Abstract
The discovery of RNAi has revolutionized loss-of-function genetic studies in mammalian systems. However, significant challenges still remain to fully exploit RNAi for mammalian genetics. For instance, genetic screens and in vivo studies could be broadly improved by methods that allow inducible and uniform gene expression control. To achieve this, we built the lentiviral pINDUCER series of expression vehicles for inducible RNAi in vivo. Using a multicistronic design, pINDUCER vehicles enable tracking of viral transduction and shRNA or cDNA induction in a broad spectrum of mammalian cell types in vivo. They achieve this uniform temporal, dose-dependent, and reversible control of gene expression across heterogenous cell populations via fluorescence-based quantification of reverse tet-transactivator expression. This feature allows isolation of cell populations that exhibit a potent, inducible target knockdown in vitro and in vivo that can be used in human xenotransplantation models to examine cancer drug targets.
Keywords: lentivirus, vector
Loss-of-function studies represent a powerful means by which to gain insight into the molecular mechanisms behind complex biological processes. Until recently, these studies were practical for only a small set of genetically tractable model organisms (1, 2). The discovery of gene silencing through RNAi has made it possible to carry out loss-of-function studies in mammals. RNAi has transformed the way in which gene function can be investigated and has quickly become a versatile tool for a wide range of applications, including reverse genetics, high-throughput screens, and therapeutics (3). The application of RNAi technology both in vitro and in vivo has tremendous potential to further our knowledge of the molecular mechanisms that underpin both normal biology and human disease.
To achieve efficient and long-term gene silencing, we have previously generated the retroviral-encoded microRNA-based shRNA PRIME (potent RNA interference using microRNA expression) vectors that are effective in suppressing expression of their intended gene targets from single proviral integrations (4). These vectors use the miR30 backbone and are expressed from RNA polymerase II promoters, which allows for exquisite regulation of shRNA expression. We have further generated genome-wide shRNA libraries in these vectors that cover large numbers of mammalian transcripts for systematic study of gene function (5, 6). These shRNA libraries, as well as shRNA libraries generated by other laboratories (7, 8), have been used to perform genetic screens in mammalian cell culture models for a variety of phenotypes, including cell transformation, synthetic lethal interactions, and resistance to chemotherapeutic treatments (9–15).
Despite the success demonstrated by the above constitutive shRNA vectors, an inducible shRNA system would have obvious advantages in many experimental settings. First, the inducible shRNA system allows for the study of essential genes. Because constitutively expressed shRNAs targeting essential genes will be toxic, the ability to control the timing and levels of shRNA expression would be extremely valuable. Second, an inducible shRNA system allows for strict isogenicity within a given experiment. Many cultured mammalian cells, in particular cancer cell lines, are genetically or epigenetically heterogeneous in a manner that can influence the expression of transgenes from a constitutive promoter. The ability to uniformly control expression of the gene of interest in the population of cells can minimize experimental variations due to clonal heterogeneity. Third, an inducible system will provide temporal and reversible control of gene expression.
Although inducible systems provide many advantages, a key problem when introducing inducible vectors into cells is the heterogeneity in transgene expression levels among cells due to differences in the position of their integration (16). Those cells carrying their own reverse tet-transactivator (rtTA) gene can also show variable inducibility due to different rtTA expression levels, depending upon location. To address these issues and broaden the applicability of shRNA vectors and libraries, we developed a third-generation inducible shRNA expression lentiviral system, pINDUCER. We designed the pINDUCER lentiviral system to have the following properties: (i) it is a single-vector system, so there is no need to use specialized cell lines or multiple vectors; (ii) it has minimal basal expression of shRNAs and high inducibility in polyclonal populations, so gene silencing can be tightly controlled; (iii) it has reporters for tracking transduced cells and the shRNA-expressing cells; (iv) it entails a cistron encoding both rtTA and a sortable marker in the same transcription unit, thus enabling cells with higher rtTA expression and shRNA inducibility to be identified and isolated in polyclonal populations; (v) it is compatible with all of our previous generations of shRNA vectors, so that the shRNAs and libraries can be easily transferred between different vectors; and (vi) it is functional both in cultured cells and in vivo. We show that these high-expressing vectors can function in live animals to limit tumor cell proliferation in xenograft experiments. Thus, the pINDUCER system will greatly facilitate functional genetic studies in mammals.
Results
Next-Generation Lentiviral Vector Series for Inducible RNAi or cDNA Expression.
