genes in the generation of B-cell lymphoma in transgenic mice. EMBO J. 1994;13:3487–3495. doi: 10.1002/j.1460-2075.1994.tb06655.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Kwek SS, Roy R, Zhou H, Climent J, Martinez-Climent JA, Fridlyand J, et al. Co-amplified genes at 8p12 and 11q13 in breast tumors cooperate with two major pathways in oncogenesis. Oncogene. 2009;28:1892–1903. doi: 10.1038/onc.2009.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Suen TC, Hung MC. c-myc reverses neu-induced transformed morphology by transcriptional repression. Mol Cell Biol. 1991;11:354–362. doi: 10.1128/mcb.11.1.354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Barsyte-Lovejoy D, Lau SK, Boutros PC, Khosravi F, Jurisica I, Andrulis IL, et al. The c-Myc oncogene directly induces the H19 noncoding RNA by allele-specific binding to potentiate tumorigenesis. Cancer Res. 2006;66:5330–5337. doi: 10.1158/0008-5472.CAN-06-0037. [DOI] [PubMed] [Google Scholar]
- 116.Barr LF, Campbell SE, Diette GB, Gabrielson EW, Kim S, Shim H, et al. c-Myc suppresses the tumorigenicity of lung cancer cells and downregulates vascular endothelial growth factor expression. Cancer Res. 2000;60:143–149. [PubMed] [Google Scholar]
- 117.Boxer RB, Jang JW, Sintasath L, Chodosh LA. Lack of sustained regression of c-MYC-induced mammary adenocarcinomas following brief or prolonged MYC inactivation. Cancer Cell. 2004;6:577–586. doi: 10.1016/j.ccr.2004.10.013. [DOI] [PubMed] [Google Scholar]
- 118.Podsypanina K, Politi K, Beverly LJ, Varmus HE. Oncogene cooperation in tumor maintenance and tumor recurrence in mouse mammary tumors induced by Myc and mutant Kras. Proc Natl Acad Sci USA. 2008;105:5242–5247. doi: 10.1073/pnas.0801197105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Tilli MT, Furth PA. Conditional mouse models demonstrate oncogene-dependent differences in tumor maintenance and recurrence. Breast Cancer Res. 2003;5:202–205. doi: 10.1186/bcr614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Tran PT, Fan AC, Bendapudi PK, Koh S, Komatsubara K, Chen J, et al. Combined Inactivation of MYC and K-Ras oncogenes reverses tumorigenesis in lung adenocarcinomas and lymphomas. PLoS One. 2008;3:2125. doi: 10.1371/journal.pone.0002125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Pelengaris S, Abouna S, Cheung L, Ifandi V, Zervou S, Khan M. Brief inactivation of c-Myc is not sufficient for sustained regression of c-Myc-induced tumors of pancreatic islets and skin epidermis. BMC Biol. 2004;2:26. doi: 10.1186/1741-7007-2-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Glockner S, Buurman H, Kleeberger W, Lehmann U, Kreipe H. Marked intratumoral heterogeneity of c-myc and cyclinD1 but not of c-erbB2 amplification in breast cancer. Lab Invest. 2002;82:1419–1426. doi: 10.1097/01.lab.0000032371.16521.40. [DOI] [PubMed] [Google Scholar]
- 123.Liao DJ, Dickson RB. Cell death in MMTV-c-myc transgenic mouse mammary tumors may not be typical apoptosis. Lab Invest. 2003;83:1437–1449. doi: 10.1097/01.lab.0000090153.13977.ae. [DOI] [PubMed] [Google Scholar]
