Abstract
Yeast cells, like mammalian cells, enlarge steadily as they age. Unabated cell growth can promote cellular senescence; however, the significance of the relationship between size and cellular lifespan is not well understood. Herein, we report a genetic link between cell size, growth rate and lifespan. Mutations that increase cell size concomitantly increase growth rate and decrease lifespan. As a result, large cells grow, divide and age dramatically faster than small cells. Conversely, small cell mutants age slowly and are long-lived. Investigation of the mechanisms involved suggests that attainment of a maximal size modulates lifespan. Indeed, cumulative results revealed that life expectancy is size-dependent, and that the rate at which cells age is determined in large part by the amount of cell growth per generation.
Key words: size, cell growth, longevity, senescence, aging
Introduction
Since S. cerevisiae proliferate by the inherently asymmetrical process of budding, larger mother cells are easily distinguished from their smaller daughter cells. Replicative lifespan assays in yeast have revealed the curious observation that while mother cells gradually age, daughter cells are generally born with a fully regenerated lifespan.1 Although, the steady accumulation of molecular damage is believed to be the major cause of cellular aging, the mechanisms whereby mother and daughters age asymmetrically are not completely understood.1 One accepted paradigm posits that the selective retention of extra-chromosomal ribosomal DNA circles (ERCs) by mother cells act as senescence factors.2 However, this theory is not without its difficulties. First, ERCs do not appear to be relevant to aging in higher eukaryotes.1 Second, recent experiments in yeast have demonstrated that ERC levels do not always correlate with lifespan.1 As such, additional theories for asymmetrical aging should be investigated.
Another theory is that cellular size limits lifespan. A correlation between size and lifespan was first observed in yeast.3 Subsequently, similar observations have been made in a wide-range of mammalian cells.4–7 For example, as cells near senescence, proliferation slows,8,9 but cell growth or the addition of cell mass continues relatively unchecked.4,5 The end result is a gradual increase in cell size with age. Indeed, old mammalian cells are often two or three times larger than young cells.4,9–11 Interestingly, the rate of cell size increase is inversely correlated with lifespan; the fastest growing cells enter senescence the soonest.5,12 Moreover, mammalian cells in vivo also steadily increase in size with age.6,7 Most importantly, recent studies have shown that stimulation of cell growth in human fibroblasts or epithelial cells in the absence of cell cycle progression leads to senescence.13 Specifically, ectopic expression of the p21 cyclin dependent kinase inhibitor (CDKI) induces hypertrophy, increases cell size and reduces the replicative lifespan of cells.13–17 Strikingly, treatment with either rapamycin or resveratrol decreases growth rate, reduces cell size and delays or prevents senescence.13,15–17 These studies suggest the potential for a direct relationship between size and replicative lifespan of cells. However, despite a long-standing correlation between increased size and decreased proliferative capacity, the significance of these observations has been historically underdeveloped.
Like mammalian cells, yeast cells also increase in size as they age.3,18–20 A potential role for cell size in the determination of lifespan was strengthened by the observation that in mating experiments, lifespan length correlated with cell size.21 Mammalian studies have also revealed that fusion of young and old cells resulted in senescent hetero-karyons.22,23 From these studies, it was concluded that lifespan length was “dominantly” inherited from the largest/shortest lived parent.21,22 Furthermore, centrifugation and cytometric techniques have revealed a correlation between cell size and lifespan.19,24,25 In addition, daughters from very old mothers are larger than normal and have a shortened lifespan.20 These results suggest that large cell size may be incompatible with viability. However, since the use of alpha-factor to produce abnormally large cells did not result in a reduced lifespan, it was concluded that cell size does not have a causative role in aging.20 Based upon these results a role for size in lifespan determination has been largely ruled out.
Recently, reports have challenged this conclusion.26–28 In addition, many genes that modulate longevity also concomitantly affect cell size. For example, deletion of SCH9, RPL31a, GPA2 and many other genes reduces cell size and extends lifespan.29–32 Moreover, several elegant studies in yeast have implicated the TOR pathway and ribosome biogenesis as major modulators of lifespan.30,33 Remarkably, the deletion of a sub-set of genes involved in 60S ribosomal function conferred a long-life phenotype.33 Like dietary restriction, inhibition of either the TOR pathway or ribosome biogenesis reduces translation rates and cell size.34 Moreover, epistasis experiments in yeast suggest that dietary restriction, TOR and ribosome biogenesis impact aging via a common pathway.30,33 However, it is currently unclear whether lifespan extension is dependent upon reduced cell growth or decreased cell size. Nonetheless, a flurry of recent papers using pharmacological inhibitors has confirmed that suppression of cell growth reduces cell size and extends lifespan in mammalian cells.13,15–17 Therefore, we have re-examined the role of growth and size in the determination of cellular lifespan in yeast.
Herein, we report that aging yeast cells grow steadily and enter senescence at a relatively constant cell size. Subsequently, lifespan is abnormally short in large cells suggesting that a maximal size limits viability. Moreover, mutations that increase cell size concomitantly increase growth per generation and decrease lifespan. Therefore, lifespan was limited because a smaller number of divisions were necessary to reach maximal cell size. Conversely, small cell mutants grew, divided and aged slowly. When cell cycle block-and-release experiments were used to enlarge the size of longevity mutants, their relative growth rate was increased and lifespan was dramatically shortened. We conclude that size and the rate of cell growth per generation have an integral role in the control of cellular lifespan.
Results
Large cells are short-lived.
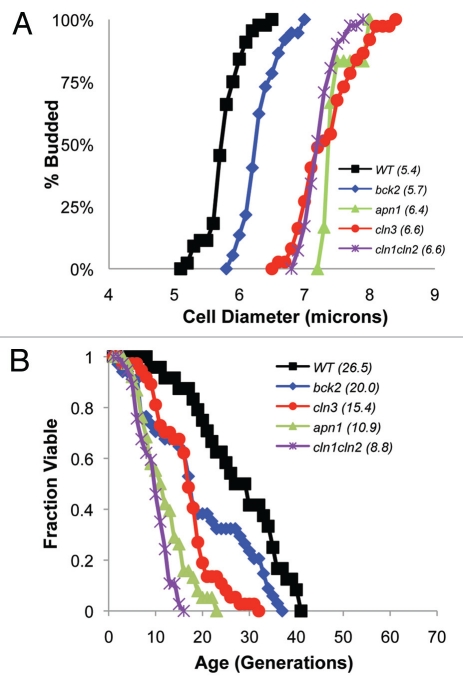
Analyses of two systematic genome-wide cell size mutant screens revealed that a number of large cell gene deletions (e.g., pph3Δ, agp17Δ, akr1Δ and others) have a shortened lifespan suggesting that size might limit replicative capacity.29–32 To follow up, we determined the lifespan of four representative large cell size mutants (bck2Δ, cln3Δ, apn1Δ and cln1cln2Δ). These deletions were chosen because despite having an enlarged size, cells from these strains had reasonably normal cell cycle times (Table 1) and were otherwise morphologically “healthy”.31,32 Log phase liquid cultures confirmed these observations (data not shown). However, since it is difficult to statistically differentiate the significance of observed size differences in populations of cells, single cells were grown on thin strips of YPD agar and followed for several generations using photomicroscopy. Moreover, since these conditions resemble those used in lifespan assays, the measurements obtained are more physiologically relevant. Using time-lapse digital photomicroscopy of 30–50 individual cells and the Axiovision multidimensional acquisition program, it was determined that the average size at birth (diameters measured in microns) of virgin daughters for bck2Δ, cln3Δ, apn1Δ and cln1cln2Δ cells were all statistically larger (p < 0.0003) than wild type cells (Fig. 1A). In addition, the size at which virgin daughters budded was determined and plotted (Fig. 1A). Analyses revealed that each of these strains budded at a size that was statistically larger (p < 0.0001) than wild type cells (Fig. 1A) and had an average lifespan that was significantly shorter than asynchronous wild type cells (Fig. 1B). The decrease in lifespan was strongly proportional to observed increases in size. For example, in bck2Δ cells, which are 13% larger than wild type cells, lifespan decreased 25% while in cln1Δcln2Δ cells (32% larger) average lifespan decreased 67% (Fig. 1B and Table 1).