Current lentiviral technology has facilitated a wide range of loss- and gain-of-function studies. However, the exploitation of these technologies for inducible control of gene function in vivo has remained a major challenge across a broad spectrum of models of mammalian development and cancer. Specifically, the power of inducible lentiviral tools in vivo has been limited by inconsistent inducibility in various cell types and across polyclonal populations. To address these limitations, we generated an inducible vector series (pINDUCER; Fig. 1A) that is broadly inducible in vitro and also elicits robust inducible cDNA and shRNA expression in vivo. Based on previous systems, this lentiviral-based system encodes rtTA3 and an interchangeable marker as a constitutive, bicistronic transcript (4, 17). Upon the addition of doxycycline (dox), transcription of the turboRFP (tRFP)-shRNA cassette or the cDNA is activated. This system has the advantage over previous vectors of easy detection of cells with activated shRNA transcription, as well as the ability to sort cells with viruses that have landed in regions optimal for expression of rtTA3, which we find increases inducibility (see below). This system allows easy cloning and transfer of shRNAs from our previous versions of the Mir30-based shRNA vectors (SM2C, MSCV-PM, GIPZ) or the cloning of cDNAs from Gateway entry vectors.
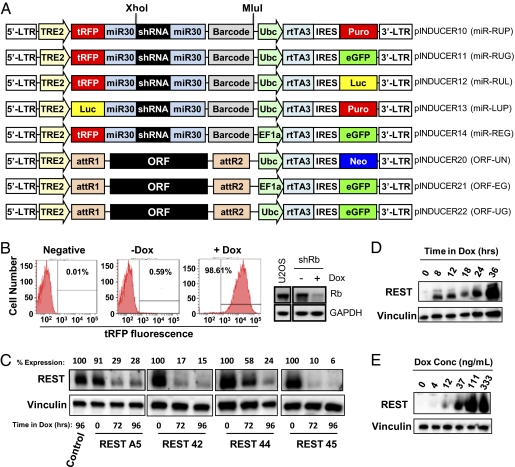
Fig. 1.
pINDUCER lentivirus elicits inducible cDNA and shRNA expression. (A) Diagrams of the pINDUCER vector series. R, tRFP; U, Ubc promoter; E, EF1-α promoter; L, luciferase; G, eGFP; P, puromycin resistance; N, neomycin resistance. (B) pINDUCER10 elicits robust cDNA expression and RNAi across polyclonal populations. U2OS osteosarcoma cells were transduced [multiplicity of infection (MOI) = 0.5] with pINDUCER10-shRB, selected for puromycin resistance, and cultured with or without dox for 4 d. Cells were analyzed for the tRFP fluorescence by flow cytometry (Left) or expression of Rb or Gapdh (loading control) proteins by Western blot (Right). (C) pINDUCER10 elicits effective RNAi in nontransformed cells. Multiple shRNAs targeting Rest were subcloned into pINDUCER10. Polyclonal populations of HMECs transduced with the indicated lentiviral shRNA were cultured with or without dox for the indicated times, and Rest or Vinculin (loading control) levels were determined by Western blot. Control indicates a nontargeting shRNA. (D) Kinetics of cDNA induction with pINDUCER20 (ORF-UN). Polyclonal populations of SW1417 colon cancer cells transduced with pINDUCER20-REST were cultured with or without dox for the indicated times, and Rest or Vinculin (loading control) protein levels were determined by Western blot. (E) Dose-dependent cDNA induction with pINDUCER20. Cells from D were cultured with the indicated dox concentration for 48 h, and Rest or Vinculin (control) levels were determined by Western blot.
To determine whether this lentiviral system yields tractable and inducible RNAi in a polyclonal population, an shRNA targeting Rb was subcloned into pINDUCER10 (miR-RUP). U2OS cells were transduced with pINDUCER10-shRB, and transduced cells that survived puromycin selection were cultured in the absence (−) or presence (+) of dox for 4 d to induce tRFP and shRNA expression, followed by flow cytometry and Western blot analysis. In the absence of dox, less than 1.0% of cells are tRFP positive, demonstrating the tight regulation of pINDUCER10. Addition of dox led to tRFP fluorescence in >98% of cells, indicating that these cells express the Rb hairpin (Fig. 1B, Left). This is confirmed by significant Rb protein depletion in the +dox population compared with the −dox population (Fig. 1B, Right). Additional Rb shRNAs also demonstrated robust depletion of Rb (Fig. S1A). We next tested whether pINDUCER10 exhibits robust inducibility in nonmalignant cells and is effective for other gene targets. Multiple shRNAs targeting the Rest tumor suppressor were cloned into pINDUCER10 and transduced into immortalized human mammary epithelial cells (HMECs). Puro-selected cells were cultured with or without dox for 72–96 h (as indicated) and harvested for Western blot analysis. Each shRNA significantly depleted Rest protein by 72 h after dox addition (Fig. 1C). pINDUCER10 shRNAs targeting ZRANB3 also elicited inducible ZRANB3 mRNA depletion (Fig. S1B). Collectively, these results indicate that the pINDUCER system produces robust target depletion using multiple shRNAs in multiple cell types.
