SUMMARY
Defining protein interactions forms the basis for discovery of biological pathways, disease mechanisms, and opportunities for therapeutic intervention. To harness the robust binding affinity and selectivity of structured peptides for interactome discovery, we engineered photoreactive stapled BH3 peptide helices that covalently capture their physiologic BCL-2 family targets. The crosslinking α-helices covalently trap both static and dynamic protein interactors, and enable rapid identification of interaction sites, providing a critical link between interactome discovery and targeted drug design.
INTRODUCTION
Protein interactions mediate innumerable cellular activities in health and disease. Like the teeth of a key that perfectly match a lock, complementary protein shape is critical to the execution of biological interactions. These contact points are embedded within a complex protein structure that provides the infrastructure to maintain the essential bioactive fold. When the biological activity of proteins is assigned to discrete subdomains, their mode of action, menu of protein targets, and explicit sites of target engagement are frequently unknown. Ideally, these evolutionarily honed substructures could be used to capture and thereby catalogue their protein targets. However, out of context from the whole protein, bioactive subdomains often unfold, resulting in loss of biological shape, potency, and specificity.
From the surface of the cell to its inner nuclear core, α-helical interactions govern such critical processes as infection (Weissenhorn et al., 1997), the immune response (Blum et al., 1993), apoptosis (Sattler et al., 1997), and transcription (Kussie et al., 1996). Importantly, the natural complexity of peptide α-helices enables them to discern among homologous targets. Whereas some protein interaction sites are remarkably similar in topography, helical peptides can function as high fidelity ligands due to the variability afforded by their amino acid composition. Indeed, we have found that chemical stabilization of pro-apoptotic BCL-2 family peptides yields reliable α-helical surrogates that bind both stable and transient physiologic interactors (Walensky et al., 2006), and even uncover novel sites of protein interaction (Gavathiotis et al., 2008). To leverage the enormous capacity of structured peptides to selectively bind and capture their protein targets, we combined chemical restoration of α-helical shape with installation of a covalent capture moiety to generate photoreactive stabilized alpha-helices of BCL-2 domains (pSAHBs) for protein interaction analysis.
RESULTS
Development of pSAHBs: Synthesis, Binding Activity, and Covalent Capture
pSAHBs are designed to incorporate through conservative aromatic residue substitution a non-natural amino acid bearing a benzophenone moiety (4-benzoylphenylalanine, Bpa), which covalently crosslinks to protein targets upon exposure to ultraviolet (UV) light (Dorman and Prestwich, 1994; Saghatelian et al., 2004; Vodovozova, 2007) (Figure 1A). To restore α-helical structure to a synthetic peptide that incorporates Bpa, we further install an i, i+4 pair of non-natural amino acids bearing olefin tethers that when subjected to ring closing metathesis yields a hydrocarbon stapled peptide (Blackwell et al., 2001; Schafmeister et al., 2000). We generated a panel of pSAHBs modeled after the BH3 death domain of the pro-apoptotic protein BAD (Yang et al., 1995) to test our α-helical interaction covalent capture strategy. Substitution of Bpa at each of the native aromatic amino acid positions contained within BAD BH3 yielded BAD pSAHBs 1–4, three of which (BAD pSAHBs 2–4) localized to the BH3-binding interface (Figure 1B). Importantly, fluorescence polarization assays performed using FITC-derivatized BAD pSAHBs and recombinant BCL-XLΔC confirmed that Bpa substitution did not disrupt target binding (Fig. 1C).
Figure 1. Transformation of a Bioactive Peptide Domain into a Structurally Stable and Photoreactive α-Helix.
(A) Photoreactive stabilized alpha-helix of BCL-2 domains (pSAHBs) for protein capture are generated by replacing native aromatic residues with 4-benzoylphenylalanine (Bpa) followed by ring closing metathesis of olefinic non-natural amino acids installed at i, i +4 positions.
(B) A panel of pSAHBs was modeled after the BH3 death domain of pro-apoptotic BAD.
(C) BAD pSAHBs retain high affinity binding to anti-apoptotic BCL-XLΔC, as measured by fluorescence polarization assay. Error bars represent mean +/− s.d. for binding assays performed in at least triplicate.
(D) Select fluorescently labeled BAD pSAHBs crosslink to BCL-XLΔC upon exposure to UV irradiation, as detected by coomassie stain and fluorescence imaging.
(E) Time-course for UV-dependent crosslinking of BAD pSAHB-2 to BCL-XLΔC as monitored by fluorescence scan and coomassie stain, and the corresponding densitometry-based calculation of crosslinking yield using ImageJ software.
FITC-BAD pSAHBs were incubated with BCL-XLΔC at a ratio of 1:1 and upon exposure to UV light, FITC-BAD pSAHB-2 and to a lesser extent FITC-BAD pSAHB-4, which contain Bpa residues predicted to localize to the BAD BH3/BCL-XLΔC binding interface (PDB ID 2BZW), underwent crosslinking to BCL-XLΔC as detected by coomassie stain and fluorescence scan (Figure 1D). Whereas the Bpa residue in FITC-BAD pSAHB-1 does not localize to the helical binding interface and correspondingly exhibited no crosslinking activity, FITC-BAD pSAHB-3, which contains an F121Bpa substitution that does reside at the binding interface, also showed no photoaffinity labeling. These data highlight that even when target binding is uniformly preserved in Bpa-substituted SAHBs (Figure 1C), generating a panel of stapled peptides containing differentially localized photoreactive moieties is advisable so that an optimal construct for protein crosslinking can be identified. Kinetic analysis of a 1:1 peptide:protein mixture of our most effective photoreactive α-helix, BAD pSAHB-2, demonstrated time-dependent production of crosslinked BCL-XLΔC, with approximately 35% conversion by 2 hours of UV exposure (Figure 1E).
