Abstract
Overcoming the inherent chemoresistance of pancreatic cancers remains a major goal of therapeutic investigations in this disease. In this study, we discovered a role for the human concentrative nucleoside transporter-1 (hCNT1; SLC28A1), a high-affinity pyrimidine nucleoside transporter, in determining the chemosensitivity of human pancreatic cancer cells to gemcitabine, the drug used presently as a standard of care. Compared with normal pancreas and pancreatic ductal epithelial cells, hCNT1 expression was frequently reduced in pancreatic tumors and tumor cell lines. In addition, hCNT1-mediated 3H-gemcitabine transport was lower in pancreatic cancer cell lines and correlated with cytotoxic IC50 estimations of gemcitabine. In contrast to gemcitabine-sensitive pancreatic cancer cell lines, MIA PaCa-2, a gemcitabine-resistant pancreatic cancer cell line exhibited relatively restrictive, cell cycle-dependent hCNT1 expression and transport. hCNT1 translation was suppressed in the late G1-enriched MIA PaCa-2 cell population possibly in an miRNA-dependent manner, which corresponded with the lowest hCNT1-mediated gemcitabine transport during this phase. While hCNT1 protein was induced during G1/S transition, increased hCNT1 trafficking resulted in maximal cell surface recruitment and transport-overshoot in the G2/M phase-enriched cell population. hCNT1 protein was directed predominantly to proteasomal or lysosomal degradation in S or G2/M phase MIA PaCa-2 cells, respectively. Pharmacological inhibition of hCNT1 degradation moderately increased cell surface hCNT1 expression and cellular gemcitabine transport in MIA PaCa-2 cells. Constitutive hCNT1 expression reduced clonogenic survival of MIA PaCa-2 cells and steeply augmented gemcitabine transport and chemosensitization. In addition to supporting a putative tumor suppressor role for hCNT1, our findings identify hCNT1 as a potential candidate to render drug-resistant pancreatic cancer cells amenable to chemotherapy.
Keywords: hCNT1, pancreatic cancer, gemcitabine-resistance, cell cycle, proliferation
Introduction
2’,2’-Difluoro-2’-deoxycytidine (dFdC; gemcitabine) is currently used as a first-line treatment against locally-advanced and metastatic adenocarcinoma of the pancreas (1, 2). Gemcitabine is phosphorylated intracellularly to its active diphosphate (dFdC-DP) and triphosphate (dFdC-TP) forms that inhibit DNA and RNA replication (3, 4, 5). In order for hydrophilic gemcitabine (log P = −1.1 to −2.0) to enter cells, membrane-bound nucleoside transporters are prerequisites (since cells deficient in nucleoside transporter are resistant to gemcitabine cytotoxicity (6)). Studies to date suggest two transporters from each of the human concentrative nucleoside transporter family (hCNT1, hCNT3) and the equilibrative nucleoside transporter family (hENT1, hENT2) are capable of translocating gemcitabine across the cell surface (3, 6, 7–10). Despite the expression of one or more of the aforementioned transporters in most cells types, hENT1 is generally considered to be predominantly involved in gemcitabine transport in tumors since its expression correlates with cellular proliferation (8,11). Clinical evidence also shows pancreatic tumor cells with high hENT1 expression exhibiting increased gemcitabine chemosensitivity (6, 12–15).
Unlike hENT1, the roles of CNTs, especially hCNT1, in governing gemcitabine cytotoxicity in tumors are not well understood. hCNT1 is normally expressed at the apical surfaces of well-differentiated epithelial tissues such as intestinal villi and hepatocytes (16,17). hCNT1 mRNA and/or protein are also detected in the pancreas, kidneys, skeletal muscles, uterus corpus, salivary glands, and placenta (16, 18). Consistent with hCNT1 expression in differentiated epithelial tissues, the rat ortholog of hCNT1, rCNT1, is regulated by differentiation-promoting agents (19–23). While the degree of tumor cell differentiation often correlates with the clinical response to chemotherapy, it is not clear whether the expression of hCNT1 itself is altered in tumors, and if so, whether such alterations influence tumor cell proliferation and drug sensitization. Earlier studies detected hCNT1 expression in solid (e.g. breast) tumors, predominantly localized in the cytoplasm and nucleus (24). Evidence for the reduction or complete loss of hCNT1 expression in solid (ovarian) tumors has also surfaced, suggesting that alterations in hCNT1 expression during tumor initiation/progression may have impacts on nucleoside chemosensitivities (25).
hCNT1’s precise involvements in pancreatic cancer cell growth, proliferation, and differentiation have not been investigated and its role in determining gemcitabine sensitivity in pancreatic cancer cells remains elusive. A previous study showed that ectopic expression of hCNT1 cDNA into pancreatic cancer cells augmented gemcitabine influx and cytotoxicity, suggesting the importance of hCNT1 in improving gemcitabine sensitivity in these cells (8). However, endogenous hCNT1 is found to be transiently expressed in sub-confluent pancreatic cancer cells (8) further obscuring its relationship to gemcitabine sensitivity. In this study, we investigated the role of hCNT1 in pancreatic cancer cell proliferation and gemcitabine chemosensitization in detail.
Materials and methods
Reagents
Gemcitabine and 3H-gemcitabine were obtained from Moravek Radiochemicals (Brea, CA) and Chemie Tek (Indianapolis, IN), respectively. Fetal bovine serum (FBS) was from PAA Laboratories Inc. (Ontario, Canada). Uridine, cytosine, inosine, crystal violet, G418, nitrobenzyl mercaptopurine riboside (NBMPR), 4’,6’-diamidino-2-phenylindole (DAPI), dimethylsulfoxide (DMSO), 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT), trypan blue, cycloheximide, propidium iodide, Z-Leu-Leu-Leu-al (MG 132) and leupeptin hemisulfate salt were obtained from Sigma-Aldrich (St. Louis, MO). Bicinchoninic acid (BCA) protein assay reagent was from Pierce Chemical (Rockford, IL). Fluorescent anti-fade mounting reagent and Vybrant DyeCycle green were obtained from Molecular Probes (Invitrogen). Plastic wares for cell culture were obtained from Corning, NY.
Pancreatic Tissues
Five pancreatic cancer tissue samples along with their matched normal adjacent tissues were procured from the National Disease Research Interchange (Philadelphia, PA). The collection and use of human tissue for research was approved by the University of Georgia Human Subjects Review Board.
Cell Culture
The pancreatic cancer cell lines (AsPC-1, BxPC-3, Capan-1, HPAF-II, MIA PaCa- 2, PANC-1, PL45, and SW1990) and Madin Darby Canine Kidney (MDCK) cell line were received from the American Type Culture Collection (ATCC; Manassas, VA) cell bank. These cell lines were propagated, expanded, and frozen immediately after arrival. The cells revived from the frozen stock were used within 10–20 passages, not exceeding a period of 2–3 months. The ATCC uses morphological, cytogenetic and DNA profile analysis for characterization of cell lines. Human pancreatic ductal epithelial cell (HPDE) (1) was kindly received from Dr. Ming Tsao, Ontario Cancer Institute (Toronto, Canada). The L3.6pl cell line (2) was kindly received from Dr. Isiah D. Fidler at The University of Texas MD Anderson Cancer Center (Houston, TX). The HPDE and L3.6pl cell lines were handled as other cell lines (described above) and were genotyped by DNA fingerprinting (PowerPlex 16, Promega, Inc.) as per the manufacturer’s instructions. AsPC-1 and BxPC-3 cells were grown in RPMI-1640 Medium supplemented with 10% FBS and subcultured at a 1:5 ratio. L3.6pl was cultured in Eagle’s Minimum Essential Medium (MEM) supplemented with 10% FBS, 1% non-essential amino acid solution 100× (Sigma-Aldrich St. Louis, MO), 1% 100 mM sodium pyruvate (Sigma-Aldrich St. Louis, MO), 1% 200mM L-glutamine, and 2% 100× vitamins, and subcultured at a 1:8 ratio. HPAF-II was cultured in MEM supplemented with 10% FBS and subcultured at a 1: 2 ratio. MIA PaCa-2 cells were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM) and supplemented with 10% FBS and 2.5% horse serum and subcultured at a 1:8 ratio. PL-45 was cultured in DMEM supplemented with 10 % FBS and subcultured at a 1:4 ratio. Capan1 was cultured in Iscove’s DMEM containing 20% FBS and subcultured at a 1:3 ratio. PANC-1 was cultured in DMEM supplemented with 10% FBS and subcultured at a 1:8 ratio. SW1990 cells were grown in Leibovitz’s L-15 Medium supplemented with 10% FBS and subcultured at a 1:3 ratio. A retroviral packaging cell line (ATCC CRL 9078) was maintained in DMEM supplemented with 5% FBS. MDCK-hCNT1HA cells were cultured in MEM containing 5% FBS and subcutured at a 1:8 ratio. All media for the above cell lines were purchased from MediaTech (Manassas, VA) and were supplemented with 100 units of penicillin/mL and 2 µg streptomycin/mL (Sigma-Aldrich, St. Louis, MO). The cells were incubated at 37°C in a 5% CO2 environment and subcultured every 48–96 h.
