Abstract
Members of the eukaryotic phylum Apicomplexa are the cause of important human diseases including malaria, toxoplasmosis, and cryptosporidiosis. These obligate intracellular parasites produce new invasive stages through a complex budding process. The budding cycle is remarkably flexible and can produce varied numbers of progeny to adapt to different host-cell niches. How this complex process is coordinated remains poorly understood. Using Toxoplasma gondii as a genetic model, we show that a key element to this coordination is the centrocone, a unique elaboration of the nuclear envelope that houses the mitotic spindle. Exploiting transgenic parasite lines expressing epitope-tagged centromeric H3 variant CenH3, we identify the centromeres of T. gondii chromosomes by hybridization of chromatin immunoprecipitations to genome-wide microarrays (ChIP-chip). We demonstrate that centromere attachment to the centrocone persists throughout the parasite cell cycle and that centromeres localize to a single apical region within the nucleus. Centromere sequestration provides a mechanism for the organization of the Toxoplasma nucleus and the maintenance of genome integrity.
Keywords: histone, mitosis
Members of the ancient eukaryotic phylum Apicomplexa are responsible for numerous important human and animal diseases, including malaria and toxoplasmosis. Apicomplexa are obligate intracellular parasites, and the infection is spread from cell to cell by a motile stage (1). Host-cell invasion results in the formation of a parasitophorous vacuole in which the parasites typically reside and develop (some species escape from the initial vacuole into the host-cell cytoplasm). With few exceptions, parasite replication is confined to the intracellular phase of the lifecycle. The timing and extent of replication is matched to the varied size and biology of the host cell, and the number of progeny can vary from two to thousands. At the core of this flexibility is the budding mechanism by which Apicomplexa replicate. This complex process encompasses mitosis of the nucleus, assembly of the cytoskeletal and membrane elements that form the pellicle, loading of the growing bud with organelles, and finally emergence of fully formed new invasive daughters from the mother cell (2). Based on this underlying scheme, Apicomplexa have evolved three distinct cell-division types (Fig. S1A). Toxoplasma tachyzoites, featuring the simplest form, endodyogeny, bud into two daughters after each round of DNA replication (3). Plasmodium, the causative agent of malaria, divides by schizogony, whereby the cell proceeds through several rounds of DNA replication and mitosis before the now multinucleate schizont gives rise to multiple zoites at once (4). Sarcocystis, a widespread tissue parasite of mammals, uses endopolygeny (5). In this division mode, multiple rounds of DNA replication occur without nuclear division, and upon budding the large polyploid nucleus is parceled out in multiple daughters.
Interestingly, these parasite species can switch from one division mode to another depending on the developmental stage, thus fine-tuning replication to different tissue and host-cell environments. For example, Toxoplasma gondii replicates by endodyogeny in intermediate hosts but replicates by schizogony within the cat intestine (6). The molecular mechanisms that regulate apicomplexan cell division and allow this remarkable flexibility of the cell and nuclear division cycle remain poorly understood (7). An important unsolved question is how apicomplexan daughter cells emerge with the complete set of chromosomes (∼10 depending on species) after passing through multinucleated and polyploid stages. In Sarcocystis neurona the mitotic spindle persists throughout the cell cycle, and small monopolar spindles housed in a specialized elaboration of the nuclear envelope, the centrocone, are evident throughout interphase (8). More recent work in T. gondii identified a molecular marker for the centrocone, the repeat protein membrane occupation and recognition nexus protein 1 (MORN1) (9, 10) that is found in a variety of apicomplexan species representing all three cell-division modes (11).
Based on these observations, we speculated that apicomplexan chromosomes remain permanently attached to the centrocone throughout the cell cycle and that this physical tethering provides a critical means to maintain genome integrity (2). We directly tested the tethering hypothesis by developing a marker to visualize the centromeres of apicomplexan chromosomes. We used an epitope-tagged T. gondii centromeric histone 3 variant (CenH3) to identify the centromeres of T. gondii chromosomes and subsequently studied the interaction of centromeres and the unique centrocone structure throughout the T. gondii cell cycle.
Results and Discussion
Identification of the T. gondii CenH3.
The biology of apicomplexan chromosomes remains poorly understood. In contrast to most eukaryotic organisms, chromosomes in Apicomplexa show only limited condensation during mitosis, thus complicating chromosome visualization. Chromosome segregation occurs through endomitosis using an intranuclear spindle with the nuclear envelope remaining intact (12–15). To develop a molecular marker for the centromere of T. gondii chromosomes, we characterized CenH3 (also referred to as “CenpA”). In most eukaryotes CenH3 replaces the canonical histone 3 in the nucleosomes of centromeric DNA and can be used to visualize the centromeres (16). Computational analysis of the T. gondii genome reveals three putative histone H3 genes. Genes TGME49_061240 and TGME49_018260 (toxodb.org version 6.2) appear to encode the canonical H3 and H3.3 variants (17, 18). TGME49_025410 encodes an H3 protein with a unique 99-amino acid insertion at the N terminus (see multiple sequence alignment in Fig. S1B). N-terminal extensions are a hallmark of CenH3 (19).
