Abstract
Uridine insertion/deletion RNA editing in kinetoplastid mitochondria corrects encoded frameshifts in mRNAs. The genetic information for editing resides in small guide RNAs (gRNAs), which form anchor duplexes just downstream of an editing site and mediate editing within a single editing “block.” Many mRNAs require multiple gRNAs; the observed overall 3′ to 5′ polarity of editing is determined by the formation of upstream mRNA anchors by downstream editing. Hel61, a mitochondrial DEAD-box protein, was previously shown to be involved in RNA editing, but the functional role was not clear. Here we report that down-regulation of Hel61 [renamed REH1 (RNA editing helicase 1)] expression in Trypanosoma brucei selectively affects editing mediated by two or more overlapping gRNAs but has no effect on editing within a single block. Down-regulation produces an increased abundance of the gRNA/edited mRNA duplex for the first editing block of the A6 mRNA. Recombinant REH1 has an ATP-dependent double strand RNA unwinding activity in vitro with a model gRNA-mRNA duplex. These data indicate that REH1 is involved in gRNA displacement either directly by unwinding the gRNA/edited mRNA duplex or indirectly, to allow the 5′ adjacent upstream gRNA to form an anchor duplex with the edited mRNA to initiate another block of editing. Purified tagged REH1 is associated with the RNA editing core complex by RNA linkers and a colocalization of REH1, REL1, and two kinetoplast ribosomal proteins with the kinetoplast DNA was observed by immunofluorescence, suggesting that editing, transcription, and translation may be functionally linked.
Keywords: Leishmania, RNAi, tandem affinity purification, streptavidin binding and protein A purification
Uridine insertion/deletion RNA editing (1, 2) in the mitochondria of kinetoplastid protists is an essential process that results in the correction of encoded frameshifts thereby rendering the mRNAs translatable. The reaction involves the interaction of “preedited” mRNAs transcribed from maxicircle kinetoplast DNA with small gRNAs that act as templates for the precise insertion and deletion of uridylyl residues at multiple sites (3). The editing reaction is mediated by a holoenzyme or “editosome” (4). The core component of the editosome is the RECC (RNA editing core complex) (5) that contains approximately 15–20 polypeptides (2, 6–9). Eight RECC proteins have been so far functionally characterized. The remaining proteins contain a variety of motifs including zinc fingers, single-strand binding (SSB), and OB folds, and the roles of these proteins in the editing reaction are not known, although several are required for structural stability of the RECC. Low-resolution cryoEM 3D structures for RECC particles from Trypanosoma brucei and Leishmania tarentolae have been published (7, 10), but these do not have sufficient resolution to shed light on the precise mechanisms involved in the editing reactions.
Several multiprotein complexes have been described that interact with the RECC via RNA in substoichiometric amounts. These include the MRP1/2 complex (11, 12) and the guide RNA binding complex (GRBC)/mitochondrial RNA binding (MRB1) complex (13–16). Heterogeneous-sized high molecular weight complexes (L* or RECC*) consisting of the RECC together with several RNA-linked complexes have been visualized by electrophoresis in blue native gels (4). These break down with RNase treatment, giving rise to the 1-MDa RECC, and we have suggested that they may represent the holoenzyme (4).
The initial editing event occurs when a gRNA forms an RNA duplex with a complementary mRNA sequence just downstream of the editing site (3). A single gRNA encodes the information for several adjacent editing sites; this constitutes an “editing block.” RNA editing has a 3′ to 5′ polarity within a single block (3) due to the fact that the gRNA first forms a duplex anchor just downstream of an editing site. Insertion and deletion of u residues occurs at the first gRNA/mRNA mismatch. This extends the mRNA-gRNA duplex in a 5′ direction and editing is then reinitiated at the next upstream editing site. However, “misedited” sequences occur that are due in some cases to the hybridization of the incorrect gRNA and in other cases apparently to stochastic errors in the editing mechanism. Alternative editing has been identified in several mRNAs that contain multiple gRNA-mediated editing domains (17). This mechanism would greatly increase the repertoire of proteins, but confirmation of this phenomenon and its generality remain to be examined.
The editing of most mRNAs is mediated by multiple overlapping gRNAs. Editing utilizing the adjacent upstream gRNA cannot proceed until the first block is completely edited because the gRNA can only form an anchor duplex with edited sequence to initiate the second editing block. This is responsible for the observed overall 3′ to 5′ polarity (18), but the mechanism of displacement of adjacent gRNAs is unknown. An RNA helicase has been suggested to be involved in this process by displacing the initial gRNA, thereby allowing the adjacent upstream gRNA to form an anchor duplex with the edited sequence (19). Two trypanosome mitochondrial DExD/H-box proteins have been identified (14, 16, 20, 21) and shown to be involved in RNA editing. RNA editing helicase 2 or REH2 is a component of GRBC or MRB1 complexes and is involved in gRNA biogenesis (13, 14, 16). Hel61, another DEAD-box protein, was shown to be involved in RNA editing by a gene disruption experiment in T. brucei, which produced a slow growth phenotype and affected editing efficiency, and ectopic reexpression of Hel61 rescued a partially restored editing phenotype (21). However, gene disruption had no effect on either an experimentally observed mitochondrial RNA unwinding activity (20) or on full cycle in vitro editing reactions (21). Furthermore, the observed RNA unwinding activity did not cosediment with Hel61 (21). The functional role of Hel61 and the mechanism of gRNA displacement were not clear.