Table 1.
Lifespan, cycle time, relative growth rate per generation in selected cell size mutants
| Size1 | ||||||
| Strain2 | Start | End | Cycle time3 | LS4 | % Change5 | RGR6 |
| whi5Δ | 5.7 | 9.1 | 108.6 | 34.7 | 31% | 0.6 |
| fob1Δ | 5.9 | 9.83 | 91.3 | 32.4 | 22% | 0.7 |
| ssf1Δ | 6.4 | 12.3 | 153.4 | 44.1 | 66% | 0.8 |
| cln3Δ7 small | 6.5 | 10.5 | 99.4 | 18.3 | −31% | 1.3 |
| rpl42aΔ | 6.7 | 11.5 | 131.6 | 40.4 | 52% | 0.7 |
| whi5Δ7 Large | 6.9 | 10.0 | ND | 27.3 | 3% | 0.7 |
| Wild type8 | 6.9 | 11.3 | 83.6 | 26.5 | - | 1.0 |
| rpl42bΔ | 6.9 | 9.7 | 107.5 | 9.7 | −64% | 1.7 |
| ssf1Δ cdc289 | 7.4 | 9.7 | 137.3 | 12.9 | −51% | 1.1 |
| Wild type8,10 | 7.5 | 11.1 | ND | 16.8 | −37% | 1.3 |
| cln3Δ whi5Δ11 | 7.6 | 12.5 | 83.9 | 20.3 | −24% | 1.5 |
| Wild type7 Large | 7.8 | 11.0 | ND | 20.9 | −21% | 0.9 |
| bck2Δ | 7.8 | 11.9 | 88.1 | 20.0 | −25% | 1.2 |
| ssf1Δ7 Large | 7.9 | 11.7 | ND | 31.6 | 19% | 0.7 |
| hxk2Δ12 | 8.0 | 12.2 | 154.2 | 37.8 | 43% | 0.7 |
| ssf1Δ nocodazole13 | 8.1 | 10.3 | 88.3 | 7.1 | −73% | 1.8 |
| cln3Δ | 8.2 | 12.0 | 125.1 | 15.4 | −42% | 1.5 |
| ssf1Δ9 Large II | 8.8 | 11.9 | 114.6 | 19.0 | −28% | 1.0 |
| Wild type7 Large II | 9.0 | 10.9 | 73.1 | 9.7 | −63% | 1.2 |
| whi5Δ7 Large II | 9.0 | 12.3 | ND | 11.1 | −58% | 1.8 |
| cln1Δ cln2Δ14 | 9.1 | 13.2 | 89.6 | 8.8 | −67% | 2.8 |
| apn1Δ | 9.2 | 11.8 | 176.4 | 10.9 | −59% | 1.4 |
| cln3Δ7, Large | 9.5 | 13.7 | 90.5 | 7.6 | −71% | 3.3 |
| cln3Δ7 Large II | 11.1 | 12.5 | 95.6 | 2.8 | −89% | 3.1 |
The cell diameter (microns) of all virgin daughter cells was determined at the start and end of the replicative lifespan assays. Numbers are the average for all cells. Due to technical complications, start sizes are not birth sizes but are closer to the size at which virgin daughters bud.
All strains are homozygous deletions in wild type BY4743 background. Specific strain genotypes, experimental conditions, or noted results are listed in superscripts. All cells were propagated on 2% glucose at all times.
Cycle time in minutes was determined by calculating the time between three subsequent buddings. Experiments were conducted using 30–50 individual virgin daughters for each sample. ND: not determined.
LS is average lifespan of each experiment.
Percent change in lifespan of each experiment compared to wild type cells.
RGR is relative growth rate per generation calculated by dividing net growth (size at end of the experiment minus size at start) by the average lifespan and normalized to wild type (set to 1).
Centrifugal elutriation was used to size-fractionate log phase YPD cultures.
WT is diploid wild type (BY4743).
An ssf1Δcdc28ts was created by mating a MATα ssf1Δ strain with a MATa strain carrying cdc28-17ts mutations in the s288c background. Heterozygotes were sporulated. Haploids with an ssf1Δcdc28ts genotype were determined by PCR and cell cycle arrests at the restrictive temperature. Subsequently, homozygous diploids were generated by mating.
The selection of virgin granddaughters and great-granddaughters from huge wild type cells resulted in smaller longer-lived cells further demonstrates that lifespan correlates with size.
cln3Δ::LEU2 whi5Δ::NAT.
The start size of these deletions was unexpectedly large despite measurements in other experiments or procedures that reproducibly demonstrated that hxk2Δ virgin daughters are small.
ssf1Δ cells were treated with nocodazole as described to produce large cells within a single generation.
cln1Δ::LEU2 cln2Δ::KANMX.
Figure 1.
Large cell mutants are short-lived. (A) Time-lapse movies of 30–50 individual cells were used to determine the birth size of virgin daughters (avg. birth size in microns shown in parentheses). In addition, the size at which each virgin daughter budded was measured. Subsequently, the percent of budded cells for large cell mutants (bck2Δ apn1Δ, cln3Δ and cln1Δ cln2Δ) was plotted as function of cell diameter and compared to wild type cells allowing the size of mothers and virgin daughters to be analyzed statistically. These analyses revealed that in all deletion strains, daughters were born and budded at a significantly larger size (p < 0.0003) as compared to wild type cells. (B) Survival curves for four large cell mutants revealed that lifespan in each case was statistically shorter than wild type (p = 0.0075 for bck2Δ and p < 0.0001 for all others). Average lifespan is shown in parentheses (Table 1).
Cells senesce at a relatively constant size.
To precisely monitor the relationship between size and longevity, we developed a replicative lifespan assay conducted in medium suspended in drops of mineral oil. This protocol enabled us to measure cell growth and lifespan in real time. Digital photomicroscopy of individual aging cells revealed a gradual increase in cell diameter with age (Sup. Mov. and Fig. 2A) and corroborated previous results.19,20 Moreover, in a standard replicative lifespan assay, most cells were relatively similar in size at the end of their lifespan (Fig. 2A), suggesting that the attainment of a maximal cell size limits longevity. Previous investigations demonstrated that old mother cells budded symmetrically to produce larger than normal daughters with a shortened lifespan.20 Nevertheless, since symmetric budding was only observed in very old mother cells and because increasing the size of daughter via alpha-factor treatment did not shorten lifespan, it was concluded cell size did not directly modulate lifespan.20 However, given that recent attempts to repeat these experiments have yielded the opposite result,26–28 we have re-examined the relationship between cell size and lifespan.