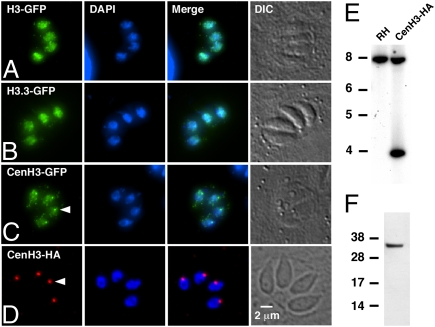
To validate these assignments, the coding sequence of each H3 gene was amplified from T. gondii mRNA, cloned into a plasmid construct fused to a C-terminal YFP tag, and expressed in tachyzoites. As expected, histones H3 and H3.3 label the entire nucleus (Fig. 1 A and B). TGME49_025410 showed faint labeling throughout the nucleus in addition to a more intense spot (Fig. 1C). We were unable to establish stable transformants for this gene using the plasmid constructs driving expression of the CenH3-YFP cassette or a CenH3-HA cassette. Expression of the transgene from these plasmids was driven by the strong α-tubulin promoter, and it seemed likely that inappropriate strength or timing of expression was detrimental to the parasite, as described for CenH3 overexpression in other organisms (20).
Fig. 1.
T. gondii histone H3 variants. (A–C) Immunofluorescence assays of tachyzoites transiently transfected with constructs expressing TGME49_061240 (H3) (A), TGME49_018260 (H3.3) (B), and TGME49_025410 (CenH3) (C) fused to the N terminus of YFP. Anti-GFP antibody is shown in green; DAPI is shown in blue. (D) To construct a strain carrying an epitope tag in the genomic locus of CenH3, a suitable cosmid was engineered by recombination in EL250 cells and transfected into T. gondii. These TgCenH3-HA lines showed a single spot of nuclear staining (red, arrowhead) in the majority of parasites in immunofluorescence. (E) Southern analysis of a stable TgCenH3-HA clone shows the presence of two bands when probed with the CenH3 coding sequence, suggesting nonhomologous insertion. (F) This clone expressed a protein of the expected size when probed in Western blot.
To provide appropriate expression, we introduced an epitope-tagging cassette into a genomic TgCenH3 locus cosmid clone by recombineering (21) and established stable clonal T. gondii TgCenH3-HA lines. When these parasites were subjected to immunofluorescence assays using an HA-specific antibody, a single nuclear spot was evident (Fig. 1D). PCR and Southern analysis of genomic DNA from these clones revealed a single nonhomologous insertion of the modified cosmid into the parasite genome, and Western blot of protein extracts indicated the presence of a tagged protein of appropriate size (Fig. 1 E and F).
Identification of Toxoplasma Centromeres by ChIP and Microarray Analysis.
The T. gondii genome is distributed into 14 chromosomes (22). CenH3 staining typically shows multiple foci reflecting the number of chromosomes (16). It therefore was surprising to observe a single TgCenH3-positive spot. To confirm the localization of TgCenH3 to the centromeres, we determined the position of the centromeres on T. gondii chromosomes by identifying TgCenH3-associated DNA using ChIP. Tachyzoites expressing TgCenH3-HA were fixed with 1% formaldehyde, and DNA was sheared by sonication. TgCenH3-HA was precipitated using an HA-specific antibody, and the coprecipitated DNA was amplified and hybridized to a tiled oligonucleotide array of the complete T. gondii genome. As a control, we immunoprecipitated and hybridized chromatin associated with the canonical H3 variant (23).
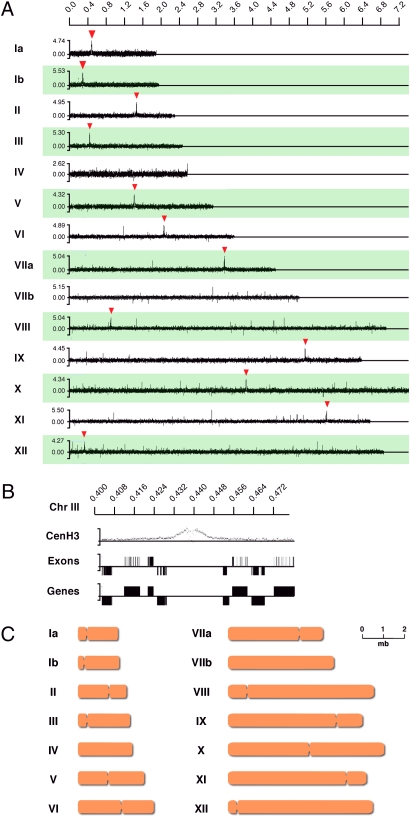
As shown in Fig. 2A, we detected significant hybridization of the CenH3-HA ChIP to the genomic chip for 12 of the 14 chromosomes (see Materials and Methods for details on peak calling and statistical analyses). Detailed analysis revealed extended hybridizing regions (16 kb ± 3.4 kb) that fall into areas of the chromosomes that are largely devoid of protein-coding genes (17 kb ± 5.6 kb). Fig. 2B shows a close-up view of chromosome III as an example, and the remaining centromere regions are shown in Fig. S2. Consistent with substitution of CenH3 for H3 at centromeres, we note a reduced signal for H3 at the center of the TgCenH3 regions of the T. gondii chromatin.
Fig. 2.
