Abstract
The safety and efficacy of viral therapies for solid tumors can be enhanced by redirecting the virus infection to tumor-specific cell-surface markers. Successful retargeting of herpes simplex virus type 1 (HSV-1) has been achieved using vectors that carry a modified envelope glycoprotein D (gD) engineered to interact directly with novel receptors. In addition, soluble bridging molecules (adapters) have been used to link gD indirectly to cell-specific receptors. Here, we describe the development of an adapter connecting gD to the common tumor antigen carcinoembryonic antigen (CEA). The adapter consisted of a CEA-specific single-chain antibody fused to the gD-binding region of the gD receptor, herpes virus entry mediator (HVEM). We used this adapter in combination with a vector that is detargeted for recognition of the widely expressed gD receptor nectin-1, but retains an intact binding region for the less common HVEM. We show that the adapter enabled infection of HSV-resistant Chinese hamster ovary (CHO) cells expressing ectopic CEA and nectin-1/CEA-bearing human gastric carcinoma cells that are resistant to the vector alone. We observed cell-to-cell spread following adapter-mediated infection in vitro and reduced tumor growth in vivo, indicating that this method of vector retargeting may provide a novel strategy for tumor-specific delivery of tumoricidal HSV.
Introduction
The goal of this study was to develop a soluble bridging molecule (adapter) to redirect herpes simplex virus type 1 (HSV-1) infection to cells that express the tumor marker carcinoembryonic antigen (CEA). Normal HSV-1 infection requires the binding of viral envelope glycoprotein D (gD) to one of several specific entry receptors, predominantly nectin-1/HveC, a widely expressed intercellular adhesion molecule, and herpes virus entry mediator (HVEM)/HveA, expressed mainly on cells of the immune system.1 Receptor binding activates the latent effector (“profusion”) function of gD, which in turn is believed to activate gB and gH, the two key mediators of viral capsid delivery to the cytoplasm via fusion of the viral envelope with cellular membranes.2 Thus, because gD binding to its natural receptors launches the infection cascade, redirecting HSV infection requires (i) the elimination of these natural receptor-binding activities and (ii) insertion of a ligand for an alternate receptor in such a manner that the new binding interaction causes activation of the gD profusion function.
Accumulating knowledge of the structure–function relationship of gD acquired over many years has recently enabled the rational construction of detargeted and retargeted versions of gD.3,4,5 These constructs hold great promise for highly specific HSV targeting to a variety of cell types and tissues although their ability to function with diverse ligands is yet unknown.
An alternate retargeting strategy that does not require target-specific engineering of viral gD involves the use of bispecific adapters to promote virus interaction with novel receptors.6,7 This strategy is based in part on the repeated demonstration that HSV infection through HVEM and nectin-1 is blocked by soluble versions of these receptors,8,9,10,11,12 suggesting that adapters composed of the gD-binding domain of either receptor combined with a targeting ligand would obviate the need to detarget gD from both receptors. Our previous study using a nectin-1-based adapter targeted by a single-chain antibody (scFv) against the human epidermal growth factor receptor demonstrated efficient adapter-mediated infection of gD receptor–deficient cells expressing ectopic epidermal growth factor receptor, but infection via nectin-1 was not blocked.13
Here, we combined a novel HVEM-based adapter with a nectin-1-detargeted virus to promote CEA-dependent infection of cancer cells. The rationale for this design was that functional HVEM expression beyond the immune system is relatively limited8,14,15,16 and thus that organs such as stomach17 should be largely resistant to nectin-1-detargeted viruses. Because these viruses remain capable of binding to HVEM, tumors in these organs may be specifically targeted with HVEM-based adapters. CEA is an attractive antigen for therapeutic targeting because it is expressed in a high percentage of certain cancers,18 but is rare in normal adult tissues. We show that the adapters promoted the CEA-dependent HSV-1 infection of HSV-resistant Chinese hamster ovary (CHO)-K1 cells and increased the infection of CEA-bearing human gastric carcinoma MKN45 cells by a nectin-1-detargeted mutant virus. Lateral virus spread was detectable in vitro following adapter-enhanced infection. In vivo experiments demonstrated an adapter-dependent infection and growth inhibition of MKN45 tumors. Our results indicate that adapters may be useful not only for the cell-specific delivery of nonreplicating HSV vectors, but also to target replication-competent vectors for specific tumor destruction.
Results
scCEA-HveA adapter-mediated HSV infection of CEA-expressing CHO cells
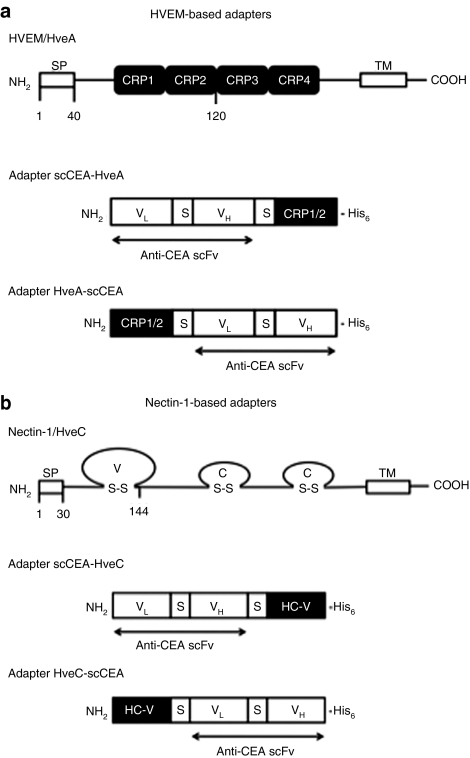
To promote HSV-1 interaction with CEA, we generated a bispecific adapter consisting of, in sequence, a scFv against human CEA, a Gly4–Ser linker, the gD-binding 102 N-terminal amino acids of HVEM/HveA [cysteine-rich pseudorepeats (CRPs) 1 and 2],19 and a C-terminal histidine (His6) tag (scCEA-HveA; Figure 1a). The coding sequence was assembled in a mammalian expression plasmid and the protein expressed by transfection of 293T cells. The adapter protein was affinity-purified from the culture media and identified by immunoblotting with His6- and HVEM-specific antibodies (data not shown), as described earlier.20
Figure 1.
HSV-1 receptors and CEA-directed adapters. Structures of full-length (a) HVEM and (b) nectin-1, including their signal peptides (SPs), and the four adapters used in this study. CEA, carcinoembryonic antigen; CRP, cysteine-rich pseudorepeat; HC-V, nectin-1 variable region; His6, histidine tag; HSV, herpes simplex virus; HVEM, herpes virus entry mediator; S–S, disulfide bond; TM, transmembrane region; VL and VH, light- and heavy-chain variable region of anti-CEA; S, (Gly)4Ser spacer.