Binding Specificity of pSAHBs
To examine the specificity of covalent crosslinking, FITC-BAD pSAHB-2 was first incubated with bovine serum albumin (BSA) alone or with mixtures of BCL-XLΔC containing increasing amounts of BSA. In each case, FITC-BAD pSAHB-2 only crosslinked to BCL-XLΔC and the quantity of added BSA had no impact on the yield of the FITC-BAD pSAHB-2/BCL-XLΔC adduct, which was equivalent across all conditions (Figure 2A). To confirm that Bpa substitution itself did not account for the absence of BSA binding, we conducted a fluorescence polarization binding assay comparing FITC-BAD SAHB and FITC-BAD pSAHB-2 (Supplementary Figure 1A). We found that wild-type and Bpa-substituted FITC-BAD SAHBs engaged albumin with similarly low affinity, highlighting that covalent capture of BCL-XLΔC by BAD pSAHB is indeed selective and that despite the abundance of BSA in the reaction mixture, no off-target crosslinking is observed.
Figure 2. pSAHBs Selectively Capture their Physiologic Targets, Trapping Both Static and Dynamic Interactors.
(A) The selectivity of BAD pSAHB covalent capture is highlighted by the absence of crosslinking to bovine serum albumin (BSA) and the inability of BSA to disrupt FITC-BAD pSAHB-2/BCL-XLΔC crosslinking.
(B) Biotinylated BAD pSAHB-2 was incubated with an equimolar mixture of homologous anti-apoptotic proteins at the indicated ratios in the presence of UV light for 1.5 hours. Western analysis of the isolated crosslinked proteins revealed high fidelity capture of physiologic interactors BCL-XLΔC, BCL-2ΔC, and BCL-wΔC, but not BFL1/A1ΔC, which lies outside the in vivo binding spectrum of the BAD protein. SA, streptavidin.
(C) A panel of pSAHBs was modeled after the BIM BH3 domain for comparative crosslinking studies with BAD pSAHBs.
(D) BAD and BIM pSAHBs covalently trap their distinct profile of anti-apoptotic binding partners within U937 cellular extracts.
(E) In addition to capturing stable protein interactors, BIM pSAHBs 2–4 crosslink to pro-apoptotic BAX, a fleeting “hit and run” target of the BH3-only protein BIM. See also Supplementary Figure 1.
Having demonstrated that BAD pSAHB-2 was grossly selective for its protein target in vitro, we next investigated whether the photoreactive stapled peptide could discriminate with high fidelity among the variety of homologous anti-apoptotic proteins and crosslink only to its physiologic binding partners. Because cells express variable levels of anti-apoptotic proteins, potentially biasing crosslinking results based on relative protein abundance, we first examined crosslinking fidelity by incubating increasing amounts of biotinylated BAD pSAHB-2 with an equimolar mixture of recombinant BCL-2ΔC, BCL-XLΔC, BCL-wΔC, BFL-1/A1ΔC, and MCL-1ΔNΔC. Following UV irradiation, the reaction mixtures were subjected to streptavidin-based affinity purification, stringent washing to eliminate non-covalent binders, elution, electrophoresis, and western analysis. Whereas exposure to BAD pSAHB-2 caused dose-responsive photoaffinity labeling of BCL-2ΔC, BCL-XLΔC, and BCL-wΔC, as reflected by a ~3.4 kDa molecular shift, no such effect was observed for BFL-1/A1ΔC (Figure 2B). Because the truncated MCL-1ΔNΔC construct was not detected by available anti-MCL-1 antibodies, we subjected the electrophoresed eluates to silver stain and MS analysis, and confirmed that BAD pSAHB-2 likewise did not crosslink to MCL-1ΔNΔC (Supplementary Figure 1B). Thus, pSAHBs can be designed to retain high affinity target binding and selectively capture their physiologic interaction partners in the context of an equimolar mixture of homologous protein targets.
pSAHBs Trap Static and Dynamic Protein Interactors
We next tested whether pSAHBs could function as reliable surrogates of BH3 domain-containing proteins and discriminate among homologous targets in a complex protein mixture. Whereas pro-apoptotic BAD exhibits a selective anti-apoptotic binding profile, engaging BCL-XL but not MCL-1, pro-apoptotic BIM broadly interacts with anti-apoptotic proteins (Chen et al., 2005). We incubated biotinylated BAD pSAHB-2 (Figure 1B) or BIM pSAHB-3 (Figure 2C) with a cellular extract from U937 myeloid leukemia cells that overexpress both BCL-XL and MCL-1 (Chen et al., 2007) in the presence or absence of UV irradiation, followed by streptavidin-based affinity purification, stringent washing to eliminate non-covalent binders, elution, electrophoresis, and western analysis. Whereas the non-irradiated samples and vehicle-treated irradiated extracts showed no crosslinked species, BAD pSAHB-2 crosslinked to endogenous BCL-XL but not MCL-1, and BIM pSAHB-3 crosslinked to both proteins (Figure 2D), mirroring the physiologic binding profile of the corresponding BH3-only proteins.