In this paper we present evidence that Hel61 is involved in the displacement of gRNAs to allow the 5′ adjacent gRNA to form an anchor duplex with the edited sequence. We also show that recombinant REH1 has ATP-dependent duplex RNA unwinding activity. We have therefore relabeled Hel61 with the functional name, REH1 (RNA editing helicase 1).
Results
Effect of Down-Regulation of REH1 Expression in T. brucei on Relative Abundance of mRNAs Edited in Block 1 Versus Those Edited in Two or More Blocks.
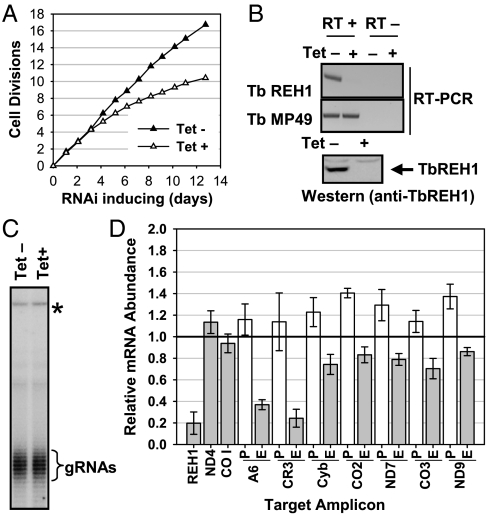
Down-regulation of expression of REH1 by conditional RNAi in T. brucei procyclic cells produced a slow growth phenotype (Fig. 1A). An 80% decrease in the abundance of REH1 mRNA by day 3 was demonstrated by RT-PCR and real-time RT-PCR (Fig. 1 B and D) and REH1 protein was not detectable by Western blot analysis (Fig. 1B). Repression of REH1 expression showed no effect on the stability or length of the gRNA 3′ oligo U tails (Fig. 1C), as was reported for the trypanosome REH2 mitochondrial RNA helicase (14, 16)
Fig. 1.
Effect of RNAi down-regulation of REH1 expression on editing in procyclic T. brucei cells. (A) Growth curve of T. brucei strain 2913 procyclic cells after induction of RNAi by addition of tetracycline. (B, Upper) RT-PCR of RNA from Tet+ and Tet− cells using primers for REH1. The RT− lanes are a control for contamination with DNA. RT-PCR of MP49 RNA was also performed as a loading control. (Lower) Western blot analysis of expression of REH1 in Tet+ and Tet− cells with anti-REH1 antibody. (C) Lack of effect of REH1 down-regulation on gRNAs. Total gRNA from control cells and cells 3 d after tetracycline addition was 5′ end labeled with [α-32P]GTP using guanylyl transferase. The gRNAs appear as ladders due to the heterogeneous 3′-oligo(U) tails. The top band marked with “*” is a cytosolic RNA that is also labeled and was used as a loading control. (D) Real-time RT-PCR analysis of relative abundance of preedited (clear bars) and mature edited (gray bars) mRNAs for several maxicircle genes from T. brucei procyclic cells after 3-d down-regulation of REH1 expression. The ND4 and COI mRNAs, which are not edited, are used as controls. The ordinate represents the relative abundance of each mRNA compared to that of the same RNA from Tet− cells (set at 1.0). The genes are indicated below each set of bars. Error bars indicate one standard deviation.
Real-time RT-PCR was also used to quantitate the effect of REH1 down-regulation on the relative abundance of several preedited and mature edited mRNAs (Fig. 1D and Table S1). The abundances of edited CR3 and A6 mRNAs were significantly reduced with down-regulation of REH1, but the effects on edited mRNAs for Cyb, ND7, CO3, and ND9 were small and probably not significant because a similar decrease was observed for the CO2 edited mRNA, which is mediated by a single in cis gRNA and does not require an overlapping gRNA. Interestingly, the abundances of preedited mRNAs for CO2 and ND9 were increased 30–40%, raising the possibility that REH1 has an effect on RNA turnover of some mRNAs. Two never-edited RNAs, ND4 and COI, were examined as controls: Neither showed a significant change in abundance. The changes in the abundances of the A6, CR3, Cyb, ND7, and CO3 preedited mRNAs were not statistically significant.