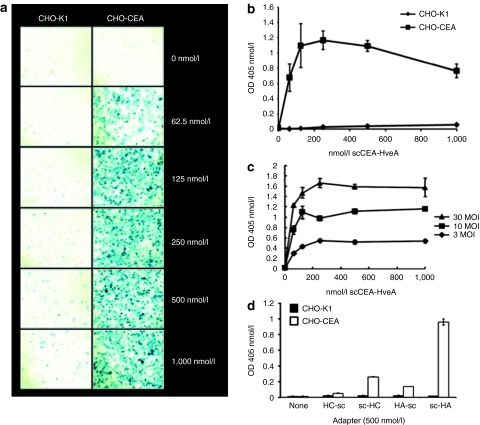
CHO-K1 cells are resistant to HSV-1 infection because of the absence of gD receptors.8,9 We tested the activity of the scCEA-HveA adapter by infection of CEA-expressing (CHO-CEA)21 and parental CHO-K1 cells with a replication-defective reporter virus, QOZHG,22 in the presence of increasing amounts of adapter. QOZHG contains a lacZ expression cassette enabling detection of infected cells by X-gal staining and quantification of β-galactosidase activity by o-nitrophenyl-β--galactopyranoside assay. As shown in Figure 2a,b, the adapters increased the infection of CHO-CEA cells at all concentrations tested, whereas infection of CHO-K1 cells remained minimal throughout. The level of adapter-mediated infection of CHO-CEA cells increased with the multiplicity of infection (MOI; established on complementing Vero-7b cells23), but reached a plateau at ~250 nmol/l adapter at MOIs of 3–30 (Figure 2c), suggesting that this concentration of adapter was saturating for the receptor. We compared the activity of scCEA-HveA to three related adapters (Figure 1a,b) that had (i) the positions of the scFv and HVEM portions of scCEA-HveA switched such that the HVEM CRPs were N-terminal to the scFv (HveA-scCEA); (ii) the HVEM portion of scCEA-HveA replaced by the gD-binding V domain of nectin-1/HveC (scCEA-HveC); or (iii) the nectin-1 V domain positioned N-terminally to scFv-His6 (HveC-scCEA). We observed little activity with the two C-terminal scFv adapters (HveA-scCEA and HveC-scCEA) and reduced activity with the N-terminal scFv-nectin-1 fusion (scCEA-HveC) (Figure 2d). These results established that the relative positions of the specificity elements of the adapter are important activity determinants and that the gD-binding region of HVEM as an adapter component is at least as effective as the previously used V domain of nectin-1.13 Together, these observations demonstrated that scCEA-HveA is an efficient mediator of CEA-targeted HSV infection.
Figure 2.
Adapter-mediated infection of Chinese hamster ovary-carcinoembryonic antigen (CHO-CEA) cells. (a,b) Infection of CHO-K1 and CHO-CEA cells with QOZHG (MOI = 10) in the presence of increasing amounts of scCEA-HveA. Cells were stained with (a) X-gal or (b) processed for o-nitrophenyl-β--galactoside (ONPG) assay at 16 hours postinfection. (c) CHO-CEA cells were infected with QOZHG at different MOIs in the presence of increasing concentrations of scCEA-HveA adapter. Virus entry was determined by ONPG assay, as above. (d) CHO-K1 (filled bars) and CHO-CEA (open bars) cells were infected with QOZHG (MOI = 10) in the absence of adapter or in the presence of HveC-scCEA, scCEA-HveC, HveA-scCEA, or scCEA-HveA, abbreviated as HC-sc, sc-HC, HA-sc, and sc-HA, respectively. Virus entry was determined at 16 hours postinfection by ONPG assay. Each value in b–d represents the mean of three determinations ± SD. MOI, multiplicity of infection.
Specificity of adapter-mediated CHO-CEA infection
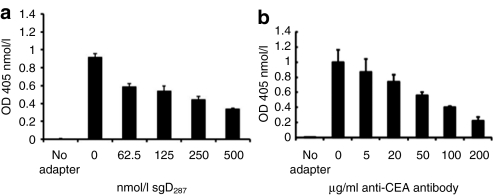
To confirm that scCEA-HveA functioned by interacting with both viral gD and cellular CEA, we performed blocking experiments with the soluble ectodomain of gD (sgD287)24 and with anti-CEA antibodies. As shown in Figure 3a, preincubation of QOZHG and scCEA-HveA (250 nmol/l) with increasing amounts of sgD287 inhibited adapter-mediated infection of CHO-CEA cells in a dose-dependent manner. Likewise, preincubation of the cells with various concentrations of CEA-specific monoclonal antibody reduced scCEA-HveA-mediated infection (Figure 3b); the antibody had no effect on QOZHG infection of Vero cells (data not shown). Together, these results indicated that the adapters acted by connecting virion gD to cell-surface CEA, thereby initiating virus entry into the cells.
Figure 3.
Competitive inhibition of scCEA-HveA-mediated infection. Chinese hamster ovary (CHO)-CEA cells were infected with QOZHG (multiplicity of infection = 10) in the presence of 250 nmol/l scCEA-HveA and increasing concentrations of (a) soluble gD ectodomain (sgD287) or (b) anti-CEA monoclonal antibody F11-39. Infection without adapter was used as a control. Virus entry at 16 hours postinfection was determined by o-nitrophenyl-β--galactoside assay. Values represent the means of three determinations ± SD. CEA, carcinoembryonic antigen.
