Abstract
Elucidating the mechanism of genetic exchange is fundamental for understanding how genes for such traits as virulence, disease phenotype, and drug resistance are transferred between pathogen strains. Genetic exchange occurs in the parasitic protists Trypanosoma brucei, T. cruzi, and Leishmania major, but the precise cellular mechanisms are unknown, because the process has not been observed directly. Here we exploit the identification of homologs of meiotic genes in the T. brucei genome and demonstrate that three functionally distinct, meiosis-specific proteins are expressed in the nucleus of a single specific cell type, defining a previously undescribed developmental stage occurring within the tsetse fly salivary gland. Expression occurs in clonal and mixed infections, indicating that the meiotic program is an intrinsic but hitherto cryptic part of the developmental cycle of trypanosomes. In experimental crosses, expression of meiosis-specific proteins usually occurred before cell fusion. This is evidence of conventional meiotic division in an excavate protist, and the functional conservation of the meiotic machinery in these divergent organisms underlines the ubiquity and basal evolution of meiosis in eukaryotes.
Keywords: African trypanosomes, Kinetoplastida, Euglenozoa, fluorescent reporter, Glossina
Elucidating the mechanism of genetic exchange is fundamental for understanding how genes for such traits as virulence, disease phenotype, and drug resistance are transferred within pathogen populations (1). The African trypanosome Trypanosoma brucei is representative of a group of kinetoplastid protozoa that are responsible for several vector-borne diseases important to human and animal health worldwide. Analysis of the inheritance of markers has been used to demonstrate genetic exchange in kinetoplastids, first in T. brucei (2) and more recently in T. cruzi (3) and Leishmania major (4). In T. brucei, the pattern of inheritance is predominantly Mendelian, indicating the occurrence of meiosis or a similar process (5), but polyploid hybrids also frequently occur (6). In contrast, T. cruzi undergoes some form of parasexual process involving cell fusion and subsequent gene loss apparently in the mammalian host (3). Current knowledge of genetic exchange in these three parasites has been recently reviewed in the context of the evolution of sexual reproduction in microbial pathogens (7).
The cellular events involved in genetic exchange in trypanosomes have long been the subject of speculation, for several reasons. First, despite the intensive work over the past 100 y on describing the complex life cycle of T. brucei, which includes at least 10 successive developmental forms (reviewed in ref. 8), cells undergoing meiosis have not been identified. Second, trypanosomes have a fixed, cage-like, microtubule cytoskeleton underlying the plasma membrane that defines the elongated cell shape and have a single flagellum subtended by a basal body that is physically linked by sets of filaments to the mitochondrion and mitochondrial genome, the kinetoplast (9–11). Inheritance of the basal body and flagellum cannot use established replication mechanisms during the reduction division of meiosis, because the generation of new organelles is linked to DNA replication (11–13), and the fixed microtubule cytoskeleton would not allow nuclear fusion without partial disassembly. Third, the process of genetic exchange in T. brucei involves mixing of mitochondrial (kinetoplast) and nuclear genomes, because hybrid progeny have hybrid kinetoplast DNA (kDNA) networks with mini-circles derived from both parents (14, 15). Plausible models for the generation of hybrid kDNA networks are limited by the complex structure and highly ordered replication of this concatenated mass of small DNA circles (16). Finally, trypanosomes belong to the Euglenozoa, a deep branch within the excavate eukaryote supergroup (17, 18). The production of four haploid gametes and subsequent fusion to reform the diploid occurring in trypanosomes would strongly suggest the presence of a typical meiosis in the last eukaryotic ancestor. The only other excavate in which a form of genetic exchange has been investigated in depth is Giardia, a distantly related diplomonad, in which genetic exchange occurs without cell fusion. Vegetative Giardia cells contain two nuclei that are maintained separately throughout the cell cycle. However, during formation of the tetranucleate cyst, the two nuclei express genes characteristic of meiosis in other eukaryotes, and may fuse and exchange genetic material (19). Whether this process is an evolutionary derivative or an alternative to classical meiosis is unclear (7). Trypanosomes have a single nucleus and must use a different strategy for genetic exchange.
The unique features of meiosis that set it apart from mitosis are the pairing of homologous chromosomes, the formation of synaptonemal complexes (SCs) and recombination between nonsister chromatids during prophase I, and the segregation of homologs in the first meiotic division. Well-characterized proteins intrinsic to these unique processes include Spo11 (20), Mnd1, Dmc1 (21) (all three of which function in processing DNA during recombination), and Hop1 (22) (a component of the SC). These four meiosis-specific genes occur widely in yeast, animals, and higher plants, and homologs have been found in the trypanosome genome (23, 24).
In T. brucei, meiosis is predicted to occur in the tsetse fly vector because this, rather than the mammalian host, is the site of genetic exchange (2). When a tsetse fly feeds on an infected host, ingested trypanosomes first differentiate and multiply in the midgut for a week or so before migrating to the salivary glands (SGs), where they differentiate into epimastigotes that attach to the SG epithelium via an elaborated flagellar membrane (25). The epimastigotes proliferate and subsequently differentiate again into infective metacyclics that are transferred into new hosts via the saliva when the fly takes a bloodmeal. The SGs have been identified as the location of mating; in experimental crosses of red and green fluorescent trypanosomes, yellow fluorescent hybrids appeared in the SGs but were not found in the midgut or among migrating trypanosomes (6).
Investigation of SG trypanosomes is technically challenging, because there is no culture system and few experimentally infected tsetse flies develop SG infection. An infected SG contains few trypanosomes, particularly in the early phase of colonization. These factors thwart most cell biology approaches, including the use of antibodies; however, fluorescent protein reporters enable detection of even a single fluorescent trypanosome (26, 27). Here, we demonstrate that three fluorescently tagged, functionally distinct, meiosis-specific proteins are expressed in the nucleus of a specific type of dividing cell in the SG, and infer that this developmental form is the meiotic stage of the trypanosome life cycle.