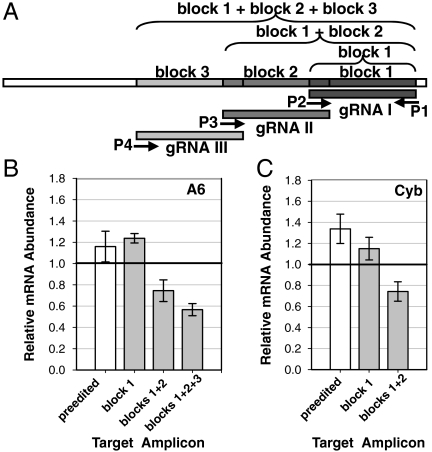
One possible mechanism for the inhibition of editing by REH1 down-regulation is that REH1 is involved with displacement of the gRNA from the edited mRNA/gRNA duplex. To test this hypothesis, we quantitatively compared the effect of down-regulation of REH1 expression on the relative abundance of mRNAs edited in block 1 versus those edited in block 1 and the adjacent upstream blocks. The experimental protocol is diagrammed in Fig. 2A (also see Table S1). The only T. brucei genes for which putative gRNAs are known for the 3′-most editing blocks are A6 and Cyb (Figs. S1 and S2) (22, 23). Real-time RT-PCR was used to measure the relative abundances of T. brucei partially edited A6 and Cyb mRNAs after 3 d of REH1 RNAi compared to these mRNAs in cells without induction of RNAi. Down-regulation caused an increase of around 20% in the A6 mRNAs completely edited in block 1 (and also the preedited A6 mRNAs). On the other hand, there was a decrease of around 20% in A6 mRNAs edited in both blocks 1 and 2 and a decrease of around 40% in A6 mRNAs edited in blocks 1, 2, and 3 (Fig. 2B). It was noted that the primers for edited A6 mRNA in Fig. 1D are located near the 5′ end of A6 gene. Repression of REH1 expression caused an increase of around 20–30% in both the preedited Cyb mRNA and Cyb mRNA completely edited in block 1 and caused a decrease of around 20% in the mature Cyb mRNA edited in both blocks 1 and 2 (there are only two editing blocks in Cyb) (Fig. 2C). Thus, down-regulation of REH1 expression in T. brucei selectively affects editing that is mediated by two or more overlapping gRNAs but does not affect editing occurring within a single gRNA-mediated editing block. This result is consistent with a role in gRNA displacement.
Fig. 2.
Effect of repression of REH1 T. brucei on relative abundances of A6 and Cyb mRNAs edited in block 1 versus mRNAs edited in both blocks 1 and 2 and 3. (A) Diagram of real-time RT-PCR analysis. Primer sets are indicated with arrows. See Table S1 for sequences. (B) Relative abundance of A6 mRNAs in T. brucei procyclic cells induced for down-regulation of expression of REH1 by RNAi. The abundance of these mRNAs from untreated cells is set at 1.0. Clear bar, preedited mRNA; gray bars, partially edited mRNAs. (C) Relative abundance of Cyb mRNAs in T. brucei procyclic cells induced for down-regulation of expression of REH1 by RNAi. See B for details. The difference between the effect of RNAi on the abundance of the A6 edited mRNA and the results in this experiment are due to the use of an upstream primer near the 5′ end of the mRNA in Fig. 1D instead of the primer used in this assay, which is close to the 3′ end.
Effect of Down-Regulation of REH1 Expression on Relative Abundance of Block 1 Edited mRNA/gRNA Duplex.
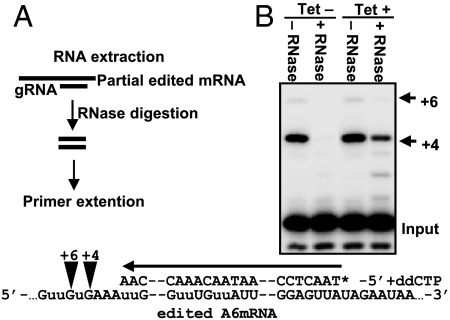
Another prediction is that REH1 down-regulation would produce an increased abundance of a block 1-fully edited mRNA/gRNA duplex by inhibiting the progression of editing from block 1 to block 2. The relative abundance of this duplex was assayed by an RNase protection experiment in which poisoned primer extension using a primer complementary to fully edited block 1 mRNA was used to measure the abundance of the RNase-protected duplex RNA. The experimental protocol is diagrammed in Fig. 3A. As shown in Fig. 3B, lanes 2 and 4, there is a significant increase in the relative abundance of the RNase-protected block 1 duplex after down-regulation of REH1 expression. Although the primer covers two U-deletion and two U-insertion sites, there is no significant decrease in the no RNase control band in lanes 1 and 3, indicating that down-regulation of REH1 has no effect on intrablock editing. We conclude that REH1 is required for releasing gRNA from A6 mRNA that is completely edited in block 1.
Fig. 3.
Relative abundance of the T. brucei gRNA I/edited A6 block 1 mRNA duplex after RNAi down-regulation of REH1 expression. (A) Diagram of assay. The abundance is assayed by poisoned primer extension of RNase-protected A6 mRNA. (B) Poisoned primer extension assay of RNase-resistant A6 block 1 duplex RNA with and without down-regulation of REH1 expression. Note the presence of a +4 labeled band only in the +Tet, +RNase lane. (Lower) Diagram of the extension reaction.
Recombinant Streptavidin-Binding Motif (SBP)-tagged REH1 has RNA-dependent ATPase and Double Strand RNA Unwinding Activities.