ChIP of CenH3-associated DNA and microarray analysis. (A) ChIP-chip analysis of DNA isolated from the transgenic CenH3-HA strain using an HA epitope-specific antibody. All 14 T. gondii chromosomes are shown. Significant hybridization peaks are indicated by red arrowheads. The x axis shows the position of probes along the chromosome; the y axis shows relative fluorescence intensity for each oligonucleotide. (B) Representative detailed view of CenH3-associated regions on chromosome III. Gene models and exon predictions for this region (version 4 annotation; www.toxodb.org) are shown below (up is sense; down is antisense). Note that CenH3-associated DNA is devoid of protein-coding genes. (C) Schematic representation of the positions of the centromeres of 12 of the 14 T. gondii chromosomes.
Biochemical Confirmation of Centromeres by Etoposide-Mediated Chromosome Breakage.
Topoisomerase II accumulates at the centromeres in metaphase and plays a critical role in kinetochore structure and sister chromatid decatenation (24). The anticancer drug etoposide binds to topoisomerase II with high specificity and locks the enzyme in an intermediate state, blocking the religation step (25). This treatment results in multiple double-strand breaks in regions of topoisomerase II accumulation and has been used to map centromeres in a variety of organisms (25–27).
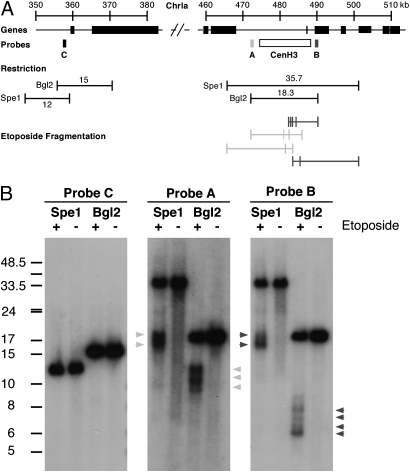
We used etoposide treatment of T. gondii tachyzoites to assess independently the validity of the array-based assignment of the centromere for chromosome Ia. T. gondii-infected cells were cultured for 2 h in the presence of 100 μM etoposide or a DMSO solvent as control. Parasite genomic DNA was extracted, digested with SpeI or BglII restriction enzymes, and separated by pulsed-field gel electrophoresis. DNA was blotted onto membrane and probed with three probes specific for chromosome Ia. Probes A and B flank the TgCenH3-binding sites, whereas probe C is localized 115,000 bp 3′ of this region and serves as control (see Fig. 3 for a map). Restriction fragments containing the putative centromere identified by probes A and B showed marked fragmentation in etoposide-treated parasites. We identified multiple breakage points that all map within 5,000 bp of each other and within the zone of TgCenH3 binding (Fig. 3 A and B). We observed no breakage in the absence of etoposide or in the control locus on the same chromosome (probe C), regardless of whether cells were drug treated. We conclude that the TgCenH3-binding regions of the T. gondii chromosomes represent the centromeres.
Fig. 3.
Etoposide mapping of the centromere of chromosome Ia. (A) Schematic map of chromosome Ia showing the CenH3-associated region as well as a control region located ∼115 kb 5′. Position of restriction fragments, probes, and mapped etoposide-induced fragments are indicated. (B) T. gondii-infected cultures were incubated for 2 h in medium containing 100 μM etoposide (+) or DMSO solvent as control (−). Parasites were purified, and genomic DNA was extracted, restricted with the indicated enzymes, and subjected to Southern analysis using the indicated probes. Note etoposide-induced fragmentation of the putative centromeric region [probes A (light gray arrowheads) and B (dark gray arrowheads)], which was not observed for the control regions (probe C).
Although TgCenH3 ChIP-microarray analysis (ChIP-chip analysis) robustly identified the centromeres of 12 chromosomes, we did not obtain conclusive hybridization data for chromosomes IV and VIIb. It is likely that probes representing these centromeres are missing from the array, either because in these regions repetitive DNA is present that was excluded from the chip design or because of incomplete or incorrect assembly of the draft genome in these regions. We scanned both chromosomes for regions that show significant gaps in probe coverage and are devoid of protein coding genes. We identified a candidate region on each chromosome (position 2,136,000–2,206,000 on chromosome IV and position 3,720,000–3,745,000 on chromosome VIIb). Note that there are significant differences between these regions among the different strain-specific assemblies in toxoDB 6.2. We tested the candidate regions by etoposide treatment and Southern blotting using a modified restriction and electrophoresis protocol to generate and analyze larger fragment encompassing the entire candidate region (Fig. S3). Etoposide treatment did not result in fragmentation, and furthermore the restriction fragments obtained did not match the genomic predictions. This result suggests that these regions are not the centromeres and that this portion of the chromosome probably is misassembled (see Fig. S3 and its legend for additional details). This misassembling may not be surprising, because centromeres and telomeres often are challenging to assemble because of repeats and nucleotide bias. The quality of the assembly of the T. gondii centromeres probably will improve with ongoing sequencing efforts.
