Abstract
Genome-wide erasure of DNA cytosine-5 methylation has been reported to occur along the paternal pronucleus in fertilized oocytes in an apparently replication-independent manner, but the mechanism of this reprogramming process has remained enigmatic. Recently, considerable amounts of 5-hydroxymethylcytosine (5hmC), most likely derived from enzymatic oxidation of 5-methylcytosine (5mC) by TET proteins, have been detected in certain mammalian tissues. 5hmC has been proposed as a potential intermediate in active DNA demethylation. Here, we show that in advanced pronuclear-stage zygotes the paternal pronucleus contains substantial amounts of 5hmC but lacks 5mC. The converse is true for the maternal pronucleus, which retains 5mC but shows little or no 5hmC signal. Importantly, 5hmC persists into mitotic one-cell, two-cell, and later cleavage-stage embryos, suggesting that 5mC oxidation is not followed immediately by genome-wide removal of 5hmC through excision repair pathways or other mechanisms. This conclusion is supported by bisulfite sequencing data, which shows only limited conversion of modified cytosines to cytosines at several gene loci. It is likely that 5mC oxidation is carried out by the Tet3 oxidase. Tet3, but not Tet1 or Tet2, was expressed at high levels in oocytes and zygotes, with rapidly declining levels at the two-cell stage. Our results show that 5mC oxidation is part of the early life cycle of mammals.
Methylation at the 5-position of cytosines is an important component of the epigenetic code (1, 2). Cell differentiation, X chromosome inactivation, reprogramming, and malignant transformation are major events characterized by remarkable changes in the epigenome and involve remodeling of DNA methylation patterns (3–10). Despite the relatively stable and heritable features of DNA methylation in somatic cells, genome-wide DNA demethylation occurs both in developing primordial germ cells and in fertilized oocytes (zygotes) (11, 12). In zygotes, a striking asymmetric DNA demethylation of the two parental genomes seems to occur within the same oocyte cytoplasm, beginning as early as 6 h after fertilization, when the paternal genome undergoes active DNA demethylation but the maternal genome resists demethylation (13–15). This process appears to be largely independent of DNA replication. The maternal genome later on undergoes passive demethylation in the absence of maintenance methyltransferase DNMT1 during DNA replication in cleavage-stage embryos (11, 13, 16).
The replication-independent DNA demethylation of the paternal genome points to the existence of a mammalian DNA demethylase activity. However, the identity of such an activity has remained enigmatic and controversial for over a decade (17, 18). Activation-induced cytidine deaminases or related activities may work in conjunction with DNA glycosylases to remove 5-methylcytosine (5mC) from DNA. After deamination of 5mC to thymine has been catalyzed by the deaminase, the mismatched thymine will be excised from the resulting G:T base pairs (19–26). The base excision repair pathway can then be further engaged to incorporate cytosine bases, resulting in replacement of 5mC with C (20). In plants, a demethylase pathway involving direct removal of 5mC by DNA glycosylase activity has been identified (27, 28), but these proteins do not have mammalian homologs. Furthermore, it was reported that the protein GADD45A promotes demethylation of CpG-methylated DNA (29), perhaps in conjunction with excision repair activities (23, 30). However, a role of GADD45A in DNA demethylation has not been confirmed (31, 32). Specifically addressing active demethylation of the paternal genome in zygotes, Okada et al. have used a siRNA knockdown strategy in oocytes followed by intracytoplasmic sperm injection to screen for candidate DNA demethylase genes. Okada et al. identified the elongator complex, and in particular its subunit Elp3, as a component required for zygotic DNA demethylation in the paternal pronucleus (33).
Taking all available information into account, perhaps the most considerable evidence suggests that cytidine deaminases work in conjunction with DNA glycosylases to remove 5mC in a DNA repair pathway (19–26). However, if not strand-specifically coordinated, excision repair would put the genome at risk for DNA double-strand breakage, and this is expected to be detrimental at those critical stages of development when the reprogramming events take place.
One plausible mechanism for demethylation of 5mC, without the need for a DNA repair process, is oxidation of the methyl group followed by secondary reactions that eventually lead to restoration of cytosine. Recently, Kriaucionis and Heintz and Tahiliani et al. made the important discovery that substantial amounts of 5-hydroxymethylcytosine (5hmC), initially thought to be only a rare DNA damage product (34), are present in mouse Purkinje and granule neurons and in embryonic stem cells (35, 36). An enzymatic activity involved in producing 5hmC from 5mC by oxidation was identified as TET1 (36). The two other mammalian homologs of TET1, TET2, and TET3, all containing a dioxygenase motif involved in Fe(II) and α-ketoglutarate binding and catalytic activity, were shown to posses similar activities as well (37).
The goal of our study was to investigate if 5mC oxidation occurs in fertilized oocytes and is part of the apparent DNA demethylation process that takes place during this early developmental stage.