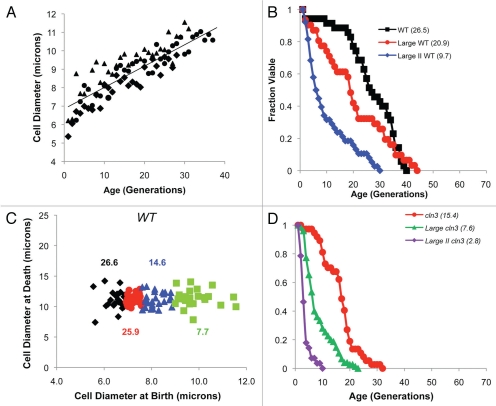
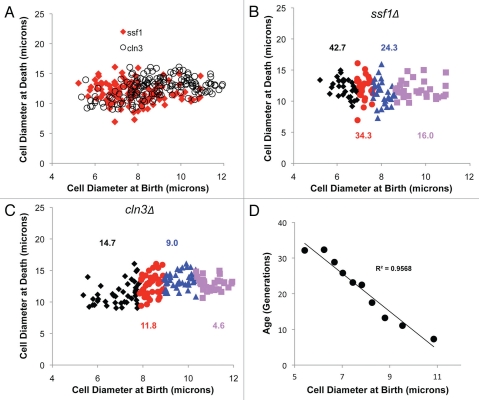
Figure 2.
Cells enlarge as they age and enter senescence at a relatively constant size. (A) The diameter of three independent (▲, ●, ♦) diploid wild type cells increases steadily with age. (B) Survival curves for large (avg. diameter 7.8 microns) and very large (Large II: avg. 9 microns) compared to asynchronous wild type (avg. 6.9 microns). Average lifespan is shown in parentheses (Table 1). (C) Diameter of cells at senescence is plotted as a function of birth size. Cells ranging in size from small to very large were selected from centrifugal elutriation fractions using a micromanipulator. Analyses of the cumulative group in quartiles revealed that virgin daughters displayed statistically different birth sizes (p < 0.0001, Table 1), but statistically similar sizes at senescence (Table 1). For example, the smallest 25% of cells (diamonds) had average size at senescence of 11.1 microns compared to 11.4 microns for the largest 25% (squares) (p = 0.47). Intermediate quartiles are shown as circles or triangles and average lifespan for all groups is shown. (D) Survival curves for large and very large cln3Δ cells (Large II) were compared to non-fractionated cln3Δ (avg. diameter 9.5, 11.1 and 8.2 microns, respectively). Average lifespan is shown in parentheses (Table 1).
To minimally perturb cell growth, centrifugal elutriation was used to size fractionate wild type cells. Subsequently, micro-manipulation of size-selected cells was used to isolate large virgin daughters. This experiment yielded two important conclusions. First, a direct correlation between size and lifespan was detected (Fig. 2B). Thus, even in only moderately larger virgin daughters, lifespan correlated with cell size (Fig. 2B). Moreover, virgin daughters that were even bigger (avg. 30% larger, p < 0.0001) had an average lifespan that was dramatically shortened (e.g., 63%, p < 0.0001) (Fig. 2B). By grouping cells in quartiles, it was found that lifespan decreased steadily with increasing birth size (Fig. 2C). Moreover, the largest 25% had an average lifespan that was 3.4 fold shorter than the smallest 25% (p <0.0001) (Fig. 2C). Importantly, these large cells were not morphologically “sick” nor did they have any obvious growth defects. In fact, the largest wild type cells had the shortest cell cycle times (Table 1). In addition, lifespan decreased well before cells began to divide symmetrically. Second, we found that cell size at senescence was relatively constant and independent of birth size (Fig. 2C). While virgin daughters were born at a range of 6–11 microns in diameter, all cells entered senescence at a relatively constant size (avg. 11.2 microns) (Fig. 2C). These results suggest that a maximum cell size may in part determine lifespan. Therefore, size fractionation of large cell mutants (e.g., cln3Δ) should yield cells with very short life expectancy. Centrifugal elutriation and micro-manipulation were used to isolate large cln3Δ virgin daughters (avg. 16% and 38%) larger in diameter than asynchronous cln3Δ or wild type cells, respectively (p < 0.0001). These cells had an average lifespan of 7Δ6 generations (50% shorter than asynchronous cln3Δ cells and 71% shorter than wild type cells, p < 0.0001) (Fig. 2D). Again, lifespan decreased before cells began to divide symmetrically. Furthermore, the lifespan of very large cln3Δ virgin daughters was extremely short (avg. 2.8 generations) (Fig. 2D). In addition, the size selected cln3Δ cells were not “sick” but in fact grew robustly with cell cycle times considerably faster than asynchronous cln3Δ cells (Table 1). These results demonstrate that reduced lifespan correlates strongly with increased cell size.
Small-sized mutants are long-lived.
If a large cell size limits lifespan, we reasoned that mutations that reduce size should increase lifespan. Indeed, many gene deletions that result in extended longevity are also small-size (whi) mutants (e.g., sch9Δ, rpl31aΔ, gpa2Δ and many others).29–32 In a classic tour-de-force, Steffen et al. determined that 14 ribosomal protein (RP) single gene deletions resulted in significantly long-lived cells.33 To determine if these RP genes had a role in cell size control, the size of long-lived cells with RP gene deletions was re-examined. Nine of the top ten longest-lived RP MATa gene deletions were found to also be whi mutants (Table 2 and Fig. 3A). In the remaining deletion, rpl20b, both the MATa and the diploid strain were abnormally small (data not shown). All ten of these RP genes, like ∼85% of RP genes, have paralogs in the genome. Importantly, for all ten genes, loss of function of its duplicate gene conferred neither a small cell size nor a phenotype long-lived.31,33 For example, rpl42aΔ cells are small and long-lived while rpl42bΔ are neither small, nor long-lived (Fig. 3B and Table 1). In addition, deletion of three additional genes involved in ribosome biogenesis, NOP12, LOC1 and SSF1, reduces cell size and dramatically increases lifespan (Fig. 3A, C and D and Tables 1 and 2).33 Importantly, mother cells lacking the FOB1 gene, an archetypal longevity gene, are also smaller than wild type cells (data not shown and Table 1). In contrast, short-lived sir2Δ strains are abnormally large (Table 2). These data suggest a causal relationship between cell size and longevity.
Table 2.
Mean cell size of log-phase haploid cultures1
| Strain | MCV (fL) |
| Wild type | 68.6 |
| rpl6bΔ | 53.3 |
| rpl22aΔ | 54.4 |
| nop12Δ | 54.9 |
| rpl23aΔ | 55.4 |
| loc1Δ | 57.4 |
| rpl31aΔ | 58.2 |
| rpl21bΔ | 58.8 |
| rpl34bΔ | 58.8 |
| ssf1Δ | 59.9 |
| sir2Δ | 85.6 |
The mean cell volume of long-lived RP MATa gene deletions is shown. Of the top ten longest-lived RP MATa gene deletions,33 five (rpl21bΔ, rpl31aΔ, rpp2bΔ, rpl13aΔ and rpl19aΔ) were previously identified as whi mutants.31 Four of the remaining five deletion strains (rpl22aΔ, rpl34bΔ, rpl23aΔ and rpl6bΔ) had mean cell volumes (MCV) considerably smaller than the wild type control (BY4741). In addition, nop12Δ, loc1Δ and ssf1Δ strains were abnormally small, and sir2Δ cells were large.
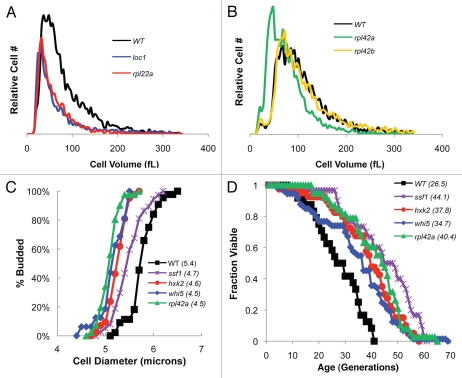
Figure 3.
whi mutants are long-lived. (A) Two long-lived haploid strains, loc1Δ and rpl22aΔ are markedly smaller than wild type cells. (B) Diploid rpl42aΔ cells are small and long-lived while rpl42bΔ cells are not (Table 1). (C) Plotting the percent of budded cells as function of cell diameter, as determined from time-lapse movies, indicated that virgin daughters of whi mutants were born (avg. birth size in microns shown in parentheses) and budded at a smaller size than wild type cells. (D) Survival curves for four long-lived whi mutants. Average lifespan is shown in parentheses (Table 1).