Results
Expression of Meiosis-Specific Genes.
Homologs of meiosis-specific genes (HMGs) were identified by phylogenomic analysis of the T. brucei genome (23, 24). Four HMGs encoding proteins that function solely during meiosis and are expressed only during meiosis in the other eukaryotes analyzed to date were selected: DMC1, Tb09.211.1210; HOP1, Tb10.70.1530; MND1, Tb11.02.3380; and SPO11, Tb927.5.3760. All four proteins function during homologous recombination during the prophase of meiosis I: SPO11 initiates recombination by introducing a double-stranded break, MND1 stabilizes heteroduplexes after double-stranded break formation, DMC1 is a homolog of RAD51 and promotes strand exchange, and HOP1 is a component of the lateral elements of the SC.
We reasoned that if the function of the HMG proteins in trypanosomes were conserved, then expression of HMGs would be restricted to meiotic cells and the proteins would be absent in other cell types. To identify any developmental stage that expressed HMGs, we prepared transgenic cell lines in which one of the two HMG alleles was modified to become an N-terminal YFP fusion (YFP::HMG). In addition to the ORF, the modification altered the 5′ UTR, but the 3′ UTR was unaltered and expression relied on endogenous transcription. The modifications were performed in cultured procyclic lines of the mating-competent T. b. brucei strain J10 (6, 28), and all cell lines were taken through at least one complete life cycle.
Expression of YFP::MND1, YFP::DMC1, and YFP::HOP1 was restricted to a subset of trypanosomes in the SGs (Fig. 1 and Movie S1). In these cells, intense fluorescence was localized to the cell nucleus in the majority of cells, with a punctate pattern for YFP::DMC1 and YFP::HOP1 (Fig. 1); some YFP::DMC1 cells also exhibited weak fluorescence in the cytoplasm. These observations agree with those for yeast; Dmc1 localizes to discrete foci in the nucleus (29), and Hop1 also should produce punctate fluorescence, because it is a component of SC lateral elements (22). YFP::MND1 was diffuse throughout the nucleus, again similar to yeast (30). YFP fluorescence was not observed in any other developmental stage, including earlier forms from the midgut or proventriculus (Table 1).
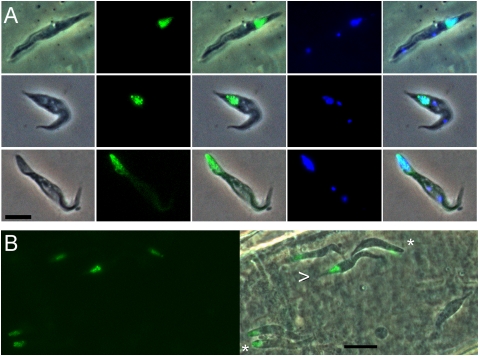
Fig. 1.
Expression of meiosis-specific YFP fusion proteins in trypanosomes from SGs of tsetse flies. (A) (Top) J10 YFP::MND1. (Middle) J10 YFP::DMC1. (Bottom) J10 YFP::HOP1. The first column shows phase contrast images of fixed trypanosomes in salivary exudate; the other columns show epifluorescence microscopy images of YFP fusion protein expression or DAPI-stained nucleus and kinetoplast, along with merged images. In all cases, YFP fluorescence colocalizes with DAPI-stained nucleus toward the posterior end of the trypanosome, and there are two kinetoplasts, (center and anterior small blue dots). (B) Live phase contrast and epifluorescence images of trypanosomes of clone J10 YFP::HOP1 inside a tsetse SG. Trypanosomes expressing the fluorescent fusion protein have a blunt (asterisk) or pointed (arrow) posterior, with the nucleus very near the posterior end. The RH nonfluorescent trypanosome is a typical epimastigote with an elongated tube-like posterior. Cell movement has compromised merge of phase and fluorescence images. (Scale bar: 5 μm.)
Table 1.
Summary of data from tsetse fly transmission of J10 transfected clones
| Cell line* | Midguts† | SGs (pair)† | |||
| Total infected | Number with fluorescent cells | Total infected | Number with fluorescent cells | Days after infection‡ | |
| J10 YFP::DMC1 clones 1 and 2 | 349/499 (70%) | 0/124 (0%) | 15/349 (4%) | 14/15 (93%) | 14–31 |
| J10 YFP::MND1 clones 1 and 2 | 219/340 (64%) | 0/110 (0%) | 9/219 (4%) | 9/9 (100%) | 17–33 |
| J10 YFP::HOP1 clones 1 and 2 | 307/339 (91%) | 0/74 (0%) | 44/307 (14%) | 35/44 (80%) | 20–28 |
| J10 YFP::SPO11 clones 1, 2, and 3 | 456/727 (63%) | 0/66 (0%) | 17/456 (4%) | 0/17 (0%) | NA |
*At least two different clones were analyzed for each fusion construct.
†The infection rates for midguts and SGs are similar to those routinely achieved for WT J10, showing that the fusion constructs did not have an adverse effect.
‡Fluorescent trypanosomes were observed in the SGs during the time window given.
Examination of two independent clones of each transgenic line revealed expression in most SGs examined at 14–33 d after infection (Table 1). The proportion of fluorescent trypanosomes per SG was variable, being most prevalent in SGs dissected 17–21 d after infection. Expressers never constituted more than an estimated 20% of the population. (Counts were approximate because nonfluorescent trypanosomes were obscured by the SG's thick walls and expressers were distributed unevenly.) Expression of YFP::SPO11 was not detected in any life cycle stage (three independent clones analyzed; Table 1). This negative result is not interpretable; it may indicate that SPO11 is not expressed, but other possible explanations are that expression was below the threshold of detection and/or very transient, or that modification of the gene interfered with regulation of expression or protein stability.