T. gondii Centromeres Are Delineated by Histone H3 Lysine K9 Di- and Trimethylation.
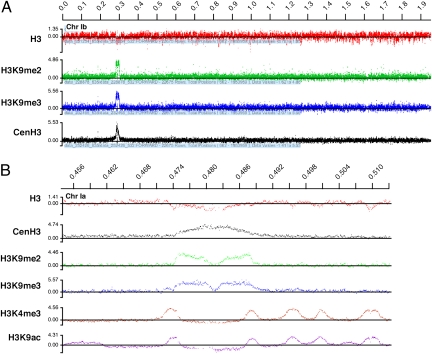
Centromeric DNA contains subdomains of CenH3 and H3 nucleosomes, and dynamic modification of H3 is believed to influence centromere assembly (28, 29). We therefore analyzed the modification status of H3 in the T. gondii centromere. As shown in Fig. 4B, centromeres are devoid of trimethylation of H3 lysine 4 (H3K4me3). In contrast, H3 lysine 9 di- and trimethylation modifications (H3K9me2 and H3K9me3, respectively), typical hallmarks of heterochromatin, were concentrated in two peaks directly flanking the center of the centromere. Interestingly, in T. gondii, the centromere appears to be the chromosomal region of most significant accumulation of both H3K9me3 and H3K9me2 (Fig. 4A, Fig. S4, and Table S1). Heterochromatin typically flanks the centromeres in metazoans but generally is clearly distinct from centromeric chromatin (29–31). In the much smaller T. gondii centromere we note considerable overlap, identifying only a singular central region as devoid of lysine 9 modification.
Fig. 4.
Histone H3K9 di- and trimethylation marks the boundaries of the T. gondii centromeres. ChIP-chip analysis using the indicated antibodies was conducted as detailed in Materials and Methods. (A) Full view of chromosome Ib. Note that centromeric H3K9me2 and H3K9me3 labeling represents the most significant accumulation of these modifications across the T. gondii chromosome. (B) Detailed view of the centromeric region of chromosome Ia. Note the absence of centromeric signal for marks previously associated with promoter regions in T. gondii [H3K4me3 and histone H3 acetylated at lysine 9 (H3K9ac) (23)].
Centromeres Are Maintained in Close Proximity to the Centrocone Throughout the Cell Cycle.
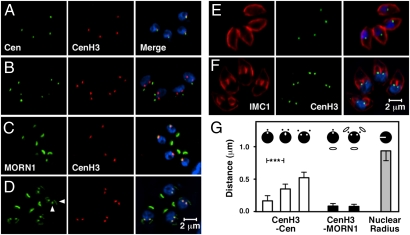
Having demonstrated that TgCenH3 marks the centromeres of at least 12 of the 14 chromosomes in T. gondii, we further characterized TgCenH3 nuclear localization by quantifying the number of spots seen in immunofluorescence assays. We observed either one (75%) or two (25%) CenH3-HA spots per parasite nucleus (n = 400). We next double-labeled parasites to compare CenH3-HA localization with that of centrin, a component of the centrosome (32, 33), with MORN1, which associates with the centrocone and rings delineating the parasite pellicle (10), and with inner membrane complex protein 1 (IMC1), a structural component of the parasite pellicle (34).
Fig. 5 shows images representing parasites during cell division (Fig. 5 B, D, and F) or during interphase (Fig. 5 A, C, and E). During division we observe two CenH3 spots that appear close to markers of the spindle, consistent with chromosome segregation during mitosis (see below). Surprisingly, in nondividing parasites this association is maintained strictly and appears even closer than during actual division. The mean distance between centromeres and centrosome during interphase was 0.17 μm (SD = 0.08 μm) compared with an average nuclear radius of 0.94 μm (SD = 0.15 μm). The distance between the centromeres and the centrocone as identified by MORN1 staining is even smaller (0.08 μm) and does not change significantly regardless of the cell-cycle phase.
Fig. 5.
The T. gondii centromeres remain proximal to the centrocone at all times. Host-cell cultures were infected with CenH3-HA transgenic parasites and fixed and processed for immunofluorescence 24 h later. Cells were stained with an antibody to HA (CenH3) and centrin (Cen) (A and B), MORN1 (C and D), and IMC1 (E and F). Representative images are shown for nondividing (A, C, and E) and dividing parasites (B, D, and F). The vertical arrowhead in D indicates the centrocone, and the horizontal arrowhead indicates the daughter-cell basal complex. (G) The distance from the center of CenH3 labeling to the center of the centrin or MORN1 label was measured by image analysis. The mean distances from centrin (54 nuclei) and MORN1 (70 nuclei) are shown. Error bars depict SD. Values were grouped by cell-cycle phase as indicated by symbols at the top of the graph. (See Fig. 6 for further details about T. gondii mitosis.) The radii of nuclei are shown for size comparison (area of DAPI-stained nuclei was measured and radii were calculated from area, assuming a circular shape for simplicity; n = 52 nuclei). ***P < 0.001.
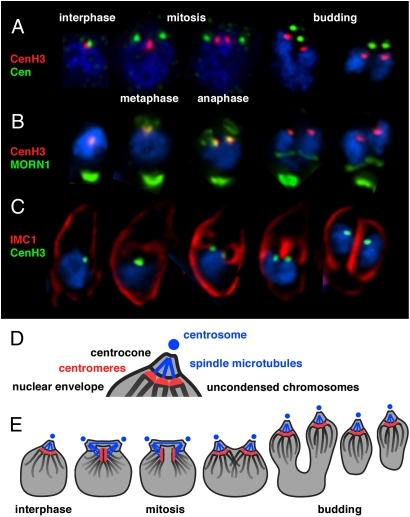
The CenH3 marker allows us to position the centromeres within the apicomplexan budding process and leads us to propose the following model for mitosis in these parasites. Despite the lack of visible chromosome condensation, the centromeres line up in a classical metaphase plate (Fig. 6 A and E). The metaphase plate remains close to the nuclear envelope and is suspended evenly between the two centrosomes. Anaphase results in two clearly separated daughter centrocones that lead the daughter nuclei into the emerging buds. Importantly, during metaphase the two daughter buds already are detectable using the MORN1 and IMC1 markers (Fig. 6 B and C). Mitosis and budding in Apicomplexa therefore are not sequential events but are closely intertwined.
