Abstract
A significant portion of the naive T-cell repertoire is capable of responding to allogeneic MHC, violating the paradigm of self-MHC restriction. Recent studies have demonstrated convincing evidence for germ-line affinity of T-cell receptors (TCR) for MHC, providing explanation for recognition of MHC not encountered during thymic development. However, although germ-line affinity proposes all TCR have inherent affinity for MHC, most T cells are not alloreactive to a given MHC. We propose that specific recognition of endogenous presented peptides, rather than inability to interact with allogeneic MHC molecules, is the primary determinant of alloreactivity. Here, we demonstrate that alloreactive and nonalloreactive TCR differ specifically in the CDR3 sequences responsible primarily for the peptide specificity of T-cell recognition. Limitations on alloreactivity imposed by a requirement for recognition of presented peptides are directly demonstrated by expansion of the alloreactive T-cell repertoire through the addition of peptide mimotopes enabling response to two distinct allogeneic MHC by otherwise nonalloreactive T cells. Responses to peptide mimotopes were specific and depended on TCR interaction with MHC. These results demonstrate that recognition of presented endogenous peptides, and not the inability to interact with allogeneic MHC, is the primary limiter on alloreactivity. This observation reconciles the concept of an inherently MHC-reactive TCR repertoire with observed frequencies of T cells responding to allogeneic stimulation and underscores the fundamental nature of TCR recognition of ligands, where both MHC and presented peptides contribute critically to T-cell recognition.
Although the diverse T-cell receptor (TCR) repertoire generated by germ-line V(D)J recombination is optimized during thymic selection for recognition of foreign antigens presented by self-MHC (1), it is hypothesized that response to MHC not encountered during thymic development is driven by inherent TCR affinity for MHC (2, 3). Recent studies have provided convincing evidence of germ-line affinity (4–8). However, it is not clear that response to allogeneic stimulation is determined primarily by the ability to make sufficient interactions between germ-line TCR elements and allogeneic MHC. Assessments of polyclonal alloreactive responses have demonstrated broad use of germ-line TCR segments (9, 10), suggesting that differential MHC affinity among germ-line elements does not account for differential alloreactivity among T cells. Conversely, analyses of non-germ-line–encoded CDR3 regions have demonstrated skewing in alloreactive responses. In a MHC-centric view of alloreactivity, CDR3 could determine alloreactivity through direct interaction with the MHC (11) or by imposing limitations on critical germ-line–encoded contacts (12).
However, placing the entirety of determination for alloreactive potential on interaction with MHC does not address clear evidence for recognition of presented peptides in alloreactivity. Alterations to peptide presentation are sufficient to provoke alloreactive T-cell responses (13–15), globally inhibited presentation of endogenous peptides decreases alloreactive responses (16, 17), and several studies have clearly demonstrated specific peptide recognition by individual alloreactive T cells (reviewed in ref. 18), including all TCR-allogeneic peptide-MHC (pMHC) cocrystals to date (19). However, these studies are contrasted by reports of peptide-degenerate recognition of allogeneic MHC (20, 21), leaving the role of peptide recognition in alloreactivity unclear. We hypothesized that T-cell alloreactive potential is defined by specific recognition of presented endogenous peptides; the repertoire of ≈2 × 104 self-peptide–MHC on a given APC (22) is several orders of magnitude less than the repertoire of 107 peripheral T-cell clones (23), providing a clear limiter on a TCR repertoire predisposed to interact with allogeneic MHC.
Results
Alloreactive and Nonreactive T Cells Have Distinct CDR3 Repertoires.
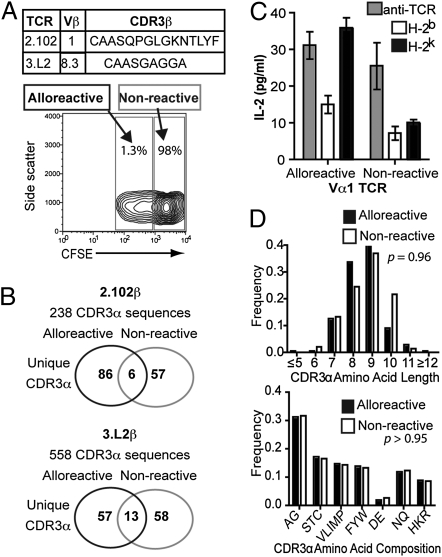
To address TCR determinants of T-cell alloreactivity at a population level, we compared TCR repertoires of CD4+ T cells responding to stimulation by allogeneic MHC with nonresponders by DNA sequence analysis. CFSE-labeled CD4+ T cells from two strains of B6.Cα+/−.fixed TCRβ (2.102β, Vβ1 and 3.L2β, Vβ8.3) mice (H-2b) were cultured in one-way mixed lymphocyte reaction (MLR) with allogeneic B6.K (H-2k) splenocytes. Alloreactive and nonalloreactive cells were FACS sorted based on CFSE dilution (Fig. 1A), and TCRα libraries were generated by using pooled primers enabling amplification of all TCRVα gene segments. Analysis of >750 TCRα DNA sequences (Table S1 and Table S2) demonstrated broad use of germ-line TCR Vα and Jα segments by alloreactive and nonalloreactive TCR, suggesting differential germ-line affinity does not define alloreactive potential. However, CDR3 sequences demonstrated minimal overlap (<10%) between alloreactive and nonalloreactive populations (Fig. 1B). The critical role for CDR3 in determining alloreactive potential was directly demonstrated in T-cell hybrids with retroviral expression of TCR identified from sequence analysis. Alloreactivity to B6.K splenocytes was seen in 0/5 TCR with CDR3α from nondividing cells compared with alloreactivity in 6/6 TCR with the same TCRVα but CDR3α from divided cells (Fig. 1B and Fig. S1).