Expression of hCNT1HA and hCNT1 siRNA, Serum Synchronization and Flow cytometry, Immunostaining and Western Blotting, MTT Cytotoxicity and Clonogenic Assay, MicroRNA Array and Lentiviral-mediated Overexpression of miRNAs
Details are given in Supplemental Materials and Methods.
Real-time PCR analysis and 3H-gemcitabine Transport
Nucleoside transport and real-time PCR analysis were performed as described previously (17).
Results
hCNT1 expression was frequently reduced in pancreatic cancer
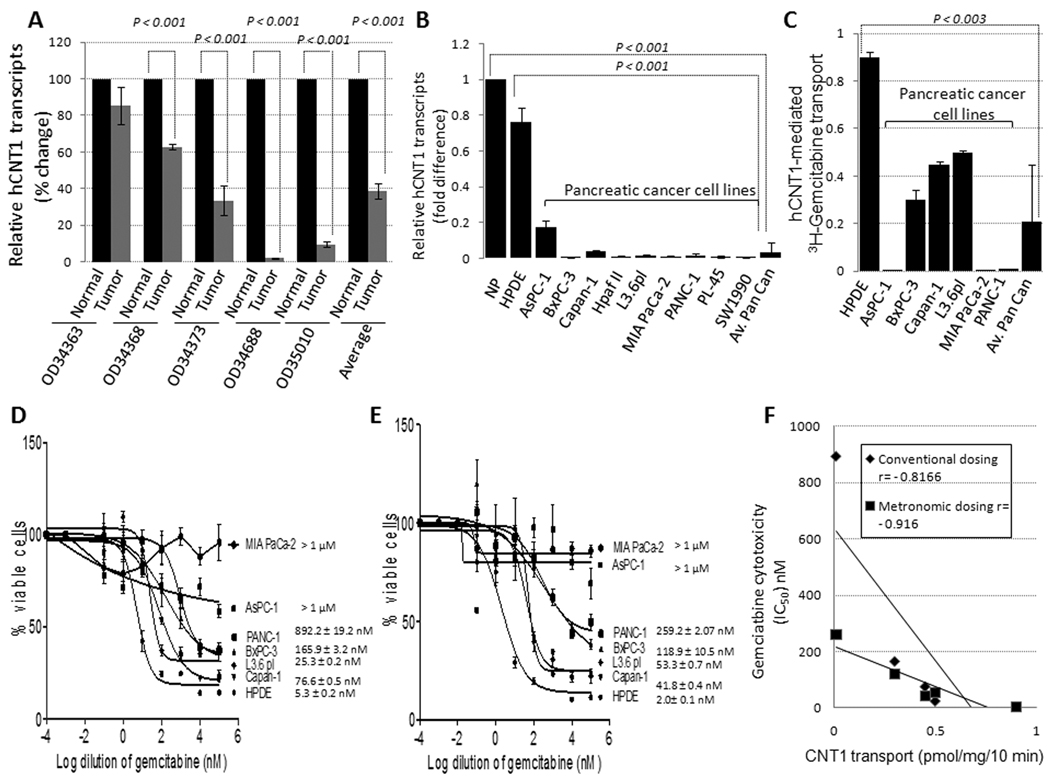
To test whether hCNT1 expression is altered in pancreatic tumors, we quantitatively assessed hCNT1 mRNA and protein levels by real-time PCR and Western blotting, respectively, in 5 pancreatic tumors and compared them with the matched (i.e. derived from the same donors) adjacent normal pancreatic tissues. hCNT1 mRNA was significantly decreased in 4 out of 5 tumor samples examined (Fig. 1A) with an average of 38.6 ± 4.2 % (p <0.001) expression in tumors. hCNT1 protein was decreased (>2-fold) in 3 out of 5 samples tested (Supplementary Fig. 1). Next we assessed hCNT1 mRNA levels in normal pancreatic ductal epithelial cells (HPDE) and in several human pancreatic cancer cell lines (AsPC-1, BxPC-3, Capan1, HpafII, L3.6pl, MIA PaCa-2, PANC-1, PL45 and SW1990). hCNT1 mRNA was decreased by 24- and 30-folds in all pancreatic cancer cell lines (average) compared with HPDE cells and normal pancreatic tissues (n=5), respectively (Fig. 1B). To further investigate the functional relevance of hCNT1 expression reductions, we chose to examine hCNT1-mediated gemcitabine transport and cytotoxicity in detail in HPDE and 6 well-characterized pancreatic cancer cell lines. hCNT1-mediated 3H-gemcitabine transport activity was significantly reduced in all pancreatic cancer cell lines examined (Fig. 1C). However, substantial relative hCNT1 transport activity was still detected in exponentially growing BxPC-3, Capan-1, and L3.6pl cell lines (~0.1–0.9 pmol/mg/10 min) but not, or only minimally, in AsPC-1, MIA PaCa-2 and PANC-1 cell lines (<0.1 pmol/mg/10 min) (Fig. 1C). Further, significant correlations were established between hCNT1-mediated 3H-gemcitabine transport influxes, and gemcitabine cytotoxic IC50 values determined by conventional (r=−0.82; p <0.05; Fig. 1D & 1F) and metronomic (r=−0.92; p <0.01; Fig. 1E & 1F) dosing schedules. Since hENT1 expression did not change significantly during the gemcitabine treatments (data not shown), the above data suggest hCNT1 transport as a determinant of gemcitabine sensitivity in pancreatic cancer.
Figure 1. hCNT1 mRNA and transport is reduced in pancreatic cancer.
A, Relative hCNT1 mRNA expression in pancreatic cancer tissues and matching adjoining normal pancreas. B, Relative levels of hCNT1 mRNA in normal pancreas tissue, HPDE, and cancerous cell lines. Columns, mean of triplicate; bars, SD. n=3. C, hCNT1-mediated 3H-gemcitabine cellular transport levels in HPDE and pancreatic cancer cell lines. Columns, mean of triplicate; bars, SE. n=3. D & E, 5 × 10 to the power 3 cells were treated with gemcitabine (0.1 nM – 100 µM) by conventional (D) or metronomic (E) dosing schedules, and percent inhibition of cellular proliferation was measured using the MTT assay. Points, mean of triplicate; bars, SE. n=3. Gemcitabine IC50 estimations are indicated. F, Gemcitabine transport levels significantly (Spearman’s correlation test; p<0.05) correlated with the cytotoxic IC50 values in those pancreatic cancer cell lines where IC50 values were estimable.