Fig. 6.
Toxoplasma mitosis and budding. (A, B, and C) Immunofluorescence micrographs showing details for individual nuclei at key points of the T. gondii cell-division cycle (note that each panel represents five individual cells consolidated and cropped from different micrographs). Three-color merged images are shown for the antibodies indicated and the DNA stained with DAPI (blue). Note that MORN1 staining is present in the centrocones of all budding stages but is not visible in the two panels showing budding in B because of stronger signal from the CenH3 (red channel). (D) Schematic view of the centrocone highlighting its components. (See ref. 10 for additional details.) (E) Model of centromere sequestration throughout the parasite cell cycle.
Conclusion.
In this study we identify the centromeres of the chromosomes of T. gondii. Unlike Plasmodium (26, 35), Toxoplasma centromeres show no nucleotide bias, and we did not identify obvious shared sequence motifs upon inspection of the primary sequence and bioinformatics motif searches. The centromeres are relatively small but have distinctive epigenetic features. In addition to the presence of CenH3, we demonstrate a pronounced accumulation of histone H3K9me2 and H3K9me3, a modification often associated with heterochromatin, flanking the core of the centromere. In our analyses these flanks appear to be the main sites of accumulation of these modifications across the chromosomes. This result contrasts with recent studies in Plasmodium falciparum where H3K9me3 and heterochromatin protein 1 (HP1) are found mainly in islands of the genome that contain transcriptionally silent members of multigene families but are not associated with centromeres. HP1 is a chromodomain protein that binds to H3K9me3 and in other species is associated with heterochromatin regions of centromeres (36). The epigenetic differences between T. gondii and Plasmodium may reflect significant differences in the biology of the centromeres or differences in the extent of gene silencing related to antigenic variation in Plasmodium (37–39). T. gondii does not undergo antigenic variation.
The TgCenH3 cellular marker offers a deeper molecular understanding of apicomplexan chromosomal and nuclear architecture during mitosis. Most remarkably T. gondii centromeres are confined to a single, apically oriented bouquet on the nuclear periphery throughout the cell cycle (Figs. 5 and 6). We propose a model of centromere sequestration (Fig. 6 D and E) in which the centrocone, a unique subcompartment of the nucleus, serves as a master organizer of chromosome location not only during mitosis but throughout the intracellular development of the parasite. Persistence of kinetochores and spindle microtubules could provide the mechanism for this association through a constant spindle (2, 8). Alternatively, centromeric heterochromatin could be anchored through protein components of the nuclear membranes (40, 41). Although the centrocone is not universally conserved among Apicomplexa, a simplified structure has been described during the gametogenesis of the malaria parasite. In a pioneering ultrastructural study on Plasmodium yoelii the authors noted what appeared to be the association of kinetochores and the nuclear envelope; during mitosis they also observed semi-spindles underlying nuclear pores (42). Whether these structures share the same molecular underpinnings as those described here for Toxoplasma awaits further investigation. Continuously “holding on” to their chromosomes may offer these parasites a means to maintain genome stability through rapid DNA replication cycles and dramatically variable budding processes.
Materials and Methods
Parasite Culture and Manipulation.
Parasites were propagated in human foreskin fibroblasts and human fibroblast reverse transcriptase (hTERT) cells and were genetically manipulated as described in detail previously (43). Transgenes were introduced by electroporation into tachyzoites of the T. gondii RH strain, and stable transformants were selected by culture in the presence of 20 μM chloramphenicol. Clonal lines were established by limiting dilution.
DNA Manipulation.
The GenBank accession number for TgCenH3 is XM 002366173. The coding sequences of T. gondii H3 variants were amplified from parasite cDNA by PCR introducing synthetic BglII or BclI (5′), and AvrII (3′) restriction sites (primers: CenH3: atcgtgatcaaaaATGGCTCGCATCAAGACGACGCC, cgatcctaggGCAAGGAAACGCATGCCGCAGTC; H3.3: atcgagatctaaaATGGCGCGAACTAAGCAGACTGC, cgatcctaggAGACCTCTCGCCTCGGATTCGCC; H3: atcgagatctaaaATGGCGCGCACCAAGCAGACC, cgatcctaggAGACCGTTCACCACGGATGCGG). The resulting amplicons were introduced into plasmid tubYFP-YFPsagCAT (44) restricted with BglII/AvrII, resulting in translational fusion to the N terminus of YFP. The genomic locus of CenH3 was epitope tagged in a cosmid clone carrying the locus by recombineering (21). Briefly, a cassette carrying the coding sequence of the HA epitope followed by a 3′ UTR and markers for the selection in bacteria and parasites was amplified by PCR introducing 50-bp flanks to direct the cassette to the 3′ end of the CenH3 gene. The cassette was introduced into EL250 E. coli cells carrying cosmid PSBM061, recombination was induced by heat shock, and tagged cosmids were identified by double selection on kanamycin and gentamycin.
ChIP and Microarray.