Fig. 1.
Alloreactive and nonreactive TCR have unique CDR3 repertoires. (A) CFSE-labeled CD4+ T cells from B6.Cα+/−.fixed TCRβ mice were cultured in 4-d MLR with irradiated splenocytes from B6.K mice. Alloreactive and nonresponsive T cells were sorted by flow cytometry based on CFSE dilution. Representative FACS data from one experiment. (B) DNA sequence analysis of TCRα libraries from sorted CD4+ T cells demonstrated nonoverlapping CDR3α between alloreactive and nonreactive T cells. (C) TCR identified from DNA sequence analysis were retrovirally expressed in TCR-deficient T-cell hybrids and alloreactivity assessed by ELISA measurement of IL-2 production after 24-h culture with irradiated splenocytes. T-cell hybrids were cultured with 0.1 μg of anti-TCRβ and 0.1 μg of anti-CD28 as positive control. Data shown are mean ± SEM of one representative of three independent experiments. (D) Analysis of CDR3α of Vα2+ 2.102β TCR revealed no differences in CDR3 length (Mann–Whitney) or amino acid use grouped by biochemical properties; small (AG), nucleophilic (STC), hydrophobic (VLIMP), aromatic (FYW), acidic (DE), amide (NQ), and basic (HKR) between alloreactive and nonreactive TCR (χ2 analysis).
To define potential mechanisms of CDR3 influence on alloreactivity, we compared CDR3α amino acid composition between alloreactive and nonreactive TCR. Additional Vα2 TCR libraries were generated from the original 2.102β cDNA libraries to eliminate possible interferences from comparing CDR3 from multiple Vα with different germ-line contributions (Table S3). Comparison of CDR3 from alloreactive and nonreactive TCR revealed no differences in CDR3 length or amino acid composition (Fig. 1D and Fig. S2). These data suggest that the CDR3 does not mediate its effects on alloreactive potential through limitations on flexibility, hydrophobicity, or charge in TCR interaction with MHC.
Increasing the Presented Peptide Repertoire Expands the Alloreactive Repertoire.
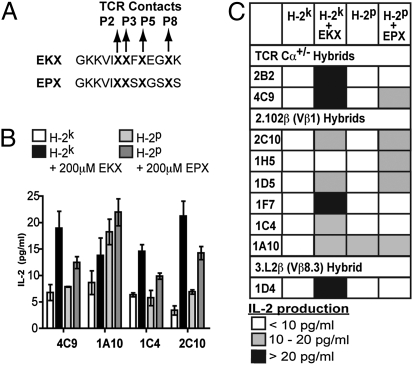
The CDR3 sequence diversity between alloreactive and nonreactive TCR supports our hypothesis that alloreactivity is determined by recognition of presented peptide antigens. We predicted that if the alloreactive T-cell repertoire is limited primarily by a requirement for specific recognition of presented endogenous peptides, addition of peptide libraries would expand alloreactivity. Peptide libraries were generated for I-Ek (EKX) and I-Ep (EPX) with defined I-Ek or I-Ep MHC anchor residues and unrestricted amino acid use (all 20 amino acids present) at the four TCR contact residues (Fig. 2A). Measurement of the effect of peptide libraries at a population level by ELISPOT analysis of IFN-γ production by naive B6 CD4+ T cells cultured with B6.K splenocytes failed to demonstrate significant expansion of the alloreactive repertoire (Fig. S3). We attributed this difficulty in analyzing polyclonal alloreactive responses to the extremely low concentration of a given peptide species (≈1 nM), competition with endogenous presented peptides, and T-cell clone diversity.
Fig. 2.
Allogeneic mimotope peptide pools expand the alloreactive T-cell repertoire. (A) Peptide mimotope pools were designed with unrestricted amino acid use at the TCR contact points of P2, P3, P5, and P8 and canonical MHC anchor residues for either I-Ek (EKX) or I-Ep (EPX). (B) EKX and EPX pools were added at a final concentration of 200 μM to nonalloreactive primary CD4+ T-cell hybrids cultured with irradiated B6.K or B6.P splenocytes. Stimulation was assessed by measurement of IL-2 production by ELISA after 24 h of culture in three independent experiments for each hybrid. Data presented are mean ± SEM from one representative experiment. (C) Addition of peptide mimotope pools enabled response to I-Ek in 8/9 otherwise nonalloreactive T-cell hybrids and response to I-Ep by 4/8 nonalloreactive T-cell hybrids. Results are means of three independent experiments for each hybrid.