To investigate a potential relationship between decreased cell size and increased lifespan, the replicative lifespan of an archetypal whi mutant, whi5Δ, was examined. In addition, we have identified a new whi RP gene deletion, rpl42aΔ (Dungrawala et al. in preparation), and its lifespan was also determined. For reference, these deletions were compared to hxk2Δ and ssf1Δ cells, two long-lived strains. Time-lapse photomicroscopy revealed that the average size at birth and budding (diameters measured in microns) of virgin daughters for whi5Δ, hxk2Δ, ssf1Δ and rpl42aΔ cells were all statistically smaller (p < 0.0001) than wild type cells (Fig. 3C). Importantly, in all four deletion strains, average lifespan was also significantly extended (p < 0.0001), ranging from 31% longer for whi5 deletions to 66% longer for ssf1Δ cells (Fig. 3D and Table 1). Finally, we have also identified the long-lived rom2 deletion as a whi mutant (Dungrawala et al. in preparation). These data suggest that mutations that reduce cell size concomitantly extend lifespan.
To test the hypothesis that reduced cell size was responsible for lifespan extension in longevity mutants, we examined the relationship between cell size and lifespan in long-lived ssf1Δ and whi5Δ cells. Centrifugal elutriation and micromanipulation were used to size fractionate ssf1Δ and whi5Δ cells (Fig. 4A and data not shown). In both cases, larger cells had a statistically shorter (p < 0.0001) lifespan as compared to small asynchronous parental cells (Fig. 4B and data not shown), suggesting that size and not the gene deletions per se modulate lifespan. Size-fractionated whi mutants were not “sick” and in fact had faster cell cycle times than parental asynchronous cultures (Table 1). To further evaluate the correlation between size and longevity, we examined the lifespan of size-matched cells. Strikingly, this comparison revealed that size-matched strains had a nearly identical lifespan. For example, while small whi5Δ cells live three times longer than apn1Δ cells (Figs. 1B and 3C), size-matched whi5Δ and apn1Δ cells had a statistically indistinguishable lifespan (avg. 11.1 and 10.9 generations, respectively p = 0.78) (Fig. 4C). This also held true when large wild type cells were matched with asynchronous bck2Δ cells (avg. lifespan 20.9 and 19.9 generations, respectively p = 0.67) (Fig. 4C).
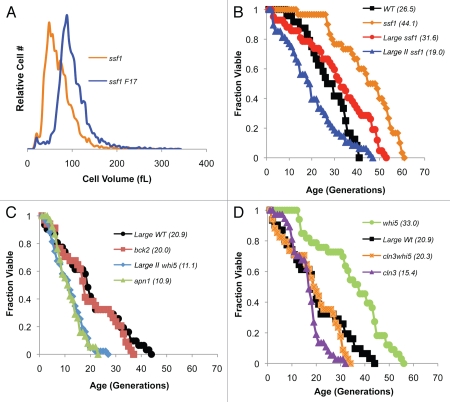
Figure 4.
Lifespan correlates with size in both large cell and whi mutants. (A) Size fractionation of a log phase ssf1Δ diploid culture yielded large cell fractions (F17). (B) Survival curves for large and very large ssf1Δ cells were compared to non-fractionated ssf1Δ or wild type cells (avg. diameter 7.9, 8.8, 6.4 and 6.9 microns, respectively). (C) Size-matching reveals that large wild type cells have a lifespan similar to bck2Δ cells (avg. diameter 7.8 and 7.8, respectively). Size-matched very large whi5Δ cells have a lifespan similar to apn1Δ cells (avg. diameter 9.0 and 9.2, respectively). (D) Survival curves demonstrate that whi5Δ is partially epistatic to cln3Δ. Average lifespan is shown in parentheses (Table 1).
Genetic complementation assays also indicated that lifespan expectancies closely mirror cell size phenotypes. For example, cln3Δ cells are large and short-lived while whi5Δ cells are small and long-lived (Figs. 1–3). Moreover, whi5Δ is partially epistatic to cln3Δ,31,35 and whi5Δcln3Δ cells have an intermediate cell size that is moderately larger than log phase wild type cells (Table 1). Importantly, the lifespan of whi5Δcln3Δ cells mirrors their size (Fig. 4D). For example, the average lifespan of whi5Δcln3Δ is longer than cln3Δ, shorter than whi5Δ, and nearly identical to size matched large wild type cells (Fig. 4D).
Wild type cells entered senescence at a relatively constant size that was independent of birth size (Fig. 2C). To determine whether this was also true in whi or large cell mutants, the size at senescence for ssf1Δ and cln3Δ cells was determined (Fig. 5B and C). Analyses revealed that like wild type cells, ssf1Δ and cln3Δ cells, entered senescence at a relatively constant size that was independent of birth size. Moreover, the ssf1Δ and cln3Δ plots were super imposable (Fig. 5A). Indeed, the average size at which all cells studied entered senescence (11.4 microns) was nearly identical to wild type cells (11.3 microns p = 0.41; Table 1). These data support the conclusion that growth to a maximal cell size limits lifespan. In addition, as was observed with wild type cells, a graded and proportional relationship between size and lifespan was seen in ssf1Δ and cln3Δ cells; increases in birth size translate into decreases in lifespan (Fig. 5B and C). Indeed, by examining the data in quartiles, the lifespan of the largest 25% of ssf1Δ cells decreased 2.7 fold as compared to the smallest 25% (p < 0.0001) (Fig. 5B). Similarly, the lifespan of the largest 25% of cln3Δ cells decreased 3.2 fold as compared to the smallest 25% (p < 0.0001) (Fig. 5C). Furthermore, if the nearly 850 individual cellular aging assays are grouped irrespective of genotype and lifespan is plotted as a function of birth size, a striking correlation between size and lifespan is observed (Fig. 5D). In this case, the largest 10% (avg. 10.9 microns at birth) has an average lifespan that is 4.4 fold less than the smallest 10% (avg. 5.8 microns) (p < 0.0001).
Figure 5.
Longevity is size-dependent. (A) The diameter of ssf1Δ (diamonds) and cln3Δ (circles) cells at senescence are plotted as a function of birth size. Individual ssf1Δ (B) or cln3Δ cells (C) ranging from small to very large were selected from centrifugal elutriation fractions using a micromanipulator. Analyses in quartiles from the smallest (diamonds) to the largest (squares) virgin daughters revealed a strong correlation between birth size and lifespan. Specifically, the size of the smallest 25% of cells was significantly smaller and longer-lived than the largest 25% (p < 0.0001 Table 1). Intermediate quartiles are shown as circles or triangles and average lifespan for each group is shown. (D) Data from 850 individual cellular aging assays were broken into ten sub-groups, from smallest to largest, independent of genotype and lifespan is plotted as a function of birth size. A linear trend line and its R2 value are shown.
Cell size is a major determinant of lifespan.