REH1 has conserved motifs found in DEAD-box proteins (Fig. 4A and Fig. S3), but there is no evidence in the literature for REH1 having RNA helicase activity. To address this question, we expressed tagged Leishmania major (Lm) REH1 (Fig. 4B) in Sf9 cells and analyzed the activity. A modified tandem affinity purification (TAP) tag (24), the streptavidin binding and protein A purification (SAP) tag (Fig. 4B), was used in which the calmodulin binding peptide motif was substituted with a high affinity streptavidin binding peptide (SBP) motif. The rREH1 was purified to homogeneity by IgG-Sepharose binding, MonoS ion-exchange chromatography, and streptavidin affinity chromatography (Fig. 4C, lane 1). The expressed band was confirmed to be SBP-tagged REH1 by Western analysis using anti-REH1 or HRP-conjugated streptavidin (SA-HRP) (Fig. 4C, lanes 2, 3). His6x-tagged REH1 was also expressed in Escherichia coli and purified to homogeneity for antibody generation.
Fig. 4.
Purification, RNA-dependent ATPase activity, and ATP-dependent double strand RNA unwinding activity of recombinant SBP-tagged REH1. (A) Diagram showing the conserved motifs of REH1. Open boxes represent the conserved helicase motifs with the indicated the amino acid sequences. The number of amino acids separating the motifs is shown. (B) Diagram of the SAP-tagged Lm REH1. SBP, streptavidin-binding peptide; PrA, protein (A). (C) Purification of SBP-tagged rREH1 expressed in sf9 insect cells using the Baculovirus expression system (Invitrogen). SDS gels stained with Sypro. Blots probed with SA-HRP or anti-REH1 antibody. (D) RNA-dependent ATPase activity of purified SBP-tagged REH1. The ATPase reactions were performed without added RNA, with added poly U, or with added poly U plus 0.05 mg/mL RNase. (E) Model gRNA/mRNA substrate with a 46-bp duplex and a 49-nt 3′ overhang. The 32P-labeled end is indicated by * in the diagram. The unwinding reaction required ATP and showed a dose response with the amount of protein. (F) Model gRNA/mRNA substrate with a 15-bp duplex and a 24-nt 3′ overhang; 5 nM rREH1. (G) 22-bp duplex with 15-nt 5′ overhang; 5 nM rREH1. (H) 15-bp blunt end duplex; 5 nM rREH1.
The purified SBP-tagged recombinant REH1 protein showed a robust poly U–stimulated ATPase activity. This activity was destroyed by RNase treatment (Fig. 4D). As a control for ATPase contamination from the Sf9 cells, SAP-tagged RET2 RECC protein was expressed and purified using the same procedure. The tagged RET2 did not show ATPase activity (Fig. 4D). The REH1 ATPase activity requires ATP or dATP (Fig. S4 D and E). The optimal pH is around 8.3, the Km for the reaction is 620 μM ATP, and the Kcat is 82.3 min-1 (Fig. S4 A–C).
Recombinant REH1 showed an ATP-dependent double strand RNA unwinding activity using a partially edited model gRNA/edited mRNA duplex (Fig. 4E and Table S2). The reaction required ATP and showed a protein dose response (Fig. 4 E and F). We also tested the unwinding of shorter RNA duplexes with 5′ or 3′ overhangs or no overhang (Fig. 4 F–H). The latter were unwound more efficiently probably due to their lower stability. It has been shown that helicase activity in vitro shows an inverse relationship between duplex stability and unwinding (25). As a control, a mutation of K143A in motif I, which is the conserved ATP binding domain of REH1, was introduced and the mutant protein lost ATPase and unwinding activities (Fig. S5). These data indicate that REH1 is an ATP-dependent RNA helicase. The REH1 helicase apparently differs from most other DEAD-box RNA helicases that exhibit very poor in vitro unwinding activity and require a 5′ or 3′ single-stranded RNA region for unwinding activity (25).
REH1, REL1, and the Kinetoplast Ribosome Proteins, L3 and S17, Show Concentration in the Kinetoplast Region of the Mitochondrion.
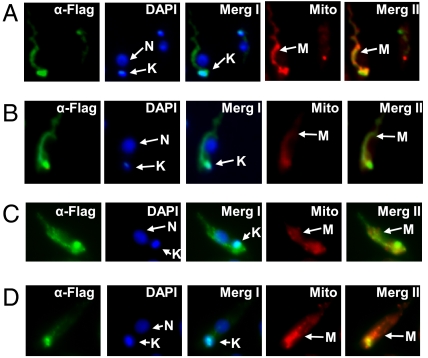
Immunolocalization of SAP-tagged REH1 within the single mitochondrion of transfected L. tarentolae cells was performed using an antibody against the C-terminal FLAG epitope (Fig. 5A). This antibody does not have cross-reactivity with any endogenous protein (Fig. S6D). The cells were stained with MitoTracker Red to visualize the single mitochondrion and costained with DAPI to detect the nuclear and kinetoplast DNA, and then treated with anti-FLAG antibody and secondary anti-IgG antibody conjugated with Alexa Fluor 488 to visualize SAP-tagged REH1. The REH1 protein colocalized with the MitoTracker mitochondrial image. Interestingly, in addition to a dispersed localization throughout the tubular mitochondrion there is an apparent concentration of REH1 protein in the kinetoplast DNA-containing region. Lower resolution images of entire fields that show that the selected fields in Fig. 5A are representative are shown in Fig. S6A. As a control, cells were analyzed for the localization of glutamate dehydrogenase, a soluble mitochondrial protein not involved in RNA editing (Fig. S6C). These cells showed a somewhat punctate distribution of immunofluorescence throughout the mitochondrion without the level of concentration in the kinetoplast region observed in the REH1 immunofluorescence, indicating that the observed kinetoplast concentration of REH1 is probably not artifactual (Fig. S6).