To overcome these limitations, we tested the ability of peptide libraries to expand T-cell alloreactivity at the clonal level. We examined the ability of peptide libraries to enable responses to allogeneic MHC by otherwise nonalloreactive T-cell hybrids. Primary CD4+ T-cell hybrids from B6.Cα+/− and B6.Cα+/−.fixed TCRβ mice generated by immunization with I-Ab–presented listeriolysin O190–201 peptide and fusion with TCR-deficient T-cell hybrids were used for increased sensitivity compared with T-cell hybrids with TCR expression reconstituted by retrovirus. T-cell hybrids were stimulated with irradiated B6.K and B6.P (H-2p) splenocytes in the presence of peptide pools, and alloreactive responses were assessed by measurement of IL-2 production. The addition of the EKX pool enabled alloreactivity to H-2k splenocytes in 8/9 not otherwise reactive hybrids (Fig. 2 B and C). Similarly, 3/7 hybrids with no reactivity to H-2p splenocytes reacted upon addition of the EPX pool. Although these libraries are not exhaustive of potential presented peptides, the enabled alloreactivity in the majority of hybrids tested indicates most TCR are predisposed to recognize allogeneic MHC but are restricted by a requirement for recognition of presented peptides.
Sensitive and Specific Peptide Recognition Enables Response to Allogeneic MHC.
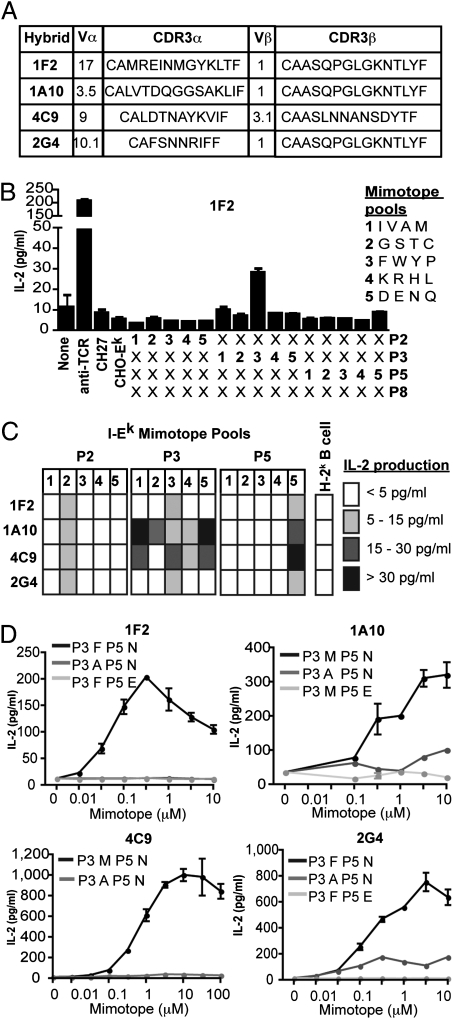
To define requirements for recognition of peptides enabling response to allogeneic MHC, we identified individual allostimulatory peptide mimotopes. Four nonalloreactive hybrids (1F2, 1A10, 4C9, and 2G4) with different TCR were tested (Fig. 3A). Single-peptide mimotopes were identified by a strategy of limited (4-aa) pool substitutions at an individual TCR contact point in combination with unrestricted amino acid use at the other TCR contacts. T-cell hybrids were cultured with Chinese hamster ovary (CHO) cells expressing I-Ek (with limited presentation of self-peptides) in the presence of the limited peptide pools (Fig. 3B). All four hybrids demonstrated reactivity against the limited peptide pools (Fig. 3C). From the patterns of reactivity, individual mimotopes were designed to evaluate the specificity of recognition. T-cell hybrids responded in a dose-dependent manner to individual peptide mimotopes, with micromolar affinities comparable with that for foreign peptides presented by self MHC (Fig. 3D). Recognition of mimotopes was eliminated by conservative amino acid substitutions at TCR contact residues (Fig. 3D), indicating the importance of peptide specificity in determining alloreactivity. Although the mimotopes shared the Hb64–76 epitope backbone and MHC anchor residues, none of the hybrids responded to the Hb64–76 epitope. None of the mimotopes corresponded to predicted murine self-peptides, supporting our contention that the lack of alloreactivity of these hybrids was due to the lack of a recognizable endogenous peptide.
Fig. 3.