- 124.Biliran H, Jr, Wang Y, Banerjee S, Xu H, Heng H, Thakur A, et al. Overexpression of cyclin D1 promotes tumor cell growth and confers resistance to cisplatin-mediated apoptosis in an elastase-myc transgene-expressing pancreatic tumor cell line. Clin Cancer Res. 2005;11:6075–6086. doi: 10.1158/1078-0432.CCR-04-2419. [DOI] [PubMed] [Google Scholar]
- 125.Biliran H, Jr, Banerjee S, Thakur A, Xu H, Heng H, Thakur A, et al. c-Myc-induced chemosensitization is mediated by suppression of cyclin D1 expression and nuclear factor-kappaB activity in pancreatic cancer cells. Clin Cancer Res. 2007;13:2811–2821. doi: 10.1158/1078-0432.CCR-06-1844. [DOI] [PubMed] [Google Scholar]
- 126.Diehl JA, Benzeno S. Cyclin D1 and pancreatic carcinoma: a proliferative agonist and chemotherapeutic antagonist. Clin Cancer Res. 2005;11:5665–5667. doi: 10.1158/1078-0432.CCR-05-1016. [DOI] [PubMed] [Google Scholar]
- 127.Robles AI, Rodriguez-Puebla ML, Glick AB, Trempus C, Hansen L, Sicinski P, et al. Reduced skin tumor development in cyclin D1-deficient mice highlights the oncogenic ras pathway in vivo. Genes Dev. 1998;12:2469–2474. doi: 10.1101/gad.12.16.2469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Rodriguez-Puebla ML, Robles AI, Conti CJ. ras activity and cyclin D1 expression: an essential mechanism of mouse skin tumor development. Mol Carcinog. 1999;24:1–6. [PubMed] [Google Scholar]
- 129.Yu Q, Geng Y, Sicinski P. Specific protection against breast cancers by cyclin D1 ablation. Nature. 2001;411:1017–1021. doi: 10.1038/35082500. [DOI] [PubMed] [Google Scholar]
- 130.Jiang W, Kahn SM, Zhou P, Zhang YJ, Cacace AM, Infante AS, et al. Overexpression of cyclin D1 in rat fibroblasts causes abnormalities in growth control, cell cycle progression and gene expression. Oncogene. 1993;8:3447–3457. [PubMed] [Google Scholar]
- 131.Michaux L, Wlodarska I, Theate I, Stul M, Scheiff JM, Deneys V, et al. Coexistence of BCL1/CCND1 and CMYC aberrations in blastoid mantle cell lymphoma: a rare finding associated with very poor outcome. Ann Hematol. 2004;83:578–583. doi: 10.1007/s00277-004-0879-2. [DOI] [PubMed] [Google Scholar]
- 132.Shiina H, Igawa M, Shigeno K, Terashima M, Deguchi M, Yamanaka M, et al. Beta-catenin mutations correlate with overexpression of C-myc and cyclin D1 Genes in bladder cancer. J Urol. 2002;168:2220–2226. doi: 10.1016/S0022-5347(05)64359-5. [DOI] [PubMed] [Google Scholar]
- 133.Butt AJ, Caldon CE, McNeil CM, Swarbrick A, Musgrove EA, Sutherland RL. Cell cycle machinery: links with genesis and treatment of breast cancer. Adv Exp Med Biol. 2008;630:189–205. doi: 10.1007/978-0-387-78818-0_12. [DOI] [PubMed] [Google Scholar]
- 134.Zinser GM, Leonis MA, Toney K, Pathrose P, Thobe M, Kader SA, et al. Mammary-specific Ron receptor overexpression induces highly metastatic mammary tumors associated with beta-catenin activation. Cancer Res. 2006;66:11967–11974. doi: 10.1158/0008-5472.CAN-06-2473. [DOI] [PubMed] [Google Scholar]
- 135.Sa G, Hitomi M, Harwalkar J, Stacey AW, GC GC, Stacey DW. Ras is active throughout the cell cycle, but is able to induce cyclin D1 only during G2 phase. Cell Cycle. 2002;1:50–58. [PubMed] [Google Scholar]
- 136.Stacey DW. Cyclin D1 serves as a cell cycle regulatory switch in actively proliferating cells. Curr Opin Cell Biol. 2003;15:158–163. doi: 10.1016/s0955-0674(03)00008-5. [DOI] [PubMed] [Google Scholar]