Heterogeneities in hCNT1 protein localization in gemcitabine-resistant pancreatic cancer cells
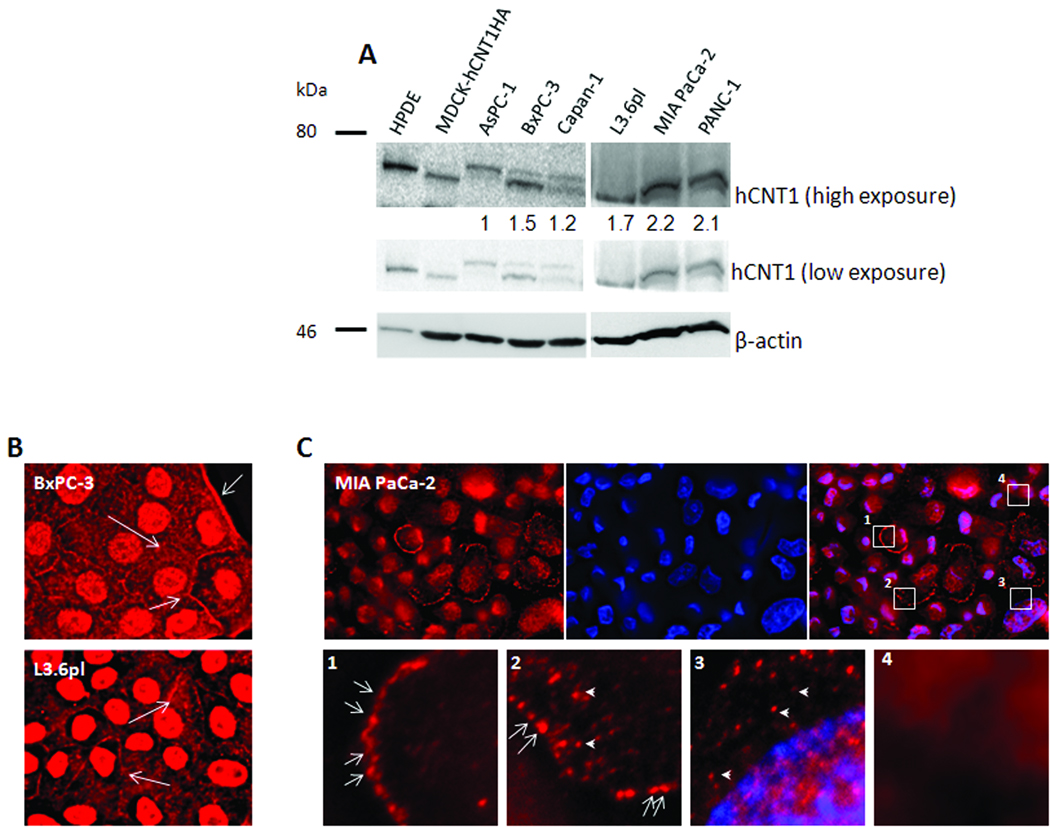
Despite similar levels of reduction in hCNT1 mRNA in all pancreatic cancer cell lines examined, significant hCNT1-mediated gemcitabine transport activities were only observed in gemcitabine-sensitive cells (HPDE, L3.6pl, BxPC-3, PANC-1, CaPan-1) and not gemcitabine-resistant cells (MIA PaCa-2, AsPC-1). To further investigate this discrepancy, we examined hCNT1 protein in these cell lines by Western blotting (Fig.2A). hCNT1 protein was detected in all cell lines as two bands (~70 kDa), most likely corresponding to its glycosylated and non-glycosylated forms (26). Although the relative abundance of these two forms varied between cell lines (Fig. 2A), the total hCNT1 protein levels (glycosylated and non-glycosylated forms) were not significantly different between gemcitabine-sensitive and gemcitabine-resistant cancer cell lines and did not account for the differences observed in hCNT1 transport activities between these groups. These results prompted an investigation as to whether aberrant cellular localization or lack of cell surface targeting of hCNT1 protein resulted in inadequate gemcitabine transport/cytotoxicity in gemcitabine-resistant cancer cells. Interestingly, immunocytochemical analyses displayed hCNT1 protein at appreciable levels in the cell periphery of most L3.6pl and BxPC-3 (gemcitabine-sensitive) cells (Fig.2B, arrows), while only a small fraction of MIA PaCa-2 (gemcitabine-resistant) cells alone displayed hCNT1 expression (Fig. 2C, top row). In addition, hCNT1-staining patterns in MIA PaCa-2 cells varied extensively with culture conditions and cell densities. Furthermore, hCNT1 immunoreactivity in MIA PaCa-2 cells was mainly intracytoplasmic (Fig. 2C, arrowheads) with only a subpopulation of cells exhibiting distinct hCNT1 cell surface expression (Fig. 2C, arrows). The observed heterogeneities in hCNT1 staining (Fig. 2C, insets) suggest that intricate cell-cycle regulation was influencing hCNT1 transport characteristics in drug-resistant MIA PaCa-2 cells.
Figure 2. hCNT1 protein expression and localization in human pancreatic cancer cell lines.
A, Western blotting analysis of hCNT1 using an anti-hCNT1 goat polyclonal antibody in whole cell lysates of HPDE and cancerous cell lines. MDCK-hCNT1HA lysate was used as a positive control (lane 2, top panel), and β-actin (45 kDa) was used as a loading control (bottom panel). hCNT1:β-actin ratios are indicated (top) B & C. Immunocytochemical analysis of exponentially-growing gemcitabine-sensitive (L3.6pl and BxPC-3; B) and gemcitabine-resistant (MIA PaCa-2; C) cells for hCNT1 (red). Nuclei stained with DAPI are blue. Original magnification X40. MIA PaCa-2 cells showing variable hCNT1 staining patterns (C, insets) were expanded (C, bottom panels). Arrows: cell surface hCNT1; Arrowheads: intracellular hCNT1. Original magnification X60 (insets).
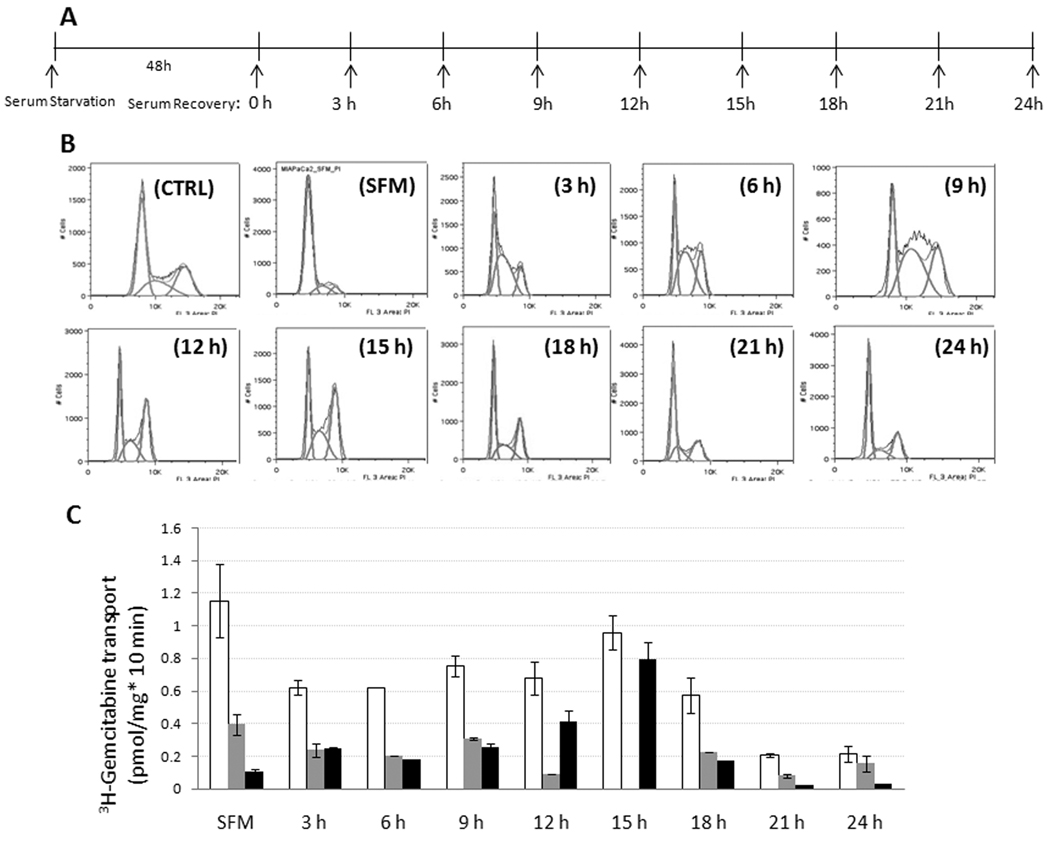
To further investigate cell cycle control of hCNT1 in MIA PaCa-2 cells, we utilized serum synchronization experiments (Fig. 3A, 3B, and Supplemental Table 1) to study alterations in hCNT1-mediated 3H-gemcitabine transport during the cell cycle. Transport studies performed in parallel-synchronized MIA PaCa-2 cell populations identified that hCNT1 contribution towards total gemcitabine cellular uptake was very low in the G1 phase (Fig. 3C; labeled SFM; Supplemental Table I). hCNT1 transport contributions significantly increased from 3 h of serum recovery in S phase-predominated cell populations (Fig. 3C; Supplemental Table I). Despite hCNT1’s implications as an S phase-inducible protein (27), the maximal hCNT1-mediated gemcitabine transport was not observed in the S phase-predominated cell population (3–9 h; 0.25–0.3 pmol/mg*10 min) but was observed in the G2/M phase-predominated cell population (~12–15 h; 0.4–0.7 pmol/mg*10min) (Fig. 3C; Supplemental Table I). Unlike hCNT1, significant proportions (30–40%) of total gemcitabine transport into G1 and S phase-enriched cell populations were mediated by hENT1 (Fig. 3C; Supplemental Table I). Compared with MIA PaCa-2 cells, hCNT1-mediated gemcitabine transport levels in G1, S, and G2/M phases of a gemcitabine-sensitive cell line (BxPC-3) were significantly higher at each of the corresponding phases (data not shown). These results suggest a cell cycle-dependent, restrictive role of hCNT1 in impeding gemcitabine transport in drug-resistant MIA PaCa-2 cells.
Figure 3. hCNT1-mediated 3H-gemcitabine transport activities varied among cell cycle synchronized MIA PaCa-2 cell populations.