Specific recognition of peptide mimotopes enables alloreactivity. (A) Four nonalloreactive T-cell hybrids with distinct TCR were examined for the specificity of recognition of mimotopes enabling alloreactivity. (B) Specificity of responses to I-Ek mimotopes was assessed by using peptide pools with defined I-Ek anchor residues and limited amino acid pools at a single TCR contact residue combined with unlimited amino acid use at other TCR contacts. Responses were assessed by ELISA measuring IL-2 production after 24-h culture of T-cell hybrids with CHO-Ek cells and 100 μM peptide pools. Results shown are mean ± SEM of one of three independent experiments. (C) T-cell hybrid recognition of limited peptide mimotope pools defined as IL-2 production in response to stimulation with CHO-Ek and 100 μM peptide pools with background IL-2 production from unstimulated T-cell hybrid cultures subtracted. Data shown are means of three independent experiments. (D) Dose–responses of T-cell hybrids to mimotopes assessed by measurement of IL-2 production from 24-h culture of T-cell hybrids with CHO-Ek. Data are mean ± SEM from one of three independent experiments.
Recognition of Different Allogeneic MHC Requires Distinct TCR Interaction with Presented Peptides.
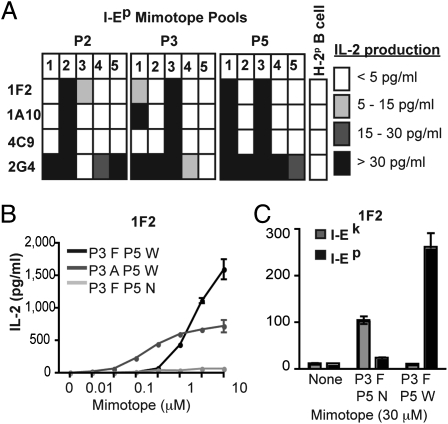
The limited peptide pool strategy was similarly used to examine specificity for peptides enabling recognition of I-Ep. All four hybrids reacted with I-Ep peptide pools, but with patterns distinct from I-Ek (Fig. 4A). Importantly, identification of a single I-Ep mimotope for the T-cell hybrid 1F2 (Fig. 4B) enabled direct comparison of mimotopes enabling reactivity to different allogeneic MHC. The presence of Phe at P3 with Asn at P5 specifically enabled alloreactivity to I-Ek, whereas Phe at P3 with Trp, but not Asn, at P5 enabled alloreactivity to I-Ep (Fig. 4C). These data indicate that T-cell alloreactivity represents a unique set of interactions for each allogeneic pMHC rather than focus on biochemical features shared between pMHC.
Fig. 4.
Recognition of different allogeneic MHC requires distinct peptide recognition. (A) Response to I-Ep peptide mimotope pools was assessed by measuring IL-2 production by ELISA after 24-h culture of T-cell hybrids with CHO-Ep cells and 100 μM peptide pools. Responses to limited mimotope pools were defined as IL-2 production in response to stimulation with CHO-Ep and 100 μM peptide pools with background IL-2 production from unstimulated T-cell hybrid cultures subtracted. Data shown are means of three independent experiments. (B) Dose–response of 1F2 T-cell hybrid to I-Ep single peptide mimotopes was measured by ELISA of 24-h culture supernatants from T-cell hybrid cells cultured with CHO-Ep plus single peptide mimotopes. Data are mean ± SEM from one of three independent experiments. (C) IL-2 production of 1F2 hybrid cells cultured 24 h with CHO-Ek or CHO-Ep in the presence of 30 μM of single peptide mimotopes. Data are mean plus SEM from one of three independent experiments.
Peptide Mimotopes Enable Interaction with MHC.
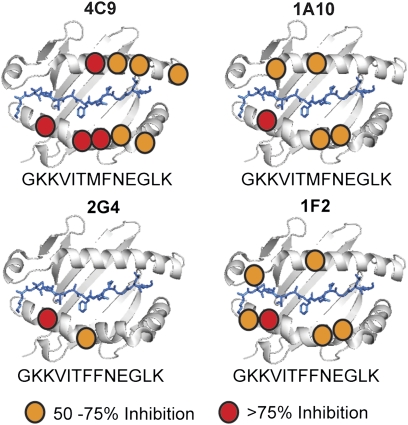
We hypothesized that alloreactivity after the addition of peptide mimotopes resulted from enabling typical interaction with allogeneic MHC rather than through an atypical peptide-focused interaction. TCR–pMHC interactions were examined by using a panel of CHO cells expressing I-Ek with disruptive mutations at the TCR interface (Fig. S4). Mimotopes enabled normal interactions between TCR and I-Ek, with critical contacts observed for both the class IIα and class IIβ chains (Fig. 5). Interestingly, although the 4C9 and 1A10 hybrids and 1F2 and 2G4 hybrids recognized the same I-Ek mimotopes, and three of the hybrids shared use of the 2.102 TCRβ, they had distinct interactions with the I-Ek molecule, demonstrating the unique molecular interactions driving TCR recognition of allogeneic pMHC.
Fig. 5.
Mimototopes enable typical interaction with allogeneic MHC. Interactions of TCR with allogeneic MHC and mimotopes were mapped by using a panel of CHO cells expressing I-Ek with individual disruptive mutations. T-cell hybrid responses were assessed by ELISA measurement of IL-2 production in 24-h culture supernatants. Inhibition of alloreactivity was defined as response to mimotopes presented by mutant I-Ek molecules divided by response to mimotopes presented by wild-type I-Ek. Data are averages of three independent experiments.