Morphology of HMG Expressers.
In trypanosomes, the mitochondrial DNA is concatenated in the kinetoplast, which appears as a small, extranuclear body in cells stained for DNA. Morphologically, cells expressing YFP::HMGs were identified as epimastigotes, because the kinetoplast was anterior to the nucleus (Fig. 1A). When live cells were observed in dissected SGs, some of the trypanosomes expressing YFP::HMGs were attached epimastigotes (Movie S1), whereas others were unattached and spilled out of the SGs with the saliva. However, cells expressing YFP::HMGs differed from the previously described SG epimastigotes, with the posterior lacking the characteristic elongated protrusion (Fig. 1B) (25) and the nucleus often occupying the posterior end of the cell rather than the usual central position (Fig. 1 A and B). Cells expressing YFP::HMGs clearly had two well-separated kinetoplasts (Fig. 1A), indicating that they were in late stages of the cell cycle. In other developmental stages, the two daughter kinetoplasts separate in G2 before mitosis (12, 13).
Trypanosome cell lines carrying YFP::HMG transgenes were further modified to express a YFP::PFR1 transgene, so that they constitutively expressed YFP fused to the paraflagellar rod (PFR) component, PFR1 (31). These cell lines were used to visualize flagella in cells expressing YFP::HMGs, which confirmed that epimastigotes expressing YFP::HMGs have two flagella as well as two kinetoplasts (Fig. 2A). In cultured trypanosomes, the new flagellum appears to contain less PFR1 per unit length compared with the old flagellum (Fig. 2B). In epimastigotes expressing YFP::HMGs, the intensity of YFP::PFR1 fluorescence was consistent with the anterior flagellum being the old flagellum (Fig. 2A). The PFR in the new flagellum was noticeably shorter in cells expressing YFP::MND1 than in cells expressing YFP::HOP1 (Movies S2 and S3). Given that PFR length is a measure of cell cycle progression (12), this observation suggests a way to ascertain the temporal order of expression of the YFP::HMGs. We could not measure the length of the new PFR directly in fixed cells, because that PFR runs alongside the old PFR. Instead, we measured the distance between the starts of the old and new PFRs and used it as a proxy for the separation of the old and new basal bodies during cell division (11–13) [Fig. 3A, (a)]. The shortest values were obtained for cells expressingYFP::MND1, indicating that MND1 is expressed before DMC1 or HOP1 (Fig. 3B; inter-PFR distance). The total cell length of YFP::HOP1 expressers was significantly greater than that of MND1 or DMC1 expressers (Fig. 3B; total cell length), indicating that a later stage of the cell cycle had been reached in the HOP1 expressers. These two parameters thus provided a temporal framework for the interpretation of other morphological changes. Although the size of the nucleus did not change significantly (Fig. 3B; nucleus area), the three YFP::HMG expressers demonstrated changes in the shape and position of the nucleus (Fig. 3 A and B). The nucleus was closer to the posterior pole in YFP::DMC1 and YFP::HOP1 expressers relative to YFP::MND1 expressers (Fig. 3B; posterior nucleus), and was both close to the posterior and elongated in YFP::HOP1 expressers (Fig. 3B; nucleus length). Whereas the distance between the two kinetoplasts remained constant (Fig. 3B; kin1–kin2), the distance between the new kinetoplast and the nucleus increased progressively (Fig. 3B; nucleus–kin1). The progression of changes in cell morphology is summarized in Fig. 3C. The order of expression of trypanosome HMGs derived here—MND1 followed by DMC1, followed by HOP1—agrees with the sequence of events in other eukaryotes (21, 22, 30).
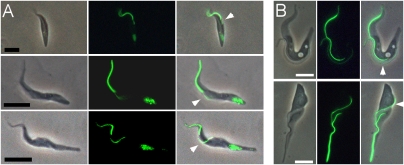
Fig. 2.
Morphology of the meiotic cell. (A) (Top) J10 YFP::MND1, YFP::PFR1. (Middle) J10 YFP::DMC1, YFP::PFR1. (Bottom) J10 YFP::HOP1, YFP::PFR1. The first column shows phase contrast images of fixed trypanosomes in salivary exudate; the other columns show epifluorescence microscopy images of YFP fusion protein expression and merged images. In trypanosomes expressing YFP::PFR1, the PFR incorporates the fusion protein and is fluorescent. The brightly fluorescent anterior PFR is old, whereas the less-bright PFR (arrow) is that of the daughter cell. (Scale bar 5 μm.) (B) Identification of the daughter flagellum in dividing procyclic cells. Previous studies of the process of cell division and timing of construction of new organelles have shown that the daughter flagellum emerges posterior to the parental flagellum (12). In the examples shown, the new PFR (arrow) is seen to fluoresce less brightly than the old PFR in the phase contrast and epifluorescence images. (Scale bar: 5 μm.)
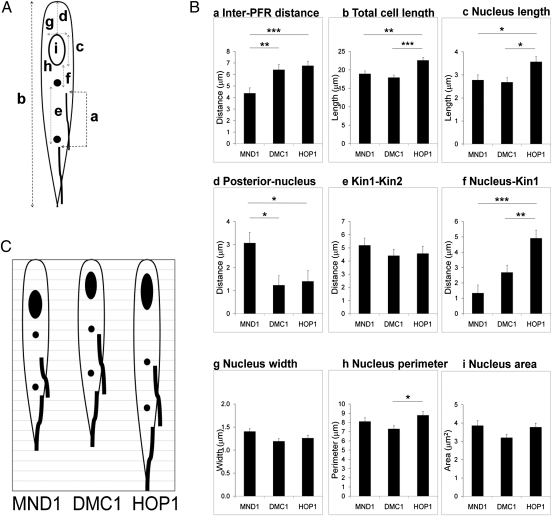
Fig. 3.