To determine the kinetics of inducible expression from pINDUCER, cDNAs encoding enhanced GFP (eGFP), Rest, p21Cip1, or Fox2 were recombined into pINDUCER20 (ORF-UN). SW1417 colon cancer cells were transduced with pINDUCER20-eGFP or pINDUCER20-REST, selected for neomycin resistance, cultured with or without dox for indicated times, and analyzed by flow cytometry (Fig. S2A) or Western blot (Fig. 1D). eGFP fluorescence and REST expression was detectable by 8–12 h after the addition of dox and continued to accumulate over 36–48 h (Fig. S2A and Fig. 1D, respectively). Likewise, robust inducibility of p21Cip1 or Fox2 expression from pINDUCER20 was also observed in human fibroblasts or murine myoblasts, respectively (Fig. S2 B and C), demonstrating the effective inducibility of pINDUCER20 across multiple genes and cell types. Additionally, Rest or Fox2 expression could be titrated with increasing dox concentrations (Fig. 1E and Fig. S2C), indicating that inducible RNAi or cDNA expression from pINDUCER can be used to create allelic series of target gene expression or depletion. Collectively, these data illustrate that the pINDUCER system elicits rapid, dox-dependent cDNA expression and RNAi across several target genes and in multiple cell types.
pINDUCER11 (miR-RUG) Lentivirus Enables Rapid Isolation of Inducible Polyclonal Populations.
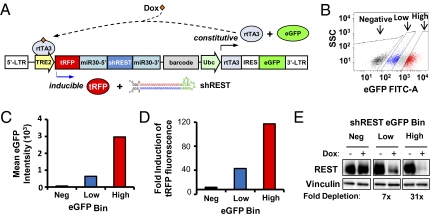
Our central goal in developing this lentiviral vector series was to facilitate ex vivo engineering of target tissues or tumors with inducible transgene expression or RNAi and subsequent transplantation. In such xenograft or allograft experiments, there are two important challenges: (i) the transduced cells must be rapidly isolated to prevent effects of prolonged in vitro manipulation, and (ii) the transduced cells must exhibit broad inducibility across a polyclonal population. We hypothesized that engineering a fluorescent reporter in a constitutive, bicistronic cassette with rtTA3 would facilitate these two objectives, because (i) transduced populations may be rapidly isolated via FACS, and (ii) quantification of cellular fluorescence during FACS would provide a surrogate measurement of rtTA3 levels, thus enabling isolation of cellular populations exhibiting high rtTA3 levels and consequent inducibility. To test this hypothesis, we transduced immortalized HMECs with pINDUCER11 (Fig. 2A) encoding a Rest shRNA, and FACS-isolated populations with different levels of eGFP fluoresence (Fig. 2B). Notably, infection could be detected (by eGFP fluorescence) within 24 h of adding virus, with maximum fluorescence detection within 48 h (Fig. S3). FACS-isolated populations with high, low, or no eGFP fluorescence were cultured in vitro in the presence or absence of dox for 72 h. As shown in Fig. 2 C and D, eGFP fluorescence correlated with tRFP inducibility as measured by flow cytometry, with “high-eGFP” populations exhibiting threefold greater inducibility than “low-eGFP” populations. Consistent with their higher tRFP-shRNA expression, high-eGFP populations also exhibited 3.5-fold greater depletion of endogenous Rest protein (Fig. 2E), indicating that isolating the population with the highest eGFP fluorescence improves target knockdown within the polyclonal population. Taken together, these data suggest that pINDUCER11 enables rapid isolation of polyclonal populations exhibiting robust inducibility and RNAi.
Fig. 2.
Dual-fluorescent pINDUCER11 (miR-RUG) system improves inducible cDNA expression and RNAi across polyclonal populations. (A) Diagram of pINDUCER11 (miR-RUG). pINDUCER11 encodes a constitutive cassette (rtTA3 and eGFP) and an inducible transcript (shRNA and tRFP). (B) Isolation of cells with differing rtTA3/eGFP expression. Polyclonal populations of HMECs were infected with pINDUCER11 (MOI = 0.7) and FACs-sorted for indicated levels of eGFP fluorescence. (C) Stable eGFP fluorescence in pINDUCER11 transduced cells. Transduced HMECs from B were measured for mean eGFP fluorescence intensity by flow cytometry 14 d after the original FACs sorting. (D) Higher rtTA3/eGFP expression increases inducibility in polyclonal cell populations. HMECs from B were cultured in Dox for 4 d and measured for tRFP fluorescence by flow cytometry. The graph shows the fold induction of tRFP fluorescence (mean +dox tRFP fluorescence intensity/mean −dox tRFP fluorescence intensity). (E) Higher rtTA3/eGFP expression increases inducible RNAi in polyclonal cell populations. HMECs transduced with pINDUCER11-shREST and FACs-sorted for various levels of eGFP fluorescence were cultured with or without dox for 3 d and analyzed by Western blot for Rest or Vinculin (control) protein levels.
pINDUCER System Elicits Robust Inducibility in the Mouse Mammary Gland and Murine and Human Cancer Xenografts in Vivo.