Discussion
This work addresses a long-standing debate regarding the relative importance of interaction with MHC versus presented peptides in determining T-cell alloreactivity. Simplistically, the MHC-centric view of alloreactivity suggests that T-cell responses to allogeneic pMHC ligands are determined primarily by the ability of some percentage of TCR to interact with MHC molecules not encountered during thymic development, whereas the peptide-centric view suggests that alloreactivity is primarily a response to a novel array of endogenous antigens presented by allogeneic MHC. We hypothesized that although germ-line TCR elements predispose interaction with allogeneic MHC, specific recognition of presented endogenous peptide antigens is the primary determinant for alloreactivity. Our data demonstrating expansion of T-cell alloreactivity through the addition of peptide mimotopes indicates that alloreactivity results from an inherently MHC-reactive T-cell repertoire limited by a requirement for recognition of presented peptide antigens, rather than an inability to interact with allogeneic MHC. This view of alloreactivity is consistent with the fundamental nature of TCR recognition of pMHC ligands, where the TCR interacts with both the MHC molecule and presented peptide to generate sufficient binding for T-cell activation.
Although our data clearly demonstrate both inherent TCR affinity for allogeneic MHC and a role for recognition of presented peptides in alloreactivity, they do not conclusively define parameters for recognition of peptides presented by allogeneic MHC. Elimination of mimotope-enabled responses to allogeneic MHC by conservative amino acid substitutions at TCR contact peptide residues suggests peptide specificity in alloreactivity is comparable with recognition of foreign antigens presented by self-MHC. This observation contrasts with reports of degenerate peptide recognition in alloreactivity (20, 21). It is possible that peptide-degenerate alloreactive T cells were present in our system but were not examined because they responded to allogeneic stimulation without the addition of exogenous peptides. However, it is thought that highly cross-reactive or degenerate TCR are eliminated during negative selection in the thymus (1). This point is relevant when examining evidence for peptide specificity, because the results presented here examine T cells generated with normal levels of thymic selection, whereas studies demonstrating peptide degenerate alloreactivity examined T cells generated in mice with a single pMHC with markedly limited thymic selection. We propose that peptide degenerate responses may be a function of thymic selection rather than a property of alloreactivity. However, alloreactivity represents interaction with MHC against which there has been no negative selection, a situation where it is quite conceivable that the TCR is not constrained by typical rules for interacting with pMHC ligands. The combination of limited thymic selection and peptide degenerate response to pMHC may be directly relevant to a discussion of alloreactivity, in light of our report of the disproportionate contribution of secondary TCRα rearrangements to the alloreactive T-cell repertoire (24). Secondary TCR are not subject to the same stringency of thymic selection (25, 26) and may have different requirements for peptide recognition. It is possible that the increased frequency of alloreactivity among T cells with secondary TCR may be related to decreased peptide specificity and warrants further investigation.
The high frequency of responses to the EKX and EPX pools, which are not exhaustive of the theoretical peptide repertoire, suggest multiple recognizable pMHC permutations likely exist for individual TCR. Although this recognition could be construed as degeneracy in ligand recognition by TCR, the existence of multiple distinct and specific pMHC ligands for a TCR has been clearly demonstrated by structural studies of individual TCR with multiple self and allogeneic pMHC (6) and by our previous finding of polyspecific recognition of allogeneic pMHC (27). It is also possible that the importance of specific recognition of presented peptides depends on the allogeneic MHC in question. It has been suggested that recognition of more similar allogeneic MHC molecules depends more on recognition of presented peptide antigens, whereas responses to more structurally distinct MHC are directed toward unique molecular motifs on the MHC (14). This hypothesis would imply that alloreactivity to structurally similar MHC molecules may be an instance of mimicry (28). Interestingly, a recent report has demonstrated a form of mimicry where TCR binding alters the pMHC to assume a topology similar to the cognate ligand (29). Conversely, peptide binding can result in conformational alterations to MHC molecules sufficient for inducing alloreactive T-cell responses, providing a distinct mechanism for the influence of presented peptides (30, 31). However, there is clear evidence from both structural and functional studies, and evidence presented here, that TCR can specifically recognize multiple distinct ligands by using unique sets of interactions. Further structural studies of individual TCR with multiple pMHC ligands are required to determine the breadth of TCR flexibility in recognition of allogeneic pMHC ligands.
In summary, the data presented clearly demonstrate an inherent affinity of TCR for allogeneic MHC limited by a requirement for recognition of presented peptides. Although the individual energetic contributions of TCR interaction with the MHC molecule and the presented peptide likely exist along a spectrum, ranging from MHC-focused to peptide-focused, these data illustrate the conserved mode of TCR recognition of pMHC ligands, even in alloreactivity. This report reconciles the inherent TCR affinity for MHC predicted by recent descriptions of germ-line affinity with the numerous observations of the importance of presented peptide antigens in alloreactivity to explain the observed frequencies of naive T cells that respond to allogeneic stimulation.
Materials and Methods
Mice.