Many critical biological interactions are transient or are of insufficient affinity to withstand the experimental conditions required to prepare cellular extracts or isolate complexes by affinity chromatography. In order to determine if pSAHBs could be applied to capture fleeting α-helical interactions, we explored whether BIM pSAHB could trap the defined “hit and run” interaction between this direct BH3 activator and pro-apoptotic BAX (Gavathiotis et al., 2008; Wei et al., 2000). Cellular extracts prepared from mouse embryo fibroblasts were incubated with a panel of biotinylated BIM pSAHBs (Figure 2C) in the presence of UV irradiation, followed by isolation and analysis of BIM pSAHB-crosslinked species. BIM pSAHB-1, the only construct containing a Bpa moiety localized to the non-interacting surface of the BIM BH3 helix, exhibited little to no crosslinking activity. However, BIM pSAHBs-3 and -4, and to a lesser extent BIM pSAHB-2, covalently captured this otherwise fleeting target (Figure 2E). Thus, by trapping transitory interactions in addition to static binding events, pSAHBs can be used to probe the mechanisms of dynamic BH3-mediated signal transduction events.
pSAHBs Localize the Site of Protein Interaction
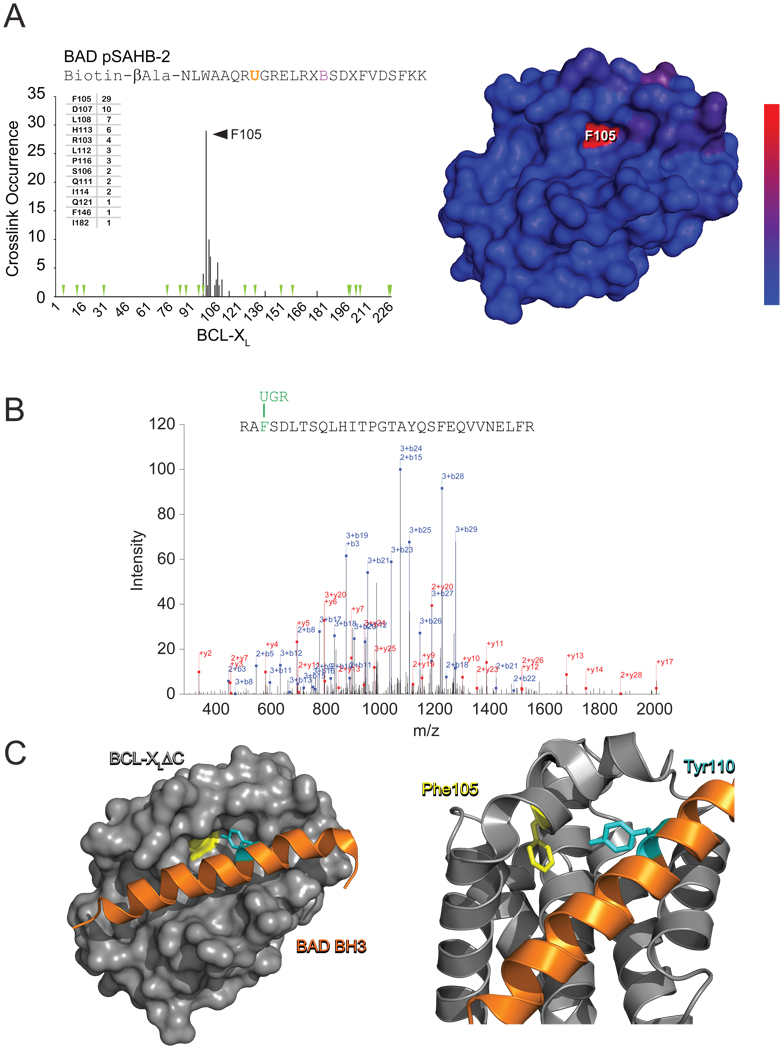
Given the importance of pharmacologic targeting of protein interactions in human disease, a rapid method for localizing helix-target binding sites would be especially advantageous for clinical translation. To explore the capacity of pSAHBs to efficiently identify explicit sites of protein engagement, we subjected a mixture of BAD pSAHB-2 and BCL-XLΔC to UV exposure, followed by SDS-PAGE electrophoresis and excision of the crosslinked species for nano liquid chromatography and mass spectrometry (nano-LC/MS) analysis. MS/MS spectra were searched using the SEQUEST algorithm (Eng et al., 1994) against a database containing BCL-XLΔC, trypsin, and common keratin contaminants. Bpa-crosslinked tryptic fragments were identified by searching for the variable modification corresponding to the mass of the tryptic BCL-XLΔC fragment plus the Bpa-containing pSAHB fragment, UGR (482.22777 Da). To obtain an estimate for false positive detection rate (FDR), the reversed protein sequences were also included in the database search (Elias and Gygi, 2010). The subset of crosslinked peptides identified by the SEQUEST search were filtered such that the maximum FDR was 5%, yielding a high confidence list of covalently modified sites, which were plotted by frequency of occurrence (Figure 3A). Strikingly, the Bpa moiety substituted at position Y110 of the BAD BH3 helix crosslinked to a discrete subset of amino acids that colocalized within the canonical BH3-binding groove of BCL-XL. Indeed, the single most frequently crosslinked BCL-XL residue, F105, represents the nearest neighbor to Y110 in the crystal structure of the BAD BH3/BCL-XLΔC complex (Figure 3B, 3C).
Figure 3. BAD pSAHB Precisely Localizes the Structurally-Defined BAD BH3/BCL-XLΔC Binding Site.
(A) BAD pSAHB-2 was incubated with BCL-XLΔC and the mixture subjected to UV irradiation (1.5 h), electrophoresis, excision of the crosslinked protein, trypsin proteolysis, and LC-MS/MS analysis. The plot depicts the frequency of crosslinked sites identified across the BCL-XLΔC polypeptide sequence. Mapping of the crosslinked residues onto the BCL-XLΔC structure revealed their striking colocalization within a circumscribed region of the canonical BH3-binding pocket, with the frequency of occurrence reflected by the color scale. Green arrowheads, trypsin digestion sites.
(B) The predominant Bpa-crosslinked tryptic fragment contained a covalent modification of BCL-XLΔC residue F105, as exemplified by the MS/MS spectrum derived from collision-induced dissociation of the (M+4H)4+ precursor (0.37 parts per million mass accuracy).
(C) The most abundant crosslink, located between BAD pSAHB-2 Bpa110 and BCL-XLΔC F105, precisely matches the structurally-defined interaction between BAD BH3 Y110 and BCL-XLΔC F105 at the BH3-binding pocket of BCL-XLΔC (PDB ID 2BZW).
