Abstract
Both the formation of long-term memory (LTM) and late-long-term potentiation (L-LTP), which is thought to represent the cellular model of learning and memory, require de novo protein synthesis. The mammalian target of Rapamycin (mTOR) complex I (mTORC1) integrates information from various synaptic inputs and its best characterized function is the regulation of translation. Although initial studies have shown that rapamycin reduces L-LTP and partially blocks LTM, recent genetic and pharmacological evidence indicating that mTORC1 promotes L-LTP and LTM is controversial. Thus, the role of mTORC1 in L-LTP and LTM is unclear. To selectively inhibit mTORC1 activity in the adult brain, we used a “pharmacogenetic” approach that relies on the synergistic action of a drug (rapamycin) and a genetic manipulation (mTOR heterozygotes, mTOR+/− mice) on the same target (mTORC1). Although L-LTP and LTM are normal in mTOR+/− mice, application of a low concentration of rapamycin—one that is subthreshold for WT mice—prevented L-LTP and LTM only in mTOR+/− mice. Furthermore, we found that mTORC1-mediated translational control is required for memory reconsolidation. We provide here direct genetic evidence supporting the role of mTORC1 in L-LTP and behavioral memory.
Keywords: behavioral learning, pharmacology, fear conditioning
A major goal of cognitive neuroscience is to identify the molecular and cellular mechanisms underlying learning and memory. Memory is essential for human experience. The loss of memory leads to the loss of lasting interactions with other human beings and ultimately the loss of self. Two general types of memory storage mechanisms have been described: short-term memory (STM), which lasts minutes, and long-term memory (LTM), which lasts days, weeks, or even a lifetime. This temporal distinction in behavior is reflected in specific forms of synaptic plasticity that underlie each type of memory, as well as specific molecular requirements. The short-term forms involve covalent modifications of preexisting proteins, whereas the long-term forms require new protein synthesis (1–4). At the cellular level, the most studied form of synaptic plasticity is long-term potentiation (LTP), which refers to long-lasting increases in synaptic strength (5, 6). Like memory, LTP occurs in two temporally distinct phases: early LTP (E-LTP), which is typically induced by a single train of high-frequency (tetanic) stimulation, lasts only 1–2 h and depends on modification of preexisting proteins; and late-LTP (L-LTP), generally induced by several (typically four) tetanic trains separated by 5–10 min, persists for many hours and requires new protein synthesis (1–4). Although a small number of genes and proteins that are important for the formation of long-lasting memories have been identified, we are far from fully understanding the precise underlying molecular steps involved in this process.
The Mammalian Target of Rapamycin (mTOR), an evolutionarily conserved Ser/Thr protein kinase that promotes translation rates, is not only regulated by nutrients, but is also the central node of a highly conserved signaling network that integrates information from various synaptic inputs (1, 7). mTOR forms two distinct protein complexes. mTOR complex 1 (mTORC1) contains Raptor (Regulatory Associated Protein of mTOR), LST8 (also known as GβL), and PRAS40 and is sensitive to the drug rapamycin (which directly binds to mTORC1 as a complex with the immunophilin FKBP12; reviewed in refs. 8–10). mTORC1 is thought to regulate mRNA translation through phosphorylation of its downstream effectors: p70 S6 kinase (S6K) and eIF4E-binding proteins (4E-BPs; refs. 8–10). In contrast, mTOR complex 2 (mTORC2), which was only recently discovered, contains SIN1 (SAPK Interacting protein I), LST8, and Rictor (Rapamycin-Insensitive Companion of mTOR), is rapamycin insensitive, and phosphorylates Akt at Serine 473 (8, 10).
Most of the evidence for mTORC1 signaling in L-LTP and LTM is based on the finding that rapamycin inhibits L-LTP in mammalian brain slices in vitro (11) and partially blocks LTM (reviewed in ref. 12). Moreover, mice lacking either the mTORC1 negative regulator Tsc2 or the downstream targets 4e-bp2 and S6k1/2 exhibit altered synaptic plasticity and memory (13–15). However, several aspects of these results are controversial. First, exceptionally high doses of rapamycin (150 mg/kg) are required to block contextual LTM in mice (15), whereas, in humans, blockage of mTORC1 signaling with the mTOR inhibitor everolimus appears to improve (not impair) cognition (16). Second, it was recently shown that rapamycin does not block L-LTP in the dentate gyrus in vivo (17). Third, translation is maintained in the presence of rapamycin (reviewed in ref. 9). Fourth, rapamycin specifically inhibits mTORC1 activity after a short treatment, but prolonged treatment with rapamycin could block the activity of mTORC2 (18). Fifth, genetic deletions that enhance mTORC1 activity, such as Tsc2+/− mice, Fkbp12−/− mice, or 4e-bp2−/− mice, generate different/contradictory phenotypes with regard to plasticity and memory (Fig. S1; refs. 14, 15, and 19). The same is true for genetic and pharmacological interventions that block mTORC1 activity (Fig. S1). Sixth, mice lacking the mTORC1 downstream target S6k1 or S6k2 exhibit normal L-LTP, suggesting that S6Ks do not control mTORC1 mediated-translation of mRNAs that underlie L-LTP (13). It is also noteworthy that mice lacking S6ks only exhibit very subtle memory phenotypes (13). Seventh, whereas mice lacking 4e-bp2, the other mTORC1 downstream target, show impaired L-LTP and LTM (14), 4E-BP2 (protein) cannot be phosphorylated (and regulated) by mTORC1 in the adult brain (20). Thus, these data suggest that mTORC1 controls protein synthesis (in the brain) in a 4E-BP2-indepenent manner. Hence, there is no direct genetic evidence supporting the role of mTORC1 in L-LTP and LTM. We integrate here genetics and pharmacology (pharmacogenetics) to selectively inhibit mTORC1, without affecting mTORC2 activity, and study long-lasting changes in synaptic strength, learning, and memory. This approach specifically targets mTORC1 function in the brain and provides strong, direct genetic evidence that mTORC1 promotes L-LTP, LTM consolidation, and reconsolidation.