Results
Using specific antibodies, we determined the levels of 5mC and 5hmC in male and female pronuclei in zygotes and in early cleavage-stage embryos. We used a recently available commercial polyclonal antibody directed against 5hmC. Initially, we verified the specificity of this antibody toward 5hmCs versus 5mCs or unmodified cytosines placed at identical positions within CpG sequences in synthetic single-stranded 76-mer oligonucleotide substrates (38). In immuno-dot blot assays, we observed that this antibody is specific for 5hmC and does not react with substrates containing only unmodified cytosines, nor does it react with substrates containing 5mC (Fig. S1A). We then tested the suitability of the anti-5hmC antibody for immunostaining experiments using human 293T cells. As initially determined by immuno-dot blot assays, this cell line contains detectable levels of 5hmC (Fig. S1B). A nuclear staining pattern was observed with the anti-5hmC antibody (Fig. S1C). To test for specificity of the staining reaction, we preincubated the antibody with synthetic oligonucleotides containing C, 5mC, or 5hmC. As shown in Fig. S1C, the nuclear staining was completely eliminated by preincubation of the antibody with 5hmC-containing oligonucleotides but not by competition with the other oligonucleotides attesting to the suitability of the antibody for immunocytochemistry.
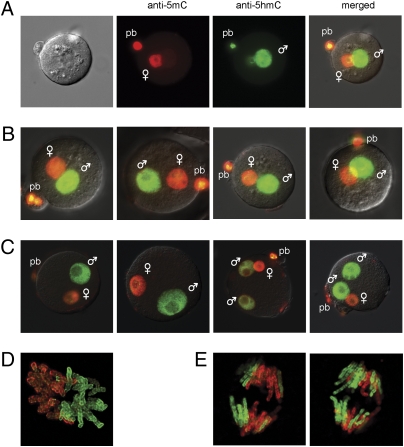
We hypothesized that 5hmC might be detectable as a potential intermediate during DNA demethylation in zygotes. Using the anti-5hmC antibody, we observed intense staining of the paternal pronucleus in mouse zygotes (Fig. 1A), whereas 5hmC staining was almost completely absent from the maternal pronucleus. To further test the specificity of the staining pattern, we carried out competition experiments with synthetic oligonucleotides and observed that the staining of zygotes for 5hmC is specific (Fig. S2). Simultaneous double-staining with an established anti-5mC antibody (16), which does not react with 5hmC (38), detected 5mC in the maternal but not in the paternal pronucleus (Fig. 1 A and B). Thus, the two staining patterns are mutually exclusive, suggesting that 5mC has been converted to 5hmC specifically in the paternal pronucleus. Fig. 1 A and B show late pronuclear stages (PN4–PN5). In vitro fertilized zygotes have similar staining patterns. Bispermic zygotes exhibit 5hmC staining in both paternal pronuclei (Fig. 1C). The maternally and paternally inherited chromosomes are localized in separate compartments at metaphase and are marked by 5mC and 5hmC staining, respectively (Fig. 1D). The condensed chromosomes at anaphase stain differentially for 5hmC or 5mC, suggesting that they are paternally or maternally derived (Fig. 1E).
Fig. 1.
5hmC is present in the male pronucleus of mouse zygotes. (A) A mouse zygote was double-stained with anti-5hmC antibody (green) and anti-5mC antibody (red). The smaller maternal pronucleus is closer to the polar body (pb). A bright-field image is shown on the far left. (B) Additional zygotes were double-stained with anti-5hmC antibody (green) and anti-5mC antibody (red). Merged images are shown. (C) Zygotes obtained by in vitro fertilization were double-stained similarly. Two polyspermic zygotes (to the right) exhibit 5hmC staining in two paternal pronuclei. (D) 5mC and 5hmC staining reveal two separate chromosome sets at metaphase of zygote division. A confocal image is shown. (E) Individual chromosomes are largely stained for either 5mC (likely originated from the maternal pronucleus) or 5hmC (likely from the paternal pronucleus) at anaphase of zygote division. Two Z sections of the same zygote are shown.
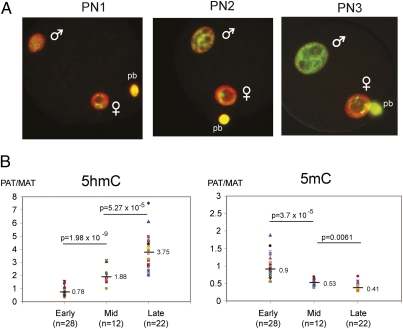
Genome-wide loss of 5mC signal by antibody staining is known to occur beginning around PN3 (15, 39), which is consistent with our observations. To determine if the timing of 5hmC appearance coincides with the loss of 5mC staining, we looked at earlier pronuclear stages, including PN1 to PN3 (Fig. 2A). The distance of the two pronuclei decreases and the size of both pronuclei increases with advancing pronuclear stages. Staining with anti-5mC antibody detected 5mC in both maternal and paternal nuclei at the earliest pronuclear stages, and this signal was decreasing in the paternal pronucleus as the zygote developed. The signal for 5hmC was visible at low levels in both pronuclei at the early stages. The level of 5hmC staining of the paternal pronucleus increased relative to the maternal pronucleus with developmental stage. At the same time, the paternal to maternal pronucleus signal decreased for 5mC staining (Fig. 2B).