Having validated the capacity of a pSAHB to correctly localize the ligand-target interface of a structurally-defined interaction, we next examined whether pSAHBs could be deployed to pinpoint and characterize a structurally-undefined binding interface. We chose the full-length BCL-2 family anti-apoptotic protein BCL-w to pilot our methodology, as BCL-w is a potential target of the BAD BH3-mimetic drug ABT-263, which is currently in clinical trials as an anti-cancer agent, yet a structural characterization of BCL-w in complex with its physiologic BAD BH3 ligand has not been reported. To obtain a pair of binding site coordinates for computational docking analysis, recombinant BCL-w was incubated with BAD pSAHB-2 or BAD pSAHB-4 as a 1:1 mixture and subjected to UV irradiation, followed by electrophoresis, tryptic digestion of the excised crosslinked species, and nano-LC/MS analysis. Of note, we created a tryptic site at the non-interacting face of BAD pSAHB-4 by D123R mutagenesis in order to ensure that the resultant Bpa-containing pSAHB-4 fragment was sufficiently small for optimal MS/MS sequencing (i.e. SUK: 484.23218 Da; SUKK: 611.34313 Da). As described above, Bpa-crosslinked tryptic fragments were identified by searching for the variable modifications corresponding to the masses of the tryptic BCL-XLΔC fragment plus the Bpa-containing pSAHB fragments, yielding a filtered list of crosslinking sites for each BAD pSAHB, plotted by frequency of occurrence per BCL-w residue (Figure 4A, 4B). As with BAD pSAHB-2/BCL-XLΔC crosslinking, one predominant amino acid site emerged with a frequency of occurrence that exceeded the average number of hits by more than two standard deviations above the mean. This top scoring hit for each pSAHB was then employed in computational docking analysis (CNS within HADDOCK 2.0) (Brunger et al., 1998; Dominguez et al., 2003) to calculate a model structure for the BAD BH3/BCL-w interaction starting with the NMR structure of full-length BCL-w (PDB ID 1O0L), the crystal structure of the BAD BH3 helix (2BZW), and the active BAD pSAHB/BCL-w residue pairs Y110/L63 and F125/R55.
Figure 4. Application of pSAHBs to Localize and Characterize a Structurally-Undefined Helix/Protein Interaction Site.
(A, B) BAD pSAHB-2 (A) or BAD pSAHB-4 (B) was incubated with full-length BCL-w and the mixture subjected to UV irradiation, electrophoresis, excision of the crosslinked protein, trypsin proteolysis, and LC-MS/MS analysis. The plots (left) and color scale (right) depict the frequency of BCL-w crosslinked sites, which map to distinct, circumscribed regions within the canonical BH3-binding pocket of BCL-w (right).
(C) The top scoring hit for each pSAHB was employed in computational docking analysis to calculate a model structure for the BAD BH3/BCL-w interaction. The BAD BH3 helix engages the canonical BH3-binding pocket of BCL-w with the N to C terminus of the peptide disposed right to left. The en face view (top) demonstrates the complementary electrostatic interactions of D119 and R115 of BAD BH3 with R94 and E84 of BCL-w, respectively. A top view of the structure (bottom) highlights the juxtaposition of crosslinked positions Y110 and F125 (cyan) of the BAD BH3 helix with L63 and R55 of BCL-w, respectively, along the core hydrophobic interface. The side chains of hydrophobic, positively charged, negatively charged, and hydrophilic residues are colored yellow, blue, red and green, respectively. To display an unobstructed view of the BH3-binding site, the BCL-w C-terminus is hidden in the images.
(D) The interaction topography of the calculated model structure of BAD BH3/BCL-w is notably similar to that of the structurally-defined BAD BH3/BCL-XLΔC complex (PDB ID 2BZW). The en face view (top) demonstrates the homologous BAD BH3/BCL-XLΔC electrostatic interaction pairs D119/R139 and R115/E129, whereas the core BAD BH3 residues A106, Y110, L114, M117, F121, and F125 of the hydrophobic interface (bottom) are remarkably similar in disposition to these same residues in the calculated model structure of BAD BH3/BCL-w.
See also Supplementary Figure 2.
The calculated model structures converged to position the BAD BH3 helix at the canonical BH3-binding pocket of BCL-w (Figure 4C, Supplementary Figure 2), with the N to C terminus directionality of the peptide disposed right to left and the interaction topography notably similar to that of the BAD BH3/BCL-XLΔC complex previously determined by X-ray crystallography (PDB ID 2BZW) (Figure 4D). The en face views of the complexes highlight the homologous complementary electrostatic interactions at the perimeter of the core hydrophobic binding interface, with charged residues D119 and R115 of BAD BH3 respectively engaging R94 and E84 of BCL-w in an analogous manner to the BAD BH3/BCL-XLΔC interaction pairs D119/R139 and R115/E129. The calculated orientation of core BAD BH3 residues A106, Y110, L114, M117, F121, and F125 at the hydrophobic interface of the BCL-w binding pocket are also remarkably similar to the disposition of these same residues in the BAD BH3/BCL-XLΔC crystal structure, highlighting both the fidelity and utility of the pSAHB approach. Thus, we establish an efficient method for combining pSAHB-derived MS/MS site identification data with computational docking analysis to rapidly localize and structurally characterize a peptide-helix/target protein binding interface.