- 137.Satyanarayana A, Kaldis P. Mammalian cell cycle regulation: several Cdks, numerous cyclins and diverse compensatory mechanisms. Oncogene. 2009;28:2925–2939. doi: 10.1038/onc.2009.170. [DOI] [PubMed] [Google Scholar]
- 138.Kehn K, Berro R, Alhaj A, Bottazzi ME, Yeh WI, Klase Z, et al. Functional consequences of cyclin D1/BRCA1 interaction in breast cancer cells. Oncogene. 2007;26:5060–5069. doi: 10.1038/sj.onc.1210319. [DOI] [PubMed] [Google Scholar]
- 139.Wang C, Li Z, Lu Y, Du R, Katiyar S, Yang J, et al. Cyclin D1 repression of nuclear respiratory factor 1 integrates nuclear DNA synthesis and mitochondrial function. Proc Natl Acad Sci USA. 2006;103:11567–11572. doi: 10.1073/pnas.0603363103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Aggarwal P, Vaites LP, Kim JK, Mellert H, Gurung B, Nakagawa H, et al. Nuclear cyclin D1/CDK4 kinase regulates CUL4 expression and triggers neoplastic growth via activation of the PRMT5 methyltransferase. Cancer Cell. 2010;18:329–340. doi: 10.1016/j.ccr.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Liu F, Matsuura I. Inhibition of Smad antiproliferative function by CDK phosphorylation. Cell Cycle. 2005;4:63–66. doi: 10.4161/cc.4.1.1366. [DOI] [PubMed] [Google Scholar]
- 142.Matsuura I, Denissova NG, Wang G, He D, Long J, Liu F. Cyclin-dependent kinases regulate the antiproliferative function of Smads. Nature. 2004;430:226–231. doi: 10.1038/nature02650. [DOI] [PubMed] [Google Scholar]
- 143.Lin HM, Zhao L, Cheng SY. Cyclin D1 Is a Ligand-independent Co-repressor for Thyroid Hormone Receptors. J Biol Chem. 2002;277:28733–28741. doi: 10.1074/jbc.M203380200. [DOI] [PubMed] [Google Scholar]
- 144.Reutens AT, Fu M, Wang C, Albanese C, McPhaul MJ, Sun Z, et al. Cyclin D1 binds the androgen receptor and regulates hormone-dependent signaling in a p300/CBP-associated factor (P/CAF)-dependent manner. Mol Endocrinol. 2001;15:797–811. doi: 10.1210/mend.15.5.0641. [DOI] [PubMed] [Google Scholar]
- 145.Fu M, Wang C, Rao M, Wu X, Bouras T, Zhang X, et al. Cyclin D1 represses p300 transactivation through a cyclin-dependent kinase-independent mechanism. J Biol Chem. 2005;280:29728–29742. doi: 10.1074/jbc.M503188200. [DOI] [PubMed] [Google Scholar]
- 146.Wang C, Pattabiraman N, Zhou JN, Fu M, Sakamaki T, Albanese C, et al. Cyclin D1 repression of peroxisome proliferator-activated receptor gamma expression and transactivation. Mol Cell Biol. 2003;23:6159–6173. doi: 10.1128/MCB.23.17.6159-6173.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Reddy HK, Mettus RV, Rane SG, Grana X, Litvin J, Reddy EP. Cyclin-dependent kinase 4 expression is essential for neu-induced breast tumorigenesis. Cancer Res. 2005;65:10174–10178. doi: 10.1158/0008-5472.CAN-05-2639. [DOI] [PubMed] [Google Scholar]
- 148.Yu Q, Sicinska E, Geng Y, Ahnstrom M, Zagozdzon A, Kong Y, et al. Requirement for CDK4 kinase function in breast cancer. Cancer Cell. 2006;9:23–32. doi: 10.1016/j.ccr.2005.12.012. [DOI] [PubMed] [Google Scholar]