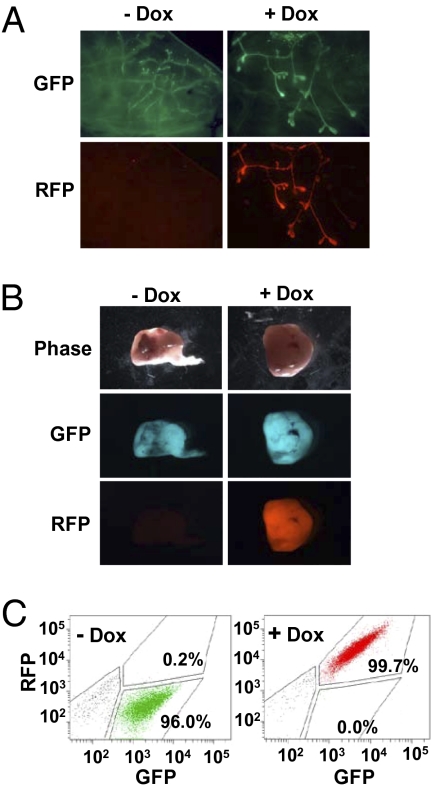
To determine whether the pINDUCER system could be used to track transduced/inducible cell populations in vivo, we tested pINDUCER11 (miR-RUG) in several well-characterized mammary development and breast cancer models. Mammary epithelial cells were isolated from mouse mammary glands, dispersed, and transduced with pINDUCER11 ex vivo and retransplanted orthotopically into the cleared fat pad of syngeneic mice in the presence or absence of dox. After 4 wk, the transduced population of cells within the mammary gland is easily identifiable by eGFP fluorescence. The inducible cells within the transduced population are also apparent by tRFP fluorescence. Importantly, tRFP fluorescence is only seen in mice treated with dox (Fig. 3A). In a similar manner, p53−/− mouse mammary tumors were transduced with pINDUCER11 ex vivo, sorted for eGFP fluorescence, and retransplanted orthotopically into syngeneic mice. Upon tumor harvest, compared to untransduced tumors, all transduced tumors are eGFP positive, and tumors from dox-treated mice are also tRFP positive (Fig. 3B and Fig. S4). Finally, the human breast cancer cell line MDA-MB-231 was transduced in vitro with pINDUCER11 and transplanted orthotopically in the absence or presence of dox. Tumors were dispersed at harvest and eGFP and tRFP fluorescence were measured by flow cytometry. The −dox tumors were eGFP positive and all +dox tumors were eGFP/tRFP positive at harvest (Fig. 3C). These data demonstrate that the pINDUCER11 vector can be used to perform tractable, inducible RNAi in multiple in vivo models.
Fig. 3.
pINDUCER11 (miR-RUG) elicits robust inducibility in the mouse mammary gland and mammary tumors in vivo. (A) Mouse mammary glands were excised, dispersed into single-cell suspension, transduced with pINDUCER11 (miR-RUG), and retransplanted orthotopically into the cleared fat pad of syngeneic mice with or without dox. After 4 wk, mammary glands were explanted and visualized by fluorescence microscopy as indicated. (B) p53−/− mouse mammary tumors were excised, dispersed into single-cell suspension, transduced with pINDUCER11, sorted for eGFP fluorescence, and retransplanted orthotopically into syngeneic mice. Upon tumor formation (>300 mm3), mice were treated with or without dox for 7 d, and tumors were explanted and visualized by phase or fluorescence microscopy as indicated. (C) MDA-MB-231 human breast cancer cells were transduced with pINDUCER11 and transplanted into the mouse mammary gland. Mice were treated with or without dox for 28 d. Tumors were dispersed into single-cell suspension and analyzed for fluorescence (eGFP and tRFP) by flow cytometry.
pINDUCER Enables Luminescence-Based Imaging of Breast Cancer Xenografts.
Luminescence-based imaging allows for tracking of luciferase-positive populations over time in live animals. To use this methodology, we engineered luciferase into the pINDUCER system to create pINDUCER12 (miR-RUL) (Fig. S5A). To determine whether luciferase is properly expressed, pINDUCER12-transduced HMECs were analyzed for luciferase activity. The pINDUCER12 cells had high levels of luciferase, whereas cells transduced with a control virus had no activity (Fig. S5B). To test whether pINDUCER12 can be used to detect tumors in live recipient animals, p53−/− mouse mammary tumors were transduced with pINDUCER12 and retransplanted orthotopically into syngeneic mice. As shown in Fig. S5C, tumors transduced with pINDUCER12 were readily detectable across independent xenografts, indicating that pINDUCER12 enables detection of live tumors in vivo.
pINDUCER Can Be Used to Validate Anticancer Targets in Human Cancer Xenografts.