A. A schematic layout for cell synchronization by serum starvation. MIA PaCa-2 cells were serum starved for 48 h and recovered with media containing 20% FBS and 5% horse serum in 3 h intervals for 24 h. B. Flow cytometric analysis of cell cycle distribution of MIA PaCa-2 cells following serum synchronization. A representative cell cycle distribution pattern is shown. C. 3H- gemcitabine transport analysis in serum synchronized MIA PaCa-2 cells. Total (open bars), hENT1 (grey bars)- and hCNT1 (black bars)-mediated gemcitabine transport levels are shown. Data are mean ± SD, n=3. SFM; serum free media, CTRL; control.
Increased trafficking of hCNT1 protein to the cell surface resulted in gemcitabine transport-overshoot in G2/M MIA PaCa-2 cells
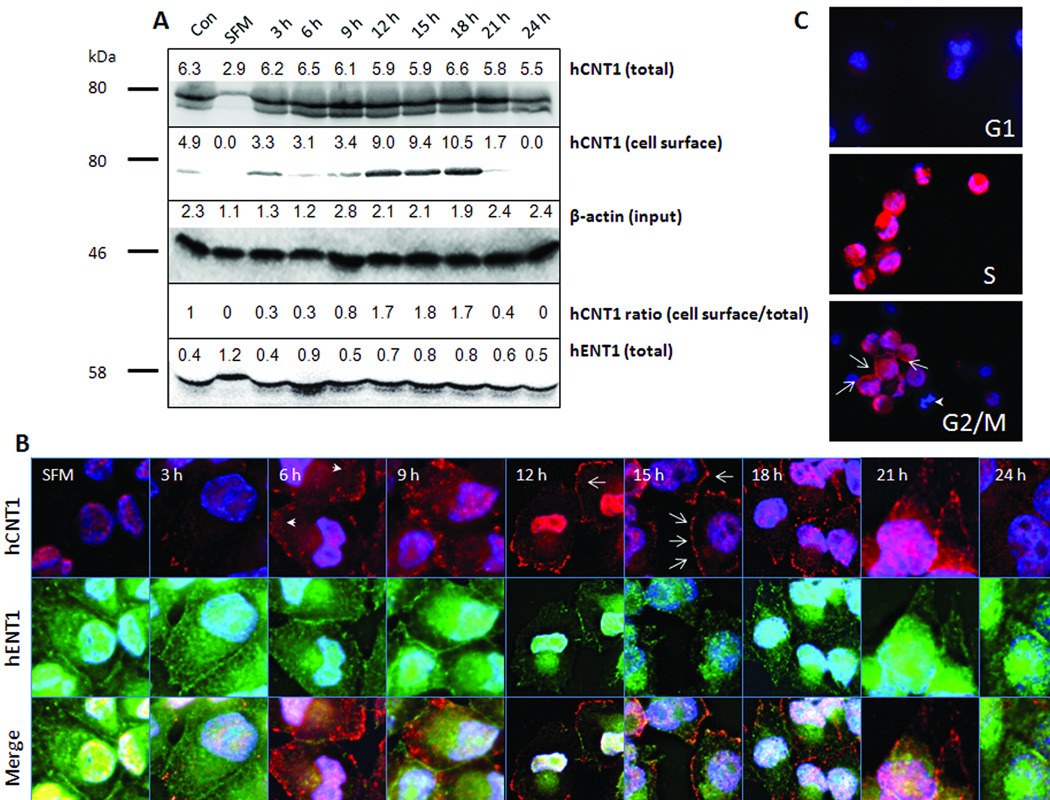
We next investigated whether the cell cycle restricted hCNT1 transport activity could be explained by alterations in hCNT1 mRNA and protein accompanying the cell cycle. Real-time PCR analysis indicated that unlike hENT1 mRNA, hCNT1 mRNA levels in G1-, S- and G2/M-enriched MIA PaCa-2 cell populations were not too different (within a 2-fold range; Supplemental Fig. 2) and did not account for the greater than 6-fold increase in hCNT1 transport activity noted between G1 and G2/M cells. Conversely, Western blotting analysis showed a highly reduced hCNT1 protein expression in G1 cells compared with that identified in cells of other cell cycle phases (Fig. 4A). hCNT1 protein expression significantly increased from 3 h of serum recovery (Fig. 4A) at which S phase cells predominated and continued to 12–18 h of serum recovery where G2/M cells predominated (Fig. 4A). However, a steep increase in hCNT1-mediated gemcitabine transport was only noticed at ~15 h of serum recovery (Fig 3C). Such a temporal difference between hCNT1 protein expression and transport activity is possible if the spatial localization of hCNT1 protein varied and if the accrual of hCNT1 protein at the cell surface ensued later to early S phase. To test this, we performed biotinylation of cell surface proteins in synchronized MIA PaCa-2 cell populations, and the cell surface pools of proteins were isolated by avidin pull-down assays (see ‘Methods’). Western blotting analysis on hCNT1 immunoprecipitated fractions identified maximal presence of hCNT1 protein at the cell surface between ~12–18 h of serum recovery (Fig. 4A), which corresponded to the preponderance of G2/M cells. Further, densitometry analysis of total and cell surface hCNT1 protein levels indicated the ratio of cell surface to total hCNT1 fraction (normalized to β-actin input) was ~3–4 folds higher in G2/M enriched cells compared with S phase cells (Fig. 4A). These data identify that increased cell surface accumulation of hCNT1 in the G2/M phase resulting in gemcitabine transport-overshoot at this phase.
Figure 4. hCNT1 expression in serum-synchronized and flow-sortedMIA PaCa-2 pancreatic cancer cells.
A. Western blotting analysis showing total (first panel) and cell surface hCNT1 (second panel) expression in serum-synchronized MIA PaCa-2 cells. The total protein blot was stripped and reprobed with anti-hENT1 goat polyclonal and anti-β-actin mouse monoclonal antibodies (third and fourth panels). SFM; serum free media. Densitometric values are indicated above the bands. B, Immunocytochemical detection of hCNT1 (arrows-cell surface hCNT1; arrowheads-cytoplasmic hCNT1) and hENT1 in serum synchronized MIA PaCa-2 cells. Original magnification X60. C, 5 ×103 cells in different phases of cell cycle were flow-sorted and immunostained for hCNT1 (arrows-cell surface hCNT1; arrowhead-mitotic cell). Nuclei were stained with DAPI are blue. Original magnification X40. SFM; serum free media.
To corroborate the data, we performed immunocytochemical analysis on various serum synchronized MIA PaCa-2 cell populations (Fig. 4B). hCNT1 immunoreactivity was drastically diminished in the G1-enriched MIA PaCa-2 cell population, with only occasional dots noticed at the periphery of some cells (Fig. 4B). hCNT1 immunoreactivity increased, beginning at 3 h of serum recovery in MIA PaCa-2 cells and was predominantly intracytoplasmic at time points 3–9 h and 21–24 h (Fig. 4B). Cell surface hCNT1 staining was readily detected in 12–18 h serum-recovered cells (Fig. 4B) with maximal cell surface hCNT1 staining identified at the 15 h time point (Fig. 4B). The increase in hCNT1 cell surface staining was accompanied by a decrease in hCNT1 cytoplasmic staining, and hCNT1 staining frequently appeared as a ‘string of beads’ at the cell surface, consistent with the docking of hCNT1-containing vesicles to the plasma membrane (Fig. 4B). In contrast to hCNT1, hENT1 was abundantly expressed in G1-enriched MIA PaCa-2 cells at cell contacts (Fig. 4B). Also, hENT1 was detected at the cell surface at 3–9 h and 21–24 h time points (Fig. 4B) when hCNT1 was predominantly intracellular. Interestingly, hENT1 staining at the cell surface diminished when hCNT1 staining at the cell surface increased at G2/M enriched cells (Fig. 4B). These data confirm that selective trafficking of hCNT1 to the cell surface in G2/M MIA PaCa-2 cells result in gemcitabine transport-overshoot at this phase.
Since an earlier study demonstrated that serum deprivation can influence hCNT1 levels (27), we alternatively validated hCNT1 expressional variations in MIA PaCa-2 cells separated into G1, S, and G2/M phases by flow cytometry. To study expression of hCNT1 at each of the designated phases, these flow-sorted cellular subpopulations were allowed only brief periods (30 min) of attachment to a substratum as drop cultures, followed by immunocytochemical analysis. The results obtained (Fig. 4C) essentially reproduced hCNT1 staining patterns obtained from the serum synchronization experiments. Mitotic cells, as judged from cells containing DAPI-stained, condensed chromatids, exhibited low cytoplasmic hCNT1 staining (Fig 4C, arrowhead). However, the cells that seemed to progress to mitotic completion (after nuclear division) began to express hCNT1 at the cell surface, suggesting reappearance in early G1. To further confirm the latter findings, we arrested MIA PaCa-2 cells in mitosis by treating with 10 µm (24 h) colchicine, a mitotic inhibitor, and then examined hCNT1 immunoreactivities as the cells progressed to G1 in the absence of the inhibitor. As observed in the flow-sorted cells, mitosis-arrested cells exhibited very low to some cytoplasmic hCNT1 staining (Supplemental Fig. 3, first and second panel), but hCNT1 was transiently observed at the cell surface in early G1 (1–2 h after inhibitor recovery; Supplemental Fig. 3, third and fourth panels) but not at late G1 cells (4 h after inhibitor recovery; Supplemental Fig. 3, fifth panel). These results identify a brief continuum of hCNT1 cell surface expression after mitosis (early G1) with a rapid loss of hCNT1 expression in late G1.