Comparison of morphological parameters in HMG expressers. (A) Measurements: (a) distance between the start of the PFR of each flagellum; (b) total cell length; (c) nucleus length; (d) posterior of th cell to posterior of the nucleus; (e) distance between kinetoplasts 1 and 2 (kin1–kin2); (f) distance between the nucleus and kinetoplast 1 (kin1); (g) nucleus width; (h), nucleus perimeter; (i) nucleus area. The open circle represents the nucleus; filled circles, kinetoplasts 1 and 2; thick lines, PFR. (B) Each parameter was measured in the three cell lines shown in Fig. 2. For PFR, n = 13 for MND1 and DMC1 and n = 18 for HOP1; for other parameters, n = 11 for MND1 and HOP1 and n = 14 for DMC1. P values were calculated by ANOVA and Tukey's post hoc test. *P < 0.05; **P < 0.01; ***P < 0.001. Error bars are SEM. (C) Comparative morphology of HMG expressers based on mean values (1-μm grid). The short inter-PFR distance places MND1 expressers at the start of the series; the cell length of HOP1 expressers places them at the end of the series. The nucleus progressively elongates and moves posteriorly. The relative position of the kinetoplasts remains constant.
Meiosis in Experimental Crosses.
The foregoing observations were obtained from clonal infections, and thus the expression of HMGs is not triggered by the presence of nonself trypanosomes. It has been suggested that interaction between different trypanosome strains promotes more frequent mating, because recombinants are found only rarely in intraclonal crosses (32–34). Our comparison of clonal transmissions of J10 YFP::DMC1 or J10 YFP::HOP1 with cotransmissions including T. b. brucei 1738 mRFP revealed no consistent difference in the numbers of trypanosomes expressing YFP::HMGs.
Cell fusion, usually involving haploids, is an integral part of mating in most eukaryotes. In trypanosomes, hybrid clones contain kinetoplast DNA from both parents, indicating that mitochondrial fusion (and hence cell fusion) has occurred (14, 15), but whether meiosis precedes cell fusion or vice versa is not known. To determine the temporal order of meiosis and cell fusion, we crossed either J10 YFP::DMC1 or J10 YFP::HOP1 with 1738 mRFP. In previous crosses of red and green fluorescent lines of J10 and 1738, yellow hybrids were observed in most SGs containing a mixed infection of both parental lines, but not in those with a single infection (6). For the J10 YFP::DMC1 × 1738 mRFP cross, none of the YFP::DMC1 expressers (21 flies with mixed SG infections) also had red fluorescence, indicating that meiosis precedes fusion (Fig. 4A). The same result was observed for the majority of YFP::HOP1 expressers in the J10 YFP::HOP1 × 1738 mRFP cross (13 flies with mixed SG infections). However, in two SGs, a minority of trypanosomes with red cytoplasm also had punctate yellow fluorescence in the nucleus (Fig. 4B), indicating that cell fusion may occasionally occur before meiosis is complete. Other than these very rare events, scrutiny of SGs with mixed infections yielded no evidence of fusion between meiotic J10 trypanosomes and 1738 mRFP. We verified the production of hybrid trypanosomes in these crosses by cloning and microsatellite genotyping progeny clones from each cross (6) (Table S1).
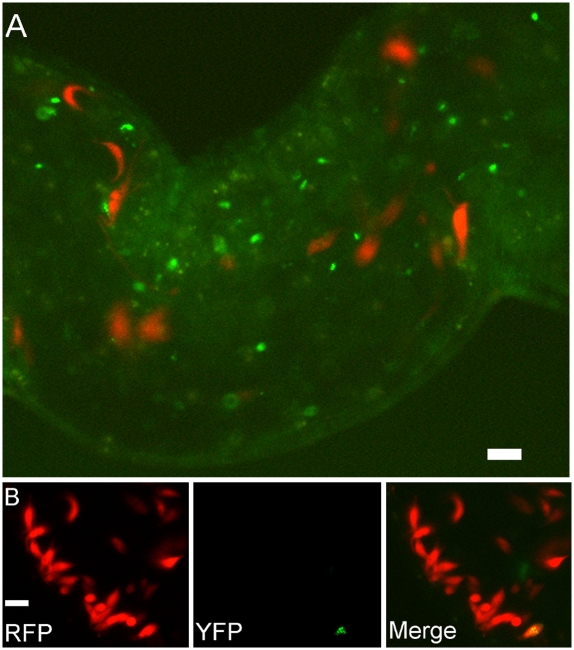
Fig. 4.
Meiosis occurs before fusion. (A) Epifluorescence image of a section of an SG infected with J10 YFP::DMC1 and 1738 mRFP. The trypanosomes expressing YFP in the nucleus (J10 YFP::DMC1) are separate from those expressing mRFP in the cytoplasm (1738 mRFP). (B) Similar image of an SG infected with J10 YFP::HOP1 and 1738 mRFP. This rare example of a trypanosome expressing both mRFP in the cytoplasm and YFP in the nucleus suggests that in this case, fusion occurred before completion of meiosis. Live images. (Scale bar: 10 μm.)
Presence of Haploids.