ChIP was performed as described (23, 45). Briefly, chromatin from CenH3-HA transgenic tachyzoites was cross-linked for 10 min with 1% formaldehyde at room temperature and purified after sonication yielding fragments of 500−1,000 bp. Chromatin was immunoprecipitated at 4 °C overnight using an HA polyclonal antibody (Abcam ab9110), an antibody to H3K9me2 (07–441; Upstate), or an antibody to H3K9me3 (ab8898; Abcam) and was washed extensively (see ref. 23 for details on additional antibodies used). The DNA was treated with proteinase K for 2 h and purified using the Qiagen PCR purification kit. Precipitated DNA was amplified using the DNA Genomeplex whole-genome amplification kit (Sigma) and subsequently labeled using random primers coupled to a fluorochrome (Cy3 or Cy5). Probes were hybridized to a tiled oligonucleotide array representing the complete T. gondii genome according to NimbleGen Systems procedures. The array was fabricated by NimbleGen Systems (http://www.nimblegen.com) and contained 740,000 oligonucleotides representing version 4 of the Me49 genome with an approximate spacing of 80 bp between each oligonucleotide. Specific information on oligonucleotides and all hybridization data have been deposited and are available on the genome browse feature of toxodb.org.
Two biological replicates of ChIP-chip were analyzed using NimbleScan software (NimbleGen Systems) to determine consensus peaks of hybridization using a 500-bp sliding window. Positions of peaks with a false-discovery rate (FDR) of <0.05 within each chromosome were compared between biological replicates using custom Perl scripts. The result of this analysis is shown in Fig. S5. Significant clustering (more than five consecutive high-confidence peaks) was readily identified for all chromosomes with the exception of IV, VIIb, and XII. Manual inspection identified a suitable region on chromosome XII base pairs 330,000–350,000. Localization of centromeres was confirmed computationally by comparing positions of consensus peaks from TgCenH3, H3K9me2, and H3K9me3 ChIP-chip experiments (>40-bp overlap was required to score). This analysis revealed 41 coincident peaks, corresponding to 10 centromeric regions (Table S1). As seen in Fig. 4, H3K9me signals flank the TgCenH3 regions of hybridization, and manual inspection of the TgCenH3, H3K9me2, and H3K9me3 hybridizing regions confirmed genomic colocalization of hybridization of TgCenH3, H3K9me2, and H3K9me3 for chromosomes III and XII.
Etoposide Treatment and Southern Mapping.
Cells were infected with T. gondii and grown for 48 h before addition of etoposide dissolved in DMSO (100 μM final concentration) (Sigma) or DMSO alone. After 2 h of treatment, cells were scraped and needle-passed, and liberated parasites were purified by filtration through a 3-μm membrane. Parasite genomic DNA was isolated by phenol-chloroform extraction, treated with restriction enzyme as indicated in Fig. 3, and separated by pulse-field electrophoresis (0.7% agarose gel, 0.25× Tris-borate-EDTA (TBE), 5V/cm over 20 h with a 120° included angle at 0.5- to 15-s switch times) and blotted onto nylon membrane. Probes A (chromosome Ia base pairs 470,412–471,063), B (chromosome Ia base pairs 488,014–488,682), and C (chromosome Ia base pairs 355,264–355,924) were amplified from parasite genomic DNA by PCR, radiolabeled by random priming, hybridized to blotted DNA, and visualized by autoradiography.
Immunostaining of Parasites and Proteins.
Immunofluorescence assays were performed as described (46) using primary antibodies to HA (rat; Roche), IMC1 (mouse; a kind gift of Gary Ward, University of Vermont, Burlington, VT), human centrin (rabbit; a kind gift of Iain Cheeseman, Whitehead Institute, Cambridge, MA), or MORN1 (rabbit) (10), respectively, at 1:1,000 dilution, followed by suitable Alexa Fluor-labeled secondary antibodies (Invitrogen) at 1:2,000 dilution. DNA was labeled by staining with DAPI. Image stacks were taken using a Deltavision microscope and an Olympus lens (UPlans APO 100×/1.40 oil). Images were subjected to deconvolution and contrast adjustment using Applied Precision software.
Supplementary Material
Acknowledgments
We thank Gary Ward, Ian Cheeseman, and Marc-Jan Gubbels for antibodies, Shipra Vaishnava and David Morrison for initial contributions, and Jean-Francois Dubremetz, Giel van Dooren, and Michael White for discussions. This work was funded by National Institutes of Health Grants AI 64671 (to B.S.); AI060496 and AI087625 (to K.K.); and AI077662 (to Michael White and B.S.). M.M.C. was supported by National Institutes of Health Grant 5T32GM007491 awarded to the Albert Einstein College of Medicine.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The microarray data reported in this paper have been deposited in the Toxoplasma Genomics Resource database, http://www.toxodb.org.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1006741108/-/DCSupplemental.