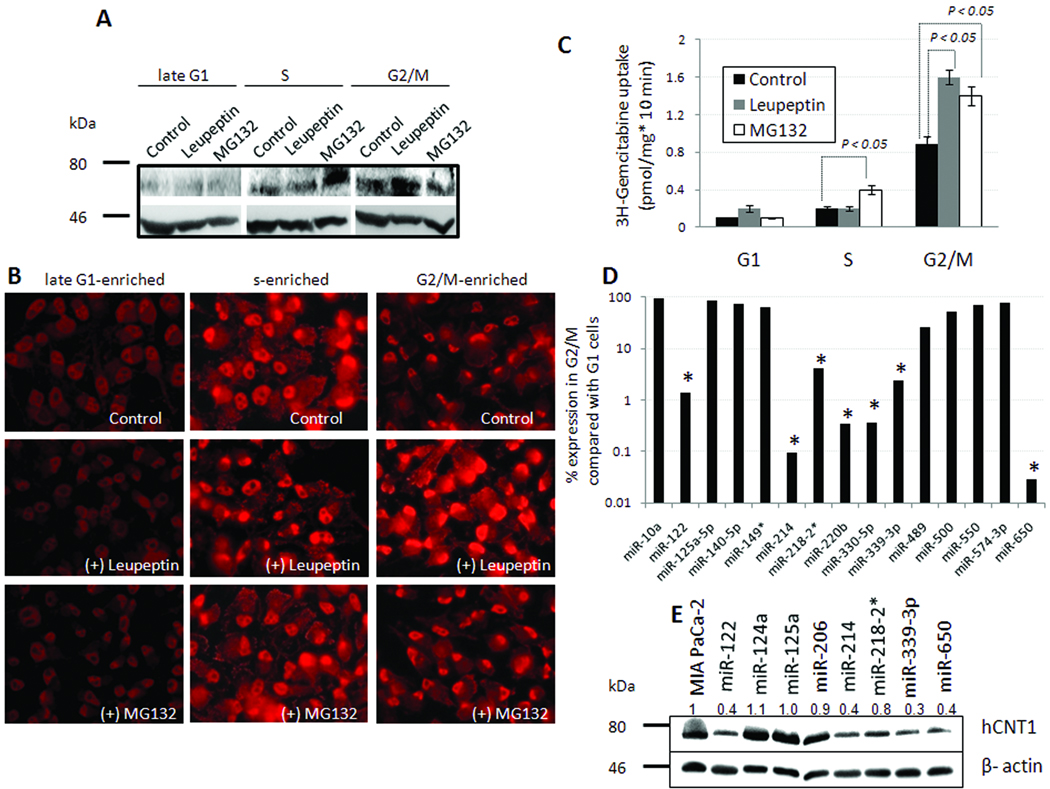
hCNT1 protein turnover by proteasomal and lysosomal pathways: miRNAs-122, 214, 339-3p, and 650 suppressed hCNT1 protein expression
The presence of steady levels of hCNT1 mRNA with varying levels of hCNT1 protein in the cell cycle suggest that alterations in hCNT1 protein synthesis or degradation controlled hCNT1 activity in MIA PaCa-2 cells. In order to test between these possibilities, we first inhibited the cellular protein degradation machineries using pharmacological inhibitors and assessed the level of hCNT1 protein accumulation at different phases of cell cycle. Pretreatment of late-G1 arrested cells with MG132 (a proteasome inhibitor) or leupeptin (a lysosome inhibitor) did not significantly increase hCNT1 protein levels (Fig.5A). Unlike in late G1 cells, hCNT1 protein was predominantly directed to proteasomal degradation in S phase-enriched cells, as judged by increased accumulation of hCNT1 protein only after MG132 treatment but not with leupeptin treatment (Fig.5A). hCNT1 in G2/M-enriched MIA PaCa-2 cells exhibited partial sensitivities to both MG132 and leupeptin treatments (Fig.5A). Immunocytochemical analysis indicated that proteasome inhibition in S and G2/M phase cells and lysosome inhibition by leupeptin in G2/M cells not only increased hCNT1 cellular staining but also appeared to augment the cell surface and nuclear expressions of hCNT1 (Fig. 5B). As a result, we tested whether MG132 and leupeptin could increase hCNT1 function and improve 3H-gemcitabine transport into drug-resistant MIA PaCa-2 cells. Our data showed that MG132 and leupeptin marginally increased 3H-gemcitabine transport by ~2- and ~1.8-folds in S and G2/M phase cells, respectively (Fig. 5C).
Figure 5. hCNT1 turnover during cell cycle.
A, Western blotting analysis showing hCNT1 turnover in serum synchronized MIA PaCa-2 cells, incubated with MG132 (20 µM) or leupeptin (20 µM) for 3 h. A β-actin antibody was used to judge input levels (bottom panel). B, Immunocytochemical analysis of hCNT1 in serum-synchronized MIA PaCa-2 cells, incubated with MG132 (20 µM) or leupeptin (20 µM) for 3 h. Nuclei stained with DAPI are blue. Original magnification X40. C, hCNT1-mediated uptake of 3H-gemcitabine (0.02 µM) in serum synchronized MIA PaCa-2 cells, incubated with MG132 (20 µM) or leupeptin (20 µM) for 3 h. hCNT1 contribution towards gemcitabine cellular uptake were plotted. Columns, mean of triplicate; bars, SE. n=3. D, Decreased expression of putative hCNT1-modulating miRNAs in G2/M MIA PaCa-2 cells compared with G1 MIA PaCa-2 cells. Asterisk indicates >10-fold reduction. E, Western blotting analysis showing hCNT1 (top panel) and β-actin (bottom panel) expression in control (first lane), and MIA PaCa-2 cells stably expressing predicted hCNT1-(miR-122, miR-125a, miR-214, miR-218-2*, miR-339-3p, miR-650), hENT1- (miR124a), or hCNT3- (miR-206) modulating miRNAs. Average hCNT1:β-actin ratios (n=2; normalized to control) are indicated (top) for comparison.
Although no reduction in hCNT1 mRNA was noticed in G1 MIA PaCa-2 cells, hCNT1 protein was almost undetectable. Further, the inhibition of protein degradation machineries did not increase hCNT1 protein, suggesting that a lack of translation and not accelerated degradation as the cause for hCNT1 loss in G1. Since recent evidence show that microRNAs (miRNAs) can regulate diverse cellular processes by binding to the 3’UTR of target mRNAs and inhibiting their translation (28, 29), we next investigated miRNA-mediated regulation of hCNT1 in G1 cells. Expression profiling of 754 miRNAs in MIA PaCa-2 cells identified a >2-fold decrease in 145 miRNAs in G2/M-enriched cells compared with G1-enriched cells (Supplemental File 1), out of which 7 predicted hCNT1-modulating miRNAs (miR-122, miR-214, miR-218-2*, miR-220b, miR-330-5p, miR-339-3p, and miR-650; Fig 5D; asterisks) (Supplemental Table 2) were decreased by more than 10 folds. Overexpression by lentiviral gene transfer of selective miRNAs identified miRNA-122, miRNA-214, miRNA-339-3p, and miRNA-650 to significantly reduce hCNT1 protein levels (Fig. 5E; Supplemental Table 3) without significantly altering hCNT1 and hENT1 mRNA levels in MIA PaCa-2 cells (Supplemental Fig. 4). In addition, these miRNAs did not significantly alter hENT1 protein levels in MIA PaCa-2 cells (data not shown). Subsequent functional analyses in L3.6pl cells identified four miRNAs (miR-122, miR-214, miR-339-3p, and miR-650) to significantly reduce both hCNT1 transport activity and gemcitabine cytotoxicity, except for miR-339-3p which did not alter gemcitabine cytotoxicity (Supplemental Fig. 5). In MIA PaCa-2 cells, despite all four miRNAs decreasing hCNT1 transport activity, only miR-122 also moderately decreased gemcitabine cytotoxicity (Supplemental Fig. 5). Together, these data suggest that miRNA-mediated control of gemcitabine chemosensitivity is governed by complex, cell line-dependent mechanisms most likely involving both the endogenous functional levels of hCNT1 as well as the other diverse targets of the respective miRNAs.