B6 (H-2b), B6.K (H-2k), and B6.P (H-2p) mice were originally purchased from The Jackson Laboratory. B6.Cα+/− mice were generated as described (24). B6.Cα+/−2.102β and B6.Cα+/−3.L2β mice were derived by crossing B6.Cα−/− mice with B6 mice transgenic for the 2.102 (32) or 3.L2 (33) TCRβ chains. Mice were typed for TCRβ transgene by PCR. All mice were bred and housed in specific pathogen-free conditions at the animal facility at Washington University. All use of laboratory animals was approved by the Washington University Division of Comparative Medicine.
Generation of TCRα Sequence Libraries.
Mixed lymphocyte reactions were performed with CD4+ T cells isolated by anti-CD4 paramagnetic bead enrichment (Miltenyi Biotech) from spleens of B6.Cα+/−2.102β and B6.Cα+/−3.L2β mice. Heterozygosity for TCRAC (Cα+/−) was used to eliminate nonproductive or noninvolved secondary TCRα rearrangements. Purified CD4+ T cells were labeled with 5 μM CFSE (Invitrogen), and 106 cells cultured at 1:1 ratio with irradiated B6.K splenocytes in 2 mL RPMI 1640 (Invitrogen) supplemented with 10% FCS, 2 mM GlutaMAX, and 50 μg/mL gentamycin for 4 d. Alloreactive and nonalloreactive T cells were sorted based on CFSE dilution by using a FACSAria (BD Biosciences). Total RNA was isolated from sorted cells by using RNEasy spin columns (Invitrogen), and cDNA libraries was generated by using SuperScript II reverse transcriptase and oligo dT primers (Invitrogen). Rearranged TCRα transcripts were amplified by nested PCR using pooled primers amplifying all TCRAV gene segments (34). TCRα libraries were bulk ligated into pCR2.1 TOPO sequencing vector (Invitrogen) and sequenced by using Big Dye Terminator v3.1 and a 3730 DNA Analyzer (Applied Biosystems) in conjunction with the Genome Sequencing Center at Washington University. DNA sequences were analyzed and protein sequences of TCRVα, TCRJα, and CDR3α were predicted by using Lasergene 7 (DNASTAR) and assigned according to standard nomenclature (35, 36). CDR3 lengths were compared nonparametrically by Mann–Whitney, and amino acid compositions were compared by χ2 analysis using Prism 4 (Graph Pad Software).
Retroviral T-Cell Hybrids.
Oligonucleotides containing TCRVα-CDR3α sequences identified by DNA sequence analysis were synthesized with 5′ EcoRI and 3′ BamHI restriction sites (Genscript) and cloned into pMIG-II-GFP retroviral vector (Addgene) containing a TCRCα (with 5′ BamHI cloning site from synonomous substitution introduced by overlap PCR)-P2A-linked 2.102TCRβ (37). Retroviruses containing TCR expression constructs were generated by transfection of Platinum-E packaging cell line (gift of T. Kitamura, University of Tokyo) and used to transfect 58TCRα−TCRβ−CD4+ T-cell hybrids. Transfected T-cell hybrids were sorted by flow cytometry for TCR expression (anti-TCRβ H57-597; Biolegend) and cultured in Iscove's Modified Dulbecco's Medium (Invitrogen) supplemented with 10% FCS, 2 mM GlutaMAX, 0.5 μM 2-ME, and 50 μg/mL gentamycin. Alloreactivity was assessed by measurement of IL-2 production by ELISA (24) following 24-h culture of 105 hybrids with 106 irradiated B6 or B6.K splenocytes in triplicate. Culture of 105 hybrids with 0.1-μg–plate-bound anti-TCRβ and anti-CD28 (37.51) mAbs (Biolegend) was used as positive control. IL-2 concentrations were calculated from concurrent standard curves by nonlinear regression using Prism 4 software.
Nonalloreactive T-Cell Hybrids.
Nonalloreactive T-cell hybrids were generated from B6.Cα+/− and B6.Cα+/−.fixed TCRβ mice by immunization with irrelevant antigen [I-Ab-restricted Listeriolysin O (LLO)190–201], fusion with BW5147TCRα−β− fusion partner, and subcloned by limiting dilution. TCR were identified by DNA sequence analysis. Alloreactivity was assessed by measuring IL-2 production by ELISA following 24-h culture of 105 T-cell hybrids with 106 irradiated B6, B6.K, or B6.P splenocytes, 106 CH27 (H-2k) or B6P.C3 (H-2p) B-cell hybrid cells, or 105 Chinese hamster ovary (CHO) cells expressing I-Ek, I-Ep, or a panel of I-Ek with single disruptive mutations along the MHC surface in triplicate.
Peptide Mimotopes.
Peptides were synthesized by Fmoc chemistry on a Rainin Symphony/Multiplex multiple peptide synthesizer (Protein Technologies). Peptide pools were generated by thr addition of multiple amino acids at the P2, P3, P5, and/or P8 positions during synthesis. Aminobutyric acid was used in substitution of cysteine for peptide synthesis. Peptide pools were used as crude preparations. Individual peptides were purified by reverse phase HPLC on a C18 column (Vydac), and peptide composition and purity were confirmed by MALDI mass spectrometry (Washington University Mass Spectrometry Facility). All peptides were sterilized by 0.45-μM filtration and suspended at stated concentrations in culture medium.