Results
Pharmacogenetic Approach to Selectively Inhibit mTORC1.
mTOR knockout mice (mTOR−/− mice) die in utero (21, 22), and brain-specific deletion of the mTORC1 up-stream regulators Tsc1 or Pten results in death within the first postnatal weeks (15, 23). Thus, the study of mTORC1 signaling in adult animals has been limited. To overcome this problem, we developed a pharmacogenetic approach that is used not only to identify genes and drug targets, but increasingly for the personalized treatment of major diseases such as epilepsy, cancer, diabetes, and autism spectrum disorders (24, 25). This approach is based on the fact that the response to a given drug is determined by a genetic variation/mutation (24–28). In addition, pharmacogenetics has been used to reveal many recessive mutant phenotypes (where the heterozygous mutant allele has no phenotype). Indeed, such heterozygotes exhibit a phenotype only when given a dose of a drug that has no effect in normal animals. Thus, pharmacogenetics relies on the synergism between a drug and a genetic manipulation on the same signaling pathway.
mTOR heterozygous mice (mTOR+/− mice) are viable, of normal size and weight, develop normally, and are phenotypically indistinguishable from their WT littermates (21). Furthermore, no gross structural abnormalities were found in mTOR+/− mouse brain, as assessed by Nissl staining and synaptic markers for the vesicular glutamate transporter 1 (VGLUT1), postsynaptic density protein 95 (PSD95), and glutamic acid decarboxylase 67 (GAD67; Fig. S2).
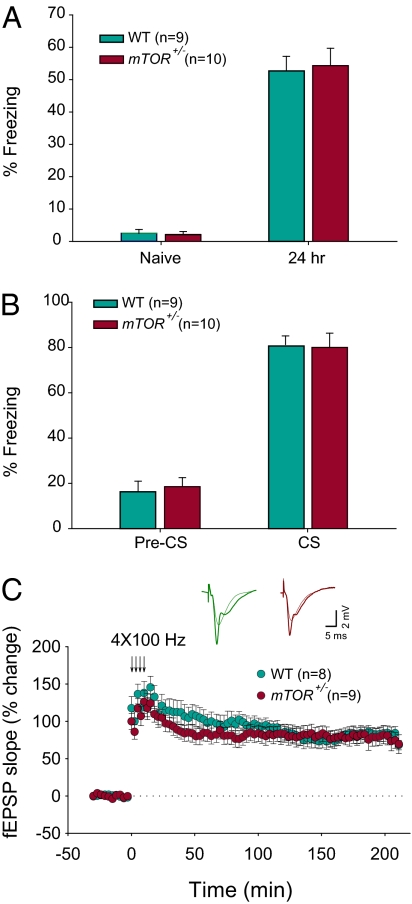
To determine whether the mTOR mutation is recessive, WT and mTOR+/− mice were analyzed in two forms of Pavlovian fear conditioning as previously described (29, 30). Contextual fear conditioning was induced by pairing a context (conditioning stimulus; CS) with a foot shock (the unconditioned stimulus, US), whereas in auditory fear conditioning the US was paired with a tone presentation (the conditioned stimulus, CS). Contextual fear conditioning involves both the hippocampus and amygdala, whereas auditory fear conditioning requires only the amygdala (31). When mice were subsequently exposed to the CS, fear responses (“freezing”) were taken as an index of the strength of the CS–US association. WT and mTOR+/− mice show similar amounts of freezing before training or when tested 24 h after training (Fig. 1A). Similarly, long-term auditory fear memory is normal in mTOR+/− mice when determined 24 h after training (Fig. 1B). Thus, the lack of one copy of mTOR does not impact auditory or contextual long-lasting fear memories.
Fig. 1.
Contextual and auditory long-term fear memories and L-LTP are normal in mTOR+/− mice. (A) Contextual fear conditioning was determined by measuring the time spent freezing (as % of total time) before the conditioning (Naïve, during 2-min period) and then 24 h after training (during a 3-min period). (B) Auditory fear memory was determined by measuring the time spent freezing (as percentage of total time) 24 h after training either before the onset of the tone (pre-CS, during a 2-min period) or during the tone presentation (during a 3-min period). (C) Similar L-LTP induced by four 100-Hz trains at 5-min intervals in slices from WT and mTOR+/− mice (LTP magnitude at 30 min, WT 110 ± 18%, mTOR+/− 97 ± 11%, P > 0.05; LTP magnitude at 220 min, WT 68 ± 6%, mTOR+/− 69 ± 12%, P > 0.05). Data are means ± SEMs.
Next, we studied synaptic transmission in slices from WT and mTOR+/− mice. Basal synaptic transmission was not altered in mTOR+/− mice as determined by the input–output relation of field excitatory postsynaptic potentials (fEPSPs; as a function of stimulus intensity), paired-pulse facilitation (PPF), and fiber volley amplitude (Fig. S3). In agreement with the behavioral data, protein-synthesis-dependent L-LTP generated by four tetanic trains of high-frequency stimulation (1, 2) was similar in CA1 hippocampal slices from mTOR+/− and WT littermates (Fig. 1C). Taken together, these data indicate that, as in their yeast and Drosophila counterparts, the TOR mutation in mammals is recessive, with respect to these forms of synaptic plasticity and memory (32, 33).
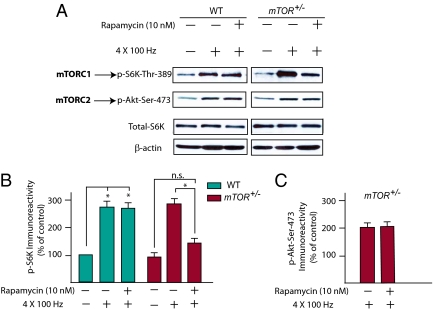
Therefore, we took advantage of the synergism between pharmacology (rapamycin administration) and genetics (mTOR+/− mice) to specifically suppress mTORC1 activity in the adult brain. Given that mTOR+/− mice lack one copy of mTOR (21), we predicted that a lower concentration of rapamycin would be required to block mTORC1 activity in these mice, compared with WT mice. Hippocampal slices from WT and mTOR+/− mice were pretreated with different concentrations of rapamycin and subsequently stimulated by four tetanic trains of high-frequency stimulation (HFS), which are known to induce a long-lasting L-LTP (1, 2) and stimulate mTORC1 signaling (1, 7). Consistent with our hypothesis, a low concentration of rapamycin (10 nM) was ineffective in CA1 slices from WT mice, but inhibited HFS-induced mTORC1 activity in CA1 slices from mTOR+/− mice (Fig. 2 A and B), as determined by measuring phosphorylation of the mTORC1 downstream target S6K1 (at Thr389), which is commonly used as readout of mTORC1 activity (8–10). As expected, rapamycin (10 nM) did not alter the activity of the rapamycin-insensitive complex mTORC2, as determined by measuring the phosphorylation of Akt-Ser-473 in mTOR+/− CA1 slices (Fig. 2 A and C), which is used as a readout mTORC2 activity (34, 35). Similar results were found when slices were incubated with forskolin, which also induces protein synthesis-dependent L-LTP (36, 37) and stimulates mTORC1 signaling (ref. 36; Fig. S4). Taken together, these data indicate that this pharmacogenetic manipulation specifically suppresses mTORC1 signaling.