- 149.Morrison BW, Leder P. neu and ras initiate murine mammary tumors that share genetic markers generally absent in c-myc and int-2-initiated tumors. Oncogene. 1994;9:3417–3426. [PubMed] [Google Scholar]
- 150.Cowling VH, Cole MD. Turning the tables: Myc activates Wnt in breast cancer. Cell Cycle. 2007;6:2625–2627. doi: 10.4161/cc.6.21.4880. [DOI] [PubMed] [Google Scholar]
- 151.Lee RJ, Albanese C, Fu M, D'Amico M, Lin B, Watanabe G, et al. Cyclin D1 is required for transformation by activated Neu and is induced through an E2F-dependent signaling pathway. Mol Cell Biol. 2000;20:672–683. doi: 10.1128/mcb.20.2.672-683.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Hinds PW, Dowdy SF, Eaton EN, Arnold A, Weinberg RA. Function of a human cyclin gene as an oncogene. Proc Natl Acad Sci USA. 1994;91:709–713. doi: 10.1073/pnas.91.2.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Landis MW, Pawlyk BS, Li T, Sicinski P, Hinds PW. Cyclin D1-dependent kinase activity in murine development and mammary tumorigenesis. Cancer Cell. 2006;9:13–22. doi: 10.1016/j.ccr.2005.12.019. [DOI] [PubMed] [Google Scholar]
- 154.Haas K, Staller P, Geisen C, Bartek J, Eilers M, Moroy T. Mutual requirement of CDK4 and Myc in malignant transformation: evidence for cyclin D1/CDK4 and p16INK4A as upstream regulators of Myc. Oncogene. 1997;15:179–192. doi: 10.1038/sj.onc.1201171. [DOI] [PubMed] [Google Scholar]
- 155.Zwicker J, Brusselbach S, Jooss KU, Sewing A, Behn M, Lucibello FC, et al. Functional domains in cyclin D1: pRb-kinase activity is not essential for transformation. Oncogene. 1999;18:19–25. doi: 10.1038/sj.onc.1202286. [DOI] [PubMed] [Google Scholar]
- 156.Leng X, Connell-Crowley L, Goodrich D, Harper JW. S-Phase entry upon ectopic expression of G1 cyclin-dependent kinases in the absence of retinoblastoma protein phosphorylation. Curr Biol. 1997;7:709–712. doi: 10.1016/s0960-9822(06)00301-0. [DOI] [PubMed] [Google Scholar]
- 157.Blain SW. Switching cyclin D-Cdk4 kinase activity on and off. Cell Cycle. 2008;7:892–898. doi: 10.4161/cc.7.7.5637. [DOI] [PubMed] [Google Scholar]
- 158.Sumrejkanchanakij P, Tamamori-Adachi M, Matsunaga Y, Eto K, Ikeda MA. Role of cyclin D1 cytoplasmic sequestration in the survival of postmitotic neurons. Oncogene. 2003;22:8723–8730. doi: 10.1038/sj.onc.1206870. [DOI] [PubMed] [Google Scholar]
- 159.Sumrejkanchanakij P, Eto K, Ikeda MA. Cytoplasmic sequestration of cyclin D1 associated with cell cycle withdrawal of neuroblastoma cells. Biochem Biophys Res Commun. 2006;340:302–308. doi: 10.1016/j.bbrc.2005.11.181. [DOI] [PubMed] [Google Scholar]
- 160.Albrecht JH, Rieland BM, Nelsen CJ, Ahonen CL. Regulation of G(1) cyclin-dependent kinases in the liver: role of nuclear localization and p27 sequestration. Am J Physiol. 1999;277:1207–1216. doi: 10.1152/ajpgi.1999.277.6.G1207. [DOI] [PubMed] [Google Scholar]
- 161.Ahamed S, Foster JS, Bukovsky A, Diehl JA, Wimalasena J. Removal of Cdk inhibitors through both sequestration and downregulation in zearalenone-treated MCF-7 breast cancer cells. Mol Carcinog. 2002;34:45–58. doi: 10.1002/mc.10048. [DOI] [PubMed] [Google Scholar]