Fig. 5.
Intracellular localization in late log phase L. tarentolae of SAP-tagged Lm REH1, TAP-tagged REL1, and the S17 and L3 TAP-tagged kinetoplast ribosome proteins. (A) Representative cells expressing SAP-tagged REH1. Cells were stained with mouse anti-FLAG antibody + Alexa Fluor 488 conjugated F(ab) fragment (α-FLAG, green) to visualize the SAP-tagged REH1. DAPI was used to visualize the nucleus and kinetoplast, and MitoTracker CMXRos was used to visualize the entire mitochondrion. A merge of the FLAG and DAPI panels is shown as MergI and a merge of the FLAG and MitoTracker panels is shown as MergII. N: nucleus, K: kinetoplast, M: mitochondrion. (B) Representative cell expressing TAP-tagged REL1. See legend in A for details. Images of entire fields are shown in Fig. S6. (C) A cell expressing TAP-tagged mitochondrial ribosomal protein S17. (D) A cell expressing TAP-tagged mitochondrial ribosomal protein L3.
The localizations of TAP-tagged REL1 RNA ligase, a RECC core component, and two TAP-tagged kinetoplast ribosome proteins (Fig. S6E), S17 and L3, were also analyzed by indirect immunofluorescence. REL1 and the two ribosome proteins showed a similar kinetoplast concentration as the SAP-tagged REH1 (Fig. 5 B–D and Fig. S6B).
REH1 Is Associated with the RECC by RNA Linkers.
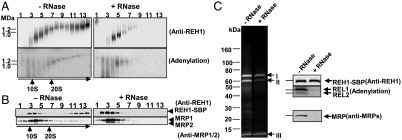
To investigate possible interactions between REH1 and the RECC, SAP-tagged Lm REH1 with a mitochondrial target signal was expressed in L. tarentolae. The tagged REH1 was found only in the whole cell and mitochondrial fractions and not in the cytosol fraction (Fig. S7A). Cell lysate was allowed to bind to IgG-Sepharose, and the bound material was released by digestion with tobacco etch virus (TEV) protease. Half of the eluted material was treated with RNase A prior to fractionation on a glycerol gradient. Gradient fractions were subjected to blue native gel electrophoresis and the blots probed with anti-REH1 antibody (Fig. 6A, Upper).
Fig. 6.
RNA-dependent association of REH1 with the RECC. (A) Clarified lysate from L. tarentolae cells expressing SAP-tagged REH1 was bound to IgG-Sepharose and the SBP-tagged REH1 released with TEV protease. Half was treated with RNase A (0.1 mg/mL). After glycerol gradient sedimentation, fractions were run in blue native gels, which were blotted and probed with anti-REH1. Aliquots of fractions were also autoadenylated with α[32P]ATP to label the REL1 RECC protein prior to blue native gel analysis. These gels were dried and exposed to a phosphoimager screen. (B) Gradient fractions from A were run in SDS gels, which were blotted and probed with anti-REH1 or anti-MRP1/2. (C) The TEV-released material in A, with or without RNase treatment, was bound to streptavidin–Sepharose and the SBP-tagged REH1 released with 2 mM biotin. The SBP-tagged REH1 was run in SDS gels, which were autoadenylated and stained with Sypro (Invitrogen), or blotted and probed with anti-REH1 or anti-MRP1/2. Note that the labeled REL1/REL2 and the MRP1/2 proteins were detected only prior to RNase treatment. The three major stained bands were subjected to mass spectrometry (data shown in Table S3).
Autoadenylation of REL1/2 with [α-32P]ATP (26) was also used to detect REL1 as a marker for the RECC (Fig. 6A, Lower). The gradient fractions were run in SDS gels that were probed with anti-REH1 antibody and with anti-MRP1/2 antibody (Fig. 6B). MRP1/2 are proteins that are known to associate with the RECC via RNA linkers (11).
The tagged REH1 sedimented as free oligomeric protein in the 5–10S region. A portion of the REH1 sedimented in the 20–25S region migrating in the blue native gel at ∼1 MDa (Fig. 6 A and B, Left) and a substantial amount sedimented in the > 25S region, migrating in the blue native gel at > 1 MDa (Fig. 6A, Left). The high molecular weight material was sensitive to RNase as were the REH1-containing RECC materials in fractions 10–13 (Fig. 6A, Right). This can also be seen in SDS gels of the same gradient fractions (Fig. 6B). The RNase-sensitive high molecular weight material is reminiscent of the previously reported RECC* particles that bound substoichiometric amounts of several other editing complexes (4). This evidence suggests that a portion of the tagged REH1 protein is associated with the RECC* particles via RNA linkers.
