Abstract
The grape is one of the earliest domesticated fruit crops and, since antiquity, it has been widely cultivated and prized for its fruit and wine. Here, we characterize genome-wide patterns of genetic variation in over 1,000 samples of the domesticated grape, Vitis vinifera subsp. vinifera, and its wild relative, V. vinifera subsp. sylvestris from the US Department of Agriculture grape germplasm collection. We find support for a Near East origin of vinifera and present evidence of introgression from local sylvestris as the grape moved into Europe. High levels of genetic diversity and rapid linkage disequilibrium (LD) decay have been maintained in vinifera, which is consistent with a weak domestication bottleneck followed by thousands of years of widespread vegetative propagation. The considerable genetic diversity within vinifera, however, is contained within a complex network of close pedigree relationships that has been generated by crosses among elite cultivars. We show that first-degree relationships are rare between wine and table grapes and among grapes from geographically distant regions. Our results suggest that although substantial genetic diversity has been maintained in the grape subsequent to domestication, there has been a limited exploration of this diversity. We propose that the adoption of vegetative propagation was a double-edged sword: Although it provided a benefit by ensuring true breeding cultivars, it also discouraged the generation of unique cultivars through crosses. The grape currently faces severe pathogen pressures, and the long-term sustainability of the grape and wine industries will rely on the exploitation of the grape's tremendous natural genetic diversity.
Keywords: genomics, SNP array, positive selection, genome-wide association
The grape is the most valuable horticultural crop in the world. The fruit from the world's ∼8 million ha of vineyard is mostly processed into wine, but some is destined for fresh consumption as table grapes, dried into raisins, processed into nonalcoholic juice, and distilled into spirits (http://faostat.fao.org/). The archaeological record suggests that cultivation of the domesticated grape, Vitis vinifera subsp. vinifera, began 6,000–8,000 y ago in the Near East from its wild progenitor, Vitis vinifera subsp. sylvestris (1). The thousands of grape cultivars in use today have been generated since then by vegetative propagation and by crosses.
Wine and table grapes currently receive intense chemical applications to combat severe pathogen pressures. This susceptibility to disease, however, is not attributable to a lack of genetic diversity. Vinifera harbors levels of genetic variation an order of magnitude greater than humans and is comparable in diversity to maize (2, 3), with polymorphism that dates back tens of millions of years (4). Thus, an environmentally sustainable grape-growing industry will rely on accessing and using the grape's tremendous genetic diversity to develop improved disease-resistant grape cultivars through marker-assisted breeding (5). Traditionally, grape breeding programs have sought genotype-phenotype associations using linkage mapping. Because of the grape's long generation time (generally 3 y), however, establishing and maintaining linkage-mapping populations is time-consuming and expensive. Thus, genome-wide association (GWA) (6) and genomic selection (GS) (7) are attractive alternatives to traditional linkage mapping in the grape and other long-lived perennial fruit crops.
Well-powered GWA and GS require a genome-wide assessment of genetic diversity, patterns of population structure, and the decay of linkage disequilibrium (LD). To this end, we recently discovered over 70,000 high-quality SNPs in the grape using next-generation DNA sequencing (4). From this SNP set, we developed and validated a 9,000-SNP genotyping array (the Vitis9kSNP array). Here, we present an analysis of genotype data from 950 vinifera and 59 sylvestris accessions using the Vitis9kSNP array as part of an effort to characterize an entire US Department of Agriculture (USDA) germplasm collection on a genome-wide scale. We provide a refined model of the domestication and breeding history of vinifera by evaluating levels of haplotype diversity, the decay of LD, and patterns of population structure in vinifera and its progenitor, sylvestris. In addition, our analyses reveal extensive clonal relationships among cultivars and a complex pedigree structure within vinifera that are the result of widespread vegetative propagation. We suggest that the last several thousand years of grape breeding explored only a small fraction of possible genetic combinations and that future marker-assisted breeding efforts therefore have tremendous diversity at their disposal to produce desirable wine and table grapes with resistance to existing and future pathogens.
Results
Pedigree Analysis Within vinifera.
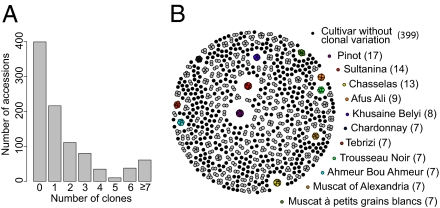
We used the Vitis9KSNP array (4) to generate 5,387 SNP genotypes from 950 vinifera accessions (451 table grape accessions, 469 wine grape accessions, and 30 accessions of unknown type) from the grape germplasm collection of the USDA, one of the most comprehensive repositories of grape diversity in the world. Currently, there are over 10,000 grape cultivar names in use worldwide (8), and their classification is often confusing because of homonyms, synonyms, scarce or incorrect historical information, and curation error. Some cultivars have been differentiated into several through the vegetative propagation of somatic mutants (9), and we expect clones derived from the same cultivar to be genetically identical at the tested marker loci. We find that 551 (58%) of the 950 vinifera accessions are clones of at least 1 other accession in the USDA grape germplasm collection. Many clonal relationships are restricted to pairs of accessions, but groups of up to 17 accessions were found to be clonally related (Fig. 1). We determined that there are 583 unique vinifera cultivars in the USDA grape germplasm collection: 399 accessions with no clonal relationships and an additional 184 accessions composed of one accession from each set of clones.