- 162.Alao JP, Gamble SC, Stavropoulou AV, Pomeranz KM, Lam EW, Coombes RC, et al. The cyclin D1 proto-oncogene is sequestered in the cytoplasm of mammalian cancer cell lines. Mol Cancer. 2006;5:7. doi: 10.1186/1476-4598-5-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Alao JP. The regulation of cyclin D1 degradation: Roles in cancer development and the potential for therapeutic invention. Mol Cancer. 2007;6:24. doi: 10.1186/1476-4598-6-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.De FM, Fedele V, De LL, Penta R, Cottone G, Cavallotti I, et al. Evaluation of cyclin D1 expression and its subcellular distribution in mouse tissues. J Anat. 2004;205:405–412. doi: 10.1111/j.0021-8782.2004.00347.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Tamamori-Adachi M, Ito H, Sumrejkanchanakij P, Adachi S, Hiroe M, Shimizu M, et al. Critical role of cyclin D1 nuclear import in cardiomyocyte proliferation. Circ Res. 2003;92:12–19. doi: 10.1161/01.res.0000049105.15329.1c. [DOI] [PubMed] [Google Scholar]
- 166.Cheng M, Olivier P, Diehl JA, Fero M, Roussel MF, Roberts JM, et al. The p21(Cip1) and p27(Kip1) CDK ‘inhibitors’ are essential activators of cyclin D-dependent kinases in murine fibroblasts. EMBO J. 1999;18:1571–1583. doi: 10.1093/emboj/18.6.1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Murakami H, Horihata M, Andojo S, Yoneda-Kato N, Kato JY. Isolation and characterization of cytoplasmic cyclin D1 mutants. FEBS Lett. 2009;583:1575–1580. doi: 10.1016/j.febslet.2009.04.036. [DOI] [PubMed] [Google Scholar]
- 168.Freedman VH, Shin SI. Cellular tumorigenicity in nude mice: correlation with cell growth in semi-solid medium. Cell. 1974;3:355–359. doi: 10.1016/0092-8674(74)90050-6. [DOI] [PubMed] [Google Scholar]
- 169.Miller FR. Xenograft models of premalignant breast disease. J Mammary Gland Biol Neoplasia. 2000;5:379–391. doi: 10.1023/a:1009577811584. [DOI] [PubMed] [Google Scholar]
- 170.Kadota M, Yang HH, Gomez B, Sato M, Clifford RJ, Meerzaman D, et al. Delineating genetic alterations for tumor progression in the MCF10A series of breast cancer cell lines. PLoS One. 2010;5:9201. doi: 10.1371/journal.pone.0009201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Podsypanina K, Du YC, Jechlinger M, Beverly LJ, Hambardzumyan D, Varmus H. Seeding and propagation of untransformed mouse mammary cells in the lung. Science. 2008;321:1841–1844. doi: 10.1126/science.1161621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Weinberg RA. Leaving home early: reexamination of the canonical models of tumor progression. Cancer Cell. 2008;14:283–284. doi: 10.1016/j.ccr.2008.09.009. [DOI] [PubMed] [Google Scholar]
- 173.Ito N, Hasegawa R, Imaida K, Hirose M, Asamoto M, Shirai T. Concepts in multistage carcinogenesis. Crit Rev Oncol Hematol. 1995;21:105–133. doi: 10.1016/1040-8428(94)00169-3. [DOI] [PubMed] [Google Scholar]
- 174.Hennings H, Glick AB, Greenhalgh DA, Morgan DL, Strickland JE, Tennenbaum T, et al. Critical aspects of initiation, promotion and progression in multistage epidermal carcinogenesis. Proc Soc Exp Biol Med. 1993;202:1–8. doi: 10.3181/00379727-202-43511a. [DOI] [PubMed] [Google Scholar]
- 175.DiGiovanni J. Multistage carcinogenesis in mouse skin. Pharmacol Ther. 1992;54:63–128. doi: 10.1016/0163-7258(92)90051-z. [DOI] [PubMed] [Google Scholar]