References
- 1.Sibley LD. Intracellular parasite invasion strategies. Science. 2004;304:248–253. doi: 10.1126/science.1094717. [DOI] [PubMed] [Google Scholar]
- 2.Striepen B, Jordan CN, Reiff S, van Dooren GG. Building the perfect parasite: Cell division in Apicomplexa. PLoS Pathog. 2007;3:e78. doi: 10.1371/journal.ppat.0030078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sheffield HG, Melton ML. The fine structure and reproduction of Toxoplasma gondii. J Parasitol. 1968;54:209–226. [PubMed] [Google Scholar]
- 4.Bannister LH, Hopkins JM, Fowler RE, Krishna S, Mitchell GH. A brief illustrated guide to the ultrastructure of Plasmodium falciparum asexual blood stages. Parasitol Today. 2000;16:427–433. doi: 10.1016/s0169-4758(00)01755-5. [DOI] [PubMed] [Google Scholar]
- 5.Speer CA, Dubey JP. Ultrastructure of shizonts and merozoites of Sarcocystis falcatula in the lungs of budgerigars (Melopsittacus undulatus) J Parasitol. 1999;85:630–637. [PubMed] [Google Scholar]
- 6.Speer CA, Dubey JP. Ultrastructural differentiation of Toxoplasma gondii schizonts (types B to E) and gamonts in the intestines of cats fed bradyzoites. Int J Parasitol. 2005;35:193–206. doi: 10.1016/j.ijpara.2004.11.005. [DOI] [PubMed] [Google Scholar]
- 7.Gubbels MJ, White M, Szatanek T. The cell cycle and Toxoplasma gondii cell division: Tightly knit or loosely stitched? Int J Parasitol. 2008;38:1343–1358. doi: 10.1016/j.ijpara.2008.06.004. [DOI] [PubMed] [Google Scholar]
- 8.Vaishnava S, et al. Plastid segregation and cell division in the apicomplexan parasite Sarcocystis neurona. J Cell Sci. 2005;118:3397–3407. doi: 10.1242/jcs.02458. [DOI] [PubMed] [Google Scholar]
- 9.Hu K, et al. Cytoskeletal components of an invasion machine—the apical complex of Toxoplasma gondii. PLoS Pathog. 2006;2:e13. doi: 10.1371/journal.ppat.0020013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gubbels MJ, Vaishnava S, Boot N, Dubremetz JF, Striepen B. A MORN-repeat protein is a dynamic component of the Toxoplasma gondii cell division apparatus. J Cell Sci. 2006;119:2236–2245. doi: 10.1242/jcs.02949. [DOI] [PubMed] [Google Scholar]
- 11.Ferguson DJ, et al. MORN1 has a conserved role in asexual and sexual development across the Apicomplexa. Eukaryot Cell. 2008;7:698–711. doi: 10.1128/EC.00021-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dubremetz JF. Ultrastructural study of schizogonic mitosis in the coccidian, Eimeria necatrix (Johnson 1930) J Ultrastruct Res. 1973;42:354–376. [PubMed] [Google Scholar]
- 13.Dubremetz JF. Genesis of merozoites in the coccidia, Eimeria necatrix. Ultrastructural study. J Protozool. 1975;22:71–84. doi: 10.1111/j.1550-7408.1975.tb00946.x. [DOI] [PubMed] [Google Scholar]
- 14.Aikawa M. The fine structure of the erythrocytic stages of three avian malarial parasites, Plasmodium fallax, P. lophurae, and P. cathemerium. Am J Trop Med Hyg. 1966;15:449–471. doi: 10.4269/ajtmh.1966.15.449. [DOI] [PubMed] [Google Scholar]
- 15.Aikawa M, Beaudoin RL. Studies on nuclear division of a malarial parasite under pyrimethamine treatment. J Cell Biol. 1968;39:749–754. doi: 10.1083/jcb.39.3.749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dalal Y, Furuyama T, Vermaak D, Henikoff S. Structure, dynamics, and evolution of centromeric nucleosomes. Proc Natl Acad Sci USA. 2007;104:15974–15981. doi: 10.1073/pnas.0707648104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sullivan WJ, Jr, Naguleswaran A, Angel SO. Histones and histone modifications in protozoan parasites. Cell Microbiol. 2006;8:1850–1861. doi: 10.1111/j.1462-5822.2006.00818.x. [DOI] [PubMed] [Google Scholar]
- 18.Sullivan WJ., Jr Histone H3 and H3.3 variants in the protozoan pathogens Plasmodium falciparum and Toxoplasma gondii. DNA Seq. 2003;14:227–231. doi: 10.1080/1042517031000089496. [DOI] [PubMed] [Google Scholar]
- 19.Malik HS, Henikoff S. Adaptive evolution of Cid, a centromere-specific histone in Drosophila. Genetics. 2001;157:1293–1298. doi: 10.1093/genetics/157.3.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heun P, et al. Mislocalization of the Drosophila centromere-specific histone CID promotes formation of functional ectopic kinetochores. Dev Cell. 2006;10:303–315. doi: 10.1016/j.devcel.2006.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brooks CF, et al. The Toxoplasma apicoplast phosphate translocator links cytosolic and apicoplast metabolism and is essential for parasite survival. Cell Host Microbe. 2010;7:62–73. doi: 10.1016/j.chom.2009.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Khan A, et al. Composite genome map and recombination parameters derived from three archetypal lineages of Toxoplasma gondii. Nucleic Acids Res. 2005;33:2980–2992. doi: 10.1093/nar/gki604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gissot M, Kelly KA, Ajioka JW, Greally JM, Kim K. Epigenomic modifications predict active promoters and gene structure in Toxoplasma gondii. PLoS Pathog. 