Fig. 1.
Clonal relationships within the USDA grape germplasm collection. (A) Number of clonal relationships was evaluated for each of the 950 vinifera accessions. Most of the accessions [551 (58%) of 950 accessions] have a clonal relationship with at least 1 other accession. (B) Degree of clonal relatedness among all 950 vinifera accessions is represented as a set of clusters. The 399 accessions that do not have a clonal relationship with another accession are shown as lone black dots. Accessions with six or fewer clonal relationships are grouped together with their clones and shown in gray. Clusters of clones with ≥7 accessions are colored, and their names are indicated in the legend. Names listed in the legend are the prime names from the Vitis International Variety Catalogue (http://www.vivc.de/).
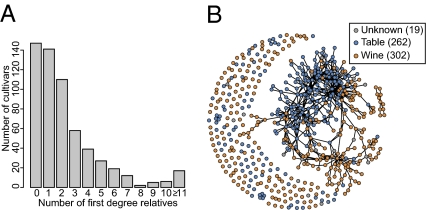
The grape is a vegetatively propagated outcrossing perennial species, which means that old cultivars propagated for hundreds or even thousands of years may coexist with cultivars generated from recent crosses. The potential for selfing and for crosses across interleaved generations, including crosses between related cultivars, makes accurate genealogical reconstruction from genomic data intractable. Nevertheless, we used patterns of identity-by-descent (IBD) and predictions of population genetic theory to infer simple pedigree relationships among the 583 unique vinifera cultivars that remained after excluding clonal relationships (Materials and Methods). We found that 74.8% of the cultivars are related to at least one other cultivar by a first-degree relationship (Fig. 2A). The resulting complex pedigree structure of our sample can be visualized as a set of networks (Fig. 2B).
Fig. 2.
First-degree relationships within the USDA grape germplasm collection. (A) Number of first-degree relationships was evaluated for each of the 583 unique vinifera cultivars. A total of 74.8% of the unique cultivars are related to at least 1 other cultivar by a first-degree relationship, and some cultivars have many first-degree relationships (i.e., >10; SI Appendix, Table S4). (B) Pedigree structure of vinifera is represented as a set of networks. Edges in the network represent inferred first-degree relationships. The vertices, or dots, represent grape cultivars and are colored by grape type (legend). The sample size of each grape type is shown in parentheses. Lone dots represent cultivars with no first-degree relatives in the dataset. Note that one single interconnected network is clearly visible and includes 384 (58.3%) of the 583 unique cultivars that are interconnected by a series of first-degree relationships.
The pedigree structure of vinifera is characterized primarily by first-degree relationships between grapes of the same type: 89.3% of edges in the network connect table grapes to table grapes or wine grapes to wine grapes (Fig. 2B). A similar trend was found for geography; only 6.1% of connections are between eastern and western cultivars (SI Appendix, Table S1). These two categories of connections in the network occur far more often than expected by chance (binomial test, P < 1 × 10−15).
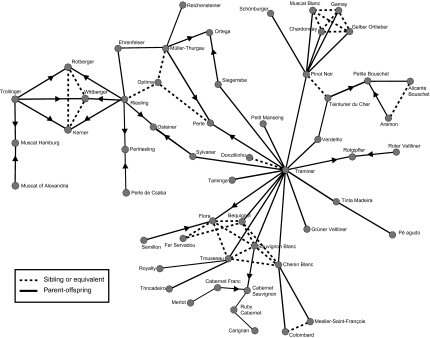
We infer that about half (47.6%) of the first-degree relationships in our sample are likely parent-offspring. The other half (52.4%) we refer to as “sibling or equivalent,” because complex crossing schemes can generate IBD values that are indistinguishable with our data from sibling relationships (SI Appendix, Fig. S1 and Table S2). By evaluating Mendelian inconsistencies, we assigned two parents to a cultivar wherever possible and thereby resolved 83 trios (SI Appendix, Fig. S1 and Table S3). A network of some well-known cultivars that includes several trios is shown in Fig. 3. Table S5 contains a list of inferred clonal and first-degree relationships for each cultivar. The assignment of clonal and pedigree relationships from the present study will be verified in the field by germplasm curators and used to improve the accuracy of the USDA grape germplasm collection.
Fig. 3.
Network of first-degree relationships among common grape cultivars. Solid vertices represent likely parent-offspring relationships. Dotted vertices represent sibling relationships or equivalent. Arrows point from parents to offspring for inferred trios (details are provided in Materials and Methods).
Haplotype Diversity and LD Decay.