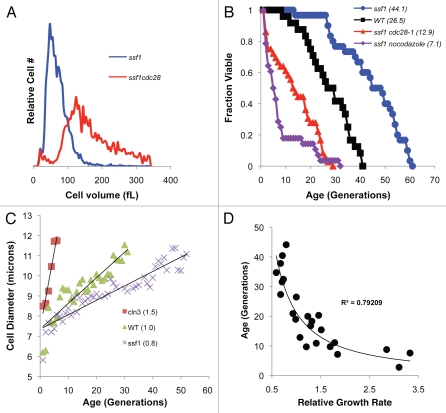
Attempts to directly determine if increasing cell size shortens longevity have had conflicting results.20,26,27 In order to clarify this discrepancy, we used two alternative approaches to artificially enlarge the size of long-lived ssf1Δ cells. Asynchronous log-phase cultures of ssf1Δ cells are dramatically smaller (MCV 66.6 fL) and longer-lived (avg. 44.1 generations) than similarly propagated wild type cells (MCV 90.1 fL and 26.5 generations). However, the generation of ssf1Δcdc28ts double mutants enabled us to rapidly modulate cell size. Incubation of ssf1Δcdc28ts cells at the restrictive temperature arrested cell cycle progression and upon release, generated greatly enlarged virgin daughters with a significantly shortened lifespan (e.g., shortened 71% from an average of 44.1 to 12.9 generations; p < 0.0001) (Fig. 6A and B and Table 1). Following a nocodazole arrest of a log phase ssf1Δ culture, the virgin daughters obtained were even larger than ssf1Δcdc28ts arrested cells (Table 1). Consequently, their average lifespan was reduced 84% to 7Δ1 generations (p < 0.0001) (Fig. 6B). Thus, artificially enlarging the size of long-lived cells within a single generation dramatically reduced their lifespan expectancy.
Figure 6.
Artificially increasing cell size dramatically reduces lifespan. (A) Cell cycle arrests induced by inactivating a cdc28ts allele at the restrictive temperature or through the addition of nocodazole dramatically increased the size of ssf1Δ cells within a single generation (Table 1 and data not shown). (B) Cell survival curves reveal that enlarged ssf1Δ cells have a significantly shortened lifespan. (C) Photomicroscopy reveals that cln3Δ cells increase in size more rapidly than wild type cells. In contrast, ssf1Δ cells enlarge more slowly. Relative growth rate per generation is shown in parentheses. (D) The relative growth rate from all experiments was normalized to wild type cells (set to 1). Plotting lifespan as a function of this rate reveals that as growth rate increases, lifespan decreases. Data from a total of 24 strains and/or experiments are plotted (Table 1) and a power trend line and its R2 value is shown.
Given the dramatic decrease in lifespan observed when the size of ssf1Δ cells was rapidly enlarged, we investigated the relationship between size and the rate at which cells aged. Specifically, since large cells divided faster than smaller isogenic cells (Table 1), perhaps large cells also increase in size faster. Time-lapse photomicroscopy of individual cells was used to determine the size of cells at the start and end of lifespan assays. Subsequently, the rate at which cells increased in size per generation was determined (Table 1). Analyses of these rates revealed that whi mutants had a growth rate per generation that was consistently lower than wild type cells (Table 1). For example, long-lived ssf1Δ cells increase in size 20% less per generation as compared to wild type cells (Fig. 6C). In contrast, short-lived cln3Δ cells increase in size 50% more per generation as compared to wild type cells (Fig. 6C). Indeed, plotting the observed lifespan from 24 different experiments reveals a strong correlation between life expectancy and the relative growth rate per generation (Fig. 6D and Table 1). Therefore, cells with the highest growth rate per generation have the shortest lifespan, and even a modest increase in growth rate greatly shortens lifespan (Fig. 6D and Table 1).
Discussion
Size and lifespan: correlative or causative?
One of the most intriguing aspects of replicative lifespan assays in yeast is the asymmetrical manner in which cells age. The lifespan of mother cells progressively shortens. In contrast, lifespan is purportedly completely regenerated in small daughter cells at birth. The gradual accumulation of ERCs in mother cells has been proposed to explain asymmetrical aging.1 This theory is supported by the observations that overexpression of the Sir2 histone deacetylase or loss of function of the Fob1 replication fork protein reduces ERC buildup and extends lifespan.1 However, this theory has several problems. First, ERC levels do not always correlate with longevity in yeast.1 Second, while Sir2 orthologues promote longevity in a wide range of organisms, ERC formation does not have a role in lifespan determination in higher eukaryotes.1 Thus, the significance of ERC accumulation in yeast needs to be re-evaluated.
Early observations in yeast also correlated increased cell size with a decreased lifespan,18 and old yeast mother cells often produce large daughters with a short lifespan.20 Indeed, a dramatic increase in cell size is one of the most demonstrable morphological changes associated with aging of a wide-range of human and other mammalian cells.4–7 Even in the fission yeast, where cell division is more symmetrical, lifespan correlates with size.36 Moreover, since cells increase in size steadily as they age, old, senescent cells are much larger than young proliferating cells.7,24,37 Furthermore, the fusion of small, young cells with large, older cells shortens lifespan suggesting a direct relationship between cell size and life expectancy.9,37,38
However, a direct relationship between cell size and lifespan has not been established leading to the conclusion that increases in cell size correlate with but are not causative of aging.2,20,39–41 Our results and the work of others have caused us to re-evaluate this conclusion.26–28,42–44
The deletion of at least 16 single or double gene deletions that result in abnormally large cells, produce short-lived cells.29–32,45 The possibility that abnormally large cells die young due to growth or proliferation defects is unlikely. In fact, in every case examined, large cells grew and divided faster than isogenic small cells. Another possibility is that the accumulation of cellular damage is size dependent. However, the observation that osmotic stabilizers extend lifespan46 suggests that cell wall integrity may limit lifespan. Moreover, the observation that at least 20 small cell mutants are long-lived strengthens the link between size and lifespan.29–31,33,34 Furthermore, our cumulative data from ∼850 individual aging assays demonstrates a very strong correlation between size and lifespan; small cells are long-lived and vice versa. Nonetheless, since all of the above results only correlate cell size with life expectancy, we sought to determine whether increasing the size of virgin daughters would directly shorten lifespan. In this manner, we found that using cdc28ts mutations or nocodazole to rapidly increase the size of small, long-lived ssf1Δ cells dramatically shortens lifespan. Importantly, enlarged ssf1Δ cells actually grew and proliferated faster than small, untreated cells demonstrating that the rapid enlargement of these cells did not have an obvious adverse affect on cell growth or division. In addition, several recent reports support the conclusion that increasing cell size limits lifespan.26,27,42,43,47 Conceptually similar experiments have shown that increasing the size of human or chicken fibroblasts also shortens lifespan.9,38 In addition, a mutation in the yeast ATP2 gene that eliminates age asymmetry (e.g., daughter cells are born old) also produces abnormally large cells.48 These results suggest that size is a major determinant of lifespan. How then was this conclusion previously ruled out? Recent results suggest that the four hour alpha-factor arrest used by previous investigators may be too short to observe size-induced senescence; longer arrests clearly yield larger cells with dramatically reduced lifespan.26,27,47 Moreover, in some strains, alpha-factor arrests induce only limited cell growth.49 In addition, since size measurements were not made in the initial study,20 it is possible that despite the alpha-factor treatment, virgin daughters from treated cells were not larger than untreated cells. Finally, it is possible that in some strain backgrounds or under certain conditions, lifespan is less constrained by size.
Growth rate per generation: a model for lifespan regulation.
Why are large yeast cells short-lived? One possibility is that accumulation of ERCs or cellular damage is size-dependent. While ERCs have no known role in mammalian cell aging, the relationship between cellular damage, cell size and senescence has recently been investigated in human cells. The results of several different experiments suggest that hypertrophy and increased size are the cause of senescence and not cellular damage.13,15–17 For example, treatment of human fibroblasts with the DNA damaging agent doxorubicin induces hypertrophy and senescence.13,17,50 However, the presence of rapamycin or decreased serum levels prevents hypertrophy. As a result, cell size is decreased and senescence partially suppressed.13,17 In addition, the ability of ectopic expression of the p21 CDKI in human cells to promote senescence is dependent upon cell growth in the absence of cell cycle progression; suppression of hypertrophy represses senescence.15,16 Thus, cellular damage may accumulate as a result of aging, but cellular damage per se may not always result in aging or senescence.