Fig. 2.
A low concentration of rapamycin specifically blocks high-frequency-induced mTORC1 activity, but not mTORC2 activity, only in mTOR+/− mice. (A) CA1-hippocampal slices from WT and mTOR+/− mice were either untetanized controls (stimulated at 0.033 Hz) or tetanized by four trains of 100 Hz (at 5-min intervals) in the presence or absence of a low concentration of rapamycin (10 nM). Extracts of CA1 regions were prepared 30 min after stimulation, and the phosphorylation of S6K (Threonine-389) and Akt (Serine-473) was determined by Western blotting. (B and C) Quantification of normalized phospho-S6K-Thr-389 (B), n = 4 mice (8 slices) per condition; and phosphor-Akt-Ser473 (C), n = 4 mice (8 slices) per conditions (*P < 0.05).
Pharmacogenetic Inhibition of mTORC1 Blocks L-LTP.
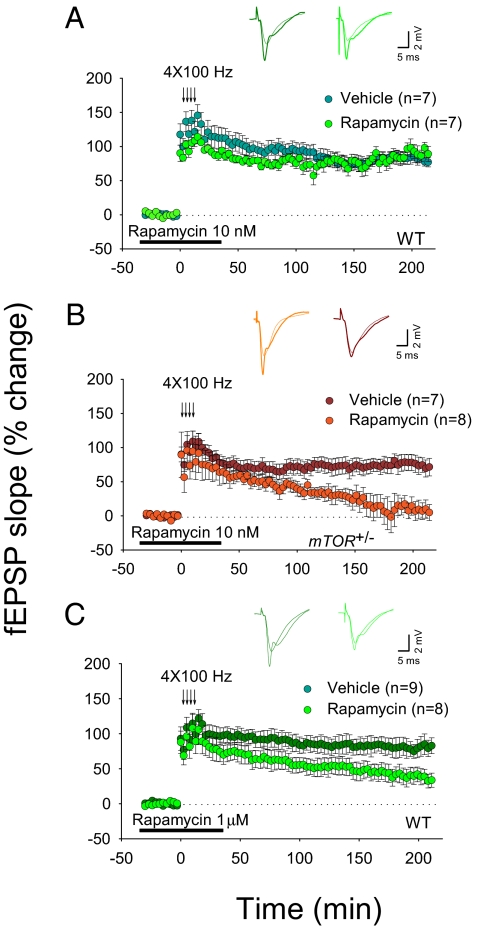
Late-LTP generated by four high-frequency trains is known to be dependent on de novo protein synthesis (1, 2). Because mTORC1 is thought to regulate translation rates (9, 10), we next examined L-LTP in slices from WT and mTOR+/− mice, as previously described (29, 30). If mTORC1 signaling is crucial for L-LTP, we predicted that a low concentration of rapamycin (10 nM) would block L-LTP in slices from mTOR+/− mice, but not in slices from WT mice. Indeed, incubation with a low concentration of rapamycin, which blocks mTORC1 signaling only in hippocampal slices from mTOR+/− mice (Fig. 2 and Fig. S4), had no effect on the L-LTP induced by four tetanic trains in slices from WT mice, but significantly blocked L-LTP in hippocampal slices from mTOR+/− mice (compare Fig. 3A to Fig. 3B). A much higher dose of rapamycin (1 μM) was necessary to reduce L-LTP in WT slices (Fig. 3C). These data show that mTORC1 activity is crucial for L-LTP and demonstrate the powerful synergistic effect of the pharmacological and genetic manipulations operating on the same target, namely, mTORC1.
Fig. 3.
A low concentration of rapamycin selectively blocks L-LTP only in slices from mTOR+/− mice. Rapamycin (10 nM) blocked L-LTP in slices from mTOR+/− mice (LTP magnitude at 30 min, vehicle 77 ± 11%, rapamycin 69 ± 18%, P > 0.05; LTP magnitude at 220 min, vehicle 76 ± 12%, rapamycin 9 ± 14%, P < 0.01; B), but not the L-LTP-induced by four tetanic trains in WT slices (LTP magnitude at 30 min, vehicle 108 ± 16%, rapamycin 90 ± 10%, P > 0.05; LTP magnitude at 220 min, vehicle 84 ± 6%, rapamycin 86 ± 8%, P > 0.05; A). (C) In WT slices, a much higher concentration of rapamcyin (1 μM) was required to block L-LTP (LTP magnitude at 20 min, vehicle 96 ± 15%, rapamycin 86 ± 18%, P < 0.05; LTP magnitude at 220 min, vehicle 83 ± 11%, rapamycin 33 ± 10%, P > 0.05). Horizontal bars indicate the period of incubation with rapamycin.
Pharmacogenetic Inhibition of mTORC1 Impairs LTM Consolidation.
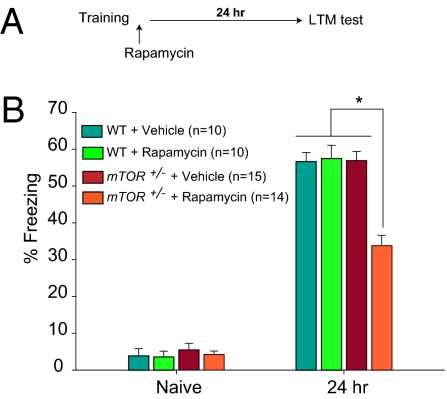
To ascertain whether mTORC1 is involved in rapid modulation of cognitive processing, we examined contextual fear conditioning in WT and mTOR+/− mice treated with rapamycin. Because memory is more susceptible to disruption immediately after training, it should be possible to block LTM only in mTOR+/− mice by administering a low concentration of rapamycin soon after training. To test this prediction, WT and mTOR+/− mice were injected acutely with a low concentration of rapamycin (40 mg/kg) immediately after Pavlovian fear conditioning training, and LTM was tested 24 h later (Fig. 4A). Accordingly, rapamycin impaired long-term contextual fear memory in mTOR+/− mice, but not in WT mice (Fig. 4B). It should be noted that this concentration of rapamycin (40 mg/kg) only blocked fear-conditioning-induced mTORC1 activity in the hippocampus (CA1 area) from mTOR+/− mice, compared with WT littermates (Fig. S5). Because translation is required only for LTM, but not STM, mice with impaired mTORC1 signaling exhibit deficient LTM consolidation, but intact STM, when tested 2 h after training (Fig. S6). These data indicate that mTORC1 positively regulates LTM consolidation.