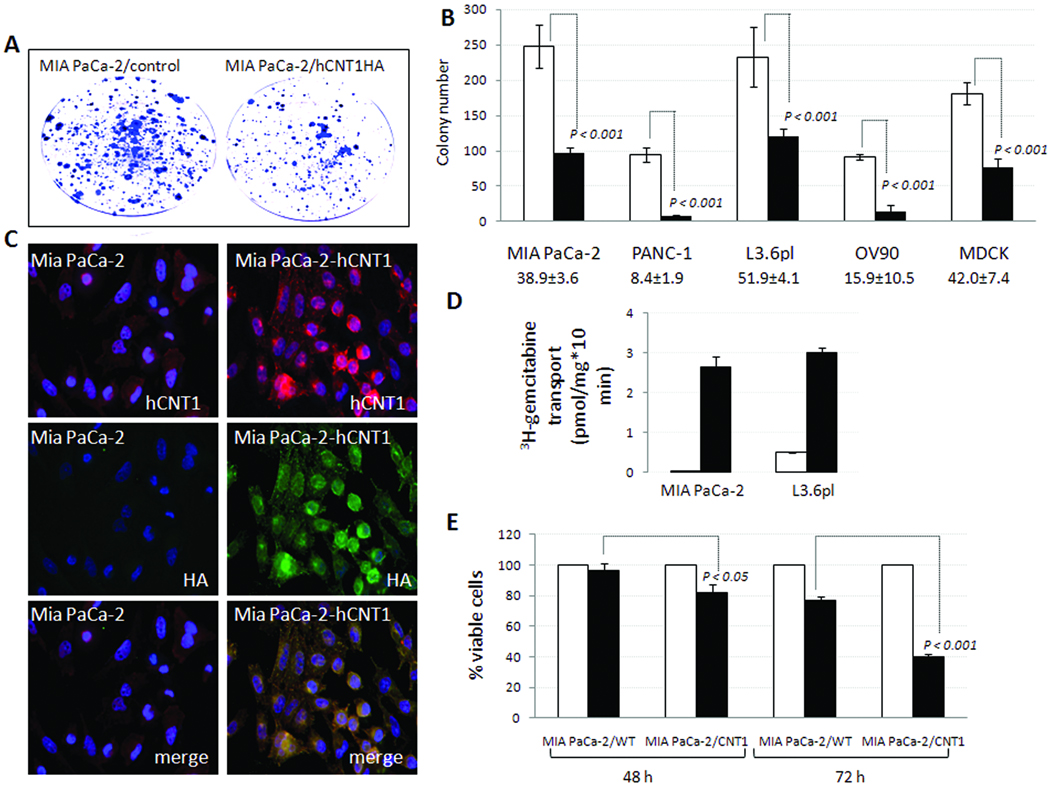
Retroviral expression of hCNT1 cDNA lacking 3’UTR in MIA PaCa-2 cells resulted in constitutive hCNT1 expression, reduced MIA PaCa-2 clonogenicity, and augmented gemcitabine chemosensitization
Since hCNT1 expression is reduced in pancreatic cancer, we next assessed whether hCNT1 overexpression could alter the growth characteristics of the MIA PaCa-2 cell line in the absence of gemcitabine. Retroviral gene transfer of the hCNT1 coding sequence alone (without 5’ and 3’ UTRs) tagged to the hemagglutinin (HA) epitope (Supplemental Fig. 6) substantially reduced colony forming abilities in MIA PaCa-2 and PANC-1 (gemcitabine-resistant), L3.6pl (gemcitabine-sensitive), OV90 (ovarian carcinoma), and MDCK (non-cancerous canine kidney cell line) cell lines (Fig. 6B). The cell lines with lower endogenous hCNT1 activities (MIA PaCa-2 and PANC-1) exhibited somewhat higher decreases in clonogenic survival (>60% and >90% reductions, respectively) than the L3.6pl cell line (<50% reduction) which possesses a relatively higher endogenous hCNT1 activity. Ectopic hCNT1HA in MIA PaCa-2 and L3.6pl clones displayed a constitutive, high hCNT1 expression at both the cell surface and intracytoplasmic vesicular structures (Fig. 6C) and steeply augmented 3H-gemcitabine transport (Fig. 6D) and chemosensitization (Fig. 6E).
Figure 6. Exogenous hCNT1 reduced clonogenic survival of MIA PaCa-2 cells and increased gemcitabine transport and chemosensitization.
A, Clonogenic assay of MIA PaCa-2 (control) and MIA PaCa-2-hCNT1HA cells. B, Clone counts made from clonogenic assays in control (open bars) and hCNT1-HA (filled bars) expressing normal and cancer cell lines were plotted. Percent survival in hCNT1 expressing cells compared with control cells are indicated (bottom). Data are mean ± SD, n=3. C, Immunocytochemical analysis of MIA PaCa-2 cells stably expressing hCNT1HA. Cells were coimmunostained with 1:1000 goat anti-hCNT1 polyclonal (red) and 1:1000 rabbit anti-HA polyclonal antibody (green) antibodies. Nuclei stained with DAPI are blue. Original magnification X40. D, 3H-gemcitabine cellular uptake in WT (open bars) and clones (filled bars) stably expressing hCNT1. Columns, mean of triplicate; bars, SE. n=3. D, 3 × 103 control (open bars) and hCNT1HA-expressing MIA PaCa-2 cells (filled bars) were treated with gemcitabine (100 µM) and percent inhibition of cellular proliferation measured by an MTT assay was plotted. Points, mean of triplicate; bars, SE; n=3.
Discussion
We provide primal evidence for the frequent downregulation of hCNT1 expression in human pancreatic tumors and that the hCNT1-mediated gemcitabine transport levels correlate with gemcitabine cytotoxicity. More importantly, our studies demonstrate that restrictive, cell cycle-dependent expression and degradation characteristics of hCNT1 act as limiting factors for a sustained and concentrative uptake of gemcitabine in drug-resistant pancreatic cancer. Our data identify novel mechanisms for nucleoside drug resistance in pancreatic cancer cells and suggest modulation of hCNT1 expression (by proteasomal inhibitors, selective miRNA antagonists, or direct hCNT1 gene transfer) as a promising approach for improving nucleoside drug sensitivity in pancreatic cancer. In addition, our data support hCNT1 as a putative tumor suppressor in pancreatic cancer.
Several lines of investigation show gemcitabine and other nucleoside-derived anticancer drugs causing cessation of tumor cell proliferation at the S phase of the cell cycle (30–34). Consistent with augmented responses at S phase, experimental evidence demonstrates parallel inductions of hCNT1 (27) and deoxycytidine kinase (dCK) (35, 36), a transporter and metabolic enzyme involved in the first and key rate-limiting gemcitabine cellular activation steps, respectively. While our findings lend credence to hCNT1 protein being induced during S phase, maximal magnitudes of hCNT1 cell surface expression and functional activity are not seen until the G2/M phase. Additionally, S phase MIA PaCa-2 cells mainly exhibit intracytoplasmic hCNT1 staining with an extended time lag between hCNT1 synthesis and hCNT1 cell surface accrual. A previous report on bone marrow leukemia cells shows the number of nitrobenzylthioinosine binding sites (a measure of ENT1 expression) to be significantly increased during S phase (37). In agreement with those observations, we observed significant cell surface hENT1 immunoreactivity and Na+-independent hENT1-mediated gemcitabine uptake in both G1 and S phases of MIA PaCa-2 cells. While these data seem consistent with the involvement of ENTs in S phase arrest of solid tumor cells, time-course studies from other laboratories have demonstrated that metabolic conversion of gemcitabine to gemcitabine triphosphate within cells peaked at 12 h (32), with a permanent block of cells in S phase noticed at ~16 h post gemcitabine treatment (38). Since G1 extends for ~10 h and S phase extends for ~5–6 h (both totaling ~15–16 h) in many cell types, hCNT1 transport-overshoot at the G2/M phase could play a key role in forcing gemcitabine-treated tumor cells to S phase arrest. In this regard, hCNT1 transport overshoot at the G2/M phase was also observed in gemcitabine-sensitive pancreatic cancer cells and in other solid tumor cell lines (e.g. ovarian tumor cells; personal communications) in which gemcitabine induces S phase arrests.
To further acquire mechanistic insights into restricted hCNT1 expression in drug-resistant MIA PaCa-2 cells, we investigated the transcriptional, translational, and post-translational (e.g. protein degradation) control of hCNT1 during the cell cycle. Our screen identified miRNAs 122, 214, 339-3p, and 650 to decrease hCNT1 protein expression with their levels cycling between G1 and S/G2/M phases to influence hCNT1 transport levels at these phases. Additionally, increased trafficking of hCNT1 protein to the cell surface facilitated the selective gemcitabine transport-overshoot in G2/M cells. hCNT1 was subjected to both proteasomal and lysosomal degradation. Interestingly, pharmacological inhibition of hCNT1 degradation not only increased hCNT1 cellular levels but also significantly increased cell surface hCNT1 expression and 3H-gemcitabine cellular influxes (in S and G2/M phases). Since several proteasomal inhibitors are now being evaluated as anti-cancer agents (39, 40), the current results suggest that combination drug regimens of these inhibitors and gemcitabine are likely to bring enhanced cytotoxicity to drug-resistant pancreatic cancer.