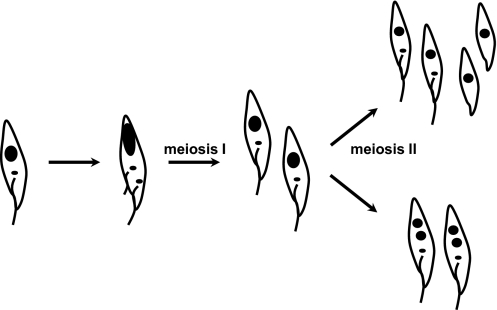
Conventionally, meiosis I ends with partitioning of the chromosomes to yield two 2N nuclei, each of which gives rise to two haploid nuclei, and this occurs without any further replication of DNA. In the trypanosome expressing HOP1, division appears to be far advanced and about to yield two trypanosome cells, each with its own kinetoplast and flagellum. Each of these cells will have a 2N nucleus, which may complete meiosis II to give two haploid nuclei within the same cell or, alternatively, go on to divide again to produce haploid gametes (Fig. 5). Another replication of the kinetoplast and flagellum is unlikely, given the tight linkage between replication of these organelles and the nuclear DNA (9–13), so two types of haploid cell may occur (Fig. 5). To search for haploid cells, we used K11, a trypanosome cell line expressing GFP under control of the Tet repressor (35). Segregation of the chromosomes carrying the GFP reporter and repressor loci during meiosis should yield fluorescent haploid trypanosomes. No fluorescent trypanosomes were seen in 11 SGs dissected at 16–30 d after infection with K11, although morphologically identifiable meiotic trypanosomes were present on days 17 and 21. In tetracycline-induced K11 from macerated SGs, GFP fluorescence was just visible after 3 h and intense by 7 h; thus, transient haploids persisting for <5 h would not have been detected in this experiment. We conclude that either no haploids were produced or any haploids produced were very transient.
Fig. 5.
Model of meiosis in trypanosomes. An epimastigote (Left) enters meiosis, and the first division results in two 2N cells. Meiosis II follows, producing haploid nuclei. Two possible outcomes are shown, assuming that replication of the kinetoplast and flagellum does not occur.
Discussion
In T. brucei, the pattern of inheritance of genetic markers is Mendelian (5), so recombination between homologs and independent assortment of chromosomes must occur. The cell biology of this process is intriguing. Despite a century of inquiry, no meiotic cells have been identified. The reduction divisions must involve a departure from the paradigm established for mitotic division, where the duplication of the basal body and flagellum is integrated into the cell cycle, and the adjustments to the subpellicular microtubule corset necessary for nuclear and mitochondrial fusion must be novel (9–13).
Here we have identified the likely meiotic stage of the trypanosome life cycle by identifying a developmental form that expresses three different HMGs characteristic of meiotic prophase I. The three YFP::HMGs exhibited a similar pattern of expression, with strong fluorescence only in the nuclei of morphologically distinct, dividing epimastigotes present in the SGs. The temporal order of expression of the three HMGs followed that seen in other eukaryotes and concurred with the functional roles of these proteins during meiosis (21, 22). MND1 was expressed first, and HOP1 was expressed last. In other eukaryotes, HOP1 is a component of the SC lateral elements, and thus we interpret the punctate fluorescence observed here for this fusion protein as the direct visualization of SC in trypanosomes. The identification of cells expressing HMGs in the SGs is consistent with this being the location of mating (6). Although meiosis is the most straightforward interpretation of the expression of these three HMGs, alternative explanations could center around DNA repair. This is unlikely for DMC1, however, given that double knockout produced no detectable phenotype in bloodstream forms (36).
After pairing of homologs, the second unique feature of meiosis is the reductive divisions to produce haploids after the separation of homologs in meiosis I and of sister chromatids in meiosis II. Proteins that could be used to mark cells during the meiotic divisions, such as Rec8 (37) or SgoI (38) in yeasts, are either not present or not readily identifiable in the trypanosome genome, and we are currently unable to specifically mark trypanosomes undergoing the meiotic divisions. Therefore, we have no information on what becomes of the trypanosome after YFP::HOP1 is degraded along with the rest of the SC at the end of pachytene. In the trypanosome expressing YFP::HOP1, the kinetoplasts are widely separated and the new and old flagella are of similar length, indicating that cell division is imminent (13). Such a division would yield two daughter cells each with a 2N nucleus. Meiosis II could then yield haploid gametes or two haploid nuclei within the same cell (Fig. 5). In the former case, it seems unlikely that each gamete would have a kinetoplast and a flagellum, given the tight control of organelle replication within the cell cycle (13). Our experiment to detect haploids using segregation of GFP and repressor genes indicates that haploid cells, if present, are transient (<5 h). Further work is needed to reveal the nature of the products of meiotic division.
The finding of HMG expression during infections initiated with both single clones and mixtures of different trypanosome strains in genetic crosses indicates that HMG expression is not triggered by the presence of nonself trypanosomes. We infer that meiosis may be a normal part of the trypanosome developmental cycle in the fly, contrary to the traditional narrative that includes only mitotic divisions. In laboratory crosses, mating (and hence meiosis) is considered to occur rarely and only when two different trypanosome strains are present, although low frequencies of intraclonal mating have been detected in both single transmissions and cotransmissions of different strains (32–34). Even though we observed fluorescence in only a small proportion of trypanosomes at any one time, it remains possible that all SG trypanosomes undergo meiosis at a certain point during establishment of the SG infection. Trypanosome invasion and colonization of the SG are not synchronous, and chance dictates that we would catch few trypanosomes in a process that probably lasts only a matter of hours. The expression of HMGs early in infection is also consistent with the timing of hybrid production, which starts as early as 13 d after fly infection (6).
Tracking the expression of fluorescently tagged genes has proven to be a powerful approach to identifying and characterizing a rare cell type. Despite the very small number of cells available for analysis, our tagging of several genes that function sequentially in the same process has elucidated details of the timing and progression of events. The parallels between meiosis in trypanosomes and meiosis in yeast underscore the ubiquity and basal evolution of meiosis in eukaryotes.