Adapter-mediated infection of CEA-expressing human cancer cells by nectin-1-detargeted HSV
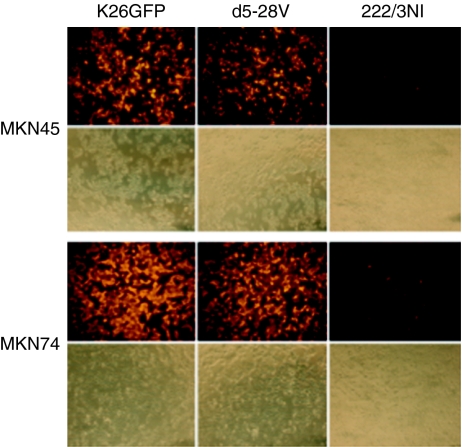
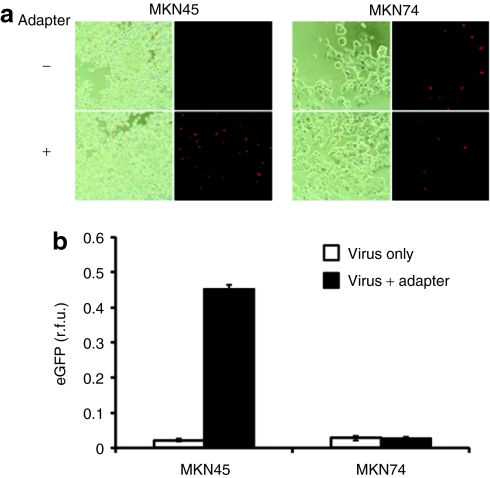
Among the principal HSV-1 entry receptors, nectin-1 is widely expressed,25 while HVEM is found predominantly on cells of the immune system.8,14,15,17 Although HVEM has also been detected on various nonimmune cell lines, several published reports have shown that its expression on these cells is often insufficient for HSV-1 infection.16,26,27 Hence, although HSV detargeting from nectin-1 is likely a universal requirement to limit infection to the target cell or tissue, preservation of the HVEM-binding activity of the virus may not dramatically increase off-target infection, depending on the site of virus application. Accordingly, we used mutant viruses detargeted for HVEM or nectin-1 to assess the contribution of HVEM to the HSV susceptibility of CEA-positive MKN45 and CEA-negative MKN74 (ref. 28) gastric carcinoma cells. K-d5-28V (K26-gD:d5-28V in ref. 27) is a mutant virus that is highly impaired for HVEM-mediated infection, while K-222/3NI (K26-gD:R222N/F223I in ref. 27) has an ~750-fold reduced ability to infect cells via nectin-1. We infected MKN45 and MKN74 cells with equal numbers of virions of the two receptor-restricted viruses and their unrestricted parent, K26GFP,29 and visualized infection at 8 hours postinfection by anti-VP16 immunostaining. As illustrated in Figure 4, both cell lines were infected efficiently by K26GFP and K-d5-28V, while infection by K-222/3NI was almost undetectable, indicating that nectin-1 is the principal HSV-1 entry receptor on these cells. We then compared K-222/3NI infection in the presence and absence of the scCEA-HveA adapter. Increased infection was observed on MKN45 cells in the presence of the adapter, while infection of MKN74 cells was essentially unaltered (Figure 5). The adapter had no effect on infection of MKN45 cells by K26GFP (data not shown).
Figure 4.
Susceptibility of carcinoembryonic antigen (CEA)-positive MKN45 and CEA-negative MKN74 cells to wild-type gD herpes simplex virus type 1 and receptor-restricted derivatives. Cells were infected with K26GFP, nectin-1-restricted K-d5-28V, or HVEM-restricted K-222/3NI at 100 genome copies/cell and stained at 8 hours postinfection with anti-VP16 antibody and Cy3-conjugated secondary antibody. The lower panels for each cell line show phase-contrast images of the same fields.
Figure 5.
Adapter-mediated K-222/3NI infection of MKN45 cells. (a) MKN45 and MKN74 cells were infected with K-222/3NI (MOI = 0.5) in the presence or absence of scEA-HveA (500 nmol/l). Infection at 8 hours postinfection was detected by anti-VP16 immunostaining (Alexa Fluor 594-conjugated secondary antibody) and fluorescence microscopy (dark panels). Phase-contrast images of the same fields are included for reference (light panels). (b) Mean fluorescent signals from triplicate wells ± SD as determined by Thermo Labsystems Fluoroskan Ascent (Thermo Scientific, Waltham, MA). eGFP, enhanced green fluorescent protein; rfu, relative fluorescent units.
Spread of HVEM-restricted virus following adapter-mediated infection
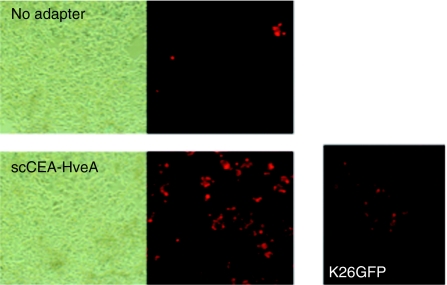
We previously showed that K-222/3NI has a residual ability to use nectin-1 for lateral spread.27 To determine whether this ability is sufficient to mediate cell-to-cell spread of the virus on MKN45 cells, we infected these cells at low MOI (0.01) in the presence of scCEA-HveA adapter, inactivated extracellular virus at 1 hour postinfection, and incubated the monolayer under 0.5% methylcellulose overlay. Anti-VP16 staining at 72 hours postinfection showed multiple clusters of infected cells indicative of lateral virus spread (Figure 6). Infection in the absence of adapter also yielded an occasional multicellular infected center (Figure 6), indicating that spread was not dependent on the presence of the adapter during the initial infection. As a control, the figure also shows a typical plaque formed by receptor-unrestricted K26GFP (MOI = 0.005). Together, these results indicated that K-222/3NI offers an attractive balance between impaired primary infection via nectin-1 and residual ability to spread via the same receptor.
Figure 6.
Lateral spread of K-222/3NI following adapter-mediated infection. MKN45 monolayers were infected with K-222/3NI (MOI = 0.01) in the presence or absence of 500 nmol/l scCEA-HveA, treated with acidic buffer to remove extracellular virus, and left to recover under fresh medium for 8 hours at 37 °C. The cells were then incubated with high-density medium for 2.5 days and stained with anti-VP16 primary antibody and Alexa Fluor 594-conjugated secondary antibody. The panel at the right shows a typical plaque formed by parallel infection with K26GFP.
Adapter-dependent infection of tumor tissue and inhibition of tumor growth
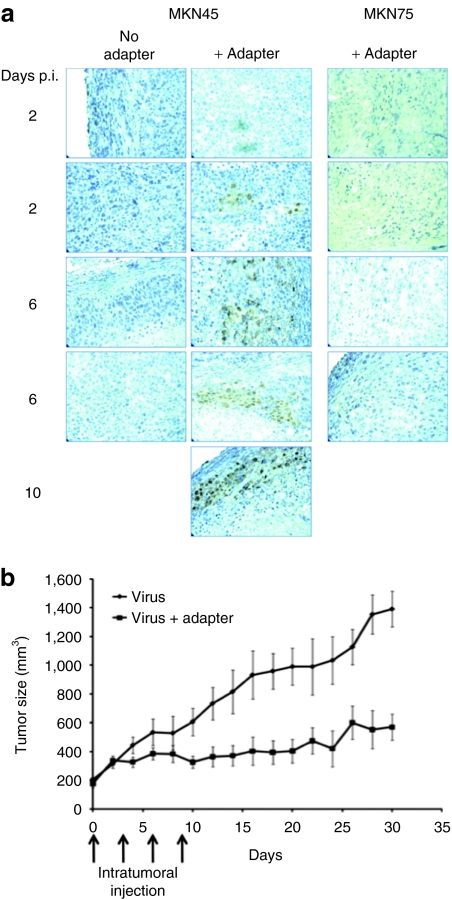
We explored the ability of the adapter to promote infection of established MKN45 tumors in nude mice. Subcutaneous tumors were injected with K-222/3NI virus with or without adapter and thin sections stained for VP16 at 2 or 6 days after virus inoculation. Representative results for two animals per time point are shown in Figure 7a. No staining was observed in MKN45 tumors without adapter or MKN74 tumors with adapter. In contrast, VP16 was detected in each of the MKN45 tumors inoculated with virus plus adapter. The infected area appeared to increase with time after vector administration, suggesting virus spread, but the sample size, including one animal examined after 10 days (Figure 7a), was too small to draw conclusions in this regard.