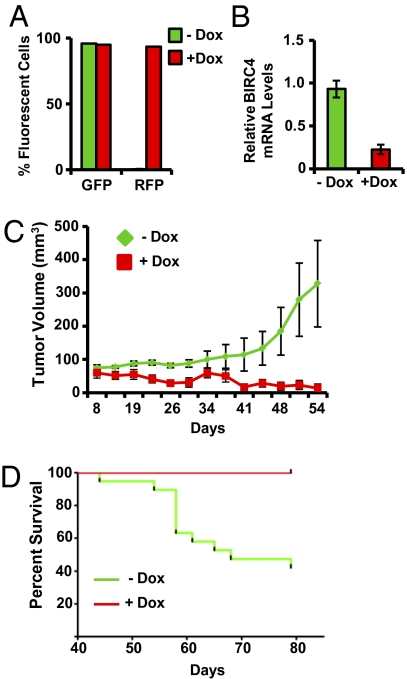
To determine whether the pINDUCER system can be used to characterize genes required for solid tumor growth in vivo, we tested the effects of inducible knockdown of BIRC4, a gene we recently identified as an essential gene in aggressive subtypes of human breast cancer. The breast cancer cell line SUM159 was transduced with pINDUCER11 (miR-RUG) encoding an shRNA targeting human BIRC4. These cells exhibited robust inducible tRFP expression and BIRC4 depletion (Fig. 4 A and B, respectively), thus confirming inducibility of pINDUCER11-shBIRC4 in this breast cancer model. These cells were transplanted orthotopically in immunocompromised mice, and mice were administered vehicle or dox and monitored for tumor growth. As shown in Fig. 4C, dox-induced BIRC4 shRNA expression resulted in a substantial diminution of tumor growth, with 69% of mice exhibiting no detectable tumor at 54 d after transplantation. This was concordant with a significant increase in animal survival upon dox administration (Fig. 4D; −dox: 40% survival; +dox: 100% survival). This suggests that pINDUCER11 can be effectively used to validate potential anticancer therapeutic targets that emerge from genomic and genetic studies of cancer. Furthermore, the effectiveness of pINDUCER11 across polyclonal populations suggests that this vector design may be used for genetic screens within solid tumors in vivo. Collectively, these data indicate that the pINDUCER vector series can be used to perform inducible gain- and loss-of-function experiments in vitro and in vivo in a wide range of experimental systems.
Fig. 4.
pINDUCER11 (miR-RUG) enables validation of tumor dependencies in vivo. (A and B) SUM159 human breast cancer cells were transduced with pINDUCER11-shBIRC4 at MOI = 3 and analyzed for (A) cellular fluorescence or (B) BIRC4 mRNA levels. (C) Cells from A were transplanted orthotopically into the mouse mammary gland. Mice were administered dox (+dox) or vehicle (−dox) and assessed for tumor volume over time. (D) Mice from C were observed for overall survival for the indicated times.
Discussion
RNAi has revolutionized the power of genetics in mammalian systems, enabling loss-of-function studies for virtually any gene in a mammalian genome. We and others have capitalized on this potential by developing genome-scale shRNA libraries and a number of methodologies for exploiting RNAi for genetic screens and candidate validation (3, 5–8). Despite these advances, the ability to study genes essential in normal development or a disease state has been hampered by the inability to effectively use RNAi to control the timing and level of gene activity. Here we describe a lentiviral toolkit (pINDUCER) for inducible RNAi and cDNA expression that exhibits robust expression in a wide range of cell types and models of tumorigenesis in vivo. The pINDUCER lentiviral system encodes a variety of markers for constitutively marking target cells with drug resistance, fluorescence, or luminescence, and further enables cells exhibiting inducible RNAi to be tracked with fluorescence or luminescence markers in vitro and in vivo. In addition, this lentiviral system encodes all elements needed for tet-inducible expression and can thus be used in a wide range of cell and tissue types without the need to deliver multiple plasmid or viral vectors. Importantly, our studies demonstrate that pINDUCER elicits robust RNAi and inducible expression in normal and malignant cells of differing tissue origins both in vitro and in vivo. Moreover, the promoter driving rtTA3 expression in the pINDUCER series can be replaced with tissue-specific promoters, thus enabling a more directed tissue- or cell-specific inducible RNAi using lentiviral methods. For example, for pINDUCER21 we have replaced the Ubc promoter with the EF1α promoter, which we have found to work more efficiently in some stem cell contexts.