Fig. 4.
A low concentration of rapamycin that is subthreshold for WT mice impairs contextual long-term fear memory in mTOR+/− mice. (A) Scheme of the experimental protocol. (B) WT and mTOR+/− mice were administered either vehicle or rapamycin (40 mg/kg) immediately after training. Rapamycin (40 mg/kg) significantly decreased contextual freezing only in mTOR+/− mice. Contextual fear conditioning was determined by measuring the time spent freezing (as percentage of total time) before the conditioning (Naïve, during a 2-min period) and then 24 h after training (during a 5-min period). Data are means ± SEMs (*P < 0.05).
Pharmacogenetic Inhibition of mTORC1 Impairs LTM Reconsolidation.
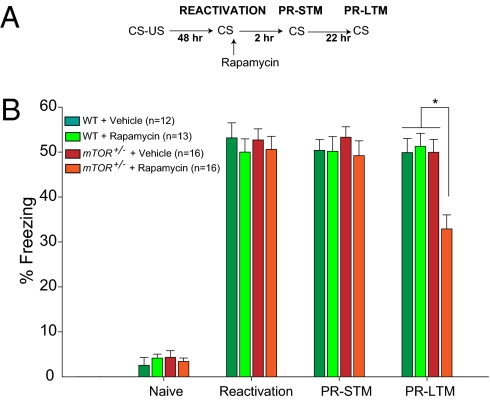
Recent reports have challenged the prevailing viewpoint that a consolidated memory is stable and immune to disruption by showing that, when a consolidated memory is reactivated, it is labile and needs to be restabilized with newly synthesized proteins (38, 39). This process, named “reconsolidation” (40, 41), has been demonstrated in species that range from sea slugs to humans (39). Because mTORC1 regulates protein synthesis, we wondered whether mTORC1 regulates the synthesis of proteins required for reconsolidation (40, 42). To this end, animals were trained using a reconsolidation protocol, which is a minor variant of common consolidation protocols (Fig. 5A). Briefly, WT and mTOR+/− mice were subjected to the standard contextual fear conditioning protocol as previously described (29, 30), and returned to their home cage for 2 d, in order for the memory trace to consolidate. Two days after training, both groups received a 5-min reactivation session in which the CS is presented, thus initiating the process of reconsolidation. Application of a low dose of rapamycin (40 mg/kg) to mTOR+/− mice immediately after the reactivation period resulted, 2 h later, in a normal protein-synthesis-independent postreactivated STM analog, called postreactivation STM (PR-STM). By contrast, the same concentration of rapamycin impaired long-term analog, defined as postreactivation LTM (PR-LTM), when tested 22 h later (Fig. 5B). It is noteworthy that rapamycin had no effect on WT mice, indicating that rapamycin specifically inhibited mTORC1 activity in mTOR+/− mice during memory reconsolidation. Importantly, in the absence of memory reactivation, rapamycin had no effect on memory in mTOR+/− mice (Fig. S7), consistent with the idea that only the reactivated memories return to a labile state and need to be reconsolidated in a mTORC1-dependent manner.
Fig. 5.
A low concentration of rapamycin that is subthreshold for WT mice impairs LTM reconsolidation in mTOR+/− mice. (A) Scheme of the experimental protocol. (B) Animals were trained as described in Fig. 4. Two days after training, memory was reactivated, and WT and mTOR+/− mice were administered either vehicle or rapamycin (40 mg/kg) immediately after reactivation. Contextual fear conditioning was determined by measuring the time spent freezing (as pecentage of total time) before the conditioning (Naïve, during a 2-min period), during reactivation (during a 5-min period), 2 h (PR-STM, during a 5-min period) and 22 h (PR-LTM, during a 5-min period) after reactivation.
Discussion
mTORC1 is activated by a variety of synaptic signals (e.g., glutamate and neurotrophins), which are thought to lead to protein-synthesis-dependent changes in synaptic strength and LTM (1, 7). Thus, mTORC1, which was identified in studies aimed at defining the molecular target of the antiproliferative drug rapamycin (10, 43, 44), serves as a “master” regulator that integrates information from various synaptic inputs to produce an appropriate translational output. However, whether mTORC1 plays a role in memory consolidation remains unclear. As recently discussed by Gkogkas et al. (45), it is virtually impossible to predict the impact of mutations which enhance or decrease mTORC1 activity on L-LTP and memory (e.g., although Fkbp12 and Tsc2 mutations both enhance mTORC1 activity, they exhibit completely different phenotypes for LTP and memory, see Fig. S1). Unfortunately, not only are the genetic data controversial, but the pharmacological data are also questionable: (i) rapamycin, like any drug, might have off-target effects; (ii) rapamycin treatment could also block mTORC2 activity (46); (iii) although rapamycin blocks L-LTP in vitro, it fails to do so in vivo in the dentate gyrus (17). Strikingly, a recent (and currently the only) study in humans shows that pharmacologically blocking mTORC1 enhances (not reduces) cognition (16). Taken together, these data demonstrate that the role of mTORC1 in synaptic plasticity and memory consolidation is far from clear. Given that mTORC1 signaling is altered in a variety of neurological disorders (12) and its inhibition appears to enhance longevity (47), it is crucial to determine whether mTORC1 enhances, impairs, or has no effect on L-LTP and LTM. Two different approaches could be used to answer this question: genetics or pharmacology. Unfortunately, the use of genetics alone is not desirable because loss of mTOR or its up-stream regulators in the brain is lethal (15, 21, 48). Furthermore, the specificity of rapamycin can only be measured in a mTOR knockout background. Thus, because the mTOR mutation is lethal and the knockout mice are not available, it is impossible to determine whether rapamycin's action in the brain is due to inhibition of mTORC1 or some other off-target action. Inspired by the work of Silva and colleagues (49), we developed a method based on the powerful synergistic action of genetics and pharmacology to “directly” and “specifically” inhibit mTORC1 activity in the brain. Several advantages are inherent to this methodology. First, we can control the specificity of the drug because, at the concentrations used, it has no effect on WT mice but blocks the activity of mTORC1, L-LTP, and LTM in mTOR+/− mice (Fig. 2 and Figs. S4 and S5). Hence, rapamycin action on the mTOR+/− mice is due to selective inhibition of mTORC1. Second, the effect of rapamycin on mTORC1 is both direct and selective because rapamycin forms a complex with FKBP12, which directly binds to and inhibits mTORC1 (50–53) but not mTORC2 signaling (Fig. 2). A selective inhibition of mTORC1 signaling is important because the other mTOR complex, mTORC2, which is thought to regulate actin dynamics, could also play a role in synaptic plasticity and memory storage. Finally, we selectively inhibited mTORC1 in the adult brain in vitro and in vivo with high temporal specificity. Thus, the use of this pharmacogenetic approach is an innovative procedure, because no other genetic manipulation that specifically and directly inhibits mTORC1 in the brain has yet been described. It is noteworthy that the increased sensitivity of mTOR mutants to a given dose of rapamycin is an evolutionarily conserved property that has also been found in Drosophila and yeast (32, 33). Therefore, by using this methodology, we conclusively determined the direct role of mTORC1 in protein synthesis-dependent plasticity and memory consolidation.