To evaluate the effects of domestication on levels of genetic diversity, haplotype diversity was measured in nonoverlapping sliding windows of varying sizes across the grape genome. Although we observe a statistically significant reduction in haplotype diversity in vinifera compared with sylvestris (P < 1 × 10−5 for eight or more SNP haplotypes), the observed reduction is relatively minor (SI Appendix, Fig. S2). Moreover, the decay of LD is very rapid in vinifera and appears unchanged between the wild ancestor and the domesticated grape (SI Appendix, Fig. S2). The identification of sylvestris is notoriously difficult because of the morphological similarity between sylvestris and vinifera and the ease with which they cross (10, 11). All studies that make use of sylvestris samples should therefore be interpreted with caution. Details of the sylvestris samples used in the present study and the impact of potential misidentification are provided in the SI Appendix.
Grape Domestication History.
Although the reduction of diversity attributable to domestication and breeding appears to be weak on a genome-wide scale [i.e., much weaker than in the tomato (12) and likely even weaker than in highly diverse maize (13)], a few notable changes in morphology have emerged since grape domestication, including perfect flowers, larger berry sizes, higher sugar content, and a wide range of berry colors (14). To identify genomic regions potentially responsible for these domestication traits, we scanned the genome for signatures of selection and identified a 5-Mb candidate domestication locus on chromosome 17 (SI Appendix, Fig. S3). Although the long-range LD generated by strong selection over a few generations prevents gene-level dissection of such loci, extended LD may also be exploited to identify genotype-phenotype associations. To test this, we performed a GWA study for berry color and identified a 5-Mb region on chromosome 2 that encompasses a group of MYB transcription factor genes known to be the major determinants of grape color (15, 16) (P = 4.8 × 10−12; SI Appendix, Fig. S4). We also observe a strong signal of positive selection for white grapes around this locus, consistent with intense breeding for lighter berry color and the rapid spread of the MYB mutations responsible for reduced pigmentation (17) (SI Appendix, Fig. S4).
Relatedness among our geographically diverse sample of vinifera and sylvestris provides strong support for an origin of vinifera in the Near East: All vinifera populations are genetically closer to eastern sylvestris than to western sylvestris (Table 1; SI Appendix, Table S1). After domestication, grape growing and winemaking expanded westward, reaching Western Europe by 2.800 y ago (1). We find that haplotype diversity in western vinifera is slightly reduced compared with eastern vinifera (SI Appendix, Fig. S5), suggesting that the grape experienced a modest reduction in genetic diversity as it was brought to Western Europe.
Table 1.
Population pairwise Fst estimates
| Sylvestris west | Sylvestris east | Vinifera west | Vinifera central | Vinifera east | |
| Sylvestris west | — | ||||
| Sylvestris east | 0.154 | — | |||
| Vinifera west | 0.120 | 0.051 | — | ||
| Vinifera central | 0.168 | 0.046 | 0.020 | ||
| Vinifera east | 0.202 | 0.035 | 0.051 | 0.031 | — |
Geographic regions are defined as follows: “east” includes locations east of Istanbul, Turkey; “west” includes locations west of Slovenia, including Austria; and “central” refers to locations between them (details are presented in SI Appendix, Table 1).
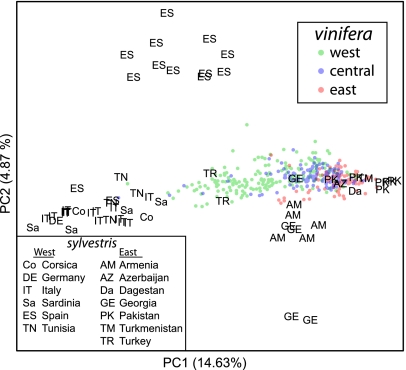
Based on morphological and genetic evidence, it has been suggested that Western European vinifera cultivars experienced introgression from local Western European sylvestris. Our finding that western vinifera are more closely related to western sylvestris than are other vinifera populations is consistent with gene flow between wild and cultivated grapes in Western Europe (Table 1). To examine this in more detail, we used principal components analysis (PCA) to visualize relationships among individual accessions. Fig. 4 shows the first two principal components (PCs) calculated from sylvestris accessions only, with vinifera cultivars subsequently projected onto the axes. Whereas PC2 differentiates a subset of geographically isolated sylvestris accessions (a subpopulation from southern Spain and two samples from Georgia), PC1 reflects a clear west-east gradient in sylvestris that is recapitulated in the vinifera that have been projected onto PC space. The observation that relationships among vinifera mirror patterns of relatedness in its wild progenitor supports a scenario in which Western European cultivars experienced introgression from local wild sylvestris grapes. Alternatively, the western sylvestris may have experienced gene flow from western vinifera. To distinguish between these two scenarios, we used a recently proposed 3-population test for admixture (18). We find strong support for a scenario in which western vinifera are a mixture of eastern vinifera and western sylvestris (f3 = −0.00481, Z score = −195.5), and we find no evidence of introgression from western vinifera into western sylvestris (f3 = 0.0268, Z score = 480.1; Materials and Methods). Thus, our data are consistent with an origin of vinifera in the Near East with subsequent introgression from wild sylvestris into vinifera in Europe.
Fig. 4.
Visualization of genetic relationships among sylvestris and vinifera. PC axis 1 (PC1) and PC2 were calculated from 59 sylvestris samples, and 570 vinifera samples were subsequently projected onto these axes. The proportion of the variance explained by each PC is shown in parentheses along each axis. The vinifera samples are represented by circles, and their origins are indicated in the legend. The countries or regions of origin of the sylvestris samples are represented by two-letter codes provided in the legend.