DISCUSSION
Here we provide the blueprints for developing a novel, versatile, and potentially expansive reagent set for protein capture based on the generation of photoreactive stapled peptide helices. By harnessing the native and complex topography of a critical protein interaction motif of the BCL-2 family, pSAHBs serve as high fidelity research tools that efficiently, selectively, and irreversibly crosslink both static and dynamic protein targets for proteomic and mechanistic analyses. In addition, the compounds can be applied to locate protein interaction sites, which may serve as templates for therapeutic targeting. This capacity to rapidly and accurately map binding interfaces using a combined MS/MS and computational docking strategy provides an alternative and complementary approach to bridging the gap between discovering protein interactions and defining biologically-relevant interfaces using definitive structural methods, which can often take years to accomplish. A unique and compelling feature of this approach is the ability to conduct rigorous validation studies for the identified targets and their sites of interaction. Because stapled peptides can penetrate intact cells and access their intracellular targets, the very reagents used for covalent capture can be applied to vet and potentially drug the interactions in cellular and in vivo studies (Bernal et al., 2007; Gavathiotis et al., 2008; Stewart et al., 2010; Walensky et al., 2004; Walensky et al., 2006). Thus, the development and application of photoreactive stapled peptide helices represents a potentially powerful approach to identifying, distinguishing, and modulating protein targets relevant to homeostatic and pathologic cellular processes.
SIGNIFICANCE
We combined peptide α-helix stabilization, photoaffinity labeling, proteomic analysis, and structure calculation to generate a new strategy for both (1) expanding our grasp of the static and dynamic α-helical interactome of the BCL-2 family and (2) localizing and targeting BH3 α-helix/protein interaction sites. Whereas hydrocarbon stapling transforms unfolded peptides into α-helices that recapitulate their native biological activities, the installation of a photoreactive moiety effectively adapts them for high fidelity covalent capture of the broad range of physiologic interactors within complex protein mixtures. To date, stapled peptides have been applied to develop a new therapeutic strategy for targeting intracellular protein interactions (Bernal et al., 2007; Moellering et al., 2009; Walensky et al., 2004; Zhang et al., 2008), uncover an unanticipated function for a death protein in metabolism (Danial et al., 2008), structurally define the elusive activation site on an essential executioner protein of the cell death pathway (Gavathiotis et al., 2008), and identify a natural α-helical peptide that can function as an exclusive inhibitor of a formidable anti-apoptotic protein linked to cancer chemoresistance (Stewart et al., 2010). We believe that the development of photoreactive stapled peptides will extend the potential for discovery of novel and unforeseen protein interactions and how they impact health and disease. Indeed, the stapled peptide constructs used to capture protein targets can be applied to interrogate them in a cellular context and provide the templates for developing novel therapeutics.
EXPERIMENTAL PROCEDURES
Peptide Synthesis
Peptide synthesis, olefin metathesis, amino terminal derivatization (e.g. FITC-βAla, biotin), reverse phase HPLC purification, and microanalysis were performed as previously described (Bird et al., 2008).
Protein Production
Glutathione S-transferase fusion proteins of BCL-XLΔC, BCL-2ΔC, BCL-wΔC, BFL1/A1ΔC, and MCL-1ΔNΔC were expressed in E. coli BL21 using pGEX2T (Pharmacia Biotech) and purified by affinity chromatography with glutathione-sepharose beads (GE Health Care), as described (Stewart et al., 2010). Tagless protein was obtained by overnight on-bead cleavage of the GST tag using 50 U thrombin protease (GE Life Sciences) in PBS buffer at room temperature followed by size-exclusion chromatography. A histidine-tagged full-length BCL-w construct for binding site analysis was expressed in BL21 competent cells and purified by Ni-NTA chromatography, followed by size exclusion chromatography. Protein concentrations were determined using the Bradford assay (Biorad).
Fluorescence Polarization Binding Assay
Fluoresceinated BAD BH3 peptides (50 nM) were incubated with BCL-XLΔC protein at the indicated concentrations in binding buffer (150 mM NaCl, 50 mM Tris-HCl pH 8.0) at room temperature. Binding activity was measured at equilibrium (20 minutes) by fluorescence polarization using a SpectraMax microplate reader (Molecular Devices). EC50 values were calculated by nonlinear regression analysis of dose-response curves using Prism software 5.0 (GraphPad).
Photoaffinity Labeling of Recombinant Proteins
Mixtures of recombinant protein and pSAHB at the indicated concentrations were vortexed, incubated for 20 minutes in the dark, and then irradiated (365 nm, Spectroline Handheld UV Lamp Model En280L, Spectronics Corp.) for 1.5 hours at room temperature. For FITC-pSAHB crosslinking experiments, the mixtures were diluted with 4X LDS load buffer (Invitrogen), electrophoresed using 12% Bis-Tris gels (Invitrogen), and analyzed by coomassie staining (SimplyBlue Safestain, Invitrogen) and fluorescence imaging (Gel Doc XR Molecular Imager and Quantity One software, Bio-Rad). Binding specificity experiments using bovine serum albumin (BSA) were conducted as above except that BSA was added at the indicated concentrations prior to addition of BAD pSAHB. For biotinylated-pSAHB crosslinking, unreacted peptide was removed from the irradiated samples by overnight dialysis at 4°C in Buffer B (200 mM NaCl, 50 mM Tris pH 7.4) using 6–8 kD molecular weight cut-off D-Tube dialyzers (EMD Biosciences). After addition of SDS to a final concentration of 0.2%, biotin capture was achieved by incubation with high capacity streptavidin agarose (50 µL 50% slurry/reaction) for 2 hours at room temperature. The streptavidin beads were successively washed at room temperature in 1% SDS in PBS, 1 M NaCl in PBS, and then 10% ethanol in PBS for 3 × 10 minutes each. Biotinylated proteins were eluted by boiling for 30 minutes in a 10% SDS solution (Promega) containing D-biotin (10 mg/mL), electrophoresed using 4%–12% gradient Bis-Tris gels (Invitrogen), and then subjected to western analysis for BCL-2 family targets (Santa Cruz Biotechnology: BCL-XL, sc-634; BCL-2, sc-492; BCL-w, sc-6172; BAX, sc-493; Abcam: BFL1/A1, ab75887). Electrophoresed samples were also subjected to silver staining, band excision, in-gel digestion, and LC/MS-based anti-apoptotic protein identification, as described below.