Additional evidence for an association of REH1 with the RECC was obtained by TAP purification, performed with or without RNase pretreatment (Fig. 6C). The REL1/REL2 internal RECC proteins and the MRP1/2 proteins were detected by Western analysis in the pull-down prior to RNase treatment but not after such treatment (Fig. 6C, Right). The coprecipitated REL1 and REH1 proteins were in substoichiometric amounts because they could not be visualized in the SDS gel by Sypro staining (Fig. 6C, Left). The three visible Sypro-stained bands in the REH1 TAP pull-down were identified by mass spectrometry as the SPB-tagged REH1 (II), a 10-kDa REH1 C-terminal breakdown product (III), and a 70-kD band (I) containing two contaminating proteins: TEV protease and a cytosolic DEAD-box protein (Fig. 6C and Table S3). The presence of this cytosolic helicase in the REH1-TAP pull-down should be investigated further.
Discussion
REH1 is essential as shown by the growth phenotype caused by down-regulation of REH1 expression. The partial growth inhibition could be due to incomplete down-regulation, a slow turnover of the protein or an enzyme redundancy. Down-regulation also causes a decrease in abundance of edited mRNAs for the six genes assayed, with editing of the A6 and CR3 mRNAs being the most affected. Possible roles of the REH1 helicase in RNA editing are (i) to directly assist in the processive editing reaction itself within a single gRNA-mediated block as editing proceeds from site to site or (ii) to be involved in gRNA displacement between adjacent editing blocks. Model 1 is ruled out by the observed lack of effect of REH1 down-regulation on intrablock editing. Model 2 is supported by the significant decrease in the relative abundance of mRNAs edited in two or more adjacent blocks by REH1 down-regulation as compared to those edited in a single block, and by the accumulation of the gRNA/block 1 edited A6 mRNA duplex. These data suggest that REH1 is involved with the progression of editing from one gRNA-mediated editing block to the next adjacent upstream block.
The mechanism of this involvement, however, is not clear. The fact that recombinant REH1 protein has gRNA/mRNA duplex unwinding activity in vitro may suggest an in vivo role in directly unwinding the mRNA/gRNA duplex formed by the editing of all the sites mediated by a single gRNA and liberating an edited mRNA single strand available for hybridization of the adjacent gRNA. However, in the case of REH1, the evidence does not distinguish between a direct effect on unwinding of the gRNA/mRNA duplex or an indirect effect, such as described for eIF4AIII, a core component of the exon junction complex, and some putative RNA helicases that are involved in snoRNA release from preribosomes (27–31). Another potential function of REH1 could be to unwind cis elements within the preedited mRNA. A dominant-negative mutation of REH1 and ectopic reexpression of wild-type REH1 might indeed help to explore the role of REH1 in vivo. In fact, the evidence that the largest effect of REH1 down-regulation on editing is on the pan-edited A6 and CR3 mRNAs suggests that the in vivo activity of REH1 is substrate-specific and perhaps is regulated by transient binding of cofactors. Along this line, no detectable in vitro unwinding activity had been observed with T. brucei gradient fractions that showed a peak of Hel61 (REH1) by blot analysis (21) nor in the Lm REH1 SAP pull-down (Fig. S7B). These data suggest the possibility of regulation of helicase activity in vivo.
There have been conflicting reports on the association of REH1 with the RECC. Tb REH1 was detected by mass spectrometry analysis in an MP63-immunoprecipitated sample and also in a RECC preparation isolated by ion-exchange chromatography (32–34). However, REH1 was not detected in REN1-, REN2-, or REN3-TAP pull-downs from T. brucei (32, 35), nor in REL1-TAP and MP44-TAP pull-downs from L. tarentolae (7, 11). These results could be explained by our finding that REH1 is associated with the RECC by RNA linkers, as are several other editing-associated complexes (6, 11), and that this linkage is easily disrupted during the isolation.
Immunofluorescence of REH1 showed a concentration in the kinetoplast DNA (kDNA) region of the mitochondrion. Interestingly, the REL1 RNA ligase, a component of the RECC, and the S17 and L3 kinetoplast ribosome proteins also exhibited a concentration in the kDNA region of the mitochondrion, suggesting the intriguing possibility of a physical linkage of kDNA transcription, translation, and editing pathways, but this remains to be investigated. Our localization results differ somewhat from several previous studies that showed that editing proteins are distributed throughout the mitochondrion with no apparent concentration (36, 37) and that the RNA-linked editing proteins, GAP1 and GAP2, are localized in discrete particles throughout the mitochondrion (14). These differences could be species-dependent or could be due to technical details.
More work is required to fully understand the precise role of the REH1 helicase in RNA editing, but it is clear from the results in this paper that REH1 is involved in the 3′ to 5′ polarity of editing in a multi-gRNA-mediated editing domain.
Materials and Methods
SI Text contains details of plasmid construction, real-time PCR, poisoned primer extension, purification of SAP-tagged Lm REH1, ATPase activity assay, immunofluorescent localization of REH1, REL1, S17, L3, and glutamate dehydrogenase, mitochondrial importation of L3-TAP and S17–TAP proteins, and sedimentation analysis of kinetoplast ribosomal complexes. The SI Text tables contain sequences of DNA primers and RNA oligonucleotides used in the assays, and sequences of peptides derived from protein bands in Fig. 3C
Supplementary Material
Acknowledgments.