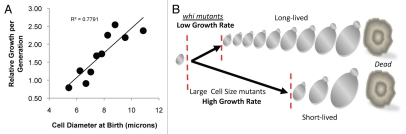
What is the relationship between size, growth rate and replicative lifespan? Our results suggest that a maximal size limits lifespan in yeast. Thus, the simplest model would predict that large cells are short-lived predominately because they are born closer to a terminal cell size than are smaller cells (Fig. 7). While our results support this model, analyses of our data enabled us to determine whether the rate at which cells aged was dependent upon size. Put simply, do large cells age faster than small cells? To examine this we determined the rate at which cells increased in size each generation. Indeed, we found that, in general, large cell mutants grew more per generation than did small cell mutants (Table 1). In addition, independent of their genotype, large cells grew proportionally faster than small cells (e.g., equivalent to exponential cell growth) (Fig. 7A). Moreover, cumulative analyses of data revealed a strong correlation between the amount of cell growth per generation and lifespan. Thus, in most cases, large cells aged faster than smaller cells. Based on these results, we propose the model that two related events, birth size and growth per generation, are major determinants of cellular lifespan (Fig. 7B).
Figure 7.
Modeling Longevity: Size and growth rate per generation impact aging. (A) Data from 850 individual cellular aging assays were broken into ten sub-groups, based on birth size from smallest to largest independent of genotype, and relative growth rate per generation (normalized to wild type cells as in Fig. 6D) is plotted as a function of birth size. A linear trend line and its R2 value are shown. (B) A model for lifespan regulation by growth rate and size. If senescence occurs at a relatively constant maximal cell size, large cell mutants are short-lived because: (1) they are born closer to terminal cell size; and (2) their intrinsically high growth rate per generation (e.g., they increase in size proportionally faster than do small cells) decreases the number of generations necessary to reach a terminal cell size (e.g., they age faster). Conversely, whi mutants are long-lived because they are born small and have an intrinsically low growth rate per generation.
Size and aging: implications, limitations and unanswered questions.
A number of elegant genetic studies have linked ribosome biogenesis with size control. Similarly, simple changes in the dynamics of cell cycle progression can have profound effects on cell size. However, this study is the first to demonstrate a genetic link between cell size control and aging. While the mechanisms that link size with aging are not known, our data suggests that the rate of cell growth per generation is the major mediator between size and lifespan. In this respect, it is not surprising that the majority of gene deletions that concomitantly alter size and lifespan are intimately involved in genetic pathways that regulate ribosome biogenesis, cell growth or cell cycle progression. For example, deletion of a large number of genes involved in ribosome biogenesis results in abnormally small long-lived cells. Specifically, many of these genes encode structural sub-units of the 60S ribosome and are present as homologous pairs in the yeast genome. Remarkably, the deletion of a specific gene of the pair results in small, long-lived cells while deletion of the other appears to have neither effect. While mechanisms responsible for these observations remain unknown, the observation that deletion of the yeast equivalent of the pRB tumor suppressor gene, WHI5, also results in small long-lived cells suggests that a link between cell cycle control and ribosome biogenesis may be integral in modulating the rate at which cells age. In this respect, it is also interesting to note that long-lived fob1Δ cells have a reduced growth rate per generation and fob1Δ mothers are smaller than wild type cells. Conversely, short-lived sir2Δ cells are considerably larger. However, clearly not all long-lived strains are whi mutants nor are all whi mutants likely to be long-lived. In addition, it remains to be determined whether decreased growth per generation is a contributing factor in most longevity mutants. For example, both dietary restriction and rapamycin might promote longevity by reducing cell growth. While the mechanisms that modulate growth per generation are not known, our observations implicate a number of cell cycle proteins. Furthermore, the factors that determine maximal cell size are also not known. While the average maximal size for all strains studied was similar to wild type cells, there were noticeable outliers, and the reasons for these differences remain to be determined. Finally, it is not clear where ERCs fit in. Currently, there is no known connection between ERC levels and size, but a tantalizing possibility is that conditions that alter lifespan without changing ERC levels (e.g., the ability of dietary restriction to increase the lifespan of fob1Δ sir2Δ cells51) might function by either reducing cell size or decreasing the rate of cell growth.
Experimental Procedures
Strains and media.
Yeast strains used were derived from the diploid BY4743 yeast ORF collection.52 Diploid strains were used in all experiments, except Figure 3A and Table 2, because size differences are more easily monitored in diploid cells and to lessen the impact of suppressors. For simplicity, homozygous diploid deletions, e.g., cln3Δ/cln3Δ, are referred to as cln3Δ. All cells were propagated in standard YPD medium.53 Standard yeast genetic techniques were used to generate deletion combinations.54 Cell cycle arrests (4–5 hours) using nocodazole or cdc28ts mutations were conducted as previously described in reference 53.
Replicative lifespan analysis, size selection and growth rate measurements.
Lifespan assays were conducted as previously described in reference 30, using standard YPD plates. Centrifugal elutriation was used for size fractionation as previously described in reference 53 and a Singer MSM Series 300 micro-manipulator was used to further fractionate cells. A Z2 Coulter Counter Channelyzer and Accucomp software (version 3.01a) was used to analyze liquid culture size data. Average cell sizes are given as the geometric mean and data were exported into 86 bins for analysis and normalized relative cell numbers are plotted as a function of cell size. Mean lifespans were calculated as previously noted30 on all cells that budded at least one time. The Wilcoxon matched pairs test (GraphPad InStat version 3.10) was used to determine statistical significance of lifespan differences using a p = 0.05 cutoff. Mann-Whitney tests (GraphPad InStat version 3.10) were used for all other statistical analyses. Cells were photographed with a Nikon Coolpix 4500 camera and a 20x objective.
Photomicroscopy and size measurement of cells in replicative lifespan assays.
To determine daughter birth and budding size, cells are propagated on thin strips of YPD agar at 30°C in an incubation chamber on a Zeiss Axiovert 200 m microscope. Zeiss's multidimensional acquisition program is used to follow 30–50 single cells during a 10–12 hr time course. An AxioCamMR3 camera using EC Plan-Neofluar 40x objective (optovar 1.6) is used to capture images every 2 minutes. The Axiovision (v. 4.17) outline spline function is used to measure and determine cell diameters from the average of three independent measurements. Plots are the diameter at which each individual cell buds. Nikon Coolpix 4,500 images captured on a MSM Series 300 micro-manipulator were used for the determination of cell size at the start or end of RLS assays. Images were imported in Axiovision and calibrated using an objective micrometer. Bright field images (Table 1) from the Singer MSM Series 300 reproducibly yielded proportionally ∼20% larger cell measurements due to the lower quality of the optics as compared to DIC images. However, cross-comparison with DIC images obtained with the Zeiss Axiovert 200 m microscope confirmed the validity of these measurements. Relative growth rate per generation was determined by dividing the net cell size increase per lifetime (size at end minus size at start of the experiment) by the average lifespan.
Conclusions
Yeast, like mammalian cells, enlarges steadily as they age. Since yeast cells enter senescence at a relatively constant cell size, small daughters have a significantly longer life expectancy. Because of this, mutations that reduce cell size extend lifespan. In contrast, mutations that increase birth size or the growth rate per generation dramatically shorten lifespan. A size-dependent entry into senescence may reflect a physical constraint. Above a maximal size, the cell wall might be too weak to support division. Indeed, growth on sorbitol extends lifespan.46 Nonetheless, increasing either cell size or growth per generation dramatically reduces lifespan even in normally long-lived mutants.