Figure 7.
Adapter effect on tumor infection and growth. (a) MKN45 and MKN74 tumors grown subcutaneously in nude mice to a size of 200 mm3 were inoculated with K-222/3NI virus (1 × 108 plaque-forming units) with or without scCEA-HveA adapter (1.2 µmol/l; n = 2/group). Tumors were removed at 2, 6, or 10 days after vector administration and sections were immunoperoxidase stained for VP16. Each image represents a separate tumor. No infection was observed in MKN74 tumors inoculated with virus alone (data not shown). (b) Established MKN45 flank tumors in nude mice were inoculated with K-222/3NI with or without adapter as above on days 0, 3, 6, and 9. Tumor diameters were measured every other day and the calculated volumes (tumor size) plotted as mean ± SD. Statistical analysis by t-test yielded a P value of 0.0033.
To determine the effect of the virus–adapter combination on tumor growth, subcutaneous MKN45 tumors were injected 4 times at 3-day intervals with K-222/3NI virus plus adapter or K-222/3NI alone. Measurement of tumor volumes every other day over a period of 30 days showed a significant reduction in tumor growth in the presence of the adapter (P < 0.05, t-test; Figure 7b). Together, these in vivo results indicated that the CEA-specific adapter combined with an unattenuated, HVEM-selective HSV vector can mediate specific infection of CEA-expressing tumor cells and reduce tumor growth.
Discussion
Research into viral therapies for solid tumors has long focused on the development of replication-impaired mutant vectors whose defects are selectively complemented by certain or all tumor cells.30 These so-called oncolytic vectors, including various mutants of HSV-1, have shown promise in preclinical studies, but continue to suffer from poor intratumoral replication and spread in part as a result of innate immune responses that are no longer suppressed by the mutated viral genes.31,32 Targeting the virus to tumor cells could alleviate the need for conditional replication defects and thereby enhance virus spread within the tumor mass.
A wide variety of adapters have been described for the targeted delivery of adenoviral and retroviral vectors, including combinations with partially or completely detargeted viruses.6,7 HSV retargeting poses unique challenges and opportunities due to the involvement of multiple glycoproteins and their receptors in virus infection. Both gB and gC bind to ubiquitous cell-surface heparan sulfate proteoglycans to mediate virus attachment to cells. Recent evidence indicates that gB interacts in addition with one or more specific receptors at a later stage of the virus entry process.33,34 Depending on the administration route, it is likely that retargeting or detargeting of the attachment functions of gB and gC will be advantageous for virus retargeting in vivo. Both have been accomplished by relatively simple genetic modifications35,36,37 and attachment-detargeted gB and gC can be used to build a standard detargeted vector as a component of a flexible, adapter-dependent system for cell-specific virus infection. Although the role of gB binding to specific cellular receptors remains to be fully defined,38 detargeting of gD from its multiple receptors is essential.2 Our previous study indicated that HSV detargeting from nectin-1 was not readily accomplished by a nectin-1-based adapter13 and in the course of this study we found that the scCEA-HveA adapter likewise does not detarget HSV from HVEM (H.B. and H.K., unpublished results). These observations are in contrast to the demonstration in other viral systems of detargeting by native receptor-based adapters.39,40,41,42 Given the well-documented inhibition of HSV infection by soluble gD receptors,8,9,10 it is conceivable that modifications to our adapter designs, such as elongation of the spacer separating the two components, will prove successful in blocking infection via the natural receptors. Ideally, however, adapters would function with a single standard virus that is detargeted from both heparan sulfate proteoglycans and the multiple gD receptors, enabling the use of readily constructed adapters of different specificities to direct the virus to applicable targets. This situation would be advantageous over genetically retargeted viruses in that the production and quality control of new adapters is simpler than that of new viruses. Kashentseva et al. have recently reported the use of a bispecific diabody recognizing c-erbB2 and His6 to promote specific infection by a detargeted, His6-tagged adenovirus.43 A similar approach may be suitable for HSV with the option of incorporating the tag in all three detargeted glycoproteins, gB, gC, and gD. Other strategies may be based on gD-specific blocking antibodies cloned as scFvs, potentially eliminating the requirement for gD detargeting, or the generation of scFvs that specifically recognize a detargeted gD and activate its profusion function.
Targeted HSV infection should reduce the need for vector mutations that prevent virus replication in normal cells, but that generally also reduce replication and spread in tumor cells. In vivo, HSV spreads by direct cell-to-cell transfer from infected to uninfected neighboring cells at cell–cell junctions in a process requiring the same entry receptors as initial infection.44,45 This understanding suggests that exogenously supplied adapters will not enable spread of detargeted viruses and are therefore more suitable for the targeted delivery of therapeutic transgenes by replication-defective HSV vectors. It is conceivable that spread can be promoted by expression of the adapter from the virus such that initial infection is mediated by exogenously provided adapter and spread facilitated by intracellular formation of virus–adapter complexes. This approach has been tested in other viral systems with mixed results.46,47,48 Instead, based on our previous observation that K-222/3NI has a greater residual ability to infect and spread via nectin-1 than the more tightly restricted K-3/38CC,27 we reasoned that the former might offer a suitable balance between selective initial infection and adapter-independent spread. We found that indeed the gD:R222N/F223I virus produced larger plaques on MKN45 cells than the gD:A3C/Y38C virus, although smaller than those generated by K26GFP, the wild-type gD parent of the two mutant viruses. Thus, although the gD:R222N/F223I virus has the potential to yield a low-level of infection of healthy tissue surrounding a CEA-positive tumor, it is expected to spread within the tumor without repeated adapter administration. By comparison, the risk of infection of healthy, CEA-negative tissue may be smaller with the gD:A3C/Y38C virus, but the tumoricidal activity of this virus will be lower due to its highly restricted spread. Because the likelihood of virus leakage beyond the tumor mass during or following intratumoral injection decreases with increasing tumor size while the need for effective virus spread increases, viruses carrying gD:R222N/F223I may be suitable for the treatment of larger tumors while gD:A3C/Y38C viruses may be preferred when considering the targeting of small tumors. These vectors can be armed to produce immunostimulatory and diffusible cytotoxic factors and can be used in combination with drugs for enhanced bystander effects.49,50 As referred to previously,27 the two gD mutant viruses require further development to stabilize their receptor-restricted phenotypes against reverting mutations, particularly for applications requiring replication.