Discussion
The use of genetic information is increasingly being used to guide breeding efforts in many crops, including the grape (5). Because establishing and evaluating linkage-mapping populations is time-consuming and costly, GWA and GS are particularly promising methods for marker-assisted breeding programs in long-lived perennial crops (19, 20). The present study provides the initial steps toward GWA and GS in the grape by providing the most comprehensive genome-wide assessment of a fruit crop to date.
Archaeological evidence suggests that grape domestication took place in the South Caucasus between the Caspian and Black Seas and that cultivated vinifera then spread south to the western side of the Fertile Crescent, the Jordan Valley, and Egypt by 5,000 y ago (1, 21). Our analyses of relatedness between vinifera and sylvestris populations are consistent with archaeological data and support a geographical origin of grape domestication in the Near East (Fig. 4 and Table 1). Grape growing and winemaking then expanded westward toward Europe, but the degree to which local wild sylvestris from Western Europe contributed genetically to Western European vinifera cultivars remains a contentious issue (1, 22–24). Our results, based on Fst (Table 1), PCA (Fig. 4), and the 3-population test for mixture, all support a model in which modern Western European cultivars experienced introgression from local wild sylvestris. Future high-resolution genetic mapping will help to reveal if specific adaptations were involved in this introgression (e.g., climate, pathogens, flavor).
Analyses of haplotype diversity and LD suggest that grape domestication involved a weak bottleneck, because present-day wine and table grapes capture much of the haplotype diversity observed in sylvestris and the decay of LD appears unchanged between vinifera and sylvestris (SI Appendix, Fig. S2). These results are in agreement with previous studies showing no reduction in genetic diversity in vinifera compared with its wild ancestor (22, 23, 25, 26) and with the relatively minor changes in morphology observed between sylvestris and vinifera (11). A recent study found a significant increase in LD in vinifera compared with sylvestris using 36 microsatellites, however (26). This result is reconcilable with our findings because the use of microsatellites, which evolve more rapidly than SNPs, will amplify the signal of the weak domestication bottleneck that we detect (SI Appendix, Fig. S2). Many of the SNPs assayed by the Vitis9kSNP array have likely been segregating for millions of years (4), and our measure of LD decay thus captures recombination events deeper into the grape's evolutionary history than LD measured from microsatellites. Although we are unable to provide precise estimates of the severity of the grape domestication bottleneck with the current dataset, it is evident that LD, as measured by r2 between SNPs, decays rapidly in vinifera, far more quickly than in humans (27), Arabidopsis (28), rice (29), and maize (30, 31). For this reason, we conclude that well-powered GWA studies in the grape will require whole-genome sequencing, as in other high-diversity plants with rapid LD decay (19). Nevertheless, for traits that were selected during domestication and/or breeding, the resulting increase in LD means that a relatively low marker density will often suffice to identify diffuse association peaks using GWA. We demonstrate this by mapping the locus responsible for lighter berry pigmentation, a trait that experienced strong artificial selection, using GWA (SI Appendix, Fig. S4).
Our weak bottleneck model for grape domestication also fits well with the recorded widespread use of vegetative propagation during the history of vinifera: Many cultivars in use today may be only a small number of generations removed from the wild progenitor (21, 24). Vegetative propagation immortalizes a cultivar by allowing numerous genetically identical copies to be produced, but it also enables clones with unique traits to be generated by propagating tissue from the mother plant, which carries somatic mutations leading to unique phenotypes (9). For example, Pinot has been extensively propagated into clones with diverse phenotypes, such as lighter berry color (e.g., Pinot Blanc, Pinot Gris) and pigmented pulp (Pinot Teinturier). We find that Pinot has the most clonal diversity in the USDA grape germplasm collection, with 17 accessions that are clonally related (Fig. 1B). Although we observe misclassification in our dataset (SI Appendix, SI Text), we mostly verify well-known clonal relationships among cultivars like Hanepoot and Muscat of Alexandria; Sultanina, Kishmish, and Thompson Seedless; and Sauvignon Blanc and Sauvignon Gris. Thus, our data suggest that a considerable proportion of the morphological diversity maintained by the USDA grape germplasm collection is the result of spontaneous somatic mutations captured through vegetative propagation rather than that of segregating polymorphism. Identifying the causal genetic variants underlying phenotypic variation among clones will be a challenging task for which deep resequencing will be required. As is the case for grape berry color (16, 32), the locus underlying a particular phenotypic difference between clones may be the same locus involved in segregating variation for that phenotype across cultivars. Thus, deep resequencing of the clones we have identified here can be used to complement future genetic mapping efforts in the grape.