A key finding in our study is that inhibition of mTORC1 signaling also blocks memory reconsolidation. It should be noted that reconsolidation studies are technically challenging in knockout or transgenic mice because these mice are usually unable to consolidate their memory. Therefore, it is impossible to determine whether an already impaired consolidated memory undergoes reconsolidation. Our pharmacogenetic approach provides a major opportunity to circumvent this problem. In addition, because most mutations are recessive, our data raise the interesting possibility that pharmacogenetics could be widely used to study memory reconsolidation in mice with recessive mutations. In addition, the finding that memories that return to a labile state and need to be reconsolidated are dependent on mTORC1 has important implications for the treatment of some mental illnesses, such as posttraumatic stress disorder.
Finally, our findings could also have implications for the study of aging. Recent studies have identified both mTORC1 and translational control as conserved longevity pathways: Reduced mTORC1 signaling and protein synthesis increase longevity (47, 54). In our experiments, administration of rapamycin (40 mg/kg) immediately after training blocked mTORC1 activity and contextual LTM in mTOR+/− mice, but not in WT mice (Fig. 4 and Fig. S5), and a much higher dose of rapamycin (150 mg/kg) appears to be required to block contextual LTM in WT mice (15). These findings are in contrast to those of Blundell et al. (55), who reported that systemic administration of rapamycin reduced LTM. The reason for the discrepancy between their data and our data is not immediately clear, but possible explanations are differences in the age of the animals, the behavioral procedure, and the timing of administration of the drug. Indeed, intra-CA1 infusion of rapamycin 15 min before training, but not immediately after training, blocks inhibitory avoidance-induced LTM, indicating that the time of injection of rapamycin is crucial (56). We also hypothesize that slow penetration of the blood–brain barrier could account for the much weaker effects observed in vivo than in vitro, as indicated by direct tests of mTOR activity in brain tissue (15). Therefore, we speculate that low systemic doses of rapamycin, which would only moderately inhibit mTORC1 in the brain, might extend life span without disturbing LTM.
In conclusion, we provide here strong direct genetic evidence that mTORC1 is crucially involved in long-term changes in synaptic strength and memory.
Materials and Methods
mTOR+/− Mice.
mTOR+/− mice were generated as previously described (21). Mice were weaned at the third postnatal week and genotyped by PCR. The mutant and corresponding WT alleles are detected by a three-primer PCR assay: Oligo1 (5′-TTCATTCCCTTGAAAGCCAGTCTCACC-3′), Oligo-2 (5′-GCTCTTGAGGCAAATGCCAGTATCACC-3′), Oligo-3 (5′-TCATTACCTTCTCATCAGCCAGCAGTT-3′). All experiments were performed on 12- to 20-wk-old mice. The mice were kept on a 12-h light/dark cycle, and the behavioral experiments were always conducted during the light phase of the cycle. The mice had access to food and water ad libitum, except during tests. Animal care and experimental procedures were approved by the Animal Care committees of Baylor College of Medicine.
Rapamycin.
For the ex vivo electrophysiological experiments, rapamycin (LC Laboratories) was initially dissolved in DMSO and used at concentrations of 10 nM and 1 μM. Hippocampal slices were perfused with rapamycin (or vehicle) for 45 min before tetanization and after at least 30 min of stable recording. For the in vivo behavioral experiments, rapamycin was first dissolved in 100% ethanol, stored at −20°C and freshly dissolved in an aqueous solution of 4% Tween 80 and 4% PEG 400 immediately before use. Rapamycin (or vehicle) was injected intraperitoneally (i.p.) immediately after fear conditioning training or reactivation, at a dose of 40 mg/kg.
Field Recordings.
Horizontal hippocampal slices (350 μm) were cut from brains of WT or and mTOR+/− mice in 4 °C artificial cerebrospinal fluid (ACSF) and kept in ACSF at room temperature for at least 1 h before recording. Slices were maintained at 28–29 °C in an interface-type chamber perfused with ACSF containing 124 mM NaCl, 2 mM KCl, 1.3 mM MgSO4, 2.5 mM CaCl2, 1.2 mM KH2PO4, 25 mM NaHCO3, and 10 mM glucose (2-3 mL/min), aerated with 95% O2 and 5% CO2. Bipolar stimulating electrodes were placed in the CA1 stratum radiatum to stimulate Schaffer collateral and commissural fibers. Field potentials were recorded using ACSF-filled micropipettes. The recoding electrodes were placed in the stratum radiatum for fEPSPs, and in the stratum pyramidale for population spikes. The stimulus strength of the 0.1-ms pulses was adjusted to evoke 30–35% of maximal response for fEPSPs. A stable baseline of responses was established for at least 30 min. Tetanic LTP was induced by applying four trains of high-frequency stimulation (100 Hz, 1 s) separated by 5-min intervals. To reduce day-to-day variability, whenever possible, simultaneous recordings (in the same chamber) were obtained from WT and mTOR+/− mice treated with rapamycin or vehicle. Statistical analysis was performed using the t test and two-way ANOVA. All data are presented as means ± SEM, and n indicates the number of slices and mice.