In summary, traditional oncolytic herpes viruses derive their tumor specificity from conditional replication defects that are generally only incompletely suppressed inside the tumor cell, resulting in reduced virus production compared to wild-type strains and limited intratumoral spread. Our approach of using a target-specific adapter to deliver receptor-restricted viruses is aimed in part at reducing the need for conditional replication defects and thereby at increasing the oncolytic potency of the virus. In addition, the combination of cell-specific adapters and receptor-restricted viruses can be applied to the delivery of therapeutic transgenes by replication-defective viruses. In these instances, target cell specificity is the key objective so that a highly receptor-restricted virus such as K-3/38CC will be advantageous over a less tightly restricted virus such as K-222/3NI. Our results show that adapters can be used to mediate HSV infection via a tumor antigen and that this method in combination with a fully replication-competent virus can accomplish a significant reduction in tumor growth. Future studies aim at both refining the adapter and virus designs, as discussed above, and defining the survival benefit and risk of off-target infection in a range of tumor models.
Materials and Methods
Cells and viruses. CHO-K1 cells (CCL-61; ATCC, Manassas, VA) and CEA-expressing CHO cells (CHO-CEA)21 were grown in Ham's F-12K medium with 10% fetal bovine serum. Human gastric adenocarcinoma MKN45 and MKN74 cells were obtained from the JCRB Cell Bank (Osaka, Japan) and maintained in RPMI 1640 medium supplemented with 10% fetal bovine serum.
Recombinant virus QOZHG, a replication-defective derivative of HSV-1 strain KOS,22 was grown on ICP4/ICP27-complementing Vero-7b cells.23 K26GFP and HVEM-restricted K-222/3NI were propagated and titered on J/A cells.27 Nectin-1-restricted K-d5-28V was propagated and titered on J/C cells.27 Physical titers in genome copies/ml were determined by real-time quantitative PCR for the ICP47 gene, as described.51
Plasmids. pCEA-HveA for expression of scCEA-HveA was constructed by ligation of two PCR products. The coding sequence for anti-CEA single-chain antibody F39scFv,52 including the signal peptide (SP), was amplified with forward primer F39scFv-VL (5′-CAAGGATCCGCCACCATGAGTGTGCCCACTCAGGTCCTGGGG-3′) and reverse primer F39scFv-VH (5′-CCCGAATTCTGAACCGCCACCACCAGCAGAGACAGTGACCAGAGTCCC-3′); the F39scFv-VH primer included a 15-nucleotide sequence (underlined) specifying a flexible linker (GGGGS) immediately following the scFv coding region. The coding region for the N-terminal 82 amino acids of mature human HVEM, corresponding to the first two CRPs,8 was amplified from pBEC14 (ref. 8; kindly provided by Patricia Spear, Northwestern University, IL) with sense primer HA4 (5′-CCCGAATTCCTGCCGTCCTGCAAGGAGGAC-3′) and antisense primer HA5 (5′-CTCTCGAGTTAGTGGTGGTGGTGGTGGTGGCCACACACGGCGTTCTC-3′); primer HA5 specified six histidine residues (His6 tag, underlined) followed by a translation termination codon. The scFv PCR product was digested with BamHI and EcoRI (bold in the corresponding primer sequences), the HVEM PCR product was digested with EcoRI and XhoI (bold in the HVEM primer sequences), and the two fragments were sequentially cloned into pcDNA3.1 (Invitrogen, Carlsbad, CA).
pHveA-CEA for expression of HveA-scCEA was similarly constructed by combining two PCR fragments in pcDNA3.1. Primers HA6 (5′-CCCAAGCTTGCCACCATGGAGCCTCCTGGAGACTGG-3′) and HA7 (5′-CCCGGATCCTGAACCGCCACCACCGCCACACACGGCGTTCTC-3′) were used to amplify the sequence for the HVEM SP, CRP1, and CRP2 on pBEC14 (GGGGS spacer underlined; restriction sites in bold). The CEA scFv coding sequence was amplified with primers F39scFv-LS (5′-CAAGGATCCGACATCCAGATGACTCAGTCT-3′) and F39scFv-His (5′-AAACTCGAGCTAGTGGTGGTGGTGGTGGTGAGCAGAGACAGTGACCAG-3′ His tag underlined). The fragments were digested with HindIII/BamHI and BamHI/XhoI, respectively, and sequentially cloned into pcDNA3.1.
pCEA-HveC and pHveC-CEA for expression of scCEA-HveC and HveC-scCEA were generated by first inserting the sequence encoding the N-terminal 114 amino acids of human nectin-1/HveC without or with SP into pcDNA3.1. Primers HC1 (5′-CAAGATATCCCCAGGTGGTCCAGGTGAACGAC-3′) and HC2 (5′-CCCCTCGAGCTAGTGGTGGTGGTGGTGGTGGGCCATCACCGTGAGATT-3′) were used for amplify the nectin-1 fragment without SP using pNec1153-his (ref. 20) as template. The sequence including the SP was amplified using primers HC3 (5′-CCCAAGCTTGCCACCATGGCTCGGATGGGGCTTGCGGGCGCC-3′) and HC4 (5′-GGCGGATCCTGAACCGCCACCACCGGCCATCACCGTGAGATTGAG-3′). The amplified fragments were digested with EcoRV/XhoI and HindIII/BamHI, respectively, and substituted for the HVEM/HveA fragments of pCEA-HveA and pHveA-CEA, respectively.
Plasmid pCEAscFv for the production of C-terminally His6-tagged anti-CEA scFv was generated by PCR with forward primer F39scFv-VL and reverse primer F39scFv-VH-His (5′-AAACTCGAGCTAGTGGTGGTGGTGGTGGTGAGCAGAGACAGTGACCAGAGTCC-3′). The PCR product was digested with BamHI and XhoI and cloned into pcDNA3.1.
All PCR-derived plasmid inserts were verified by complete DNA sequencing.
Production and purification of soluble proteins. Soluble proteins were expressed by LipofectAMINE-Plus (Invitrogen)-mediated transfection of 293T cells, as described,20 and affinity-purified from the culture media by passage over Probond resin (Invitrogen) according to the manufacturer's instructions. The concentration of each purified protein was determined by Bradford assay (Bio-Rad, Hercules, CA).
Infection assays. Adapter and QOZHG virus were incubated for 30 minutes at 4 °C before infection of CHO-K1 and CHO-CEA cells (3 × 104 cells/well in a 96-well plate) for 1 hour at 37 °C. The culture media were replaced with fresh media and the cells were cultured for 16 hours at 37 °C. Virus entry was determined by X-gal staining or o-nitrophenyl-β--galactoside assay, as described previously.13
Adapter-free infection of gastric carcinoma cells with parental and receptor-restricted viruses was performed and detected at 8 hours postinfection as described previously for other cells.27 A virus input of 100 genome copies/cell was used.