The present study identified diminution of hCNT1 expression, possibly at multiple levels, in pancreatic cancer. hCNT1 expression was frequently diminished in pancreatic tumors, and restriction of hCNT1 activity to specific phases of the cell cycle further limited gemcitabine transport in drug-resistant MIA PaCa-2 pancreatic cancer cells. Unlike in MIA PaCa-2 cells, hCNT1 expression is continuously observed at the cell periphery of gemcitabine-sensitive L3.6pl and BxPC-3 cells. In this regard, the latter cell lines exhibit well-differentiated, epithelial characteristics (41) with high levels of epithelial markers (e.g. E-cadherin) expression (42–44), whereas MIA PaCa-2 cells show poorly-differentiated characteristics (41) with a mesenchymal morphology (45) and express mesenchymal markers (e.g. vimentin, ZEB1) (42, 46). Since our categorization of pancreatic cancer cell lines, based on hCNT1 transport and gemcitabine sensitivity, to some extent reflect the differentiation state of these pancreatic cancer cells, the observed hCNT1 heterogeneities are a likely representation of the tumor dedifferentiation events in vivo and may not be mere artifacts of the cellular transformation process. Our data also identify reduction in the proliferative capacity of pancreatic cancerous cells upon constitutive expression of hCNT1. While hCNT1 was widely characterized for its transport properties, it is not yet clear whether non-transport related cellular functions are involved in its growth suppressive properties. We believe that hCNT1 brings growth control by inducing differentiation (or reverting dedifferentiation) in pancreatic tumor cells. This hypothesis is based on earlier investigations (19, 21, 22), including our own studies (16, 17), supporting hCNT1’s association with cellular differentiation. In addition, we found that unlike the control MIA PaCa-2 clones that were larger and irregular with corrugated edges, hCNT1-expressing MIA PaCa-2 clones were relatively smaller and compact with differentiated epithelial characteristics (Fig. 6A). Moreover, the ability to induce differentiation might be sufficient enough to promote growth control in pancreatic cancer since pro-differentiation activity has been shown to act as a tumor suppressor mechanism (47, 48). Overall, our data support that hCNT1’s involvement in cellular transport, proliferation, and differentiation could synergistically impact tumor growth control in drug-resistant pancreatic cancer.
Supplementary Material
Acknowledgments
Grant Support: The work was supported by the National Cancer Institute award 5 P50 CA128613 SPORE (RG) and by the Department of Pharmaceutical and Biomedical Sciences, University of Georgia.
Footnotes
Supplementary data for this article are available at Cancer Research Online
Potential Conflict of Interest: No potential conflicts of interest were disclosed.
References
- 1.Wong A, Soo RA, Yong WP, Innocenti F. Clinical pharmacology and pharmacogenetics of gemcitabine. Drug Metab Rev. 2009;41:77–88. doi: 10.1080/03602530902741828. [DOI] [PubMed] [Google Scholar]
- 2.Bayraktar S, Bayraktar UD, Rocha-Lima CM. Recent developments in palliative chemotherapy for locally advanced and metastatic pancreas cancer. World J Gastroenterol. 2010;16:673–682. doi: 10.3748/wjg.v16.i6.673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huber-Ruano I, Pastor-Anglada M. Transport of nucleoside analogs across the plasma membrane: a clue to understanding drug-induced cytotoxicity. Curr Drug Metab. 2009;10:347–358. doi: 10.2174/138920009788499030. [DOI] [PubMed] [Google Scholar]
- 4.Hidalgo M. Pancreatic cancer. N Engl J Med. 2010;362:1605–1617. doi: 10.1056/NEJMra0901557. [DOI] [PubMed] [Google Scholar]
- 5.Veltkamp SA, Pluim D, van Eijndhoven MA, et al. New insights into the pharmacology and cytotoxicity of gemcitabine and 2',2'-difluorodeoxyuridine. Mol Cancer Ther. 2008;7:2415–2425. doi: 10.1158/1535-7163.MCT-08-0137. [DOI] [PubMed] [Google Scholar]
- 6.Mackey JR, Mani RS, Selner M, et al. Functional nucleoside transporters are required for gemcitabine influx and manifestation of toxicity in cancer cell lines. Cancer Res. 1998;58:4349–4357. [PubMed] [Google Scholar]
- 7.Lostao MP, Mata JF, Larrayoz IM, Inzillo SM, Casado FJ, Pastor-Anglada M. Electrogenic uptake of nucleosides and nucleoside-derived drugs by the human nucleoside transporter 1 (hCNT1) expressed in Xenopus laevis oocytes. FEBS Lett. 2000;481:137–140. doi: 10.1016/s0014-5793(00)01983-9. [DOI] [PubMed] [Google Scholar]
- 8.Garcia-Manteiga J, Molina-Arcas M, Casado FJ, Mazo A, Pastor-Anglada M. Nucleoside transporter profiles in human pancreatic cancer cells: role of hCNT1 in 2',2'-difluorodeoxycytidine- induced cytotoxicity. Clin Cancer Res. 2003;9:5000–5008. [PubMed] [Google Scholar]
- 9.Hu H, Endres CJ, Chang C, et al. Electrophysiological characterization and modeling of the structure activity relationship of the human concentrative nucleoside transporter 3 (hCNT3) Mol Pharmacol. 2006;69:1542–1553. doi: 10.1124/mol.105.018945. [DOI] [PubMed] [Google Scholar]
- 10.Mackey JR, Yao SY, Smith KM, et al. Gemcitabine transport in xenopus oocytes expressing recombinant plasma membrane mammalian nucleoside transporters. J Natl Cancer Inst. 1999;91:1876–1881. doi: 10.1093/jnci/91.21.1876. [DOI] [PubMed] [Google Scholar]
- 11.Andersson R, Aho U, Nilsson BI, et al. Gemcitabine chemoresistance in pancreatic cancer: molecular mechanisms and potential solutions. Scand J Gastroenterol. 2009;44:782–786. doi: 10.1080/00365520902745039. [DOI] [PubMed] [Google Scholar]
- 12.Farrell JJ, Elsaleh H, Garcia M, et al. Human equilibrative nucleoside transporter 1 levels predict response to gemcitabine in patients with pancreatic cancer. Gastroenterology. 2009;136:187–195. doi: 10.1053/j.gastro.2008.09.067. [DOI] [PubMed] [Google Scholar]
- 13.Giovannetti E, Del Tacca M, Mey V, et al. Transcription analysis of human equilibrative nucleoside transporter-1 predicts survival in pancreas cancer patients treated with gemcitabine. Cancer Res. 2006;66:3928–3935. doi: 10.1158/0008-5472.CAN-05-4203. [DOI] [PubMed] [Google Scholar]
- 14.Marechal R, Mackey JR, Lai R, et al. Human equilibrative nucleoside transporter 1 and human concentrative nucleoside transporter 3 predict survival after adjuvant gemcitabine therapy in resected pancreatic adenocarcinoma. Clin Cancer Res. 2009;15:2913–2919. doi: 10.1158/1078-0432.CCR-08-2080. [DOI] [PubMed] [Google Scholar]
- 15.Spratlin J, Sangha R, Glubrecht D, et al. The absence of human equilibrative nucleoside transporter 1 is associated with reduced survival in patients with gemcitabine-treated pancreas adenocarcinoma. Clin Cancer Res. 2004;10:6956–6961. doi: 10.1158/1078-0432.CCR-04-0224. [DOI] [PubMed] [Google Scholar]
- 16.Govindarajan R, Bakken AH, Hudkins KL, et al. In situ hybridization and immunolocalization of concentrative and equilibrative nucleoside transporters in the human intestine, liver, kidneys, and placenta. Am J Physiol Regul Integr Comp Physiol. 2007;293:R1809–R1822. doi: 10.1152/ajpregu.00293.2007. [DOI] [PubMed] [Google Scholar]