Acknowledgements
We would like to thank Sandra Whelly, Jim Hutson and Colette Schneider for critical insights. B.L.S. has been supported by grants from the NIH R01GM077874 and R01GM077874-04S1 and the Ted Nash Long Life Foundation.
Supplementary Material
References
- 1.Kaeberlein M. Lessons on longevity from budding yeast. Nature. 2010;464:513–519. doi: 10.1038/nature08981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Henderson KA, Gottschling DE. A mother's sacrifice: What is she keeping for herself? Curr Opin Cell Biol. 2008;20:723–728. doi: 10.1016/j.ceb.2008.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mortimer RaJ., JR Life span of individual yeast cells. Nature. 1959;183:1751–1752. doi: 10.1038/1831751a0. [DOI] [PubMed] [Google Scholar]
- 4.Cristofalo VJ, Kritchevsky D. Cell size and nucleic acid content in the diploid human cell line WI-38 during aging. Med Exp Int J Exp Med. 1969;19:313–320. doi: 10.1159/000137216. [DOI] [PubMed] [Google Scholar]
- 5.Simons JW. The use of frequency distributions of cell diameters to characterize cell populations in tissue culture. Exp Cell Res. 1967;45:336–350. doi: 10.1016/0014-4827(67)90184-x. [DOI] [PubMed] [Google Scholar]
- 6.Adolphe M, Ronot X, Jaffray P, Hecquet C, Fontagne J, Lechat P. Effects of donor's age on growth kinetics of rabbit articular chondrocytes in culture. Mech Ageing Dev. 1983;23:191–198. doi: 10.1016/0047-6374(83)90067-2. [DOI] [PubMed] [Google Scholar]
- 7.Treton JA, Courtois Y. Evolution of the distribution, proliferation and ultraviolet repair capacity of rat lens epithelial cells as a function of maturation and aging. Mech Ageing Dev. 1981;15:251–267. doi: 10.1016/0047-6374(81)90134-2. [DOI] [PubMed] [Google Scholar]
- 8.Hayflick L, Moorhead PS. The serial cultivation of human diploid cell strains. Exp Cell Res. 1961;25:585–621. doi: 10.1016/0014-4827(61)90192-6. [DOI] [PubMed] [Google Scholar]
- 9.Pendergrass W, Angello J, Norwood TH. The relationship between cell size, the activity of DNA polymerase alpha and proliferative activity in human diploid fibroblast-like cell cultures. Exp Gerontol. 1989;24:383–393. doi: 10.1016/0531-5565(89)90046-6. [DOI] [PubMed] [Google Scholar]
- 10.Greenberg SB, Grove GL, Cristofalo VJ. Cell size in aging monolayer cultures. In Vitro. 1977;13:297–300. doi: 10.1007/BF02616174. [DOI] [PubMed] [Google Scholar]
- 11.Schneider EL, Fowlkes BJ. Measurement of DNA content and cell volume in senescent human fibroblasts utilizing flow multiparameter single cell analysis. Exp Cell Res. 1976;98:298–302. doi: 10.1016/0014-4827(76)90441-9. [DOI] [PubMed] [Google Scholar]
- 12.Absher PM, Absher RG. Clonal variation and aging of diploid fibroblasts. Cinematographic studies of cell pedigrees. Exp Cell Res. 1976;103:247–255. doi: 10.1016/0014-4827(76)90261-5. [DOI] [PubMed] [Google Scholar]
- 13.Demidenko ZN, Blagosklonny MV. Growth stimulation leads to cellular senescence when the cell cycle is blocked. Cell Cycle. 2008;7:3355–3361. doi: 10.4161/cc.7.21.6919. [DOI] [PubMed] [Google Scholar]
- 14.Chang BD, Xuan Y, Broude EV, Zhu H, Schott B, Fang J, et al. Role of p53 and p21waf1/cip1 in senescence-like terminal proliferation arrest induced in human tumor cells by chemotherapeutic drugs. Oncogene. 1999;18:4808–4818. doi: 10.1038/sj.onc.1203078. [DOI] [PubMed] [Google Scholar]
- 15.Demidenko ZN, Blagosklonny MV. At concentrations that inhibit mTOR, resveratrol suppresses cellular senescence. Cell Cycle. 2009;8:1901–1904. doi: 10.4161/cc.8.12.8810. [DOI] [PubMed] [Google Scholar]
- 16.Demidenko ZN, Shtutman M, Blagosklonny MV. Pharmacologic inhibition of MEK and PI-3K converges on the mTOR/S6 pathway to decelerate cellular senescence. Cell Cycle. 2009;8:1896–1900. doi: 10.4161/cc.8.12.8809. [DOI] [PubMed] [Google Scholar]
- 17.Demidenko ZN, Zubova SG, Bukreeva EI, Pospelov VA, Pospelova TV, Blagosklonny MV. Rapamycin decelerates cellular senescence. Cell Cycle. 2009;8:1888–1895. doi: 10.4161/cc.8.12.8606. [DOI] [PubMed] [Google Scholar]
- 18.Johnston JR. Reproductive capacity and mode of death of yeast cells. Antonie Van Leeuwenhoek. 1966;32:94–98. doi: 10.1007/BF02097448. [DOI] [PubMed] [Google Scholar]
- 19.Egilmez NK, Chen JB, Jazwinski SM. Preparation and partial characterization of old yeast cells. Journal of gerontology. 1990;45:9–17. doi: 10.1093/geronj/45.1.b9. [DOI] [PubMed] [Google Scholar]
- 20.Kennedy BK, Austriaco NR, Jr, Guarente L. Daughter cells of Saccharomyces cerevisiae from old mothers display a reduced life span. J Cell Biol. 1994;127:1985–1993. doi: 10.1083/jcb.127.6.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Muller I. Parental age and the life-span of zygotes of Saccharomyces cerevisiae. Antonie van Leeuwenhoek. 1985;51:1–10. doi: 10.1007/BF00444223. [DOI] [PubMed] [Google Scholar]
- 22.Norwood TH, Pendergrass WR, Sprague CA, Martin GM. Dominance of the senescent phenotype in heterokaryons between replicative and post-replicative human fibroblast-like cells. Proc Natl Acad Sci USA. 1974;71:2231–2235. doi: 10.1073/pnas.71.6.2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pereira-Smith OM, Stein GH, Robetorye S, Meyer-Demarest S. Immortal phenotype of the HeLa variant D98 is recessive in hybrids formed with normal human fibroblasts. J Cell Physiol. 1990;143:222–225. doi: 10.1002/jcp.1041430204. [DOI] [PubMed] [Google Scholar]
- 24.Angello JC, Pendergrass WR, Norwood TH, Prothero J. Proliferative potential of human fibroblasts: an inverse dependence on cell size. J Cell Physiol. 1987;132:125–130. doi: 10.1002/jcp.1041320117. [DOI] [PubMed] [Google Scholar]
- 25.Woldringh CL, Huls PG, Vischer NO. Volume growth of daughter and parent cells during the cell cycle of Saccharomyces cerevisiae a/alpha as determined by image cytometry. J Bacteriol. 1993;175:3174–3181. doi: 10.1128/jb.175.10.3174-3181.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zadrag R, Kwolek-Mirek M, Bartosz G, Bilinski T. Relationship between the replicative age and cell volume in Saccharomyces cerevisiae. Acta Biochim Pol. 2006;53:747–751. [PubMed] [Google Scholar]
- 27.Zadrag-Tecza R, Kwolek-Mirek M, Bartosz G, Bilinski T. Cell volume as a factor limiting the replicative lifespan of the yeast Saccharomyces cerevisiae. Biogerontology. 2008 doi: 10.1007/s10522-008-9192-0. [DOI] [PubMed] [Google Scholar]