Materials and Methods
Trypanosomes.
Two tsetse-transmissible strains of Trypanosoma brucei brucei were used: J10 (MCRO/ZM/73/J10 CLONE 1) (39) and 1738 (MOVS/KE/70/1738) (40). The 1738 mRFP carried a transgene for monomeric red fluorescent protein (6, 41). Strain K11 is derived from T. b. gambiense group 2 [MHOM/CI/78/TH2 (78E)] (42) and contains a GFP gene driven by a procyclin promoter under control of the tet repressor (35). Procyclic form (PF) trypanosomes were grown in Cunningham's medium (CM) (43) supplemented with 10% vol/vol heat-inactivated FCS, 5 μg/mL of hemin, and 10 μg/mL of gentamycin at 27 °C.
Transfection.
Fusion constructs were assembled in plasmid vectors (44). For the HMGs (DMC1, Tb09.211.1210; HOP1, Tb10.70.1530; MND1, Tb11.02.3380; SPO11, Tb927.5.3760), the gene for enhanced YFP was fused to the N terminus of the endogenous ORF, whereas for PFR1 (Tb927.3.4290), a C-terminal fusion was used. Expression of the modified gene and selectable marker gene relied on endogenous transcription; the 3′ UTR of the modified gene was unaltered, because this was the most likely location of determinants of developmentally regulated expression (45). PF trypanosomes were transfected by electroporation using two pulses of 1.5 kV, 25 μF, and transfectants were selected at 16 h or 24 h postelectroporation by the addition of appropriate antibiotics. Clones were obtained by limiting dilution of PF trypanosomes in CM in 96-well plates incubated at 27 °C in 5% CO2. Correct integration was verified by PCR on genomic DNA templates using primers spanning the integration site (the left primer complementary to the YFP gene, and the right primer within the original 3′ end of the targeted gene). None of the HMG transfectants exhibited expression of the fluorescent fusion gene as cultured PF trypanosomes.
Tsetse Transmission.
Tsetse flies were offered a bloodmeal containing bloodstream form (BF) or PF trypanosomes at 24–48 h posteclosion as their first feed. The bloodmeal consisted of ∼8 × 106 BF trypanosomes mL−1 in sterile horse blood or ∼107 PF trypanosomes per mL of washed horse red blood cells resuspended in HBSS, supplemented with 10 mM l-glutathione (46). Infected flies were maintained on sterile horse blood supplemented with 2.5% wt/vol BSA (47) and 1 mM dATP (48) until dissection. Flies were dissected up to 9 wk after the infective feed. Metacyclics from infected SGs were inoculated into mice; if required, BF trypanosomes were subsequently transformed to PF trypanosomes by incubation in CM at 27 °C.
Tsetse Dissection.
Fly organs (SGs and alimentary tract from the proventriculus to the hindgut) were dissected in a drop of PBS and examined for the presence of fluorescent trypanosomes using a DMRB microscope (Leica) equipped with a Retiga EXi camera (QImaging) and Volocity imaging software (PerkinElmer). Cells were fixed in 2% wt/vol paraformaldehyde at room temperature for 20 min and stained with DAPI in VECTASHIELD mounting medium (Vector Laboratories) to visualize the nucleus and kinetoplast. PF trypanosomes grown in culture were washed in PBS before fixation, improving preservation of their morphology; this was not possible for the small number of SG trypanosomes available.
Genotype Analysis.
Genomic DNA was prepared from PF trypanosomes using a spin column DNA purification kit (Qiagen). Microsatellite analysis was performed by PCR as described previously (6) using loci on a total of four chromosomes (5). Products were resolved by electrophoresis in 1× TAE buffer through 3–5% MetaPhor agarose (Lonza) gels.
Statistical Analysis.
ImageJ software (http://rsb.info.nih.gov/ij) was used to analyze digital images of fixed, DAPI-stained trypanosomes expressing HMGs; measurements are indicated in Fig. 3A. The distance between the start of the new and old PFRs was measured in trypanosomes coexpressing YFP::HMG and YFP::PFR1 by following the new PFR from its start to the start of the old PFR. Because the PFR begins at a short, fixed distance from the basal body (9), this serves as a proxy for the position of the basal body. Data were analyzed using the SPSS statistical package (SPSS).
Supplementary Material
Acknowledgments
We thank the International Atomic Energy Agency, Vienna for supplying tsetse fly pupae and the Wellcome Trust (http://www.wellcome.ac.uk/) for providing funding to both laboratories.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1019423108/-/DCSupplemental.