Fig. 2.
5hmC and 5mC in early pronuclear stage zygotes. (A) Zygotes at pronuclear stages PN1, PN2, and PN3 were double-stained with anti-5hmC antibody (green) and anti-5mC antibody (red). Merged images are shown. (B) The levels of 5hmC and 5mC in paternal and maternal pronuclei were quantitated. The ratio of staining signal between the paternal and maternal pronucleus is plotted. The number of zgotes analyzed in PN1/PN2 (Early), in PN3 (Mid), and in PN4/PN5 (Late) are indicated with n values. The median value is indicated by a horizontal line and a number. The difference between each two datasets is statistically significant, as seen in the P values of t-tests.
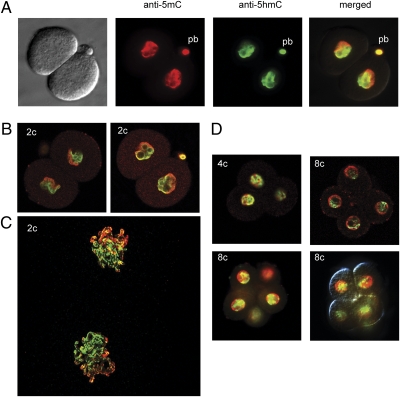
The asymmetrical staining pattern is still observed in two-cell–stage embryos (Fig. 3 A and B), in which different compartments of the nuclei are strikingly enriched for 5mC or 5hmC, respectively. These nuclear compartments are derived from paternal and maternal chromosomes, respectively, which occupy distinct territories (13, 40). Confocal microscopy images give a clear example of the asymmetric distribution of 5hmC and 5mC along the two-chromosome sets in two-cell–stage embryos entering mitosis (Fig. 3C). We further observed that the asymmetrical 5hmC signal persists toward the four- and eight-cell stages (Fig. 3D). These findings suggest that 5hmC is maintained for a considerable amount of time after it was initially formed in the paternal pronucleus at the one-cell stage by 5mC oxidation.
Fig. 3.
5hmC and 5mC in early cleavage-stage embryos. (A) Two-cell stage embryos were double-stained with anti-5hmC antibody (green) and anti-5mC antibody (red). pb, polar body. A bright-field image is shown on the far left. (B) Two-cell–stage embryos double-stained with anti-5hmC antibody (green) and anti-5mC antibody (red). These images were obtained by confocal microscopy. (C) Confocal microscopy image of a two-cell (2c) stage embryo entering mitosis. The condensed chromosomes are labeled with anti-5mC antibody (red) and anti-5hmC antibody (green). (D) 5hmC and 5mC in four- (4c) and eight-cell (8c) –stage embryos. Four-cell (Upper Left) and eight-cell (remaining images) embryos were double-stained with anti-5hmC antibody (green) and anti-5mC antibody (red). A confocal image is shown in the upper right image.
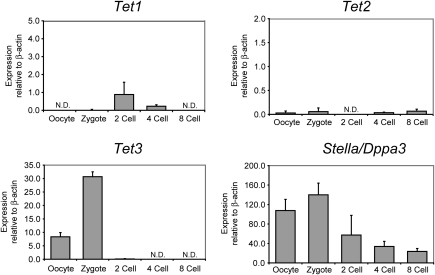
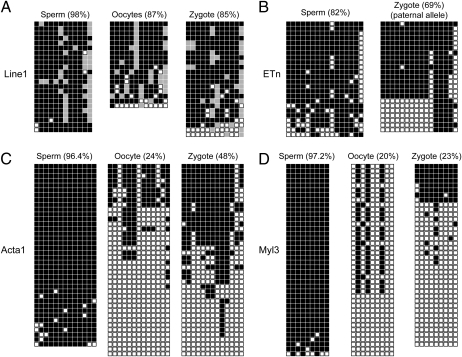
There are three mammalian proteins with known 5mC oxidase activities: Tet1, Tet2, and Tet3 (36, 37). One strategy employed previously in the search for mammalian DNA demethylases is that this activity should be expressed at high levels and specifically in oocytes and zygotes (31, 41). Therefore, we examined the expression of the three Tet genes in mouse oocytes, zygotes, two-, four-, and eight-cell–stage embryos by quantitative real-time PCR (Fig. 4) using primers, as indicated in Fig. S3. We found that Tet3 is expressed at high levels in oocytes and zygotes, but its expression is drastically down-regulated at the two-cell stage and at later cleavage stages. On the other hand, Tet1 and Tet2 were not expressed at substantial levels in oocytes and zygotes. Tet1 was expressed at moderate to low levels at the two- and four-cell stages. As a control, we measured the expression of the Stella/Dppa3 transcript encoding a protein that protects the maternal genome from active DNA demethylation (42). As expected, Stella/Dppa3 was expressed at high levels in oocytes and zygotes, and its level of expression gradually declined toward the eight-cell stage (Fig. 4). Although expression of Tet3 has been demonstrated in other tissues by RT-PCR (37), our data for oocyte- and zygote-specific expression of Tet3 are consistent with a set of published microarray data, which show almost complete absence of the Tet3 transcript in all somatic mouse tissues tested but high expression of Tet3 in oocytes and fertilized eggs, in a pattern similar to that of Stella/Dppa3 (Fig. S4). Likely, there are differentially spliced isoforms of Tet3, which give rise to the different expression patterns. To determine if 5hmC in zygotes is further converted to cytosine, we conducted sodium bisulfite sequencing analysis of DNA from mouse sperm, oocytes, and zygotes (PN4–PN5). We analyzed the methylation pattern of the Line1 (long interspersed element-1) 5′ region and of the ETn (early transposon) repetitive elements (Fig. 5 A and B). The Line1 sequences were highly methylated in sperm DNA (98%) and in oocytes (87%). This level was 85% in zygotes, indicating only a rather limited conversion of 5mC or 5hmC to C, although the difference between sperm and oocyte combined and zygotes is statistically significant (P = 0.0016; Fisher's exact test, two-tailed). Our data are showing less demethylation than reported in previous studies in which the same sequences were analyzed (33, 39). ETn sequences were methylated at a level of 82% in sperm DNA. The level of ETn methylation in the paternal genome, identified by sequence polymorphism of the mouse strains used, was 69% in zygotes (P = 0.0001; Fisher's exact test, two-tailed), again indicating limited conversion of modified cytosines to cytosines relative to sperm DNA. We then analyzed two single-copy genes, Myl3 and Acta1, coding for myosin light-chain-C and α-actin, respectively, by bisulfite sequencing (Fig. 5 C and D). Both genes were highly methylated in sperm DNA (96–97%). Myl3 was methylated at a level of 20% in oocytes and 23% in zygotes indicating significant demethylation (i.e., conversion of 5mC/5hmC to C). Acta1 was methylated at a level of 24% in oocytes and 48% in zygotes, indicating some demethylation in zygotes.