- 28.Bilinski T, Bartosz G. Hypothesis: cell volume limits cell divisions. Acta Biochim Pol. 2006;53:833–835. [PubMed] [Google Scholar]
- 29.Kaeberlein M, Kirkland KT, Fields S, Kennedy BK. Genes determining yeast replicative life span in a long-lived genetic background. Mech Ageing Dev. 2005;126:491–504. doi: 10.1016/j.mad.2004.10.007. [DOI] [PubMed] [Google Scholar]
- 30.Kaeberlein M, Powers RW, 3rd, Steffen KK, Westman EA, Hu D, Dang N, et al. Regulation of yeast replicative life span by TOR and Sch9 in response to nutrients. Science. 2005;310:1193–1196. doi: 10.1126/science.1115535. [DOI] [PubMed] [Google Scholar]
- 31.Jorgensen P, Nishikawa JL, Breitkreutz BJ, Tyers M. Systematic identification of pathways that couple cell growth and division in yeast. Science. 2002;297:395–400. doi: 10.1126/science.1070850. [DOI] [PubMed] [Google Scholar]
- 32.Zhang J, Schneider C, Ottmers L, Rodriguez R, Day A, Markwardt J, et al. Genomic Scale Mutant Hunt Identifies Cell Size Homeostasis Genes in S. cerevisiae. Curr Biol. 2002;12:1992–2001. doi: 10.1016/s0960-9822(02)01305-2. [DOI] [PubMed] [Google Scholar]
- 33.Steffen KK, MacKay VL, Kerr EO, Tsuchiya M, Hu D, Fox LA, et al. Yeast life span extension by depletion of 60s ribosomal subunits is mediated by Gcn4. Cell. 2008;133:292–302. doi: 10.1016/j.cell.2008.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jorgensen P, Rupes I, Sharom JR, Schneper L, Broach JR, Tyers M. A dynamic transcriptional network communicates growth potential to ribosome synthesis and critical cell size. Genes Dev. 2004;18:2491–2505. doi: 10.1101/gad.1228804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Costanzo M, Nishikawa JL, Tang X, Millman JS, Schub O, Breitkreuz K, et al. CDK activity antagonizes Whi5, an inhibitor of G1/S transcription in yeast. Cell. 2004;117:899–913. doi: 10.1016/j.cell.2004.05.024. [DOI] [PubMed] [Google Scholar]
- 36.Barker MG, Walmsley RM. Replicative ageing in the fission yeast Schizosaccharomyces pombe. Yeast (Chichester, England) 1999;15:1511–1518. doi: 10.1002/(sici)1097-0061(199910)15:14<1511::aid-yea482>3.3.co;2-p. [DOI] [PubMed] [Google Scholar]
- 37.Mitsui Y, Schneider EL. Relationship between cell replication and volume in senescent human diploid fibroblasts. Mech Ageing Dev. 1976;5:45–56. doi: 10.1016/0047-6374(76)90007-5. [DOI] [PubMed] [Google Scholar]
- 38.Angello JC, Pendergrass WR, Norwood TH, Prothero J. Cell enlargement: one possible mechanism underlying cellular senescence. J Cell Physiol. 1989;140:288–294. doi: 10.1002/jcp.1041400214. [DOI] [PubMed] [Google Scholar]
- 39.Sinclair D, Mills K, Guarente L. Aging in Saccharomyces cerevisiae. Annu Rev Microbiol. 1998;52:533–560. doi: 10.1146/annurev.micro.52.1.533. [DOI] [PubMed] [Google Scholar]
- 40.Sinclair DA, Mills K, Guarente L. Molecular mechanisms of yeast aging. Trends Biochem Sci. 1998;23:131–134. doi: 10.1016/s0968-0004(98)01188-8. [DOI] [PubMed] [Google Scholar]
- 41.Lin SJ, Sinclair D. Molecular mechansisms of aging: Insights from budding yeast. Cold Spring Harbor, NY: Cold Spring Harbor Press; 2008. [Google Scholar]
- 42.Demidenko ZN, Blagosklonny MV. Quantifying pharmacologic suppression of cellular senescence: prevention of cellular hypertrophy versus preservation of proliferative potential. Aging. 2009;1:1008–1016. doi: 10.18632/aging.100115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Blagosklonny MV, Hall MN. Growth and aging: a common molecular mechanism. Aging. 2009;1:357–362. doi: 10.18632/aging.100040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chiocchetti A, Zhou J, Zhu H, Karl T, Haubenreisser O, Rinnerthaler M, et al. Ribosomal proteins Rpl10 and Rps6 are potent regulators of yeast replicative life span. Exp Gerontol. 2007;42:275–286. doi: 10.1016/j.exger.2006.11.002. [DOI] [PubMed] [Google Scholar]
- 45.Kaeberlein M, Guarente L. Saccharomyces cerevisiae MPT5 and SSD1 function in parallel pathways to promote cell wall integrity. Genetics. 2002;160:83–95. doi: 10.1093/genetics/160.1.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kaeberlein M, Andalis AA, Fink GR, Guarente L. High osmolarity extends life span in Saccharomyces cerevisiae by a mechanism related to calorie restriction. Mol Cell Biol. 2002;22:8056–8066. doi: 10.1128/MCB.22.22.8056-8066.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zadrag R, Bartosz G, Bilinski T. Is the yeast a relevant model for aging of multicellular organisms? An insight from the total lifespan of Saccharomyces cerevisiae. Curr Aging Sci. 2008;1:159–165. doi: 10.2174/1874609810801030159. [DOI] [PubMed] [Google Scholar]
- 48.Lai CY, Jaruga E, Borghouts C, Jazwinski SM. A mutation in the ATP2 gene abrogates the age asymmetry between mother and daughter cells of the yeast Saccharomyces cerevisiae. Genetics. 2002;162:73–87. doi: 10.1093/genetics/162.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Goranov AI, Cook M, Ricicova M, Ben-Ari G, Gonzalez C, Hansen C, et al. The rate of cell growth is governed by cell cycle stage. Genes Dev. 2009;23:1408–1422. doi: 10.1101/gad.1777309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chang BD, Broude EV, Dokmanovic M, Zhu H, Ruth A, Xuan Y, et al. A senescence-like phenotype distinguishes tumor cells that undergo terminal proliferation arrest after exposure to anticancer agents. Cancer Res. 1999;59:3761–3767. [PubMed] [Google Scholar]
- 51.Kaeberlein M, Kirkland KT, Fields S, Kennedy BK. Sir2-independent life span extension by calorie restriction in yeast. PLoS Biol. 2004;2:296. doi: 10.1371/journal.pbio.0020296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Winzeler EA, Shoemaker DD, Astromoff A, Liang H, Anderson K, Andre B, et al. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science. 1999;285:901–906. doi: 10.1126/science.285.5429.901. [DOI] [PubMed] [Google Scholar]
- 53.Day A, Schneider C, Schneider BL. Yeast cell synchronization. Methods Mol Biol. 2004;241:55–76. doi: 10.1385/1-59259-646-0:55. [DOI] [PubMed] [Google Scholar]
- 54.Amberg DC, Burke D, Strathern JN. Cold Spring Harbor Laboratory. Methods in yeast genetics: a Cold Spring Harbor Laboratory course manual. Cold Spring Harbor NY: Cold Spring Harbor Laboratory Press; 2005. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.