2007;3:e77. doi: 10.1371/journal.ppat.0030077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carpenter AJ, Porter AC. Construction, characterization, and complementation of a conditional-lethal DNA topoisomerase IIalpha mutant human cell line. Mol Biol Cell. 2004;15:5700–5711. doi: 10.1091/mbc.E04-08-0732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Obado SO, Bot C, Nilsson D, Andersson B, Kelly JM. Repetitive DNA is associated with centromeric domains in Trypanosoma brucei but not Trypanosoma cruzi. Genome Biol. 2007;8:R37. doi: 10.1186/gb-2007-8-3-r37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kelly JM, McRobert L, Baker DA. Evidence on the chromosomal location of centromeric DNA in Plasmodium falciparum from etoposide-mediated topoisomerase-II cleavage. Proc Natl Acad Sci USA. 2006;103:6706–6711. doi: 10.1073/pnas.0510363103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Floridia G, Zatterale A, Zuffardi O, Tyler-Smith C. Mapping of a human centromere onto the DNA by topoisomerase II cleavage. EMBO Rep. 2000;1:489–493. doi: 10.1093/embo-reports/kvd110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Turner BM. Reading signals on the nucleosome with a new nomenclature for modified histones. Nat Struct Mol Biol. 2005;12:110–112. doi: 10.1038/nsmb0205-110. [DOI] [PubMed] [Google Scholar]
- 29.Sullivan BA, Karpen GH. Centromeric chromatin exhibits a histone modification pattern that is distinct from both euchromatin and heterochromatin. Nat Struct Mol Biol. 2004;11:1076–1083. doi: 10.1038/nsmb845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Partridge JF, Borgstrøm B, Allshire RC. Distinct protein interaction domains and protein spreading in a complex centromere. Genes Dev. 2000;14:783–791. [PMC free article] [PubMed] [Google Scholar]
- 31.Blower MD, Karpen GH. The role of Drosophila CID in kinetochore formation, cell-cycle progression and heterochromatin interactions. Nat Cell Biol. 2001;3:730–739. doi: 10.1038/35087045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Salisbury JL. Centrin, centrosomes, and mitotic spindle poles. Curr Opin Cell Biol. 1995;7:39–45. doi: 10.1016/0955-0674(95)80043-3. [DOI] [PubMed] [Google Scholar]
- 33.Striepen B, et al. The plastid of Toxoplasma gondii is divided by association with the centrosomes. J Cell Biol. 2000;151:1423–1434. doi: 10.1083/jcb.151.7.1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mann T, Beckers C. Characterization of the subpellicular network, a filamentous membrane skeletal component in the parasite Toxoplasma gondii. Mol Biochem Parasitol. 2001;115:257–268. doi: 10.1016/s0166-6851(01)00289-4. [DOI] [PubMed] [Google Scholar]
- 35.Iwanaga S, et al. Functional identification of the Plasmodium centromere and generation of a Plasmodium artificial chromosome. Cell Host Microbe. 2010;7:245–255. doi: 10.1016/j.chom.2010.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fischer T, et al. Diverse roles of HP1 proteins in heterochromatin assembly and functions in fission yeast. Proc Natl Acad Sci USA. 2009;106:8998–9003. doi: 10.1073/pnas.0813063106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Flueck C, et al. Plasmodium falciparum heterochromatin protein 1 marks genomic loci linked to phenotypic variation of exported virulence factors. PLoS Pathog. 2009;5:e1000569. doi: 10.1371/journal.ppat.1000569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bougdour A, Braun L, Cannella D, Hakimi MA. Chromatin modifications: Implications in the regulation of gene expression in Toxoplasma gondii. Cell Microbiol. 2010;12:413–423. doi: 10.1111/j.1462-5822.2010.01446.x. [DOI] [PubMed] [Google Scholar]
- 39.Dzikowski R, Deitsch KW. Genetics of antigenic variation in Plasmodium falciparum. Curr Genet. 2009;55:103–110. doi: 10.1007/s00294-009-0233-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.King MC, Drivas TG, Blobel G. A network of nuclear envelope membrane proteins linking centromeres to microtubules. Cell. 2008;134:427–438. doi: 10.1016/j.cell.2008.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bhaud Y, et al. Morphology and behaviour of dinoflagellate chromosomes during the cell cycle and mitosis. J Cell Sci. 2000;113:1231–1239. doi: 10.1242/jcs.113.7.1231. [DOI] [PubMed] [Google Scholar]
- 42.Sinden RE, Canning EU, Spain B. Gametogenesis and fertilization in Plasmodium yoelii nigeriensis: A transmission electron microscope study. Proc R Soc Lond B Biol Sci. 1976;193:55–76. doi: 10.1098/rspb.1976.0031. [DOI] [PubMed] [Google Scholar]
- 43.Striepen B, Soldati D. Genetic manipulation of Toxoplasma gondii. In: Weiss LD, Kim K, editors. Toxoplasma gondii. The Model Apicomplexan—Perspectives and Methods. London: Elsevier; 2007. pp. 391–415. [Google Scholar]
- 44.Gubbels MJ, Li C, Striepen B. High-throughput growth assay for Toxoplasma gondii using yellow fluorescent protein. Antimicrob Agents Chemother. 2003;47:309–316. doi: 10.1128/AAC.47.1.309-316.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wells J, Farnham PJ. Characterizing transcription factor binding sites using formaldehyde crosslinking and immunoprecipitation. Methods. 2002;26:48–56. doi: 10.1016/S1046-2023(02)00007-5. [DOI] [PubMed] [Google Scholar]
- 46.Striepen B, Soldati D, Garcia-Reguet N, Dubremetz JF, Roos DS. Targeting of soluble proteins to the rhoptries and micronemes in Toxoplasma gondii. Mol Biochem Parasitol. 2001;113:45–53. doi: 10.1016/s0166-6851(00)00379-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.