Adapter activity on gastric carcinoma cells was determined as follows. MKN45 and MKN74 cells were plated in 48-well culture plates at 1.5 × 105 cells per well. Virus was incubated with adapter protein in a total volume of 150 µl for 30 minutes at 4 °C. The culture medium was removed from each well and the virus/adapter mixture added to the cell monolayer. After 1 hour incubation at 37 °C, infected cells were washed with 0.1 mol/l glycine (pH 3.0) for 30 seconds, washed twice with phosphate-buffered saline, and incubated with complete media for 8 hours. Infected cells were immunostained with anti-VP16 antibody 1-21 (Santa Cruz Biotechnology, Santa Cruz, CA), essentially as described.27 Secondary antibody was Alexa Fluor 594 goat anti-mouse IgG (1:500; Molecular Probes, Eugene, OR).
Infection inhibition. QOZHG (MOI = 10) was incubated with 250 nmol/l scCEA-HveA protein and 0–500 nmol/l sgD287 protein for 30 minutes at 4 °C. CHO-CEA cells were infected with the preincubation mixtures as above and virus entry was determined by o-nitrophenyl-β--galactoside assay.
Antibody-mediated inhibition was determined by preincubation of CHO-CEA cells with 0–200 µg/ml anti-CEA antibody F11-39 (ref. 53) at 37 °C for 30 minutes, addition of a preincubated (30 minutes, 4 °C) mixture of QOZHG (MOI = 10) and scCEA-HveA (250 nmol/l), infection as above, and measurement of virus entry by o-nitrophenyl-β--galactoside assay.
Lateral spread. Following infection, cells were treated with acidic glycine buffer, as above, and incubated with fresh media for 8 hours at 37 °C. The cells were then overlaid with 0.5% methylcellulose (Sigma, St Louis, MO)-containing medium and incubated for 72 hours at 37 °C. Infected clusters of cells were visualized by staining with anti-VP16 antibody.
Tumor infection. Eight-week-old male BALB/c nude mice were injected with 5 × 106 MKN45 or MKN74 cells into the right flank (n = 2/group). When the tumor volumes reached 200 mm3, HVEM-restricted K-222/3NI virus (1 × 108 plaque-forming units/tumor) with or without 1.2 µmol/l adapter in a total volume of 50 µl was inoculated into the tumor. Animals were killed and tumors removed at 2, 6, or 10 days following vector injection. Tumors were fixed in 4% formaldehyde, embedded in paraffin, and sectioned at 3 µm. Sections were sequentially incubated with anti-VP16 antibody, biotinylated anti-mouse IgG, and acidin–peroxidase complex (VECTASTAIN ABC Elite Kit; Vector Laboratories, Burlingame, CA). The slides were developed with the peroxidase substrate 3,3′-diaminobenzidine (KPL, Gaithersburg, MD) and hematoxylin counterstain (Merck, Darmstadt, Germany).
Tumor growth. Subcutaneous MKN45 tumors at 200 mm3 (n = 4/group) were injected with K-222/3NI virus (1 × 108 plaque-forming units) with or without adapter (1.2 µmol/l) on day 0. Repeat injections were performed on days 3, 6, and 9. Tumor diameters were measured every 2 days and the volume (size) calculated as (length × width2)/2. Statistical analysis was performed by t-test and a P value below 0.05 was considered significant. All animal experiments were performed in accordance with the Korean National Institutes of Health guidelines as approved by the Institutional Animal Care and Use Committee of the Korea Institute of Radiological and Medical Sciences.
Acknowledgments
We thank Patricia Spear (Northwestern University) for pBEC14 and William Goins (University of Pittsburgh) for helpful discussions. This work was supported by grants 0065410, 0002125 and 0093737 to H.K. from the Korea Science and Engineering Foundation (KOSEF) funded by the Korean government (MEST), and by NIH grants CA119298, NS40923, and DK044935 to J.C.G. J.C.G. is an inventor of patents related to HSV technology and owns equity in a publicly traded company, Diamyd Medical AB based in Stockholm, Sweden, that is evaluating HSV gene therapy applications for the treatment of chronic pain.
REFERENCES
- Spear PG. Herpes simplex virus: receptors and ligands for cell entry. Cell Microbiol. 2004;6:401–410. doi: 10.1111/j.1462-5822.2004.00389.x. [DOI] [PubMed] [Google Scholar]
- Campadelli-Fiume G, Amasio M, Avitabile E, Cerretani A, Forghieri C, Gianni T, et al. The multipartite system that mediates entry of herpes simplex virus into the cell. Rev Med Virol. 2007;17:313–326. doi: 10.1002/rmv.546. [DOI] [PubMed] [Google Scholar]
- Zhou G., and, Roizman B. Construction and properties of a herpes simplex virus 1 designed to enter cells solely via the IL-13α2 receptor. Proc Natl Acad Sci USA. 2006;103:5508–5513. doi: 10.1073/pnas.0601258103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menotti L, Cerretani A, Hengel H., and, Campadelli-Fiume G. Construction of a fully retargeted herpes simplex virus 1 recombinant capable of entering cells solely via human epidermal growth factor receptor 2. J Virol. 2008;82:10153–10161. doi: 10.1128/JVI.01133-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menotti L, Nicoletti G, Gatta V, Croci S, Landuzzi L, De Giovanni C, et al. Inhibition of human tumor growth in mice by an oncolytic herpes simplex virus designed to target solely HER-2-positive cells. Proc Natl Acad Sci USA. 2009;106:9039–9044. doi: 10.1073/pnas.0812268106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waehler R, Russell SJ., and, Curiel DT. Engineering targeted viral vectors for gene therapy. Nat Rev Genet. 2007;8:573–587. doi: 10.1038/nrg2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffer DV, Koerber JT., and, Lim KI. Molecular engineering of viral gene delivery vehicles. Annu Rev Biomed Eng. 2008;10:169–194. doi: 10.1146/annurev.bioeng.10.061807.160514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery RI, Warner MS, Lum BJ., and, Spear PG. Herpes simplex virus-1 entry into cells mediated by a novel member of the TNF/NGF receptor family. Cell. 1996;87:427–436. doi: 10.1016/s0092-8674(00)81363-x. [DOI] [PubMed] [Google Scholar]
- Geraghty RJ, Krummenacher C, Cohen GH, Eisenberg RJ., and, Spear PG. Entry of α-herpesviruses mediated by poliovirus receptor-related protein 1 and poliovirus receptor. Science. 1998;280:1618–1620. doi: 10.1126/science.280.5369.1618. [DOI] [PubMed] [Google Scholar]