References
- 1.Heitman J. Sexual reproduction and the evolution of microbial pathogens. Curr Biol. 2006;16:R711–R725. doi: 10.1016/j.cub.2006.07.064. [DOI] [PubMed] [Google Scholar]
- 2.Jenni L, et al. Hybrid formation between African trypanosomes during cyclical transmission. Nature. 1986;322:173–175. doi: 10.1038/322173a0. [DOI] [PubMed] [Google Scholar]
- 3.Gaunt MW, et al. Mechanism of genetic exchange in American trypanosomes. Nature. 2003;421:936–939. doi: 10.1038/nature01438. [DOI] [PubMed] [Google Scholar]
- 4.Akopyants NS, et al. Demonstration of genetic exchange during cyclical development of Leishmania in the sand fly vector. Science. 2009;324:265–268. doi: 10.1126/science.1169464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.MacLeod A, et al. Allelic segregation and independent assortment in T. brucei crosses: Proof that the genetic system is Mendelian and involves meiosis. Mol Biochem Parasitol. 2005;143:12–19. doi: 10.1016/j.molbiopara.2005.04.009. [DOI] [PubMed] [Google Scholar]
- 6.Gibson W, Peacock L, Ferris V, Williams K, Bailey M. The use of yellow fluorescent hybrids to indicate mating in Trypanosoma brucei. Parasit Vectors. 2008;1:4. doi: 10.1186/1756-3305-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Heitman J. Evolution of eukaryotic microbial pathogens via covert sexual reproduction. Cell Host Microbe. 2010;8:86–99. doi: 10.1016/j.chom.2010.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sharma R, et al. The heart of darkness: Growth and form of Trypanosoma brucei in the tsetse fly. Trends Parasitol. 2009;25:517–524. doi: 10.1016/j.pt.2009.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gull K. The cytoskeleton of trypanosomatid parasites. Annu Rev Microbiol. 1999;53:629–655. doi: 10.1146/annurev.micro.53.1.629. [DOI] [PubMed] [Google Scholar]
- 10.Robinson DR, Gull K. Basal body movements as a mechanism for mitochondrial genome segregation in the trypanosome cell cycle. Nature. 1991;352:731–733. doi: 10.1038/352731a0. [DOI] [PubMed] [Google Scholar]
- 11.Ogbadoyi EO, Robinson DR, Gull K. A high-order trans-membrane structural linkage is responsible for mitochondrial genome positioning and segregation by flagellar basal bodies in trypanosomes. Mol Biol Cell. 2003;14:1769–1779. doi: 10.1091/mbc.E02-08-0525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sherwin T, Gull K. The cell division cycle of Trypanosoma brucei brucei: Timing of event markers and cytoskeletal modulations. Philos Trans R Soc Lond B Biol Sci. 1989;323:573–588. doi: 10.1098/rstb.1989.0037. [DOI] [PubMed] [Google Scholar]
- 13.Woodward R, Gull K. Timing of nuclear and kinetoplast DNA replication and early morphological events in the cell cycle of Trypanosoma brucei. J Cell Sci. 1990;95:49–57. doi: 10.1242/jcs.95.1.49. [DOI] [PubMed] [Google Scholar]
- 14.Gibson W, Garside L. Kinetoplast DNA minicircles are inherited from both parents in genetic hybrids of Trypanosoma brucei. Mol Biochem Parasitol. 1990;42:45–53. doi: 10.1016/0166-6851(90)90111-x. [DOI] [PubMed] [Google Scholar]
- 15.Gibson W, Crow M, Kearns J. Kinetoplast DNA minicircles are inherited from both parents in genetic crosses of Trypanosoma brucei. Parasitol Res. 1997;83:483–488. doi: 10.1007/s004360050284. [DOI] [PubMed] [Google Scholar]
- 16.Shapiro TA, Englund PT. The structure and replication of kinetoplast DNA. Annu Rev Microbiol. 1995;49:117–143. doi: 10.1146/annurev.mi.49.100195.001001. [DOI] [PubMed] [Google Scholar]
- 17.Keeling PJ, et al. The tree of eukaryotes. Trends Ecol Evol. 2005;20:670–676. doi: 10.1016/j.tree.2005.09.005. [DOI] [PubMed] [Google Scholar]
- 18.Hampl V, et al. Phylogenomic analyses support the monophyly of Excavata and resolve relationships among eukaryotic “supergroups.”. Proc Natl Acad Sci USA. 2009;106:3859–3864. doi: 10.1073/pnas.0807880106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Poxleitner MK, et al. Evidence for karyogamy and exchange of genetic material in the binucleate intestinal parasite Giardia intestinalis. Science. 2008;319:1530–1533. doi: 10.1126/science.1153752. [DOI] [PubMed] [Google Scholar]
- 20.Keeney S, Giroux CN, Kleckner N. Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell. 1997;88:375–384. doi: 10.1016/s0092-8674(00)81876-0. [DOI] [PubMed] [Google Scholar]
- 21.Petukhova GV, et al. The Hop2 and Mnd1 proteins act in concert with Rad51 and Dmc1 in meiotic recombination. Nat Struct Mol Biol. 2005;12:449–453. doi: 10.1038/nsmb923. [DOI] [PubMed] [Google Scholar]
- 22.Penkina MV, Karpova OI, Bogdanov YF. Synaptonemal complex proteins: Specific proteins of meiotic chromosomes. Mol Biol. 2002;36:304–313. [PubMed] [Google Scholar]
- 23.Ramesh MA, Malik SB, Logsdon JM., Jr. A phylogenomic inventory of meiotic genes: Evidence for sex in Giardia and an early eukaryotic origin of meiosis. Curr Biol. 2005;15:185–191. doi: 10.1016/j.cub.2005.01.003. [DOI] [PubMed] [Google Scholar]
- 24.El-Sayed NM, et al. The genome sequence of Trypanosoma cruzi, etiologic agent of Chagas disease. Science. 2005;309:409–415. doi: 10.1126/science.1112631. [DOI] [PubMed] [Google Scholar]
- 25.Tetley L, Vickerman K. Differentiation in Trypanosoma brucei: Host–parasite cell junctions and their persistence during acquisition of the variable antigen coat. J Cell Sci. 1985;74:1–19. doi: 10.1242/jcs.74.1.1. [DOI] [PubMed] [Google Scholar]