Fig. 4.
Expression of Tet and Stella/Dppa3 genes in oocytes, zygotes, and early cleavage-stage embryos. RNA was isolated from oocytes, zygotes, two-, four-, and eight-cell–stage embryos. Real-time PCR was used to assess the expression of the three Tet genes and Stella/Dppa3. Data were normalized relative to expression of β-actin. N.D., no detectable signal in real-time PCR. Expression of Tet1 in the zygote and of Tet3 at the two-cell stage has a detectable signal, which is close to zero.
Fig. 5.
Sodium bisulfite sequencing of Line1, ETn, Mylc, and Acta1 sequences in sperm, oocytes, and zygotes. DNA was isolated from mouse oocytes, sperm, or zygotes (PN4–PN5) and subjected to sodium bisulfite conversion. (A) Line1 5′ end sequences were amplified, cloned, and sequenced. Open squares, unmethylated CpGs; black squares, methylated CpGs; gray squares, not analyzable/mutated CpG site. Each row represents an individual sequenced DNA strand. (B) ETn sequences were amplified, cloned, and sequenced. The sequences from zygotes represent the paternal allele distinguishable by a sequence polymorphism. (C) Acta1 sequences. (D) Myl3 sequences. The percentage of methylated CpGs is indicated.
Discussion
Our data explain previous observations of asymmetrical staining of maternal and paternal pronuclei by anti-5mC antibodies (13, 15, 39). This antibody does not recognize 5hmC (38), which is formed in the paternal pronucleus by genome-wide 5mC oxidation, leading to lack of staining of the male pronucleus by anti-5mC antibody. Sodium bisulfite sequencing, which cannot distinguish between 5mC and 5hmC (38, 43), has been used by several laboratories to demonstrate active DNA demethylation—that is, conversion of 5mC to C—of certain sequences in zygotes. Based on this technique, active DNA demethylation has been inferred for a few genomic loci, including the repetitive Line1 and ETn elements (14, 33, 39), although ETn demethylation in the zygote occurred only to a very small extent or not at all (33, 39). Our bisulfite sequencing data confirmed very limited demethylation (that is, conversion of 5mC or 5hmC to C) for ETn and Line1 sequences, as well as for the single-copy gene Acta1. We did see substantial conversion of modified to unmodified cytosines for the Myl3 gene in zygotes. However, there remains the possibility that the apparent conversion of 5hmC to C may have occurred during DNA replication, which begins in the late PN3 stage (39). Some DNA demethylation (conversion to C) may occur in the prereplicative phase, but it is not very pronounced (39). In any event, beyond the few loci examined by us and by others to date, we lack information about the fate of 5mC and 5hmC in most sequences of the zygotic genome. Clearly, our data show that conversion of 5mC to C in the zygote cannot be a genome-wide event because considerable amounts of 5hmC are formed and this base modification would be indistinguishable from 5mC using bisulfite conversion-based techniques, consistent with our results. Furthermore, 5hmC is formed in the zygote and persists into the two-cell stage and later cleavage stages in an asymmetrical manner (Figs. 1 D and E and 3), suggesting that it is not formed de novo by 5mC oxidase activity at the two-, four-, and eight-cell stages. Such an activity should operate on all (paternal and maternal) chromosomes at these stages. Combined, our data on 5hmC levels in zygotes and in early cleavage stage embryos and data from sodium bisulfite sequencing suggest that 5hmC conversion to C may occur only to a limited extent and perhaps at specific sequences. Our data are thus arguing against the possibility that 5hmC is efficiently removed by DNA repair-mediated processes at a genome-wide level. Although initially reported in 1988 (44), the nature of a protein or enzymatic activity that would excise 5hmC from DNA has not been determined. It is of note, however, that excision repair processes do take place in paternal pronuclei in mammalian zygotes, as indicated by the occurrence of γ-H2AX–marked DNA strand breaks and base excision repair proteins at this developmental stage (39, 45), although it is not clear what DNA base or lesion is being removed. Our results also argue against the possibility that most 5hmC may be further oxidized and potentially decarboxylated to form C, this being one possible mechanism for 5hmC processing and demethylation that has been proposed (18). However, our data do not exclude the possibility that 5hmC is processed into C at certain sequences. Alternatively, multiple mechanisms may be at work to reprogram paternal genome methylation patterns that include, for example, deamination of 5mC followed by excision repair, in addition to 5mC oxidation.