To date, a major challenge in achieving effective inducible RNAi via lentiviral delivery has been the heterogeneity of inducible shRNA expression across polyclonal cell populations. This heterogeneity can result from differing activity of lentiviral integration sites or from the varying activity of the lentiviral promoter within differing cell types (16, 18). Here we use a fluorescence-based approach to rapidly identify and isolate polyclonal populations with high rtTA3 expression and inducibility. We show that this strategy enables effective in vitro inducibility in many cell types. Furthermore, and perhaps more importantly, it enables robust in vivo inducibility in the mouse mammary gland and in murine and human orthotopic models of breast cancer. In addition to such ex vivo/in vivo applications, we note that the pINDUCER lentivirus may also enable direct in vivo infection and monitoring of inducible populations, thus providing a potential experimental approach for tissues (e.g., brain) for which ex vivo manipulation and transplantation is not possible.
In cancer research, the discovery and validation of therapeutic targets for human cancer is especially dependent on genetic tools for manipulating gene function in vivo. Specifically, although orthotopic xenograft models of human cancer have become a widespread approach in cancer research, the inability to interfere with the level and timing of gene function across tumor models and cancer cell types has limited the identification of gene programs that are essential for human tumor growth and metastasis in vivo. To date, most studies have relied upon clonally derived isolates (9). This loses the heterogeneity of the actual tumor and may be less representative because of well-recognized variability in cell types within individual tumors and tumor models. Although inducible RNAi systems have begun to address this challenge in murine hematopoietic cancers (19), tools for inducible RNAi have remained elusive for in vivo models of epithelial cancers, which are the predominant malignancies affecting human health. In the present study, we show that the pINDUCER lentiviral system is broadly inducible in multiple models of epithelial cancer and mammary development. This system provides robust polyclonal inducibility, and we show that depletion of a potential anticancer target (BIRC4) elicits dramatic effects on tumor growth and host survival. The polyclonal efficacy of pINDUCER also suggests that the pINDUCER system will facilitate large-scale in vivo genetic screening in models of human epithelial cancers.
Methods
Vectors and Virus Production.
pINDUCER10 (miR-RUP) was first made by creating a linker by annealing two oligos: 5′-CTAGGCTAGCATTGGCACCGGTATTGCCGCGGCCGCATTGCCACGCGTATTGCCGGATCCATTGCCGGCGCGCCATTGCCTTAATTAAG and 5′-CGCGTTAATTAAGGCAATGGCGCGCCGGCAATGGATCCGGCAATACGCGTGGCAATGCGGCCGCGGCAATACCGGTGGCAATGCTAGC. The annealed oligos were cloned into the XbaI- and MluI-digested pGIPZ vector from Open Biosystems to create the pGIPZ-linker.
The TRE2 promoter was PCR amplified from pSLIK-Neo-TGmiR-Luc (17) using primers 5′-ATTGGCGCTAGCTTAAAGGAACCAATTCAGTCGACTGG and 5′-ATTGGCACCGGTAGGCTGGATCGGTCCCGGTG. The PCR product was TA-cloned into the pCR2.1-TOPO vector from Invitrogen and sequence verified. The TRE2 promoter was then cloned from the pCR2.1-TOPO vector into the pGIPZ-linker with NheI and AgeI to create pGIPZ-TRE2. The turboRFP was cloned from turboRFP-N from Evrogen into the pGIPZ-TRE2 with AgeI and NotI to create pGIPZ-TRE2-RFP. The Mir30-shRNA was cloned from pPRIME-CMV-GFP (4) into pGIPZ-TRE2-RFP with NotI and MluI to create pGIPZ-TRE2-RFP-Mir30. The Ubc promoter was PCR amplified from pSLIK-Neo-TGmiR-Luc using primers 5′-ATTGGCACGCGTAAGATCTGGCCTCCGCGCCG and 5′-ATTGGCGGATCCGTCTAACAAAAAAGCCAAAAACGGCC, TA-cloned and sequenced, and cloned into pGIPZ-TRE2-RFP-Mir30 with MluI and BamHI to create pGIPZ-TRE2-RFP-Mir30-Ubc. The rtTA3 was PCR amplified from pSLIK-Neo-TGmiR-Luc using primers 5′-ATTGGCGGATCCATAACTTCGTATAATGTATGCTATACGAAGTTATGCCACCATGTCTAGACTGGACAAGAGCAAAGTC and 5′-ATTGGCGGCGCGCC ATAACTTCGTATAATGTATGCTATACGAAGTTATTTACCCGGGGAGCATGTCAAGGTC, TA-cloned and sequenced, and cloned into pGIPZ-TRE2-RFP-Mir30-Ubc with BamHI and AscI to create pGIPZ-TRE2-RFP-Mir30-Ubc-rtTA3. The IRES-Puro was PCR amplified from pGIPZ using primers 5′-ATTGGCGCGCGCCAATTCCGCCCCTCTCCCTCCC and 5′-ATTGGCTTAATTAATCAGGCACCGGGCTTGCGGG, TA-cloned and sequenced, and cloned into pGIPZ-TRE2-RFP-Mir30-Ubc-rtTA3 with AscI and PacI. The final product was sequence verified.