Photoaffinity Labeling of Cellular Lysates
Cellular lysates were generated from U937 cells or mouse embryo fibroblasts (MEFs) grown under standard culture conditions (U937: RPMI 1640 [Invitrogen] supplemented with 10% fetal bovine serum, 100 U/mL penicillin, 100 µg/mL streptomycin, 2 mM L-glutamine, 50 mM HEPES and 50 µM β-mercaptoethanol; MEF: DMEM [Invitrogen] supplemented with 10% fetal bovine serum, 100 U/mL penicillin, 100 µg/mL streptomycin, 1 mM sodium pyruvate, 50 µM β-mercaptoethanol, and 2 mM L-glutamine). Cells were treated with ice cold Buffer A (50 mM Tris pH 7.4, 150 mM NaCl, 1% CHAPS, 1mM EDTA, 1.5 mM MgCl2, EDTA-free complete protease inhibitor cocktail [Roche]), and the supernatant isolated by centrifugation. The supernatant was maintained at 4°C and pre-cleared for 2 hours in high capacity streptavidin agarose (Pierce) that was pre-equilibrated in lysis buffer (35 µL 50% slurry/mg lysate). Pre-cleared lysates were diluted to a final concentration of 4.5 mg/mL with cold lysis buffer and incubated with biotinylated pSAHB (10 µM for U937 experiments [500 µL total volume], and 100 µM for MEF experiments [200 µL total volume]), and then irradiated, subjected to streptavidin-based isolation (as described above), and the electrophoresed pSAHB-crosslinked proteins analyzed by anti-BCL-XL and anti-MCL-1 western blotting (sc-634 and sc-819 antibodies, respectively; Santa Cruz Biotechnology).
pSAHB Interaction Site Analysis
For interaction site analysis, pSAHBs (20 µM) were mixed with BCL-XLΔC or BCL-w (20 µM) in PBS for 20 minutes followed by irradiation or incubation in the dark (i.e. non-irradiated control) for 1.5 hour. Reaction samples were subjected to gel electrophoresis and then control and crosslinked bands were excised and destained at 37°C in a 50 mM ammonium bicarbonate aqueous solution containing 50% acetonitrile, followed by dehydration using two successive cycles of acetonitrile incubation (5 minutes), solvent removal, and elimination of residual solvent by speedvac (Thermo Scientific). Gel slices were rehydrated in reducing buffer (25 mM dithiothreitol in 100 mM ammonium bicarbonate) and incubated at 55°C for 30 minutes. Samples were then cooled to room temperature, and partially dehydrated by incubation with acetonitrile for 5 minutes. Acetonitrile was removed and replaced with alkylation buffer (10 mM iodoacetamide in 50 mM ammonium bicarbonate), and samples were incubated at room temperature in the dark for 1 hour. After removal of alkylation buffer, gel slices were washed three times with 100 mM ammonium bicarbonate for 15 minutes in the dark, dehydrated, and residual solution eliminated by speedvac. For in-gel digestion, gel slices were rehydrated with sufficient digestion buffer (12.5 ng/mL sequencing grade modified-trypsin [Promega] in 50 mM ammonium bicarbonate) to completely cover each slice, and incubated for 16 hours at 37°C overnight. After removal of the digestion buffer, gel slices were washed twice for 15 minutes with 5% formic acid in 50% acetonitrile. Digestion and wash solutions were then combined and evaporated by speedvac, and the samples resuspended in 40 µL of 5% formic acid in 5% acetonitrile and desalted using C18 STAGE tips (Rappsilber et al., 2007). Desalted samples were subjected to LC-MS/MS in an LTQ Orbitrap XL hybrid mass spectrometer (ThermoFisher, San Jose, CA). The instrument was operated in data-dependent mode using a setup similar to that described previously (Haas et al., 2006). MS/MS spectra were searched using the SEQUEST algorithm (Eng et al., 1994) against a partially tryptic database containing BCL-XLΔC or BCL-w, trypsin, and common keratin contaminants. Bpa-crosslinked tryptic fragments were identified by searching for the variable modification corresponding to the mass of the tryptic BCL-XLΔC or BCL-w fragment plus the Bpa-containing pSAHB fragment. The reversed protein sequences were also included in the database to generate an estimate of false positive detection rate (FDR). The subset of crosslinked peptides identified by the SEQUEST search were filtered based on tryptic state, charge state, mass accuracy, peptide length, and number of correct matches per protein, such that the maximum FDR was 5%. SEQUEST results containing more than one instance of photoaffinity labeling were excluded from the dataset, as the crosslinking stoichiometry was assumed to be 1:1. The resultant list of covalently modified sites was plotted by frequency of occurrence across the polypeptide sequence of BCL-XLΔC or BCL-w. To depict the crosslinking “hot spots” on the protein structure, a red to blue color scale was assigned by converting crosslinking frequencies to percent of maximum occurrence and then colored by groups of ten percentiles.