Although the adoption of vegetative propagation contributed to the maintenance of high levels of genetic diversity in vinifera and enabled the generation of clones with unique traits, we suggest that it also reduced grape growers’ motivation to undertake extensive crossing and to breed new cultivars. In support of this scenario, we find that 75% of the vinifera cultivars in the USDA grape germplasm collection are related to at least one other cultivar by a first-degree relationship (Fig. 2). In fact, 384 (58.3%) of the 583 cultivars form a single-pedigree network: Over half of the cultivars in the USDA grape germplasm collection are interconnected by a series of first-degree relationships (Fig. 2B). With more extensive sampling of vinifera, lone vertices and disjoint networks from Fig. 2B are likely to become increasingly interconnected. Moreover, the pairwise IBD distribution (SI Appendix, Fig. S1) suggests that there are a vast number of higher order pedigree relationships (e.g., second and third degree) within the present set of vinifera cultivars. Thus, the genetic structure of vinifera can be largely understood as one large complex pedigree. We propose that this pedigree structure is the result of a limited number of crosses made among elite cultivars that were immortalized and sometimes vegetatively propagated for centuries.
Most cultivars have only one or two first-degree relatives in our sample, but a small number of cultivars are highly connected in the network, and thus likely represent ancient cultivars widely used during grape breeding (SI Appendix, Table S4). This is consistent with the previous finding that Pinot and Gouais Blanc are the parents of 16 common French cultivars (33). Most of the highly connected cultivars are table grapes, including Muscat of Alexandria and the world's preeminent raisin grape, Sultanina (Thompson's Seedless). Table grapes have more first-degree relationships than wine grapes (Mann–Whitney U test, P = 3.72 × 10−8), and the high degree of connectivity among table grapes suggests that there was more intense breeding in table grapes than in wine grapes. This result follows from the ease with which table grapes are evaluated compared with wine grapes. The most highly connected wine grape in the present sample is Traminer, which has 20 first-degree relatives and is believed to be an ancient cultivar widely used during the history of grape breeding (34). Fig. 3 provides a more detailed view of relationships among some well-known cultivars and includes several inferred trios. Particularly noteworthy is our discovery that Chenin Blanc and Sauvignon Blanc are likely siblings and both share a parent-offspring relationship with Traminer. Also, we find that two of the most common cultivars of the Rhône Valley in France, Viognier and Syrah, are likely siblings. Not only do we find that elite cultivars were often reused in different crosses but that most first-degree relationships are between grapes of the same type (i.e., wine, table) and between grapes from the same geographic region (i.e., east, west). Together, these observations suggest that grape breeding has been restricted to a relatively small number of cultivars and that only a small number of the possible genetic combinations within vinifera have been explored.
We propose that the adoption and widespread use of vegetative propagation has been a double-edged sword during grape breeding. Although the production of fine wine would be impossible without the control over genetic variability that vegetative propagation offers, vegetative propagation has also discouraged the breeding of new cultivars and is at least partially responsible for a worldwide grape industry dominated by cultivars sharing extensive coancestry. Other factors that have contributed to the small number of cultivars in use today include the devastation of European vineyards in the second half of the 19th century by mildews and phylloxera and the development of the global wine industry. Currently, grapes face intense pathogen pressures, and are thus intensely chemically treated. There are numerous examples of sources of resistance to these pathogens, both from wild Vitis species and from vinifera cultivars that are often found in marginal areas of cultivation and remain largely unexploited (5, 35). The grape is clearly exceptional in terms of its domestication and breeding history compared with most crops studied to date. The vinifera grape has retained high levels of genetic diversity since its domestication ∼7,000 y ago, yet its genetic variation remains relatively unshuffled within an extended pedigree. Developing an environmentally sustainable wine and grape industry will rely on tapping into this tremendous diversity by genetically characterizing the world's germplasm collections and using marker-assisted breeding approaches to generate improved cultivars.
Materials and Methods
Sample Collection and Genotype Calling.
Leaf tissue was collected from the USDA grape germplasm collections in Davis, California and Geneva, New York, and DNA was extracted using standard protocols. Some sylvestris DNA samples were provided by F.G. Genotype data were generated from the custom Illumina Vitis9KSNP array, which assays 8,898 SNPs (4). After quality filters, 5,387 SNPs genotyped in 950 vinifera and 59 sylvestris samples remained for analysis (SI Appendix, SI Materials and Methods).
Pedigree Construction.
We calculated IBD for all pairwise comparisons among the 950 vinifera accessions using PLINK (36). We considered pairs of accessions to be genetically identical (i.e., clones, sports) if they had an IBD >95%. For inferences of first-degree relationships based on IBD, we reduced the samples to a set of “unique cultivars,” which included 399 accessions without clonal relationships and an additional 184 accessions composed of one randomly chosen accession from each set of clones. We refer to this set of samples as 583 unique vinifera cultivars. Our power to estimate IBD reliably is reduced by our relatively small number of SNPs and the ascertainment bias introduced during our SNP discovery procedure. We therefore used known pedigree relationships to calibrate our IBD values. From 43 confirmed parent-offspring relationships (SI Appendix, Table S2), the lowest pairwise IBD value was 0.466; therefore, we considered all pairwise relationships in our data to be likely first-degree relatives if they had an IBD value ≥0.466 (SI Appendix, Fig. S1). To differentiate between parent-offspring and other pedigree relationships, we used the Z0 values and Z1 values observed from our confirmed parent-offspring pairs as thresholds for defining other parent-offspring pairs in the data (SI Appendix, Fig. S1 and Table S2). For each cultivar that was related to at least 2 other cultivars with IBD, Z0 and Z1 scores consistent with a parent-offspring relationship, we calculated the proportion of SNPs consistent with Mendelian inheritance for each possible pair of parents. We defined trios in the data by assigning two parents to a cultivar when <1.35% of SNPs were inconsistent with Mendelian inheritance (SI Appendix, Fig. S1 and Table S3). Adjacency matrices and network images were generated using the “network” package in R (37).