pINDUCER11 (miR-RUG) was made by first digesting pINDUCER10 with XbaI and PacI to remove the rtTA through the puromycin resistance gene. The rtTA-IRES segment was removed from pIndmir3Luc-miR by XbaI and NotI digest. eGFP was PCR amplified from MSCV-N-eGFP-ENTR using primers 5′-TAATACATGT ATCGATCCACCATGGTGAGCAAGGGC and 5′-GATCTTAATTAA TTACTTGTACAGCTCGTCCATGCCG. The PCR product was digested with PciI and PacI. The vector, rtTA-IRES, and eGFP were then ligated and sequence verified.
pINDUCER12 (miR-RUL) was made by digesting pINDUCER10 with XbaI and PacI. The IRES was isolated from the same vector by digesting with XbaI and NcoI. Luciferase was PCR amplified from pEF1-Luc-IRES-neo using primers 5′- TACTACCGGTCGCCACCATGGAAGACGCCAAAAACATAAA and 5′-TAGAGATTAGGATTAGGATCTTAATTAAGCGGCCGCTTACACGGCGATCTTTCCGCC. The PCR product was then digested with NcoI and PacI, ligated to the vector and IRES, and sequence verified.
shRNAs targeting RB, REST (A5, V2LHS_57042, V2LHS_57044, V2LHS_57045), BIRC4 (V2LHS_94578), ZRANB3 sh1 AATTCTCTTCTACTAATTACT, sh2 CAGAAACTAAGATACAATACT, sh3 CGCAATTATGTTTCAGCAATG, sh4 GATTCAGACTCGCAATTATGT, and FF (firefly luciferase) were subcloned from pGIPZ Hannon-Elledge shRNA collection (Open Biosystems) or synthesized DNA oligos into pINDUCER10 or pINDUCER11 with XhoI and MluI.
pINDUCER20 (ORF-UN) was made by digesting TRE2 out of the pSLIK-Neo-TGmiR-Luc vector with BamHI and SpeI and blunt-end cloned into the backbone of SpeI and XhoI digested pHAGE-CAGS-W vector (Dr. Richard Mulligan, Harvard Institute of Medicine, Boston) to generate the pHAGE-TRE vector. A fragment containing the gateway recipient cassette was cut out of the pHAGE-EF-DEST vector (G.H.) with SpeI and inserted into pHAGE-TRE to generate the pHAGE-TRE-DEST vector. The Ubc-rtTA3-IRES-Neo fragment was cut out of the pSLIK-Neo-TGmiR-Luc vector with PacI and ClaI and blunt-end cloned into the backbone of SphI and ClaI digested pHAGE-TRE-DEST.
pINDUCER21 (ORF-UG) was made by digesting pINDUCER20 with NdeI and SphI. eGFP was PCR amplified with AgeI and SphI on the 5′ and 3′ ends, respectively, and then digested with AgeI and SphI. The vector and eGFP were ligated and sequence verified.
The eGFP, REST, and p21 cDNAs were recombined into pINDUCER20.
Lentiviral supernatants were generated by transient transfection of 293T cells according to Mirus Bio's TransIT transfection protocols and harvested 48 h after transfection.
Cell Culture.
HMECs expressing hTERT and SV40 LT (TLM-HMECs) (20, 21) were cultured in mammary epithelial growth medium (Lonza). U2OS osteosarcoma cells and MDA-MB-231 human breast cancer cells were cultured in DMEM (Gibco) supplemented with 10% FBS. C2C12 myoblasts (ATCC) were maintained in high-glucose DMEM, supplemented with 10% FBS, 1% penicillin/streptomycin, and 1% l-glutamine (Invitrogen). SW1417 human colon cancer cells were cultured in RMPI 1640 (ATCC) supplemented with 10% FBS. SUM159 human breast cancer cells were cultured in F12 (Gibco) media supplemented with 5% FBS, 10mM Hepes (Gibco), 5 μg/mL insulin (Invitrogen) and 1 μg/mL hydrocortisone. All cell lines were incubated at 37 °C and 5% CO2. Stable cell lines expressing indicated shRNAs or cDNAs were generated by lentiviral transduction in the presence of 8 μg/mL polybrene followed by selection with appropriate antibiotic resistance markers. Cell fluorescence was analyzed by flow cytometry using a BD Biosystems LSRII analyzer. FACsorting was performed on a BD Biosystems AriaII.
Supplementary Material
Acknowledgments
T.F.W. is a scholar of The V Foundation and Mary Kay Ash Foundation for Cancer Research. This work was supported by Susan G. Komen for the Cure Career Catalyst Award KG090355 (to T.F.W.) and Specialized Program of Research Excellence Developmental Grant P50 CA058183 (to T.F.W. and R.S.), Department of Defense Breast Cancer Research Program Predoctoral Fellowship BC094077 (to K.L.M.), National Institutes of Health (NIH) Grant R01GM076493 (to T.A.C.), NIH Grant R37 CA16303 (to J.M.R.), NIH and National Institute of Environmental Health Sciences Intramural Research Program Z01ES102745 (to G.H.), and US Army Innovator Award W81XWH0410197 (to S.J.E.). S.J.E. is an Investigator with the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1019736108/-/DCSupplemental.