Structure Calculation
Model structures of BAD BH3/BCL-w complexes were calculated using crystallography and NMR system solve (CNS) (Brunger et al., 1998) within HADDOCK 2.0 (Dominguez et al., 2003). Starting structures were: (1) the NMR structure of BCL-w (PDB ID 1O0L) with the C-terminal helix displaced from the BH3 pocket and energy minimized using the molecular dynamics software GROMOS (Van Der Spoel et al., 2005) and (2) the X-ray structure of the BAD BH3 helix derived from the BAD BH3/BCL-XLΔC complex (PDB ID 2BZW). The docking calculations generated models of the BAD BH3/BCL-w complex based on interaction restraints derived from the crosslinking experiments between BAD pSAHBs and BCL-w, and electrostatic and van der Waals interactions were optimized based on a combination of molecular dynamics and energy minimization protocols. Interacting residues designated by the crosslinking experiments were more than 50% solvent accessible as determined by NACCESS (Hubbard and Thornton, 1993). The two sets of residues based on the top scoring hits from MS/MS analysis were (1) BAD BH3 Y110 and BCL-w L63 and (2) BAD BH3 F125 and BCL-w R55. The force constants for the restraints were set to 10 kcal mol−1 Å−2 for the rigid-body docking stage and then set to a final value of 50 kcal mol−1 Å−2 during the semi-flexible simulated annealing stage. The restraints were set to be satisfied as soon as any one of the distances entering the sum average ([Σ1/r6]−1/6), over all individual pairwise combinations, were within the defined cut-off distance of 2 Å. The docking calculations were performed with standard HADDOCK protocols (Arnesano et al., 2004). Initially, 2000 orientations/structures of the complex were generated by rigid-body docking energy minimization of the individual structures. At this stage, structures were docked based on the restraints, van der Waals, and electrostatic energy terms. The 100 better-scored structures were semi-flexibly refined in torsion angle space and then refined in explicit water (Linge et al., 2003). Final structures were calculated following water refinement and energy minimization. During the simulated annealing and water refinement, all BAD BH3 amino acids (side chains and backbone) were allowed to move to optimize the interface packing. In addition, at this stage, specific segments of the BCL-w structure that comprise the BH3 pocket, including α2 (aa 40–57), α3 (aa 58–69), α4 (aa 70–89), α5 (aa 90–113), and α9 (aa 151–172) were fully flexible (side chains and backbone). The 20 lowest-scored structures from the water refinement step were clustered into one group using pairwise backbone root mean square deviation (r.m.s.d.) of 3 Å. The final structure ensemble was evaluated for chirality and stereochemistry with the programs WHATCHECK (Hooft et al., 1996) and PROCHECK_NMR (Laskowski et al., 1996), and then displayed and analyzed using PYMOL (DeLano, 2002).
Supplementary Material
ACKNOWLEDGMENTS
We thank E. Smith for figure design and editorial assistance. This work was supported by NIH grant 5P01CA92625, 1R01GM090299, and a Stand Up to Cancer Innovative Research Grant to L.D.W., and a National Sciences and Engineering Research Council of Canada postgraduate scholarship to C.R.B.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Arnesano F, Banci L, Bertini I, Bonvin AM. A docking approach to the study of copper trafficking proteins; interaction between metallochaperones and soluble domains of copper ATPases. Structure. 2004;12:669–676. doi: 10.1016/j.str.2004.03.006. [DOI] [PubMed] [Google Scholar]
- Bernal F, Tyler AF, Korsmeyer SJ, Walensky LD, Verdine GL. Reactivation of the p53 tumor suppressor pathway by a stapled p53 peptide. J Am Chem Soc. 2007;129:2456–2457. doi: 10.1021/ja0693587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird GH, Bernal F, Pitter K, Walensky LD. Chapter 22 Synthesis and Biophysical Characterization of Stabilized alpha-Helices of BCL-2 Domains. Methods Enzymol. 2008;446:369–386. doi: 10.1016/S0076-6879(08)01622-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackwell HE, Sadowsky JD, Howard RJ, Sampson JN, Chao JA, Steinmetz WE, O'Leary DJ, Grubbs RH. Ring-closing metathesis of olefinic peptides: design, synthesis, and structural characterization of macrocyclic helical peptides. J Org Chem. 2001;66:5291–5302. doi: 10.1021/jo015533k. [DOI] [PubMed] [Google Scholar]
- Blum JH, Stevens TL, DeFranco AL. Role of the mu immunoglobulin heavy chain transmembrane and cytoplasmic domains in B cell antigen receptor expression and signal transduction. J Biol Chem. 1993;268:27236–27245. [PubMed] [Google Scholar]
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- Chen L, Willis SN, Wei A, Smith BJ, Fletcher JI, Hinds MG, Colman PM, Day CL, Adams JM, Huang DC. Differential targeting of prosurvival Bcl-2 proteins by their BH3-only ligands allows complementary apoptotic function. Mol Cell. 2005;17:393–403. doi: 10.1016/j.molcel.2004.12.030. [DOI] [PubMed] [Google Scholar]
- Chen S, Dai Y, Harada H, Dent P, Grant S. Mcl-1 downregulation potentiates ABT-737 lethality by cooperatively inducing Bak activation and Bax translocation. Cancer Res. 2007;67:782–791. doi: 10.1158/0008-5472.CAN-06-3964. [DOI] [PubMed] [Google Scholar]
- Danial NN, Walensky LD, Zhang CY, Choi CS, Fisher JK, Molina AJ, Datta SR, Pitter KL, Bird GH, Wikstrom JD, et al. Dual role of proapoptotic BAD in insulin secretion and beta cell survival. Nat Med. 2008;14:144–153. doi: 10.1038/nm1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLano WL. The PyMOL Molecular Graphics System. San Carlos: DeLano Scientific; 2002. http://www.pymol.org. edn. [Google Scholar]