Peptide-I-Ek Surface Model.
A representative model of the surface of the I-Ek molecule in complex with presented peptide antigen was generated by using PyMOL (DeLano Scientific) based on the crystal structure of I-Ek complexed with hemoglobin64–76 peptide (Protein Data Bank accession code 1FNG). MHC surface helices are represented as ribbons, with the panel of disruptive mutations represented by coloring of the residues.
Supplementary Material
Acknowledgments
We thank S. Horvath for peptide synthesis; D. Donermeyer for assistance in retroviral expression of TCR, D. Kreamalmeyer for maintenance of our mouse colony; and T. H. Hansen, C. S. Hsieh, S. C. Morley, E. R. Unanue, and K. S. Weber for comments. This research was supported by National Institutes of Health Grant AI-061173 (to P.M.A.)
Footnotes
*This Direct Submission article had a prearranged editor.
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1017015108/-/DCSupplemental.
References
- 1.Hogquist KA, Baldwin TA, Jameson SC. Central tolerance: learning self-control in the thymus. Nat Rev Immunol. 2005;5:772–782. doi: 10.1038/nri1707. [DOI] [PubMed] [Google Scholar]
- 2.Jerne NK. The somatic generation of immune recognition. Eur J Immunol. 1971;1:1–9. doi: 10.1002/eji.1830010102. [DOI] [PubMed] [Google Scholar]
- 3.Garcia KC, Adams JJ, Feng D, Ely LK. The molecular basis of TCR germline bias for MHC is surprisingly simple. Nat Immunol. 2009;10:143–147. doi: 10.1038/ni.f.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ignatowicz L, Kappler J, Marrack P. The repertoire of T cells shaped by a single MHC/peptide ligand. Cell. 1996;84:521–529. doi: 10.1016/s0092-8674(00)81028-4. [DOI] [PubMed] [Google Scholar]
- 5.Zerrahn J, Held W, Raulet DH. The MHC reactivity of the T cell repertoire prior to positive and negative selection. Cell. 1997;88:627–636. doi: 10.1016/s0092-8674(00)81905-4. [DOI] [PubMed] [Google Scholar]
- 6.Feng D, Bond CJ, Ely LK, Maynard J, Garcia KC. Structural evidence for a germline-encoded T cell receptor-major histocompatibility complex interaction ‘codon’. Nat Immunol. 2007;8:975–983. doi: 10.1038/ni1502. [DOI] [PubMed] [Google Scholar]
- 7.Dai S, et al. Crossreactive T Cells spotlight the germline rules for alphabeta T cell-receptor interactions with MHC molecules. Immunity. 2008;28:324–334. doi: 10.1016/j.immuni.2008.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Scott-Browne JP, White J, Kappler JW, Gapin L, Marrack P. Germline-encoded amino acids in the alphabeta T-cell receptor control thymic selection. Nature. 2009;458:1043–1046. doi: 10.1038/nature07812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garman RD, Ko JL, Vulpe CD, Raulet DH. T-cell receptor variable region gene usage in T-cell populations. Proc Natl Acad Sci USA. 1986;83:3987–3991. doi: 10.1073/pnas.83.11.3987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tjoa B, Kranz DM. Diversity of T cell receptor-α chain transcripts from hyperimmune alloreactive T cells. J Immunol. 1992;149:253–259. [PubMed] [Google Scholar]
- 11.Burrows SR, et al. Hard wiring of T cell receptor specificity for the major histocompatibility complex is underpinned by TCR adaptability. Proc Natl Acad Sci USA. 2010;107:10608–10613. doi: 10.1073/pnas.1004926107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rubtsova K, et al. Many different Vbeta CDR3s can reveal the inherent MHC reactivity of germline-encoded TCR V regions. Proc Natl Acad Sci USA. 2009;106:7951–7956. doi: 10.1073/pnas.0902728106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pierres M, et al. I-A α polymorphic residues that determine alloreactive T cell recognition. J Exp Med. 1989;169:1655–1668. doi: 10.1084/jem.169.5.1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Obst R, Netuschil N, Klopfer K, Stevanović S, Rammensee HG. The role of peptides in T cell alloreactivity is determined by self-major histocompatibility complex molecules. J Exp Med. 2000;191:805–812. doi: 10.1084/jem.191.5.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rudolph MG, et al. The crystal structures of K(bm1) and K(bm8) reveal that subtle changes in the peptide environment impact thermostability and alloreactivity. Immunity. 2001;14:231–242. doi: 10.1016/s1074-7613(01)00105-4. [DOI] [PubMed] [Google Scholar]
- 16.Weber DA, et al. Requirement for peptide in alloreactive CD4+ T cell recognition of class II MHC molecules. J Immunol. 1995;154:5153–5164. [PubMed] [Google Scholar]