Population Structure Analyses.
Samples with >20% missing data were removed, and we ensured that no two samples had an Identity-By-State >0.95, which resulted in 570 vinifera cultivars and 59 sylvestris accessions included in the analysis. Average population-pairwise Fst estimates were calculated from all 5,387 SNPs weighted by allele frequency [equation 10 in (38)]. For PCA, SNPs with >20% missing data and minor allele frequency (MAF) <0.05 were excluded. We then pruned the SNPs for LD using PLINK (36) by considering a window of 10 SNPs, removing 1 of a pair of SNPs if LD >0.5, and then shifting the window by 3 SNPs and repeating the procedure. After these filters, 2,958 SNPs remained. PCA was performed using SMARTPCA (39).
3-Population Test for Mixture.
We used the 3-population test for mixture (18) to compare a focal population X with two putatively parental populations Y and W to determine whether X, Y, and W are related in a simple tree or whether X is a mixture of Y and W. The f3 statistic, f3(X;Y,W), is defined as the normalized product of the frequency difference between populations X and Y and the frequency difference between populations X and W averaged over the full 5,387 SNPs (18). If there is no mixture, such that groups X, Y and W are related by a simple unrooted tree, the expected value of the f3 statistic is positive. If X is the result of mixture between Y and W, the expected value of the f3 statistic is negative. We calculated the f3 statistics for the following scenarios: f3 (vinifera west; vinifera east, sylvestris west) and f3 (sylvestris west; vinifera west, sylvestris east). We generated SEs of the f3 statistics by a Block Jackknife procedure: The data were divided into 289 nonoverlapping blocks of 20 contiguous SNPs, each block was dropped in turn, and the f3 statistic was calculated to generate a SE estimate. The SE was used to generate Z scores, which provides a measure of confidence in the f3 statistics. Large Z scores should be viewed as statistically significant but not simply convertible to P values (18).
Haplotype Analyses.
To infer haplotypes, we used fastPHASE (40) and included SNPs with a MAF >0.05 and <10% missing data and excluded individuals with >20% missing data. We investigated imputation error rate across different numbers of clusters (K) using 343 SNPs from chromosome 8 in 570 vinifera cultivars and 59 sylvestris accessions with the following fastPHASE options: -T10 -C20 -KL1 -KU20 -H-4 -Ki1 -Ks5 -Kp.05. We chose K = 10 as values above this threshold provided little improvement in performance. This gives an imputation error rate of 0.0638, which is comparable to values reported when phasing SNP data in humans (40). We phased each of the 19 chromosomes using fastPHASE with the K = 10 option and the default settings for the remaining options. The analysis included 3,397 SNPs from the 19 assembled chromosomes. Haplotype diversity was calculated as (n/(n − 1))(1 − ∑xi2), where xi is the haplotype frequency of each haplotype and n is the sample size (41). Chromosomes with larger numbers of SNPs had lower imputation error rates, but results remain unchanged when haplotype diversity was calculated using all chromosomes vs. only the 6 chromosomes with imputation error rates <0.08. Haplotype diversity was evaluated across a range of window sizes (SI Appendix, Fig. S2 and Fig. S5).
LD.
Samples with >20% missing data were removed, and we ensured that no two samples had an IBS >0.95, which resulted in 570 vinifera cultivars and 59 sylvestris accessions included in the analysis. We only considered SNPs from the 19 assembled chromosomes with MAF >0.05 and <20% missing data, which resulted in 3,558 SNPs in vinifera and 3,349 SNPs in sylvestris. LD, as measured by r2, was calculated using PLINK (36).
Grape Color Association.
Grape color was recorded on September 8–15, 2008, using the USDA grape germplasm in Davis, California. The following scale was used: 1 = gray/white, 2 = yellow, 3 = green, 4 = rose, 5 = red, 6 = red/black, and 7 = blue/black. We performed a GWA study for grape color in 289 vinifera accessions for which genotype and phenotype data were available. After excluding SNPs with MAF <0.05 and >10% missing data, 5,110 SNPs remained for analysis. GWA was performed using the mixed model (42) implemented in EMMA (43), with the IBS matrix from PLINK (36) as a random effect. Signatures of selection were evaluated by dividing cultivars into white (scores = 1, 2, and 3; n = 139) and red (scores = 5, 6, and 7; n = 112).