- Krummenacher C, Rux AH, Whitbeck JC, Ponce-de-Leon M, Lou H, Baribaud I, et al. The first immunoglobulin-like domain of HveC is sufficient to bind herpes simplex virus gD with full affinity, while the third domain is involved in oligomerization of HveC. J Virol. 1999;73:8127–8137. doi: 10.1128/jvi.73.10.8127-8137.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez M, Cocchi F, Avitabile E, Leclerc A, Adelaide J, Campadelli-Fiume G, et al. Novel, soluble isoform of the herpes simplex virus (HSV) receptor nectin1 (or PRR1-HIgR-HveC) modulates positively and negatively susceptibility to HSV infection. J Virol. 2001;75:5684–5691. doi: 10.1128/JVI.75.12.5684-5691.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitbeck JC, Peng C, Lou H, Xu R, Willis SH, Ponce de Leon M, et al. Glycoprotein D of herpes simplex virus (HSV) binds directly to HVEM, a member of the tumor necrosis factor receptor superfamily and a mediator of HSV entry. J Virol. 1997;71:6083–6093. doi: 10.1128/jvi.71.8.6083-6093.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano K, Asano R, Tsumoto K, Kwon H, Goins WF, Kumagai I, et al. Herpes simplex virus targeting to the EGF receptor by a gD-specific soluble bridging molecule. Mol Ther. 2005;11:617–626. doi: 10.1016/j.ymthe.2004.12.012. [DOI] [PubMed] [Google Scholar]
- Kwon BS, Tan KB, Ni J, Oh KO, Lee ZH, Kim KK, et al. A newly identified member of the tumor necrosis factor receptor superfamily with a wide tissue distribution and involvement in lymphocyte activation. J Biol Chem. 1997;272:14272–14276. doi: 10.1074/jbc.272.22.14272. [DOI] [PubMed] [Google Scholar]
- Marsters SA, Ayres TM, Skubatch M, Gray CL, Rothe M., and, Ashkenazi A. Herpesvirus entry mediator, a member of the tumor necrosis factor receptor (TNFR) family, interacts with members of the TNFR-associated factor family and activates the transcription factors NF-κB and AP-1. J Biol Chem. 1997;272:14029–14032. doi: 10.1074/jbc.272.22.14029. [DOI] [PubMed] [Google Scholar]
- Manoj S, Jogger CR, Myscofski D, Yoon M., and, Spear PG. Mutations in herpes simplex virus glycoprotein D that prevent cell entry via nectins and alter cell tropism. Proc Natl Acad Sci USA. 2004;101:12414–12421. doi: 10.1073/pnas.0404211101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu H, Solovyev I, Colombero A, Elliott R, Kelley M., and, Boyle WJ. ATAR, a novel tumor necrosis factor receptor family member, signals through TRAF2 and TRAF5. J Biol Chem. 1997;272:13471–13474. doi: 10.1074/jbc.272.21.13471. [DOI] [PubMed] [Google Scholar]
- Hammarström S. The carcinoembryonic antigen (CEA) family: structures, suggested functions and expression in normal and malignant tissues. Semin Cancer Biol. 1999;9:67–81. doi: 10.1006/scbi.1998.0119. [DOI] [PubMed] [Google Scholar]
- Whitbeck JC, Connolly SA, Willis SH, Hou W, Krummenacher C, Ponce de Leon M, et al. Localization of the gD-binding region of the human herpes simplex virus receptor, HveA. J Virol. 2001;75:171–180. doi: 10.1128/JVI.75.1.171-180.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon H, Bai Q, Baek HJ, Felmet K, Burton EA, Goins WF, et al. Soluble V domain of Nectin-1/HveC enables entry of herpes simplex virus type 1 (HSV-1) into HSV-resistant cells by binding to viral glycoprotein D. J Virol. 2006;80:138–148. doi: 10.1128/JVI.80.1.138-148.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroki M, Arakawa F, Matsuo Y, Oikawa S, Misumi Y, Nakazato H, et al. Molecular cloning of nonspecific cross-reacting antigens in human granulocytes. J Biol Chem. 1991;266:11810–11817. [PubMed] [Google Scholar]
- Chen X, Li J, Mata M, Goss J, Wolfe D, Glorioso JC, et al. Herpes simplex virus type 1 ICP0 protein does not accumulate in the nucleus of primary neurons in culture. J Virol. 2000;74:10132–10141. doi: 10.1128/jvi.74.21.10132-10141.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krisky DM, Marconi PC, Oligino T, Rouse RJ, Fink DJ., and, Glorioso JC. Rapid method for construction of recombinant HSV gene transfer vectors. Gene Ther. 1997;4:1120–1125. doi: 10.1038/sj.gt.3300497. [DOI] [PubMed] [Google Scholar]
- Tsvitov M, Frampton AR, Jr, Shah WA, Wendell SK, Ozuer A, Kapacee Z, et al. Characterization of soluble glycoprotein D-mediated herpes simplex virus type 1 infection. Virology. 2007;360:477–491. doi: 10.1016/j.virol.2006.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takai Y., and, Nakanishi H. Nectin and afadin: novel organizers of intercellular junctions. J Cell Sci. 2003;116:17–27. doi: 10.1242/jcs.00167. [DOI] [PubMed] [Google Scholar]
- Simpson SA, Manchak MD, Hager EJ, Krummenacher C, Whitbeck JC, Levin MJ, et al. Nectin-1/HveC mediates herpes simplex virus type 1 entry into primary human sensory neurons and fibroblasts. J Neurovirol. 2005;11:208–218. doi: 10.1080/13550280590924214. [DOI] [PubMed] [Google Scholar]
- Uchida H, Shah WA, Ozuer A, Frampton AR, Jr, Goins WF, Grandi P, et al. Generation of herpesvirus entry mediator (HVEM)-restricted herpes simplex virus type 1 mutant viruses: resistance of HVEM-expressing cells and identification of mutations that rescue nectin-1 recognition. J Virol. 2009;83:2951–2961. doi: 10.1128/JVI.01449-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T, Huang J, Hirai S, Kuroki M, Kuroki M, Watanabe N, et al. Carcinoembryonic antigen-targeted selective gene therapy for gastric cancer through FZ33 fiber-modified adenovirus vectors. Clin Cancer Res. 2006;12:3803–3813. doi: 10.1158/1078-0432.CCR-06-0024. [DOI] [PubMed] [Google Scholar]
- Desai P., and, Person S. Incorporation of the green fluorescent protein into the herpes simplex virus type 1 capsid. J Virol. 1998;72:7563–7568. doi: 10.1128/jvi.72.9.7563-7568.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu TC, Galanis E., and, Kirn D. Clinical trial results with oncolytic virotherapy: a century of promise, a decade of progress. Nat Clin Pract Oncol. 2007;4:101–117. doi: 10.1038/ncponc0736. [DOI] [PubMed] [Google Scholar]
- Wakimoto H, Johnson PR, Knipe DM., and, Chiocca EA. Effects of innate immunity on herpes simplex virus and its ability to kill tumor cells. Gene Ther. 2003;10:983–990. doi: 10.1038/sj.gt.3302038. [DOI] [PubMed] [Google Scholar]