Behavioral Learning.
The experimenter was blind to the genotype for all behavioral tests. Mice were first handled for 5–10 min for 2 d and then habituated to the conditioning chamber for 20 min twice a day for 2 d. On the training day, after 2 min in the conditioning chamber, mice received two pairings of a tone (2,800 Hz, 85 db, 30 s) with a coterminating foot-shock (0.7 mA, 2 s). Mice remained in the chamber for 1 additional min and then were returned to their home cages. Mice were tested 2 and 24 h after training for “freezing” (immobility with the exception of respiration) in response to the tone (in a chamber to which they had not been conditioned) and to the training context (training chamber). For auditory fear conditioning, mice were placed in the chamber and freezing responses were recorded during the initial 2 min (pre-CS period) and during the last 3 min when the tone was played. Mice were returned to their cages 30 s after the end of the tone. For testing contextual fear conditioning, mice were returned to the conditioning chamber for 5 min.
For reconsolidation experiments, mice were handled and habituated as described above. After a 2-min acclimatizing period, they received two pairings of a tone (2,800 Hz, 85 db, 30 s) with a coterminating foot-shock (0.7 mA, 2 s) and were returned to their home cages. Two days later, mice were placed into the same context and given a 5-min presentation of the CS (reactivation) to measure the expression of their consolidated memory. Animals were injected with either rapamycin (40 mg/kg) or vehicle immediately after the retention test and tested 2 h (PR-STM) and 22 h (PR-LTM) after memory reactivation for “freezing” during a 5-min period. The percentage of total time during which freezing occurred was taken as an index of learning and memory. Statistical analysis was based on repeated-measures ANOVA, and between-group comparisons by Tukey's Test.
Immunohistochemistry and Western Blotting.
For Western blotting, slices were cut as above and incubated for at least 1 h at room temperature in oxygenated (95% O2/5% CO2) ACSF. Hippocampal slices were then separated into groups and equilibrated for 1 h at 28–29 °C in ACSF before treatment. Slices were pretreated with rapamycin (10 nM) for 10 min and then treated with Forskolin (Sigma) at a final concentration of 50 μM for 10 min before being snap-frozen over dry ice. In all instances, slices were treated with vehicle as a control. Frozen slices were briefly thawed for microdissection of area CA1 and then suspended into homogenizing buffer containing (40 mM Tris·HCl/275 mM NaCl/20% glycerol/2% Igepal/1% Triton X-100/50 mM NaF/2 mM Na3VO4/25 mM β-glycerophosphate/1 mM PMSF/20 μg/mL pepstatin/20 μg/mL aprotinin). A total of 50 μg of protein per sample were resolved on SDS/PAGE (15%) and transfered onto nitrocellulose membranes (Millipore). Membranes were blocked in 5% powdered milk in Tris-buffered saline with 0.1% Tween 20 (TBS-T) for 1 h, followed by incubation with primary antibodies overnight at 4 °C. Antibodies against phospho S6K1-Thr-389, phospho-Akt-Ser-473, Total-S6K, and β-actin were purchased from Cell Signaling Technology. Membranes were then washed with TBS-T three times and incubated with horseradish peroxidase-coupled secondary antibody for 1–2 h at room temperature, washed again three times, and treated with enhanced chemiluminescence before detection on X-ray film.
For immunohistochemistry, mice were deeply anesthetized and perfused intracardially with 4% paraformaldehyde (PFA) in an ice-cold 0.1 M phosphatase buffer solution (PBS). Brains were removed from the skull and stored in a 4% PFA solution overnight (at 4 °C). Horizontal sections (40 μm) were cut on a microtome (Leica VFT1000S, Germany). Free-floating method was used while rinsing between steps. Sections were first placed in a blocking solution (5% BSA/0.3% Triton/ 4% normal goat serum in PBS) at room temperature for 1 h, incubated with primary antibodies [PKR (Santa Cruz Biotechnology, CA), GAD67 (Millipore, Billerica, MA), V-Glut 1 (Synaptic Systems, Goettingen, Germany) and PSD95 (NeuroMab, CA)] overnight and then rinsed four times (for 20 min) with PBS before incubation with the secondary antibody (for 4 h). After four washes (each for 20 min) with PBS, the sections were mounted on Superfrost Plus slides (VWR, West Chester, PA). Finally, the sections were placed on cover slips with VECTASHIELD Hard Set mounting medium (Vector Lab, Burlingame, CA). Digital photos were taken with a Zeiss LSM 510 laser confocal microscope.
Supplementary Material
Acknowledgments
We thank Jingming Chen for assisting in the maintenance of the mouse colony and Andon Placzek for comments on the manuscript. This work was supported by Searle Award 09-SSP-211, the Whitehall Foundation, and the George and Cynthia Mitchell Foundation (to M.C.-M.).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1014715108/-/DCSupplemental.