Supplementary Material
Acknowledgments
We thank Heidi Schwaninger, Dianne Velasco, Amy Szewc-McFadden, and Chuck Simon for DNA preparation and distribution; Bill Srmack, Dave Beckhorn, Greg Noden, Robert Martens, and Dawn Dellefave for germplasm maintenance and distribution; Gan-Yuan Zhong, Lance Cadle-Davidson, Jean-Luc Jannink, Peter Bradbury, and Martha Hamblin for discussion; Mehmet Somel, Elliot Heffner, and Aaron Lorenz for comments on drafts of the manuscript; and Kristy Goodwin for assistance with figures. C.D.B. and A.R.B. were partially supported by National Science Foundation Grant 0701382 and A.R.B. by National Science Foundation Grant 0948510.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
See Commentary on page 3457.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1009363108/-/DCSupplemental.
References
- 1.McGovern PE. Ancient Wine: The Search for the Origins of Viniculture. Princeton: Princeton Univ Press; 2003. [Google Scholar]
- 2.Lijavetzky D, Cabezas JA, Ibáñez A, Rodríguez V, Martínez-Zapater JM. High throughput SNP discovery and genotyping in grapevine (Vitis vinifera L.) by combining a re-sequencing approach and SNPlex technology. BMC Genomics. 2007;8:424–435. doi: 10.1186/1471-2164-8-424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buckler ES, Gaut BS, McMullen MD. Molecular and functional diversity of maize. Curr Opin Plant Biol. 2006;9:172–176. doi: 10.1016/j.pbi.2006.01.013. [DOI] [PubMed] [Google Scholar]
- 4.Myles S, et al. Rapid genomic characterization of the genus vitis. PLoS ONE. 2010;5:e8219. doi: 10.1371/journal.pone.0008219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.di Gaspero G, Cattonaro F. Application of genomics to grapevine improvement. Aust J Grape Wine Res. 2010;16:122–130. [Google Scholar]
- 6.McCarthy MI, et al. Genome-wide association studies for complex traits: Consensus, uncertainty and challenges. Nat Rev Genet. 2008;9:356–369. doi: 10.1038/nrg2344. [DOI] [PubMed] [Google Scholar]
- 7.Heffner EL, Sorrells ME, Jannink J-L. Genomic selection for crop improvement. Crop Sci. 2009;49:1–12. [Google Scholar]
- 8.Alleweldt G. Genetics of grapevine breeding. Prog Bot. 1997;58:441–454. [Google Scholar]
- 9.Pelsy F. Molecular and cellular mechanisms of diversity within grapevine varieties. Heredity. 2010;104:331–340. doi: 10.1038/hdy.2009.161. [DOI] [PubMed] [Google Scholar]
- 10.Di Vecchi-Staraz M, et al. Low level of pollen-mediated gene flow from cultivated to wild grapevine: Consequences for the evolution of the endangered subspecies Vitis vinifera L. subsp. silvestris. J Hered. 2009;100:66–75. doi: 10.1093/jhered/esn084. [DOI] [PubMed] [Google Scholar]
- 11.This P, Lacombe T, Thomas MR. Historical origins and genetic diversity of wine grapes. Trends Genet. 2006;22:511–519. doi: 10.1016/j.tig.2006.07.008. [DOI] [PubMed] [Google Scholar]
- 12.Miller JC, Tanksley SD. RFLP analysis of phylogenetic relationships and genetic variation in the genus Lycopersicon. Theor Appl Genet. 1990;80:437–448. doi: 10.1007/BF00226743. [DOI] [PubMed] [Google Scholar]
- 13.Eyre-Walker A, Gaut RL, Hilton H, Feldman DL, Gaut BS. Investigation of the bottleneck leading to the domestication of maize. Proc Natl Acad Sci USA. 1998;95:4441–4446. doi: 10.1073/pnas.95.8.4441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Olmo HP. The origin and domestication of the Vinifera grape. In: McGovern PE, editor. The Origins and Ancient History of Wine. Amsterdam: Gordon and Breach; 1995. pp. 31–43. [Google Scholar]
- 15.This P, Lacombe T, Cadle-Davidson M, Owens CL. Wine grape (Vitis vinifera L.) color associates with allelic variation in the domestication gene VvmybA1. Theor Appl Genet. 2007;114:723–730. doi: 10.1007/s00122-006-0472-2. [DOI] [PubMed] [Google Scholar]
- 16.Fournier-Level A, et al. Quantitative genetic bases of anthocyanin variation in grape (Vitis vinifera L. ssp. sativa) berry: A quantitative trait locus to quantitative trait nucleotide integrated study. Genetics. 2009;183:1127–1139. doi: 10.1534/genetics.109.103929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fournier-Level A, Lacombe T, Le Cunff L, Boursiquot JM, This P. Evolution of the VvMybA gene family, the major determinant of berry colour in cultivated grapevine (Vitis vinifera L.) Heredity. 2010;104:351–362. doi: 10.1038/hdy.2009.148. [DOI] [PubMed] [Google Scholar]
- 18.Reich D, Thangaraj K, Patterson N, Price AL, Singh L. Reconstructing Indian population history. Nature. 2009;461:489–494. doi: 10.1038/nature08365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Myles S, et al. Association mapping: Critical considerations shift from genotyping to experimental design. Plant Cell. 2009;21:2194–2202. doi: 10.1105/tpc.109.068437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nordborg M, Weigel D. Next-generation genetics in plants. Nature. 2008;456:720–723. doi: 10.1038/nature07629. [DOI] [PubMed] [Google Scholar]