References
- 1.Hannon GJ, Rossi JJ. Unlocking the potential of the human genome with RNA interference. Nature. 2004;431:371–378. doi: 10.1038/nature02870. [DOI] [PubMed] [Google Scholar]
- 2.Elbashir SM, et al. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 3.Schlabach MR, et al. Cancer proliferation gene discovery through functional genomics. Science. 2008;319:620–624. doi: 10.1126/science.1149200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stegmeier F, Hu G, Rickles RJ, Hannon GJ, Elledge SJ. A lentiviral microRNA-based system for single-copy polymerase II-regulated RNA interference in mammalian cells. Proc Natl Acad Sci USA. 2005;102:13212–13217. doi: 10.1073/pnas.0506306102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Paddison PJ, et al. A resource for large-scale RNA-interference-based screens in mammals. Nature. 2004;428:427–431. doi: 10.1038/nature02370. [DOI] [PubMed] [Google Scholar]
- 6.Silva JM, et al. Second-generation shRNA libraries covering the mouse and human genomes. Nat Genet. 2005;37:1281–1288. doi: 10.1038/ng1650. [DOI] [PubMed] [Google Scholar]
- 7.Berns K, et al. A large-scale RNAi screen in human cells identifies new components of the p53 pathway. Nature. 2004;428:431–437. doi: 10.1038/nature02371. [DOI] [PubMed] [Google Scholar]
- 8.Moffat J, et al. A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high-content screen. Cell. 2006;124:1283–1298. doi: 10.1016/j.cell.2006.01.040. [DOI] [PubMed] [Google Scholar]
- 9.Luo J, et al. A genome-wide RNAi screen identifies multiple synthetic lethal interactions with the Ras oncogene. Cell. 2009;137:835–848. doi: 10.1016/j.cell.2009.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Westbrook TF, et al. A genetic screen for candidate tumor suppressors identifies REST. Cell. 2005;121:837–848. doi: 10.1016/j.cell.2005.03.033. [DOI] [PubMed] [Google Scholar]
- 11.Zender L, et al. An oncogenomics-based in vivo RNAi screen identifies tumor suppressors in liver cancer. Cell. 2008;135:793–795. doi: 10.1016/j.cell.2008.09.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Scholl C, et al. Synthetic lethal interaction between oncogenic KRAS dependency and STK33 suppression in human cancer cells. Cell. 2009;137:821–834. doi: 10.1016/j.cell.2009.03.017. [DOI] [PubMed] [Google Scholar]
- 13.Ngo VN, et al. A loss-of-function RNA interference screen for molecular targets in cancer. Nature. 2006;441:106–110. doi: 10.1038/nature04687. [DOI] [PubMed] [Google Scholar]
- 14.Barbie DA, et al. Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature. 2009;462:108–112. doi: 10.1038/nature08460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Meacham CE, Ho EE, Dubrovsky E, Gertler FB, Hemann MT. In vivo RNAi screening identifies regulators of actin dynamics as key determinants of lymphoma progression. Nat Genet. 2009;41:1133–1137. doi: 10.1038/ng.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Levis R, Hazelrigg T, Rubin GM. Effects of genomic position on the expression of transduced copies of the white gene of Drosophila. Science. 1985;229:558–561. doi: 10.1126/science.2992080. [DOI] [PubMed] [Google Scholar]
- 17.Shin KJ, et al. A single lentiviral vector platform for microRNA-based conditional RNA interference and coordinated transgene expression. Proc Natl Acad Sci USA. 2006;103:13759–13764. doi: 10.1073/pnas.0606179103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pikaart MJ, Recillas-Targa F, Felsenfeld G. Loss of transcriptional activity of a transgene is accompanied by DNA methylation and histone deacetylation and is prevented by insulators. Genes Dev. 1998;12:2852–2862. doi: 10.1101/gad.12.18.2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zuber J, et al. Toolkit for evaluating genes required for proliferation and survival using tetracycline-regulated RNAi. Nat Biotechnol. doi: 10.1038/nbt.1720. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao JJ, et al. Human mammary epithelial cell transformation through the activation of phosphatidylinositol 3-kinase. Cancer Cell. 2003;3:483–495. doi: 10.1016/s1535-6108(03)00088-6. [DOI] [PubMed] [Google Scholar]
- 21.Elenbaas B, et al. Human breast cancer cells generated by oncogenic transformation of primary mammary epithelial cells. Genes Dev. 2001;15:50–65. doi: 10.1101/gad.828901. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.