The role of 5mC oxidation in the paternal pronucleus is currently unknown. One immediate effect of this oxidation step should be the neutralization of the functional role of 5mC in gene suppression. Embryonic genome activation in the mouse takes place at the two-cell stage and it is expected that many genes that are methylation-suppressed during spermatogenesis (e.g., Oct4 and Nanog) will need to be activated to allow development to proceed. After oxidation of 5mC, the 5hmC-containing sequences will no longer be capable of interacting with repressor proteins that are known to bind to 5mC (34, 38). DNA sequences containing 5hmC in place of 5mC are not substrates for the maintenance methyltransferase activity of DNMT1 (46). This finding means that the formation of 5hmC may serve to dilute DNA CpG methylation during replication in early embryos, even in the presence of any nuclear DNMT activity. Interestingly, we did not observe much signal for 5hmC in the presumably maternally derived chromosome domains of two-, four-, and eight-cell nuclei (Fig. 3). This finding is consistent with the assumption that the maternal genome undergoes passive, replication-dependent demethylation in early cleavage-stage embryos in a manner that is not dependent on 5mC oxidation but may simply the consequence of replication in absence of DNMT1 maintenance methylation activity.
The most likely candidate for 5mC oxidation in the paternal pronucleus is Tet3, which is specifically expressed in oocytes and zygotes but not in two-cell–stage embryos (Fig. 4). We attempted to knock-down Tet3 expression in oocytes by siRNA before in vitro fertilization but were unable to achieve efficient knockdown. Mouse models are under construction to prove that Tet3 is the activity that converts 5mC to 5hmC in fertilized oocytes. In conclusion, our data show that 5mC oxidation is one initial step in reprogramming of the paternal genome upon fertilization, suggesting that this event is an important part of the early mammalian life cycle.
Experimental Procedures
Derivation and Immunostaining of Oocytes, Zygotes, and Early Embryos.
Animal handling was done in accordance with institutional guidelines and was approved by the City of Hope Institutional Animal Care and Use Committee. Preimplantation embryonic stages (one to eight cells) were collected from 6- to 8-wk-old female FVB mice. Pronuclear stages were identified as described (15). Cumulus cells were removed from zygotes with 1% hyaluronidase treatment. The zona pellucida was removed by using acidic tyrode solution. After washing in M2 medium + 0.3% BSA, zygotes or embryos were fixed in 3.7% paraformaldehyde in PBS at room temperature for 20 min. Embryos were permeabilized in 0.2% Triton-X 100 in PBS at room temperature for 10 min. Permeabilized embryos were incubated in 4 N HCl solution at room temperature for 10 min followed by neutralization in Tris-Cl, pH 8.0, for 10 min. The embryos were blocked overnight at 4 °C in 1% BSA, 0.2% Triton X-100 in PBS. Embryos were incubated with anti-5hmC (rabbit polyclonal; Active Motif) and anti-5mC antibodies (mouse monoclonal; Eurogentec) in blocking solution for 1 h at room temperature. The embryos were washed several times in 0.05% Tween 20 in PBS (PBST), transferred to secondary antibody mixture of Alexa Fluor 568 goat anti-mouse and Alexa Fluor 488 goat anti-rabbit (Molecular Probes), and incubated at room temperature for 1 h. The embryos were washed several times with PBST before mounting on slides with ProLong Gold antifade reagent with DAPI (Molecular Probes). Fluorescence images were acquired using an Olympus AX70 upright microscope with Image Pro Plus version 6.3 software. Confocal images were acquired using a Zeiss LSM 510 upright microscope and processed using the Zeiss LSM image browser. Quantitative analysis of pronuclei was done using Image-pro plus version 6.3 (Media Cybernetics Inc.). All software settings for intensity and saturation were maintained constant across all experimental groups. A region of interest was drawn around each pronucleus in zygotes and the mean optical density was calculated within the region of interest. The median 5hmC intensity in the male pronucleus was divided by the median 5hmC intensity in the female pronucleus to obtain the PAT/MAT ratio for 5hmC. The PAT/MAT ratio for the control 5mC was obtained from the respective 5mC values.