We thank Bob Nelson for the original construction of the SAP vector in our laboratory, Kent Hill for use of a fluorescence microscope, Stephen Smale and Jian Xu for the use of a Real-Time PCR apparatus, and Agda Simpson and Kestrel Rogers for generation of antibodies. The antibody against Tb REH1 was provided by Ulrich Göringer (Technical University of Darmstadt, Darmstadt, Germany) and the 2913 strain of T. brucei was provided by George Cross (Rockfeller University, New York). We thank David King for mass spectrometry of gel bands to identify proteins. This work was supported by National Institutes of Health Grant AI09102 (to L.S.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1014152108/-/DCSupplemental.
References
- 1.Simpson L, Sbicego S, Aphasizhev R. Uridine insertion/deletion RNA editing in trypanosome mitochondria: A complex business. RNA. 2003;9:265–276. doi: 10.1261/rna.2178403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stuart KD, Schnaufer A, Ernst NL, Panigrahi AK. Complex management: RNA editing in trypanosomes. Trends Biochem Sci. 2005;30:97–105. doi: 10.1016/j.tibs.2004.12.006. [DOI] [PubMed] [Google Scholar]
- 3.Blum B, Bakalara N, Simpson L. A model for RNA editing in kinetoplastid mitochondria: “Guide” RNA molecules transcribed from maxicircle DNA provide the edited information. Cell. 1990;60:189–198. doi: 10.1016/0092-8674(90)90735-w. [DOI] [PubMed] [Google Scholar]
- 4.Osato D, et al. Uridine insertion/deletion RNA editing in trypanosomatid mitochondria: In search of the editosome. RNA. 2009;15:1338–1344. doi: 10.1261/rna.1642809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Simpson L, Aphasizhev R, Lukes J, Cruz-Reyes J. Guide to the nomenclature of kinetoplastid RNA editing: A proposal. Protist. 2010;161:2–6. doi: 10.1016/j.protis.2009.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aphasizhev R, et al. Isolation of a U-insertion/deletion editing complex from Leishmania tarentolae mitochondria. EMBO J. 2003;22:913–924. doi: 10.1093/emboj/cdg083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li F, et al. Structure of the core editing complex (L-complex) involved in uridine insertion/deletion RNA editing in trypanosomatid mitochondria. Proc Natl Acad Sci USA. 2009;106:12306–12310. doi: 10.1073/pnas.0901754106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Simpson L, Aphasizhev R, Gao G, Kang X. Mitochondrial proteins and complexes in Leishmania and Trypanosoma involved in U-insertion/deletion RNA editing. RNA. 2004;10:159–170. doi: 10.1261/rna.5170704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Worthey EA, Schnaufer A, Mian IS, Stuart K, Salavati R. Comparative analysis of editosome proteins in trypanosomatids. Nucleic Acids Res. 2003;31:6392–6408. doi: 10.1093/nar/gkg870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Golas MM, et al. Snapshots of the RNA editing machine in trypanosomes captured at different assembly stages in vivo. EMBO J. 2009;28:766–778. doi: 10.1038/emboj.2009.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aphasizhev R, Aphasizheva I, Nelson RE, Simpson L. A 100-kD complex of two RNA-binding proteins from mitochondria of Leishmania tarentolae catalyzes RNA annealing and interacts with several RNA editing components. RNA. 2003;9:62–76. doi: 10.1261/rna.2134303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schumacher MA, Karamooz E, Zikova A, Trantirek L, Lukes J. Crystal structures of T. brucei MRP1/MRP2 guide-RNA binding complex reveal RNA matchmaking mechanism. Cell. 2006;126:701–711. doi: 10.1016/j.cell.2006.06.047. [DOI] [PubMed] [Google Scholar]
- 13.Weng J, et al. Guide RNA-binding complex from mitochondria of trypanosomatids. Mol Cell. 2008;32:198–209. doi: 10.1016/j.molcel.2008.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hashimi H, Čičová Z, Novotná L, Wen YZ, Lukeš J. Kinetoplastid guide RNA biogenesis is dependent on subunits of the mitochondrial RNA binding complex 1 and mitochondrial RNA polymerase. RNA. 2009;15:588–599. doi: 10.1261/rna.1411809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Acestor N, Panigrahi AK, Carnes J, Zikova A, Stuart KD. The MRB1 complex functions in kinetoplastid RNA processing. RNA. 2009;15:277–286. doi: 10.1261/rna.1353209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hernandez A, et al. REH2 RNA helicase in kinetoplastid mitochondria: Ribonucleoprotein complexes and essential motifs for unwinding and guide RNA (gRNA) binding. J Biol Chem. 2010;285:1220–1228. doi: 10.1074/jbc.M109.051862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ochsenreiter T, Hajduk SL. Alternative editing of cytochrome c oxidase III mRNA in trypanosome mitochondria generates protein diversity. EMBO Rep. 2006;7:1128–1133. doi: 10.1038/sj.embor.7400817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Feagin JE, Abraham J, Stuart K. Extensive editing of the cytochrome c oxidase III transcript in Trypanosoma brucei. Cell. 1988;53:413–422. doi: 10.1016/0092-8674(88)90161-4. [DOI] [PubMed] [Google Scholar]