References
- 1.Costa-Mattioli M, Sossin WS, Klann E, Sonenberg N. Translational control of long-lasting synaptic plasticity and memory. Neuron. 2009;61:10–26. doi: 10.1016/j.neuron.2008.10.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kandel ER. The molecular biology of memory storage: A dialogue between genes and synapses. Science. 2001;294:1030–1038. doi: 10.1126/science.1067020. [DOI] [PubMed] [Google Scholar]
- 3.Kelleher RJ, 3rd, Govindarajan A, Tonegawa S. Translational regulatory mechanisms in persistent forms of synaptic plasticity. Neuron. 2004;44:59–73. doi: 10.1016/j.neuron.2004.09.013. [DOI] [PubMed] [Google Scholar]
- 4.Klann E, Dever TE. Biochemical mechanisms for translational regulation in synaptic plasticity. Nat Rev Neurosci. 2004;5:931–942. doi: 10.1038/nrn1557. [DOI] [PubMed] [Google Scholar]
- 5.Malenka RC, Bear MF. LTP and LTD: An embarrassment of riches. Neuron. 2004;44:5–21. doi: 10.1016/j.neuron.2004.09.012. [DOI] [PubMed] [Google Scholar]
- 6.Neves G, Cooke SF, Bliss TV. Synaptic plasticity, memory and the hippocampus: A neural network approach to causality. Nat Rev Neurosci. 2008;9:65–75. doi: 10.1038/nrn2303. [DOI] [PubMed] [Google Scholar]
- 7.Richter JD, Klann E. Making synaptic plasticity and memory last: Mechanisms of translational regulation. Genes Dev. 2009;23:1–11. doi: 10.1101/gad.1735809. [DOI] [PubMed] [Google Scholar]
- 8.Guertin DA, Sabatini DM. Defining the role of mTOR in cancer. Cancer Cell. 2007;12:9–22. doi: 10.1016/j.ccr.2007.05.008. [DOI] [PubMed] [Google Scholar]
- 9.Ma XM, Blenis J. Molecular mechanisms of mTOR-mediated translational control. Nat Rev Mol Cell Biol. 2009;10:307–318. doi: 10.1038/nrm2672. [DOI] [PubMed] [Google Scholar]
- 10.Wullschleger S, Loewith R, Hall MN. TOR signaling in growth and metabolism. Cell. 2006;124:471–484. doi: 10.1016/j.cell.2006.01.016. [DOI] [PubMed] [Google Scholar]
- 11.Tang SJ, et al. A rapamycin-sensitive signaling pathway contributes to long-term synaptic plasticity in the hippocampus. Proc Natl Acad Sci USA. 2002;99:467–472. doi: 10.1073/pnas.012605299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hoeffer CA, Klann E. mTOR signaling: At the crossroads of plasticity, memory and disease. Trends Neurosci. 2010;33:67–75. doi: 10.1016/j.tins.2009.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Antion MD, et al. Removal of S6K1 and S6K2 leads to divergent alterations in learning, memory, and synaptic plasticity. Learn Mem. 2008;15:29–38. doi: 10.1101/lm.661908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Banko JL, et al. The translation repressor 4E-BP2 is critical for eIF4F complex formation, synaptic plasticity, and memory in the hippocampus. J Neurosci. 2005;25:9581–9590. doi: 10.1523/JNEUROSCI.2423-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ehninger D, et al. Reversal of learning deficits in a Tsc2+/- mouse model of tuberous sclerosis. Nat Med. 2008;14:843–848. doi: 10.1038/nm1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lang UE, et al. Immunosuppression using the mammalian target of rapamycin (mTOR) inhibitor everolimus: Pilot study shows significant cognitive and affective improvement. Transplant Proc. 2009;41:4285–4288. doi: 10.1016/j.transproceed.2009.08.050. [DOI] [PubMed] [Google Scholar]
- 17.Panja D, et al. Novel translational control in Arc-dependent long term potentiation consolidation in vivo. J Biol Chem. 2009;284:31498–31511. doi: 10.1074/jbc.M109.056077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sarbassov DD, et al. Prolonged rapamycin treatment inhibits mTORC2 assembly and Akt/PKB. Mol Cell. 2006;22:159–168. doi: 10.1016/j.molcel.2006.03.029. [DOI] [PubMed] [Google Scholar]
- 19.Hoeffer CA, et al. Removal of FKBP12 enhances mTOR-Raptor interactions, LTP, memory, and perseverative/repetitive behavior. Neuron. 2008;60:832–845. doi: 10.1016/j.neuron.2008.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bidinosti M, et al. Postnatal deamidation of 4E-BP2 in brain enhances its association with raptor and alters kinetics of excitatory synaptic transmission. Mol Cell. 2010;37:797–808. doi: 10.1016/j.molcel.2010.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gangloff YG, et al. Disruption of the mouse mTOR gene leads to early postimplantation lethality and prohibits embryonic stem cell development. Mol Cell Biol. 2004;24:9508–9516. doi: 10.1128/MCB.24.21.9508-9516.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Murakami M, et al. mTOR is essential for growth and proliferation in early mouse embryos and embryonic stem cells. Mol Cell Biol. 2004;24:6710–6718. doi: 10.1128/MCB.24.15.6710-6718.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kwon CH, et al. Pten haploinsufficiency accelerates formation of high-grade astrocytomas. Cancer Res. 2008;68:3286–3294. doi: 10.1158/0008-5472.CAN-07-6867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roses AD. Pharmacogenetics in drug discovery and development: A translational perspective. Nat Rev Drug Discov. 2008;7:807–817. doi: 10.1038/nrd2593. [DOI] [PubMed] [Google Scholar]
- 25.Spurr NK. Pharmacogenetic studies of epilepsy drugs: are we there yet? Trends Genet. 2006;22:250–252. doi: 10.1016/j.tig.2006.03.004. [DOI] [PubMed] [Google Scholar]
- 26.Eichelbaum M, Ingelman-Sundberg M, Evans WE. Pharmacogenomics and individualized drug therapy. Annu Rev Med. 2006;57:119–137. doi: 10.1146/annurev.med.56.082103.104724. [DOI] [PubMed] [Google Scholar]
- 27.Need AC, Motulsky AG, Goldstein DB. Priorities and standards in pharmacogenetic research. Nat Genet. 2005;37:671–681. doi: 10.1038/ng1593. [DOI] [PubMed] [Google Scholar]
- 28.Weinshilboum RM, Wang L. Pharmacogenetics and pharmacogenomics: Development, science, and translation. Annu Rev Genomics Hum Genet. 2006;7:223–245. doi: 10.1146/annurev.genom.6.080604.162315. [DOI] [PubMed] [Google Scholar]