- 26.Peacock L, Ferris V, Bailey M, Gibson W. Dynamics of infection and competition between two strains of Trypanosoma brucei brucei in the tsetse fly observed using fluorescent markers. Kinetoplastid Biol Dis. 2007;6:4. doi: 10.1186/1475-9292-6-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gibson W, Bailey M. The development of Trypanosoma brucei within the tsetse fly midgut observed using green fluorescent trypanosomes. Kinetoplastid Biol Dis. 2003;2:1. doi: 10.1186/1475-9292-2-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gibson WC, Garside LH. Genetic exchange in Trypanosoma brucei brucei: Variable chromosomal location of housekeeping genes in different trypanosome stocks. Mol Biochem Parasitol. 1991;45:77–89. doi: 10.1016/0166-6851(91)90029-6. [DOI] [PubMed] [Google Scholar]
- 29.Bishop DK. RecA homologs Dmc1 and Rad51 interact to form multiple nuclear complexes prior to meiotic chromosome synapsis. Cell. 1994;79:1081–1092. doi: 10.1016/0092-8674(94)90038-8. [DOI] [PubMed] [Google Scholar]
- 30.Tsubouchi H, Roeder GS. The Mnd1 protein forms a complex with hop2 to promote homologous chromosome pairing and meiotic double-strand break repair. Mol Cell Biol. 2002;22:3078–3088. doi: 10.1128/MCB.22.9.3078-3088.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bastin P, Sherwin T, Gull K. Paraflagellar rod is vital for trypanosome motility. Nature. 1998;391:548. doi: 10.1038/35300. [DOI] [PubMed] [Google Scholar]
- 32.Tait A, Buchanan N, Hide G, Turner CM. Self-fertilisation in Trypanosoma brucei. Mol Biochem Parasitol. 1996;76:31–42. doi: 10.1016/0166-6851(95)02528-6. [DOI] [PubMed] [Google Scholar]
- 33.Gibson W, Winters K, Mizen G, Kearns J, Bailey M. Intraclonal mating in Trypanosoma brucei is associated with out-crossing. Microbiology. 1997;143:909–920. doi: 10.1099/00221287-143-3-909. [DOI] [PubMed] [Google Scholar]
- 34.Peacock L, Ferris V, Bailey M, Gibson W. Intraclonal mating occurs during tsetse transmission of Trypanosoma brucei. Parasit Vectors. 2009;2:43. doi: 10.1186/1756-3305-2-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bingle LEH, Eastlake JL, Bailey M, Gibson WC. A novel GFP approach for the analysis of genetic exchange in trypanosomes allowing the in situ detection of mating events. Microbiology. 2001;147:3231–3240. doi: 10.1099/00221287-147-12-3231. [DOI] [PubMed] [Google Scholar]
- 36.Proudfoot C, McCulloch R. Trypanosoma brucei DMC1 does not act in DNA recombination, repair or antigenic variation in bloodstream stage cells. Mol Biochem Parasitol. 2006;145:245–253. doi: 10.1016/j.molbiopara.2005.10.007. [DOI] [PubMed] [Google Scholar]
- 37.Watanabe Y, Nurse P. Cohesin Rec8 is required for reductional chromosome segregation at meiosis. Nature. 1999;400:461–464. doi: 10.1038/22774. [DOI] [PubMed] [Google Scholar]
- 38.Kitajima TS, Kawashima SA, Watanabe Y. The conserved kinetochore protein shugoshin protects centromeric cohesion during meiosis. Nature. 2004;427:510–517. doi: 10.1038/nature02312. [DOI] [PubMed] [Google Scholar]
- 39.Gibson WC, Marshall TF de C, Godfrey DG. Numerical analysis of enzyme polymorphism: A new approach to the epidemiology and taxonomy of trypanosomes of the subgenus Trypanozoon. Adv Parasitol. 1980;18:175–246. doi: 10.1016/s0065-308x(08)60400-5. [DOI] [PubMed] [Google Scholar]
- 40.Gibson WC, Borst P, Fase-Fowler F. Further analysis of intraspecific variation in Trypanosoma brucei using restriction site polymorphisms in the maxi-circle of kinetoplast DNA. Mol Biochem Parasitol. 1985;15:21–36. doi: 10.1016/0166-6851(85)90026-x. [DOI] [PubMed] [Google Scholar]
- 41.Campbell RE, et al. A monomeric red fluorescent protein. Proc Natl Acad Sci USA. 2002;99:7877–7882. doi: 10.1073/pnas.082243699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mehlitz D, Zillmann U, Scott CM, Godfrey DG. Epidemiological studies on the animal reservoir of Gambiense sleeping sickness, part III: Characterization of trypanozoon stocks by isoenzymes and sensitivity to human serum. Tropenmed Parasitol. 1982;33:113–118. [PubMed] [Google Scholar]
- 43.Cunningham I. New culture medium for maintenance of tsetse tissues and growth of trypanosomatids. J Protozool. 1977;24:325–329. doi: 10.1111/j.1550-7408.1977.tb00987.x. [DOI] [PubMed] [Google Scholar]
- 44.Kelly S, et al. Functional genomics in Trypanosoma brucei: A collection of vectors for the expression of tagged proteins from endogenous and ectopic gene loci. Mol Biochem Parasitol. 2007;154:103–109. doi: 10.1016/j.molbiopara.2007.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Clayton C, Shapira M. Post-transcriptional regulation of gene expression in trypanosomes and leishmanias. Mol Biochem Parasitol. 2007;156:93–101. doi: 10.1016/j.molbiopara.2007.07.007. [DOI] [PubMed] [Google Scholar]
- 46.MacLeod ET, Maudlin I, Darby AC, Welburn SC. Antioxidants promote establishment of trypanosome infections in tsetse. Parasitology. 2007;134:827–831. doi: 10.1017/S0031182007002247. [DOI] [PubMed] [Google Scholar]
- 47.Kabayo JP. The nature of the nutritional importance of serum albumin to Glossina morsitans. J Insect Physiol. 1982;28:917–923. [Google Scholar]
- 48.Galun R, Margalit J. Adenine nucleotides as feeding stimulants of the tsetse fly Glossina austeni Newst. Nature. 1969;222:583–584. doi: 10.1038/222583a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.