- 21.Olmo H. Grapes. In: Smartt J, Simmonds N, editors. Evolution of Crop Plants. 2nd Ed. New York: Longman; 1995. pp. 485–490. [Google Scholar]
- 22.Grassi F, et al. Evidence of a secondary grapevine domestication centre detected by SSR analysis. Theor Appl Genet. 2003;107:1315–1320. doi: 10.1007/s00122-003-1321-1. [DOI] [PubMed] [Google Scholar]
- 23.Aradhya MK, et al. Genetic structure and differentiation in cultivated grape, Vitis vinifera L. Genet Res. 2003;81:179–192. doi: 10.1017/s0016672303006177. [DOI] [PubMed] [Google Scholar]
- 24.Arroyo-García R, et al. Multiple origins of cultivated grapevine (Vitis vinifera L. ssp. sativa) based on chloroplast DNA polymorphisms. Mol Ecol. 2006;15:3707–3714. doi: 10.1111/j.1365-294X.2006.03049.x. [DOI] [PubMed] [Google Scholar]
- 25.Barnaud A, Lacombe T, Doligez A. Linkage disequilibrium in cultivated grapevine, Vitis vinifera L. Theor Appl Genet. 2006;112:708–716. doi: 10.1007/s00122-005-0174-1. [DOI] [PubMed] [Google Scholar]
- 26.Barnaud A, Laucou V, This P, Lacombe T, Doligez A. Linkage disequilibrium in wild French grapevine, Vitis vinifera L. subsp. silvestris. Heredity. 2010;104:431–437. doi: 10.1038/hdy.2009.143. [DOI] [PubMed] [Google Scholar]
- 27.Pritchard JK, Przeworski M. Linkage disequilibrium in humans: Models and data. Am J Hum Genet. 2001;69:1–14. doi: 10.1086/321275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim S, et al. Recombination and linkage disequilibrium in Arabidopsis thaliana. Nat Genet. 2007;39:1151–1155. doi: 10.1038/ng2115. [DOI] [PubMed] [Google Scholar]
- 29.Mather KA, et al. The extent of linkage disequilibrium in rice (Oryza sativa L.) Genetics. 2007;177:2223–2232. doi: 10.1534/genetics.107.079616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Remington DL, et al. Structure of linkage disequilibrium and phenotypic associations in the maize genome. Proc Natl Acad Sci USA. 2001;98:11479–11484. doi: 10.1073/pnas.201394398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tenaillon MI, et al. Patterns of DNA sequence polymorphism along chromosome 1 of maize (Zea mays ssp. mays L.) Proc Natl Acad Sci USA. 2001;98:9161–9166. doi: 10.1073/pnas.151244298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kobayashi S, Goto-Yamamoto N, Hirochika H. Retrotransposon-induced mutations in grape skin color. Science. 2004;304:982. doi: 10.1126/science.1095011. [DOI] [PubMed] [Google Scholar]
- 33.Bowers J, et al. Historical Genetics: The Parentage of Chardonnay, Gamay, and Other Wine Grapes of Northeastern France. Science. 1999;285:1562–1565. doi: 10.1126/science.285.5433.1562. [DOI] [PubMed] [Google Scholar]
- 34.Regner F, Stadlhuber A, Eisenheld C, Kaserer HAHI. Considerations about the evolution of grapevine and the role of Traminer. Acta Hortic. 2000;528:179–184. [Google Scholar]
- 35.Coleman C, et al. The powdery mildew resistance gene REN1 co-segregates with an NBS-LRR gene cluster in two Central Asian grapevines. BMC Genet. 2009;10:89–109. doi: 10.1186/1471-2156-10-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Purcell S, et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Butts C. Network: A package for managing telational fata in R. J Stat Softw. 2008;24:1–36. [Google Scholar]
- 38.Weir BS, Cockerham CC. Estimating F-statistics for the analysis of population structure. Evolution. 1984;38:1358–1370. doi: 10.1111/j.1558-5646.1984.tb05657.x. [DOI] [PubMed] [Google Scholar]
- 39.Patterson N, Price AL, Reich D. Population structure and eigenanalysis. PLoS Genet. 2006;2:e190. doi: 10.1371/journal.pgen.0020190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Scheet P, Stephens M. A fast and flexible statistical model for large-scale population genotype data: Applications to inferring missing genotypes and haplotypic phase. Am J Hum Genet. 2006;78:629–644. doi: 10.1086/502802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nei M. Molecular Evolutionary Genetics. New York: Columbia Univ Press; 1987. [Google Scholar]
- 42.Yu J, et al. A unified mixed-model method for association mapping that accounts for multiple levels of relatedness. Nat Genet. 2006;38:203–208. doi: 10.1038/ng1702. [DOI] [PubMed] [Google Scholar]
- 43.Kang HM, et al. Efficient control of population structure in model organism association mapping. Genetics. 2008;178:1709–1723. doi: 10.1534/genetics.107.080101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.