- 29.Costa-Mattioli M, et al. Translational control of hippocampal synaptic plasticity and memory by the eIF2alpha kinase GCN2. Nature. 2005;436:1166–1173. doi: 10.1038/nature03897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Costa-Mattioli M, et al. eIF2alpha phosphorylation bidirectionally regulates the switch from short- to long-term synaptic plasticity and memory. Cell. 2007;129:195–206. doi: 10.1016/j.cell.2007.01.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.LeDoux JE. Emotion circuits in the brain. Annu Rev Neurosci. 2000;23:155–184. doi: 10.1146/annurev.neuro.23.1.155. [DOI] [PubMed] [Google Scholar]
- 32.Heitman J, Movva NR, Hall MN. Targets for cell cycle arrest by the immunosuppressant rapamycin in yeast. Science. 1991;253:905–909. doi: 10.1126/science.1715094. [DOI] [PubMed] [Google Scholar]
- 33.Zhang H, Stallock JP, Ng JC, Reinhard C, Neufeld TP. Regulation of cellular growth by the Drosophila target of rapamycin dTOR. Genes Dev. 2000;14:2712–2724. doi: 10.1101/gad.835000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jacinto E, et al. Mammalian TOR complex 2 controls the actin cytoskeleton and is rapamycin insensitive. Nat Cell Biol. 2004;6:1122–1128. doi: 10.1038/ncb1183. [DOI] [PubMed] [Google Scholar]
- 35.Sarbassov DD, et al. Rictor, a novel binding partner of mTOR, defines a rapamycin-insensitive and raptor-independent pathway that regulates the cytoskeleton. Curr Biol. 2004;14:1296–1302. doi: 10.1016/j.cub.2004.06.054. [DOI] [PubMed] [Google Scholar]
- 36.Gobert D, et al. Forskolin induction of late-LTP and up-regulation of 5′ TOP mRNAs translation via mTOR, ERK, and PI3K in hippocampal pyramidal cells. J Neurochem. 2008;106:1160–1174. doi: 10.1111/j.1471-4159.2008.05470.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Impey S, et al. Induction of CRE-mediated gene expression by stimuli that generate long-lasting LTP in area CA1 of the hippocampus. Neuron. 1996;16:973–982. doi: 10.1016/s0896-6273(00)80120-8. [DOI] [PubMed] [Google Scholar]
- 38.Alberini CM. The role of protein synthesis during the labile phases of memory: Revisiting the skepticism. Neurobiol Learn Mem. 2008;89:234–246. doi: 10.1016/j.nlm.2007.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nader K, Hardt O. A single standard for memory: The case for reconsolidation. Nat Rev Neurosci. 2009;10:224–234. doi: 10.1038/nrn2590. [DOI] [PubMed] [Google Scholar]
- 40.Nader K, Schafe GE, Le Doux JE. Fear memories require protein synthesis in the amygdala for reconsolidation after retrieval. Nature. 2000;406:722–726. doi: 10.1038/35021052. [DOI] [PubMed] [Google Scholar]
- 41.Sara SJ. Strengthening the shaky trace through retrieval. Nat Rev Neurosci. 2000;1:212–213. doi: 10.1038/35044575. [DOI] [PubMed] [Google Scholar]
- 42.Milekic MH, Alberini CM. Temporally graded requirement for protein synthesis following memory reactivation. Neuron. 2002;36:521–525. doi: 10.1016/s0896-6273(02)00976-5. [DOI] [PubMed] [Google Scholar]
- 43.Sehgal SN, Baker H, Vézina C. Rapamycin (AY-22,989), a new antifungal antibiotic. II. Fermentation, isolation and characterization. J Antibiot (Tokyo) 1975;28:727–732. doi: 10.7164/antibiotics.28.727. [DOI] [PubMed] [Google Scholar]
- 44.Vézina C, Kudelski A, Sehgal SN. Rapamycin (AY-22,989), a new antifungal antibiotic. I. Taxonomy of the producing streptomycete and isolation of the active principle. J Antibiot (Tokyo) 1975;28:721–726. doi: 10.7164/antibiotics.28.721. [DOI] [PubMed] [Google Scholar]
- 45.Gkogkas C, Sonenberg N, Costa-Mattioli M. Translational control mechanisms in long-lasting synaptic plasticity and memory. J Biol Chem. 2010;285:31913–31917. doi: 10.1074/jbc.R110.154476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sarbassov DD, Guertin DA, Ali SM, Sabatini DM. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science. 2005;307:1098–1101. doi: 10.1126/science.1106148. [DOI] [PubMed] [Google Scholar]
- 47.Kaeberlein M. Lessons on longevity from budding yeast. Nature. 2010;464:513–519. doi: 10.1038/nature08981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kwon CH, et al. Pten regulates neuronal arborization and social interaction in mice. Neuron. 2006;50:377–388. doi: 10.1016/j.neuron.2006.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ohno M, Frankland PW, Chen AP, Costa RM, Silva AJ. Inducible, pharmacogenetic approaches to the study of learning and memory. Nat Neurosci. 2001;4:1238–1243. doi: 10.1038/nn771. [DOI] [PubMed] [Google Scholar]
- 50.Brown EJ, et al. A mammalian protein targeted by G1-arresting rapamycin-receptor complex. Nature. 1994;369:756–758. doi: 10.1038/369756a0. [DOI] [PubMed] [Google Scholar]
- 51.Cafferkey R, et al. Dominant missense mutations in a novel yeast protein related to mammalian phosphatidylinositol 3-kinase and VPS34 abrogate rapamycin cytotoxicity. Mol Cell Biol. 1993;13:6012–6023. doi: 10.1128/mcb.13.10.6012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kunz J, et al. Target of rapamycin in yeast, TOR2, is an essential phosphatidylinositol kinase homolog required for G1 progression. Cell. 1993;73:585–596. doi: 10.1016/0092-8674(93)90144-f. [DOI] [PubMed] [Google Scholar]
- 53.Sabatini DM, Erdjument-Bromage H, Lui M, Tempst P, Snyder SH. RAFT1: A mammalian protein that binds to FKBP12 in a rapamycin-dependent fashion and is homologous to yeast TORs. Cell. 1994;78:35–43. doi: 10.1016/0092-8674(94)90570-3. [DOI] [PubMed] [Google Scholar]
- 54.Kaeberlein M, Kennedy BK. Protein translation, 2008. Aging Cell. 2008;7:777–782. doi: 10.1111/j.1474-9726.2008.00439.x. [DOI] [PubMed] [Google Scholar]
- 55.Blundell J, Kouser M, Powell CM. Systemic inhibition of mammalian target of rapamycin inhibits fear memory reconsolidation. Neurobiol Learn Mem. 2008;90:28–35. doi: 10.1016/j.nlm.2007.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bekinschtein P, et al. mTOR signaling in the hippocampus is necessary for memory formation. Neurobiol Learn Mem. 2007;87:303–307. doi: 10.1016/j.nlm.2006.08.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.