Antibody Specificity Test.
Synthetic oligonucleotides containing cytosine, 5mC, or 5hmC bases were prepared as described previously (38) and were used in antibody dot-blot assays. The 76-mer oligonucleotide sequence was 5′-CCTCACCATCTCAACCAATATTATATTAXGXGTATATXGXGTATTTXGXGTTATAATATTGAGGGAGAAGTGGTGA-3′, where X is 5hmC, C, or 5mC. For competition immunocytochemistry, we preincubated the anti-5hmC antibody (1:6,000; Active Motif) with 0.5 μg/mL of single-stranded 38-mer oligonucleotides (C38R for normal cytosine; 5mC38R for 5mC; 5hmC38R for 5hmC) in 0.05% PBST at room temperature for 1 h, then incubated with the fixed cells for immunostaining. The sequence of the 38-mers was 5′-ATTATAAXGXGAAATAXGXGATATAXGXGTAATATAAT-3′ where X is either 5hmC (5hmC38R), C (C38R), or 5mC (5mC38R).
Real-Time PCR.
Poly(A) mRNA was isolated from MII oocytes (n = 8), zygotes (n = 8), two-cell (n = 4), four-cell (n = 2), and eight-cell (n = 1) embryos by using the Dynabeads mRNA DIRECT Micro Kit (Invitrogen). Oligo (dT)25-coupled Dynabeads and mRNA complexes were immediately used for reverse transcription using the SuperScript III reverse transcriptase (Invitrogen), according to the manufacturer's instructions. Real-time quantitative PCR reactions were performed at 50 °C for 2 min and 95 °C for 10 min followed by 50 cycles at 95 °C for 15 s and 60 °C for 1 min using TaqMan Gene Expression Master Mix (Applied Biosystems) on an iQ5 real-time PCR cycler (Biorad). PCR was performed with the TaqMan MGB primer with 6FAM-based probes (Applied Biosystems) using the following assay ID numbers: Tet1 (Mm01169088_m1), Tet2 (Mm01312907_m1), Tet3 (Mm00805754_m1), and Stella/Dppa3 (Mm01184198_g1). The cDNA levels of target genes were analyzed using comparative Ct methods and normalized to internal standard, β-actin.
Bisulfite Sequencing.
For bisulfite sequencing, cells were directly subjected to bisulfite conversion by using the EZ DNA Methylation Direct kit (Zymo Research). Bisulfite-modified DNAs were amplified using the following PCR primers: Line1-5′ region, for the first PCR, the forward primer 5′-GTTAGAGAATTTGATAGTTTTTGGAATAGG-3′ and reverse primer 5′-CCAAAACAAAACCTTTCTCAAACACTATAT-3′, and for the second PCR, the forward primer 5′-TAGGAAATTAGTTTGAATAGGTGAGAGGT-3′ and reverse primer 5′-TCAAACACTATATTACTTTAACAATTCCCA-3′, were used.
For ETn elements, for the first PCR, the forward primer 5′-CTTAACTACATTTCTTCTTTT-3′ and reverse primer 5′-AGTTAGYGTTAGTATGTGTATTTGTACC-3′, and for the second PCR, the forward primer 5′-TCTAAATTCCTCTCTTACAACT-3′ and reverse primer 5′-AGTTAGYGTTAGTATGTGTATTTGTACC-3′ were used. For α-actin (Acta1) promoter amplification, for the first PCR, the forward primer 5′-AAGTAGTGATTTTTGGTTTAGTATAGT- 3′ and reverse primer 5′-ACTCAATAACTTTCTTTACTAAATCTCCAAA-3′, and for the second PCR, the forward primer 5′-GGGGTAGATAGTTGGGGATATTTTT-3′ and reverse primer 5′-CCTACTACTCTAACTCTACCCTAAATA-3′ were used.
For Myl3 promoter amplification, for the first PCR, the forward primer 5′-GTATAATAAATTTGGATAGGTAAAGGTTAG- 3′ and reverse primer 5′-AAACCTAAAACACTAATCTTAAAAATTTTA′, and for the second PCR, the forward primer 5′-ATATTATAGTAGGGGTTGGAATGATTAAAG-3′ and reverse primer 5′-CCTATTAAACTAATCTAAAAAACAATCCTC-3′ were used.
The reaction buffer contained dNTPs and 1.25 U of PfuTurbo Cx Hotstart DNA Polymerase (Stratagene) and the samples were incubated at 95 °C for 3 min, and then 36 cycles of PCR at 95 °C for 20 s, 50 °C for 30 s, and 72 °C for 45 s were performed, followed by a final extension step at 72 °C for 5 min. The second-round PCR was carried out with Platinum Taq polymerase (Invitrogen), and the samples were incubated at 95 °C for 2 min, and then 36 cycles of PCR at 94 °C for 30 s, 50 °C for 30 s, and 72 °C for 1 min were performed, followed by a final extension step at 72 °C for 5 min. The PCR products were then ligated into the pCR2.1 TA cloning vector (Invitrogen). The cloned samples were sequenced using the M13 reverse sequencing primer and analyzed.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health Grants AG036041 (to G.P.P.) and ES015185 and GM064378 (to P.E.S.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. P.A.J. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1014033108/-/DCSupplemental.