- 19.Maslov DA, Simpson L. The polarity of editing within a multiple gRNA-mediated domain is due to formation of anchors for upstream gRNAs by downstream editing. Cell. 1992;70:459–467. doi: 10.1016/0092-8674(92)90170-h. [DOI] [PubMed] [Google Scholar]
- 20.Missel A, Goringer HU. Trypanosoma brucei mitochondria contain RNA helicase activity. Nucleic Acids Res. 1994;22:4050–4056. doi: 10.1093/nar/22.20.4050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Missel A, Souza AE, Norskau G, Goringer HU. Disruption of a gene encoding a novel mitochondrial DEAD-box protein in Trypanosoma brucei affects edited mRNAs. Mol Cell Biol. 1997;17:4895–4903. doi: 10.1128/mcb.17.9.4895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hong M, Simpson L. Genomic organization of Trypanosoma brucei kinetoplast DNA minicircles. Protist. 2003;154:265–279. doi: 10.1078/143446103322166554. [DOI] [PubMed] [Google Scholar]
- 23.Ochsenreiter T, Cipriano M, Hajduk SL. KISS: The kinetoplastid RNA editing sequence search tool. RNA. 2007;13:1–4. doi: 10.1261/rna.232907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Puig O, et al. The tandem affinity purification (TAP) method: A general procedure of protein complex purification. Methods. 2001;24:218–229. doi: 10.1006/meth.2001.1183. [DOI] [PubMed] [Google Scholar]
- 25.Cordin O, Banroques J, Tanner NK, Linder P. The DEAD-box protein family of RNA helicases. Gene. 2006;367:17–37. doi: 10.1016/j.gene.2005.10.019. [DOI] [PubMed] [Google Scholar]
- 26.Peris M, et al. Characterization of two classes of ribonucleoprotein complexes possibly involved in RNA editing from Leishmania tarentolae mitochondria. EMBO J. 1994;13:1664–1672. doi: 10.1002/j.1460-2075.1994.tb06430.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Andersen CB, et al. Structure of the exon junction core complex with a trapped DEAD-box ATPase bound to RNA. Science. 2006;313(5795):1968–1972. doi: 10.1126/science.1131981. [DOI] [PubMed] [Google Scholar]
- 28.Bono F, Ebert J, Lorentzen E, Conti E. The crystal structure of the exon junction complex reveals how it maintains a stable grip on mRNA. Cell. 2006;126:713–725. doi: 10.1016/j.cell.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 29.Kos M, Tollervey D. The putative RNA helicase Dbp4p is required for release of the U14 snoRNA from preribosomes in saccharomyces cerevisiae. Mol Cell. 2005;20:53–64. doi: 10.1016/j.molcel.2005.08.022. [DOI] [PubMed] [Google Scholar]
- 30.Liang XH, Fournier MJ. The helicase Has1p is required for snoRNA release from pre-rRNA. Mol Cell Biol. 2006;26:7437–7450. doi: 10.1128/MCB.00664-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bohnsack MT, Kos M, Tollervey D. Quantitative analysis of snoRNA association with pre-ribosomes and release of snR30 by Rok1 helicase. EMBO Rep. 2008;9(12):1230–1236. doi: 10.1038/embor.2008.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Panigrahi AK, et al. Compositionally and functionally distinct editosomes in Trypanosoma brucei. RNA. 2006;12:1038–1049. doi: 10.1261/rna.45506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Panigrahi AK, Allen TE, Stuart K, Haynes PA, Gygi SP. Mass spectrometric analysis of the editosome and other multiprotein complexes in Trypanosoma brucei. J Am Soc Mass Spectr. 2003;14:728–735. doi: 10.1016/S1044-0305(03)00126-0. [DOI] [PubMed] [Google Scholar]
- 34.Stuart K, Panigrahi AK, Schnaufer A. Identification and characterization of trypanosome RNA-editing complex components. Methods Mol Biol. 2004;265:273–291. doi: 10.1385/1-59259-775-0:273. [DOI] [PubMed] [Google Scholar]
- 35.Carnes J, Trotter JR, Peltan A, Fleck M, Stuart K. RNA editing in Trypanosoma brucei requires three different editosomes. Mol Cell Biol. 2008;28:122–130. doi: 10.1128/MCB.01374-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Oberholzer M, Morand S, Kunz S, Seebeck T. A vector series for rapid PCR-mediated C-terminal in situ tagging of Trypanosoma brucei genes. Mol Biochem Parasitol. 2006;145:117–120. doi: 10.1016/j.molbiopara.2005.09.002. [DOI] [PubMed] [Google Scholar]
- 37.Panigrahi AK, et al. Association of two novel proteins, TbMP52 and TbMP48, with the Trypanosoma brucei RNA editing complex. Mol Cell Biol. 2001;21:380–389. doi: 10.1128/MCB.21.2.380-389.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.