- 176.Miller EC, Miller JA. Mechanisms of chemical carcinogenesis. Cancer. 1981;47:1055–1064. doi: 10.1002/1097-0142(19810301)47:5+<1055::aid-cncr2820471302>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 177.Briand P. Hormone-dependent mammary tumors in mice and rats as a model for human breast cancer (review) Anticancer Res. 1983;3:273–281. [PubMed] [Google Scholar]
- 178.Cutts JH. Estrogen-induced breast cancer in the rat. Proc Can Cancer Conf. 1966;6:50–68. [PubMed] [Google Scholar]
- 179.Jordan VC. Laboratory models of breast cancer to aid the elucidation of antiestrogen action. J Lab Clin Med. 1987;109:267–277. [PubMed] [Google Scholar]
- 180.Liao DJ, Dickson RB. Roles of androgens in the development, growth and carcinogenesis of the mammary gland. J Steroid Biochem Mol Biol. 2002;80:175–189. doi: 10.1016/s0960-0760(01)00185-6. [DOI] [PubMed] [Google Scholar]
- 181.McGuire WL, Chamness GC, Costlow ME, Shepherd RE. Hormone dependence in breast cancer. Metabolism. 1974;23:75–100. doi: 10.1016/0026-0495(74)90106-1. [DOI] [PubMed] [Google Scholar]
- 182.Noble RL, Cutts JH. Mammary tumors of the rat: a review. Cancer Res. 1959;19:1125–1139. [PubMed] [Google Scholar]
- 183.Noble RL. Prostate carcinoma of the Nb rat in relation to hormones. Int Rev Exp Pathol. 1982;23:113–159. [PubMed] [Google Scholar]
- 184.Cutts JH. Enzyme activities in regressing estrone-induced mammary tumors of the rat. Cancer Res. 1973;33:1235–1237. [PubMed] [Google Scholar]
- 185.Cutts JH, Froude GC. Regression of estrone-induced mammary tumors in the rat. Cancer Res. 1968;28:2413–2418. [PubMed] [Google Scholar]
- 186.Huggins C. Endocrine-induced regression of cancers. Am J Surg. 1978;136:233–238. doi: 10.1016/0002-9610(78)90235-0. [DOI] [PubMed] [Google Scholar]
- 187.Huggins C, Oka H. Regression of stem-cell erythroblastic leukemia after hypophysectomy. Cancer Res. 1972;32:239–242. [PubMed] [Google Scholar]
- 188.Huggins CB, Ueda N. Regression of myelocytic leukemia in rats after hypophysectomy. Proc Natl Acad Sci USA. 1984;81:598–601. doi: 10.1073/pnas.81.2.598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Huggins C. Endocrine-induced regression of cancers. Cancer Res. 1967;27:1925–1930. [PubMed] [Google Scholar]
- 190.Shvemberger IN. Conversion of malignant cells into normal ones. Int Rev Cytol. 1986;103:341–386. doi: 10.1016/s0074-7696(08)60840-2. [DOI] [PubMed] [Google Scholar]
- 191.Felsher DW. Tumor dormancy and oncogene addiction. APMIS. 2008;116:629–637. doi: 10.1111/j.1600-0463.2008.01037.x. [DOI] [PubMed] [Google Scholar]
- 192.Hengstler JG, Bockamp EO, Hermes M, et al. Oncogene-blocking therapies: new insights from conditional mouse tumor models. Curr Cancer Drug Targets. 2006;6:603–612. doi: 10.2174/156800906778742488. [DOI] [PubMed] [Google Scholar]
- 193.Felsher DW. Tumor dormancy: death and resurrection of cancer as seen through transgenic mouse models. Cell Cycle. 2006;5:1808–1811. doi: 10.4161/cc.5.16.3111. [DOI] [PubMed] [Google Scholar]
- 194.Aguirre-Ghiso JA. Models, mechanisms and clinical evidence for cancer dormancy. Nat Rev Cancer. 2007;7:834–846. doi: 10.1038/nrc2256. [DOI] [PMC free article] [PubMed] [Google Scholar]