- Dominguez C, Boelens R, Bonvin AM. HADDOCK: a protein-protein docking approach based on biochemical or biophysical information. J Am Chem Soc. 2003;125:1731–1737. doi: 10.1021/ja026939x. [DOI] [PubMed] [Google Scholar]
- Dorman G, Prestwich GD. Benzophenone photophores in biochemistry. Biochemistry. 1994;33:5661–5673. doi: 10.1021/bi00185a001. [DOI] [PubMed] [Google Scholar]
- Elias JE, Gygi SP. Target-decoy search strategy for mass spectrometry-based proteomics. Methods Mol Biol. 2010;604:55–71. doi: 10.1007/978-1-60761-444-9_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eng JK, McCormack AL, Yates JR. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J Am Soc Mass Spectrom. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- Gavathiotis E, Suzuki M, Davis ML, Pitter K, Bird GH, Katz SG, Tu HC, Kim H, Cheng EH, Tjandra N, et al. BAX activation is initiated at a novel interaction site. Nature. 2008;455:1076–1081. doi: 10.1038/nature07396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas W, Faherty BK, Gerber SA, Elias JE, Beausoleil SA, Bakalarski CE, Li X, Villen J, Gygi SP. Optimization and use of peptide mass measurement accuracy in shotgun proteomics. Mol Cell Proteomics. 2006;5:1326–1337. doi: 10.1074/mcp.M500339-MCP200. [DOI] [PubMed] [Google Scholar]
- Hooft RW, Vriend G, Sander C, Abola EE. Errors in protein structures. Nature. 1996;381:272. doi: 10.1038/381272a0. [DOI] [PubMed] [Google Scholar]
- Hubbard SJ, Thornton JM. NACCESS. London: University College; 1993. [Google Scholar]
- Kussie PH, Gorina S, Marechal V, Elenbaas B, Moreau J, Levine AJ, Pavletich NP. Structure of the MDM2 oncoprotein bound to the p53 tumor suppressor transactivation domain. Science. 1996;274:948–953. doi: 10.1126/science.274.5289.948. [DOI] [PubMed] [Google Scholar]
- Laskowski RA, Rullmannn JA, MacArthur MW, Kaptein R, Thornton JM. AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J Biomol NMR. 1996;8:477–486. doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
- Linge JP, Habeck M, Rieping W, Nilges M. ARIA: automated NOE assignment and NMR structure calculation. Bioinformatics. 2003;19:315–316. doi: 10.1093/bioinformatics/19.2.315. [DOI] [PubMed] [Google Scholar]
- Moellering RE, Cornejo M, Davis TN, Del Bianco C, Aster JC, Blacklow SC, Kung AL, Gilliland DG, Verdine GL, Bradner JE. Direct inhibition of the NOTCH transcription factor complex. Nature. 2009;462:182–188. doi: 10.1038/nature08543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappsilber J, Mann M, Ishihama Y. Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat Protoc. 2007;2:1896–1906. doi: 10.1038/nprot.2007.261. [DOI] [PubMed] [Google Scholar]
- Saghatelian A, Jessani N, Joseph A, Humphrey M, Cravatt BF. Activity-based probes for the proteomic profiling of metalloproteases. Proc Natl Acad Sci U S A. 2004;101:10000–10005. doi: 10.1073/pnas.0402784101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sattler M, Liang H, Nettesheim D, Meadows RP, Harlan JE, Eberstadt M, Yoon HS, Shuker SB, Chang BS, Minn AJ, et al. Structure of Bcl-xL-Bak peptide complex: recognition between regulators of apoptosis. Science. 1997;275:983–986. doi: 10.1126/science.275.5302.983. [DOI] [PubMed] [Google Scholar]
- Schafmeister C, Po J, Verdine G. An all-hydrocarbon crosslinking system for enhancing the helicity and metabolic stability of peptides. J Am Chem Soc. 2000;122:5891–5892. [Google Scholar]
- Stewart ML, Fire E, Keating AE, Walensky LD. The MCL-1 BH3 helix is an exclusive MCL-1 inhibitor and apoptosis sensitizer. Nat Chem Biol. 2010 doi: 10.1038/nchembio.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Der Spoel D, Lindahl E, Hess B, Groenhof G, Mark AE, Berendsen HJ. GROMACS: fast, flexible, and free. J Comput Chem. 2005;26:1701–1718. doi: 10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]
- Vodovozova EL. Photoaffinity labeling and its application in structural biology. Biochemistry (Mosc) 2007;72:1–20. doi: 10.1134/s0006297907010014. [DOI] [PubMed] [Google Scholar]
- Walensky LD, Kung AL, Escher I, Malia TJ, Barbuto S, Wright RD, Wagner G, Verdine GL, Korsmeyer SJ. Activation of apoptosis in vivo by a hydrocarbon-stapled BH3 helix. Science. 2004;305:1466–1470. doi: 10.1126/science.1099191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walensky LD, Pitter K, Morash J, Oh KJ, Barbuto S, Fisher J, Smith E, Verdine GL, Korsmeyer SJ. A stapled BID BH3 helix directly binds and activates BAX. Mol Cell. 2006;24:199–210. doi: 10.1016/j.molcel.2006.08.020. [DOI] [PubMed] [Google Scholar]
- Wei MC, Lindsten T, Mootha VK, Weiler S, Gross A, Ashiya M, Thompson CB, Korsmeye SJ. tBID, a membrane-targeted death ligand, oligomerizes BAK to release cytochrome c. Genes Dev. 2000;14:2060–2071. [PMC free article] [PubMed] [Google Scholar]
- Weissenhorn W, Dessen A, Harrison SC, Skehel JJ, Wiley DC. Atomic structure of the ectodomain from HIV-1 gp41. Nature. 1997;387:426–430. doi: 10.1038/387426a0. [DOI] [PubMed] [Google Scholar]
- Yang E, Zha J, Jockel J, Boise LH, Thompson CB, Korsmeyer SJ. Bad, a heterodimeric partner for Bcl-XL and Bcl-2, displaces Bax and promotes cell death. Cell. 1995;80:285–291. doi: 10.1016/0092-8674(95)90411-5. [DOI] [PubMed] [Google Scholar]
- Zhang H, Zhao Q, Bhattacharya S, Waheed AA, Tong X, Hong A, Heck S, Curreli F, Goger M, Cowburn D, et al. A cell-penetrating helical peptide as a potential HIV-1 inhibitor. J Mol Biol. 2008;378:565–580. doi: 10.1016/j.jmb.2008.02.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.