References
- 1.Holliday R, Pugh JE. DNA modification mechanisms and gene activity during development. Science. 1975;187:226–232. [PubMed] [Google Scholar]
- 2.Riggs AD. X inactivation, differentiation, and DNA methylation. Cytogenet Cell Genet. 1975;14:9–25. doi: 10.1159/000130315. [DOI] [PubMed] [Google Scholar]
- 3.Hochedlinger K, Jaenisch R. Nuclear reprogramming and pluripotency. Nature. 2006;441:1061–1067. doi: 10.1038/nature04955. [DOI] [PubMed] [Google Scholar]
- 4.Huang K, Fan G. DNA methylation in cell differentiation and reprogramming: An emerging systematic view. Regen Med. 2010;5:531–544. doi: 10.2217/rme.10.35. [DOI] [PubMed] [Google Scholar]
- 5.Straussman R, et al. Developmental programming of CpG island methylation profiles in the human genome. Nat Struct Mol Biol. 2009;16:564–571. doi: 10.1038/nsmb.1594. [DOI] [PubMed] [Google Scholar]
- 6.De Carvalho DD, You JS, Jones PA. DNA methylation and cellular reprogramming. Trends Cell Biol. 2010;20:609–617. doi: 10.1016/j.tcb.2010.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128:683–692. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen T, Li E. Establishment and maintenance of DNA methylation patterns in mammals. Curr Top Microbiol Immunol. 2006;301:179–201. doi: 10.1007/3-540-31390-7_6. [DOI] [PubMed] [Google Scholar]
- 9.Suzuki MM, Bird A. DNA methylation landscapes: Provocative insights from epigenomics. Nat Rev Genet. 2008;9:465–476. doi: 10.1038/nrg2341. [DOI] [PubMed] [Google Scholar]
- 10.Gartler SM, Riggs AD. Mammalian X-chromosome inactivation. Annu Rev Genet. 1983;17:155–190. doi: 10.1146/annurev.ge.17.120183.001103. [DOI] [PubMed] [Google Scholar]
- 11.Reik W. Stability and flexibility of epigenetic gene regulation in mammalian development. Nature. 2007;447:425–432. doi: 10.1038/nature05918. [DOI] [PubMed] [Google Scholar]
- 12.Surani MA, Hayashi K, Hajkova P. Genetic and epigenetic regulators of pluripotency. Cell. 2007;128:747–762. doi: 10.1016/j.cell.2007.02.010. [DOI] [PubMed] [Google Scholar]
- 13.Mayer W, Niveleau A, Walter J, Fundele R, Haaf T. Demethylation of the zygotic paternal genome. Nature. 2000;403:501–502. doi: 10.1038/35000656. [DOI] [PubMed] [Google Scholar]
- 14.Oswald J, et al. Active demethylation of the paternal genome in the mouse zygote. Curr Biol. 2000;10:475–478. doi: 10.1016/s0960-9822(00)00448-6. [DOI] [PubMed] [Google Scholar]
- 15.Santos F, Hendrich B, Reik W, Dean W. Dynamic reprogramming of DNA methylation in the early mouse embryo. Dev Biol. 2002;241:172–182. doi: 10.1006/dbio.2001.0501. [DOI] [PubMed] [Google Scholar]
- 16.Rougier N, et al. Chromosome methylation patterns during mammalian preimplantation development. Genes Dev. 1998;12:2108–2113. doi: 10.1101/gad.12.14.2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ooi SK, Bestor TH. The colorful history of active DNA demethylation. Cell. 2008;133:1145–1148. doi: 10.1016/j.cell.2008.06.009. [DOI] [PubMed] [Google Scholar]
- 18.Wu SC, Zhang Y. Active DNA demethylation: Many roads lead to Rome. Nat Rev Mol Cell Biol. 2010;11:607–620. doi: 10.1038/nrm2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Popp C, et al. Genome-wide erasure of DNA methylation in mouse primordial germ cells is affected by AID deficiency. Nature. 2010;463:1101–1105. doi: 10.1038/nature08829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gehring M, Reik W, Henikoff S. DNA demethylation by DNA repair. Trends Genet. 2009;25:82–90. doi: 10.1016/j.tig.2008.12.001. [DOI] [PubMed] [Google Scholar]
- 21.Morgan HD, Dean W, Coker HA, Reik W, Petersen-Mahrt SK. Activation-induced cytidine deaminase deaminates 5-methylcytosine in DNA and is expressed in pluripotent tissues: Implications for epigenetic reprogramming. J Biol Chem. 2004;279:52353–52360. doi: 10.1074/jbc.M407695200. [DOI] [PubMed] [Google Scholar]
- 22.Métivier R, et al. Cyclical DNA methylation of a transcriptionally active promoter. Nature. 2008;452:45–50. doi: 10.1038/nature06544. [DOI] [PubMed] [Google Scholar]
- 23.Rai K, et al. DNA demethylation in zebrafish involves the coupling of a deaminase, a glycosylase, and gadd45. Cell. 2008;135:1201–1212. doi: 10.1016/j.cell.2008.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bhutani N, et al. Reprogramming towards pluripotency requires AID-dependent DNA demethylation. Nature. 2010;463:1042–1047. doi: 10.1038/nature08752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim MS, et al. DNA demethylation in hormone-induced transcriptional derepression. Nature. 2009;461:1007–1012. doi: 10.1038/nature08456. [DOI] [PubMed] [Google Scholar]