- 17.Felix NJ, et al. H2-DMalpha(-/-) mice show the importance of major histocompatibility complex-bound peptide in cardiac allograft rejection. J Exp Med. 2000;192:31–40. doi: 10.1084/jem.192.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Felix NJ, Allen PM. Specificity of T-cell alloreactivity. Nat Rev Immunol. 2007;7:942–953. doi: 10.1038/nri2200. [DOI] [PubMed] [Google Scholar]
- 19.Rudolph MG, Stanfield RL, Wilson IA. How TCRs bind MHCs, peptides, and coreceptors. Annu Rev Immunol. 2006;24:419–466. doi: 10.1146/annurev.immunol.23.021704.115658. [DOI] [PubMed] [Google Scholar]
- 20.Huseby ES, Crawford F, White J, Kappler J, Marrack P. Negative selection imparts peptide specificity to the mature T cell repertoire. Proc Natl Acad Sci USA. 2003;100:11565–11570. doi: 10.1073/pnas.1934636100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huseby ES, et al. How the T cell repertoire becomes peptide and MHC specific. Cell. 2005;122:247–260. doi: 10.1016/j.cell.2005.05.013. [DOI] [PubMed] [Google Scholar]
- 22.Suri A, et al. In APCs, the autologous peptides selected by the diabetogenic I-Ag7 molecule are unique and determined by the amino acid changes in the P9 pocket. J Immunol. 2002;168:1235–1243. doi: 10.4049/jimmunol.168.3.1235. [DOI] [PubMed] [Google Scholar]
- 23.Casrouge A, et al. Size estimate of the α β TCR repertoire of naive mouse splenocytes. J Immunol. 2000;164:5782–5787. doi: 10.4049/jimmunol.164.11.5782. [DOI] [PubMed] [Google Scholar]
- 24.Morris GP, Allen PM. Cutting edge: Highly alloreactive dual TCR T cells play a dominant role in graft-versus-host disease. J Immunol. 2009;182:6639–6643. doi: 10.4049/jimmunol.0900638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.He X, et al. Dual receptor T cells extend the immune repertoire for foreign antigens. Nat Immunol. 2002;3:127–134. doi: 10.1038/ni751. [DOI] [PubMed] [Google Scholar]
- 26.Zal T, Weiss S, Mellor A, Stockinger B. Expression of a second receptor rescues self-specific T cells from thymic deletion and allows activation of autoreactive effector function. Proc Natl Acad Sci USA. 1996;93:9102–9107. doi: 10.1073/pnas.93.17.9102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Felix NJ, et al. Alloreactive T cells respond specifically to multiple distinct peptide-MHC complexes. Nat Immunol. 2007;8:388–397. doi: 10.1038/ni1446. [DOI] [PubMed] [Google Scholar]
- 28.Archbold JK, et al. Natural micropolymorphism in human leukocyte antigens provides a basis for genetic control of antigen recognition. J Exp Med. 2009;206:209–219. doi: 10.1084/jem.20082136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Macdonald WA, et al. T cell allorecognition via molecular mimicry. Immunity. 2009;31:897–908. doi: 10.1016/j.immuni.2009.09.025. [DOI] [PubMed] [Google Scholar]
- 30.Bluestone JA, Kaliyaperumal A, Jameson S, Miller S, Dick R., 2nd Peptide-induced changes in class I heavy chains alter allorecognition. J Immunol. 1993;151:3943–3953. [PubMed] [Google Scholar]
- 31.Chattopadhyay S, Theobald M, Biggs J, Sherman LA. Conformational differences in major histocompatibility complex-peptide complexes can result in alloreactivity. J Exp Med. 1994;179:213–219. doi: 10.1084/jem.179.1.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hsu BL, Evavold BD, Allen PM. Modulation of T cell development by an endogenous altered peptide ligand. J Exp Med. 1995;181:805–810. doi: 10.1084/jem.181.2.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kersh GJ, et al. TCR transgenic mice in which usage of transgenic α- and β-chains is highly dependent on the level of selecting ligand. J Immunol. 1998;161:585–593. [PubMed] [Google Scholar]
- 34.Baker FJ, Lee M, Chien YH, Davis MM. Restricted islet-cell reactive T cell repertoire of early pancreatic islet infiltrates in NOD mice. Proc Natl Acad Sci USA. 2002;99:9374–9379. doi: 10.1073/pnas.142284899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Arden B, Clark SP, Kabelitz D, Mak TW. Mouse T-cell receptor variable gene segment families. Immunogenetics. 1995;42:501–530. doi: 10.1007/BF00172177. [DOI] [PubMed] [Google Scholar]
- 36.Al-Lazikani B, Lesk AM, Chothia C. Canonical structures for the hypervariable regions of T cell alphabeta receptors. J Mol Biol. 2000;295:979–995. doi: 10.1006/jmbi.1999.3358. [DOI] [PubMed] [Google Scholar]
- 37.Holst J, et al. Generation of T-cell receptor retrogenic mice. Nat Protoc. 2006;1:406–417. doi: 10.1038/nprot.2006.61. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.