- 17.Govindarajan R, Endres CJ, Whittington D, et al. Expression and hepatobiliary transport characteristics of the concentrative and equilibrative nucleoside transporters in sandwich-cultured human hepatocytes. Am J Physiol Gastrointest Liver Physiol. 2008;295:G570–G580. doi: 10.1152/ajpgi.00542.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu C, Orozco C, Boyer J, et al. BioGPS: an extensible and customizable portal for querying and organizing gene annotation resources. Genome Biol. 2009;10:R130. doi: 10.1186/gb-2009-10-11-r130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aymerich I, Pastor-Anglada M, Casado FJ. Long term endocrine regulation of nucleoside transporters in rat intestinal epithelial cells. J Gen Physiol. 2004;124:505–512. doi: 10.1085/jgp.200409086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Valdes R, Fernandez-Veledo S, Aymerich I, Casado FJ, Pastor-Anglada M. TGF-beta transcriptionally activates the gene encoding the high-affinity adenosine transporter CNT2 in rat liver parenchymal cells. Cell Mol Life Sci. 2006;63:2527–2537. doi: 10.1007/s00018-006-6240-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Soler C, Garcia-Manteiga J, Valdes R, et al. Macrophages require different nucleoside transport systems for proliferation and activation. FASEB J. 2001;15:1979–1988. doi: 10.1096/fj.01-0022com. [DOI] [PubMed] [Google Scholar]
- 22.Soler C, Felipe A, Garcia-Manteiga J, et al. Interferon-gamma regulates nucleoside transport systems in macrophages through signal transduction and activator of transduction factor 1 (STAT1)-dependent and -independent signalling pathways. Biochem J. 2003;375:777–783. doi: 10.1042/BJ20030260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Klein K, Kullak-Ublick GA, Wagner M, Trauner M, Eloranta JJ. Hepatocyte nuclear factor-4alpha and bile acids regulate human concentrative nucleoside transporter-1 gene expression. Am J Physiol Gastrointest Liver Physiol. 2009;296:G936–G947. doi: 10.1152/ajpgi.90678.2008. [DOI] [PubMed] [Google Scholar]
- 24.Gloeckner-Hofmann K, Guillen-Gomez E, Schmidtgen C, et al. Expression of the high-affinity fluoropyrimidine-preferring nucleoside transporter hCNT1 correlates with decreased disease-free survival in breast cancer. Oncology. 2006;70:238–244. doi: 10.1159/000094541. [DOI] [PubMed] [Google Scholar]
- 25.Farre X, Guillen-Gomez E, Sanchez L, et al. Expression of the nucleoside-derived drug transporters hCNT1, hENT1 and hENT2 in gynecologic tumors. Int J Cancer. 2004;112:959–966. doi: 10.1002/ijc.20524. [DOI] [PubMed] [Google Scholar]
- 26.Hamilton SR, Yao SY, Ingram JC, et al. Subcellular distribution and membrane topology of the mammalian concentrative Na+nucleoside cotransporter rCNT1. J Biol Chem. 2001;276:27981–27988. doi: 10.1074/jbc.M100518200. [DOI] [PubMed] [Google Scholar]
- 27.Valdes R, Casado FJ, Pastor-Anglada M. Cell-cycle-dependent regulation of CNT1, a concentrative nucleoside transporter involved in the uptake of cell-cycle-dependent nucleoside-derived anticancer drugs. Biochem Biophys Res Commun. 2002;296:575–579. doi: 10.1016/s0006-291x(02)00919-1. [DOI] [PubMed] [Google Scholar]
- 28.Schickel R, Boyerinas B, Park SM, Peter ME. MicroRNAs: key players in the immune system, differentiation, tumorigenesis and cell death. Oncogene. 2008;27:5959–5974. doi: 10.1038/onc.2008.274. [DOI] [PubMed] [Google Scholar]
- 29.Fabian MR, Sonenberg N, Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Annu Rev Biochem. 2010;79:351–379. doi: 10.1146/annurev-biochem-060308-103103. [DOI] [PubMed] [Google Scholar]
- 30.Karnitz LM, Flatten KS, Wagner JM, et al. Gemcitabine-induced activation of checkpoint signaling pathways that affect tumor cell survival. Mol Pharmacol. 2005;68:1636–1644. doi: 10.1124/mol.105.012716. [DOI] [PubMed] [Google Scholar]
- 31.Morgan MA, Parsels LA, Parsels JD, Mesiwala AK, Maybaum J, Lawrence TS. Role of checkpoint kinase 1 in preventing premature mitosis in response to gemcitabine. Cancer Res. 2005;65:6835–6842. doi: 10.1158/0008-5472.CAN-04-2246. [DOI] [PubMed] [Google Scholar]
- 32.Ostruszka LJ, Shewach DS. The role of cell cycle progression in radiosensitization by 2',2'-difluoro-2'-deoxycytidine. Cancer Res. 2000;60:6080–6088. [PubMed] [Google Scholar]
- 33.Rauchwerger DR, Firby PS, Hedley DW, Moore MJ. Equilibrative-sensitive nucleoside transporter and its role in gemcitabine sensitivity. Cancer Res. 2000;60:6075–6079. [PubMed] [Google Scholar]
- 34.Shi Z, Azuma A, Sampath D, Li YX, Huang P, Plunkett W. S-Phase arrest by nucleoside analogues and abrogation of survival without cell cycle progression by 7-hydroxystaurosporine. Cancer Res. 2001;61:1065–1072. [PubMed] [Google Scholar]
- 35.Pegoraro L, Bernengo MG. Thymidine kinase, deoxycytidine kinase and deoxycytidylate deaminase activities in phytohaemagglutinin stimulated human lymphocytes. Exp Cell Res. 1971;68:283–290. doi: 10.1016/0014-4827(71)90152-2. [DOI] [PubMed] [Google Scholar]
- 36.Wan CW, Mak TW. Deoxycytidine kinase and cytosine nucleoside deaminase activities in synchronized cultures of normal rat kidney cells. Cancer Res. 1978;38:2768–2772. [PubMed] [Google Scholar]
- 37.Powell BL, Gregory BW, Evans JK, et al. Leukapheresis induced changes in cell cycle distribution and nucleoside transporters in patients with untreated acute myeloid leukemia. Leukemia. 1991;5:1037–1042. [PubMed] [Google Scholar]
- 38.Hertel LW, Boder GB, Kroin JS, et al. Evaluation of the antitumor activity of gemcitabine (2',2'-difluoro-2'-deoxycytidine) Cancer Res. 1990;50:4417–4422. [PubMed] [Google Scholar]
- 39.Adams J. The development of proteasome inhibitors as anticancer drugs. Cancer Cell. 2004;5:417–421. doi: 10.1016/s1535-6108(04)00120-5. [DOI] [PubMed] [Google Scholar]
- 40.Fahy BN, Schlieman MG, Virudachalam S, Bold RJ. Schedule-dependent molecular effects of the proteasome inhibitor bortezomib and gemcitabine in pancreatic cancer. J Surg Res. 2003;113:88–95. doi: 10.1016/s0022-4804(03)00201-4. [DOI] [PubMed] [Google Scholar]
- 41.Winter JM, Ting AH, Vilardell F, et al. Absence of E-cadherin expression distinguishes noncohesive from cohesive pancreatic cancer. Clin Cancer Res. 2008;14:412–418. doi: 10.1158/1078-0432.CCR-07-0487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Arumugam T, Ramachandran V, Fournier KF, et al. Epithelial to mesenchymal transition contributes to drug resistance in pancreatic cancer. Cancer Res. 2009;69:5820–5828. doi: 10.1158/0008-5472.CAN-08-2819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Song Y, Washington MK, Crawford HC. Loss of FOXA1/2 is essential for the epithelial-to-mesenchymal transition in pancreatic cancer. Cancer Res. 2010;70:2115–2125. doi: 10.1158/0008-5472.CAN-09-2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nishioka R, Itoh S, Gui T, et al. SNAIL induces epithelial-to-mesenchymal transition in a human pancreatic cancer cell line (BxPC3) and promotes distant metastasis and invasiveness in vivo. Exp Mol Pathol. doi: 10.1016/j.yexmp.2010.05.008. [DOI] [PubMed] [Google Scholar]
- 45.Seidel B, Braeg S, Adler G, Wedlich D, Menke A. E- and N-cadherin differ with respect to their associated p120ctn isoforms and their ability to suppress invasive growth in pancreatic cancer cells. Oncogene. 2004;23:5532–5542. doi: 10.1038/sj.onc.1207718. [DOI] [PubMed] [Google Scholar]
- 46.Li Y, VandenBoom TG, 2nd, Kong D, et al. Up-regulation of miR-200 and let-7 by natural agents leads to the reversal of epithelial-to-mesenchymal transition in gemcitabine-resistant pancreatic cancer cells. Cancer Res. 2009;69:6704–6712. doi: 10.1158/0008-5472.CAN-09-1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sellers WR, Novitch BG, Miyake S, et al. Stable binding to E2F is not required for the retinoblastoma protein to activate transcription, promote differentiation, and suppress tumor cell growth. Genes Dev. 1998;12(1):95–106. doi: 10.1101/gad.12.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thomas DM, Carty SA, Piscopo DM, et al. The retinoblastoma protein acts as a transcriptional coactivator required for osteogenic differentiation. Mol Cell. 2001;8(2):303–316. doi: 10.1016/s1097-2765(01)00327-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.