- 26.Hu XV, et al. Identification of RING finger protein 4 (RNF4) as a modulator of DNA demethylation through a functional genomics screen. Proc Natl Acad Sci USA. 2010;107:15087–15092. doi: 10.1073/pnas.1009025107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Agius F, Kapoor A, Zhu JK. Role of the Arabidopsis DNA glycosylase/lyase ROS1 in active DNA demethylation. Proc Natl Acad Sci USA. 2006;103:11796–11801. doi: 10.1073/pnas.0603563103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gehring M, et al. DEMETER DNA glycosylase establishes MEDEA polycomb gene self-imprinting by allele-specific demethylation. Cell. 2006;124:495–506. doi: 10.1016/j.cell.2005.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barreto G, et al. Gadd45a promotes epigenetic gene activation by repair-mediated DNA demethylation. Nature. 2007;445:671–675. doi: 10.1038/nature05515. [DOI] [PubMed] [Google Scholar]
- 30.Schmitz KM, et al. TAF12 recruits Gadd45a and the nucleotide excision repair complex to the promoter of rRNA genes leading to active DNA demethylation. Mol Cell. 2009;33:344–353. doi: 10.1016/j.molcel.2009.01.015. [DOI] [PubMed] [Google Scholar]
- 31.Jin SG, Guo C, Pfeifer GP. GADD45A does not promote DNA demethylation. PLoS Genet. 2008;4:e1000013. doi: 10.1371/journal.pgen.1000013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Engel N, et al. Conserved DNA methylation in Gadd45a(-/-) mice. Epigenetics. 2009;4:98–99. doi: 10.4161/epi.4.2.7858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Okada Y, Yamagata K, Hong K, Wakayama T, Zhang Y. A role for the elongator complex in zygotic paternal genome demethylation. Nature. 2010;463:554–558. doi: 10.1038/nature08732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Valinluck V, et al. Oxidative damage to methyl-CpG sequences inhibits the binding of the methyl-CpG binding domain (MBD) of methyl-CpG binding protein 2 (MeCP2) Nucleic Acids Res. 2004;32:4100–4108. doi: 10.1093/nar/gkh739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kriaucionis S, Heintz N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science. 2009;324:929–930. doi: 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tahiliani M, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ito S, et al. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature. 2010;466:1129–1133. doi: 10.1038/nature09303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jin SG, Kadam S, Pfeifer GP. Examination of the specificity of DNA methylation profiling techniques towards 5-methylcytosine and 5-hydroxymethylcytosine. Nucleic Acids Res. 2010;38:e125. doi: 10.1093/nar/gkq223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wossidlo M, et al. Dynamic link of DNA demethylation, DNA strand breaks and repair in mouse zygotes. EMBO J. 2010;29:1877–1888. doi: 10.1038/emboj.2010.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mayer W, Smith A, Fundele R, Haaf T. Spatial separation of parental genomes in preimplantation mouse embryos. J Cell Biol. 2000;148:629–634. doi: 10.1083/jcb.148.4.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jin SG, Tsark W, Szabó PE, Pfeifer GP. Haploid male germ cell- and oocyte-specific Mbd3l1 and Mbd3l2 genes are dispensable for early development, fertility, and zygotic DNA demethylation in the mouse. Dev Dyn. 2008;237:3435–3443. doi: 10.1002/dvdy.21767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nakamura T, et al. PGC7/Stella protects against DNA demethylation in early embryogenesis. Nat Cell Biol. 2007;9:64–71. doi: 10.1038/ncb1519. [DOI] [PubMed] [Google Scholar]
- 43.Huang Y, et al. The behaviour of 5-hydroxymethylcytosine in bisulfite sequencing. PLoS ONE. 2010;5:e8888. doi: 10.1371/journal.pone.0008888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cannon SV, Cummings A, Teebor GW. 5-Hydroxymethylcytosine DNA glycosylase activity in mammalian tissue. Biochem Biophys Res Commun. 1988;151:1173–1179. doi: 10.1016/s0006-291x(88)80489-3. [DOI] [PubMed] [Google Scholar]
- 45.Hajkova P, et al. Genome-wide reprogramming in the mouse germ line entails the base excision repair pathway. Science. 2010;329:78–82. doi: 10.1126/science.1187945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Valinluck V, Sowers LC. Endogenous cytosine damage products alter the site selectivity of human DNA maintenance methyltransferase DNMT1. Cancer Res. 2007;67:946–950. doi: 10.1158/0008-5472.CAN-06-3123. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.