- Ikeda K, Ichikawa T, Wakimoto H, Silver JS, Deisboeck TS, Finkelstein D, et al. Oncolytic virus therapy of multiple tumors in the brain requires suppression of innate and elicited antiviral responses. Nat Med. 1999;5:881–887. doi: 10.1038/11320. [DOI] [PubMed] [Google Scholar]
- Bender FC, Whitbeck JC, Lou H, Cohen GH., and, Eisenberg RJ. Herpes simplex virus glycoprotein B binds to cell surfaces independently of heparan sulfate and blocks virus entry. J Virol. 2005;79:11588–11597. doi: 10.1128/JVI.79.18.11588-11597.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh T, Arii J, Suenaga T, Wang J, Kogure A, Uehori J, et al. PILRα is a herpes simplex virus-1 entry coreceptor that associates with glycoprotein B. Cell. 2008;132:935–944. doi: 10.1016/j.cell.2008.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laquerre S, Anderson DB, Stolz DB., and, Glorioso JC. Recombinant herpes simplex virus type 1 engineered for targeted binding to erythropoietin receptor-bearing cells. J Virol. 1998;72:9683–9697. doi: 10.1128/jvi.72.12.9683-9697.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argnani R, Boccafogli L, Marconi PC., and, Manservigi R. Specific targeted binding of herpes simplex virus type 1 to hepatocytes via the human hepatitis B virus preS1 peptide. Gene Ther. 2004;11:1087–1098. doi: 10.1038/sj.gt.3302266. [DOI] [PubMed] [Google Scholar]
- Zhou G, Ye GJ, Debinski W., and, Roizman B. Engineered herpes simplex virus 1 is dependent on IL13Rα 2 receptor for cell entry and independent of glycoprotein D receptor interaction. Proc Natl Acad Sci USA. 2002;99:15124–15129. doi: 10.1073/pnas.232588699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Q, Lin E, Satoh T, Arase H., and, Spear PG. Differential effects on cell fusion activity of mutations in herpes simplex virus 1 glycoprotein B (gB) dependent on whether a gD receptor or a gB receptor is overexpressed. J Virol. 2009;83:7384–7390. doi: 10.1128/JVI.00087-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li HJ, Everts M, Pereboeva L, Komarova S, Idan A, Curiel DT, et al. Adenovirus tumor targeting and hepatic untargeting by a coxsackie/adenovirus receptor ectodomain anti-carcinoembryonic antigen bispecific adapter. Cancer Res. 2007;67:5354–5361. doi: 10.1158/0008-5472.CAN-06-4679. [DOI] [PubMed] [Google Scholar]
- Würdinger T, Verheije MH, Broen K, Bosch BJ, Haijema BJ, de Haan CA, et al. Soluble receptor-mediated targeting of mouse hepatitis coronavirus to the human epidermal growth factor receptor. J Virol. 2005;79:15314–15322. doi: 10.1128/JVI.79.24.15314-15322.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebbinghaus C, Al-Jaibaji A, Operschall E, Schöffel A, Peter I, Greber UF, et al. Functional and selective targeting of adenovirus to high-affinity Fcγ receptor I-positive cells by using a bispecific hybrid adapter. J Virol. 2001;75:480–489. doi: 10.1128/JVI.75.1.480-489.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dmitriev I, Kashentseva E, Rogers BE, Krasnykh V., and, Curiel DT. Ectodomain of coxsackievirus and adenovirus receptor genetically fused to epidermal growth factor mediates adenovirus targeting to epidermal growth factor receptor-positive cells. J Virol. 2000;74:6875–6884. doi: 10.1128/jvi.74.15.6875-6884.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashentseva EA, Douglas JT, Zinn KR, Curiel DT., and, Dmitriev IP. Targeting of adenovirus serotype 5 pseudotyped with short fiber from serotype 41 to c-erbB2-positive cells using bispecific single-chain diabody. J Mol Biol. 2009;388:443–461. doi: 10.1016/j.jmb.2009.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roller RJ., and, Rauch D. Herpesvirus entry mediator HVEM mediates cell-cell spread in BHK(TK-) cell clones. J Virol. 1998;72:1411–1417. doi: 10.1128/jvi.72.2.1411-1417.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cocchi F, Menotti L, Dubreuil P, Lopez M., and, Campadelli-Fiume G. Cell-to-cell spread of wild-type herpes simplex virus type 1, but not of syncytial strains, is mediated by the immunoglobulin-like receptors that mediate virion entry, nectin1 (PRR1/HveC/HIgR) and nectin2 (PRR2/HveB) J Virol. 2000;74:3909–3917. doi: 10.1128/jvi.74.8.3909-3917.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemminki A, Wang M, Hakkarainen T, Desmond RA, Wahlfors J., and, Curiel DT. Production of an EGFR targeting molecule from a conditionally replicating adenovirus impairs its oncolytic potential. Cancer Gene Ther. 2003;10:583–588. doi: 10.1038/sj.cgt.7700606. [DOI] [PubMed] [Google Scholar]
- Verheije MH, Würdinger T, van Beusechem VW, de Haan CA, Gerritsen WR., and, Rottier PJ. Redirecting coronavirus to a nonnative receptor through a virus-encoded targeting adapter. J Virol. 2006;80:1250–1260. doi: 10.1128/JVI.80.3.1250-1260.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carette JE, Graat HC, Schagen FH, Mastenbroek DC, Rots MG, Haisma HJ, et al. A conditionally replicating adenovirus with strict selectivity in killing cells expressing epidermal growth factor receptor. Virology. 2007;361:56–67. doi: 10.1016/j.virol.2006.11.011. [DOI] [PubMed] [Google Scholar]
- Niranjan A, Wolfe D, Tamura M, Soares MK, Krisky DM, Lunsford LD, et al. Treatment of rat gliosarcoma brain tumors by HSV-based multigene therapy combined with radiosurgery. Mol Ther. 2003;8:530–542. doi: 10.1016/s1525-0016(03)00232-6. [DOI] [PubMed] [Google Scholar]
- Krisky DM, Marconi PC, Oligino TJ, Rouse RJ, Fink DJ, Cohen JB, et al. Development of herpes simplex virus replication-defective multigene vectors for combination gene therapy applications. Gene Ther. 1998;5:1517–1530. doi: 10.1038/sj.gt.3300755. [DOI] [PubMed] [Google Scholar]
- Jiang C, Ataai M, Ozuer A, Krisky D, Wechuck J, Pornsuwan S, et al. Inactivation of herpes simplex type 1 gene vector on immobilized metal affinity chromatography: oxidative damage by hydroxyl free radicals and its prevention. Biotechnol Bioeng. 2006;95:48–57. doi: 10.1002/bit.20943. [DOI] [PubMed] [Google Scholar]
- Liao S, Khare PD, Arakawa F, Kuroki M, Hirose Y, Fujimura S, et al. Targeting of LAK activity to CEA-expressing tumor cells with an anti-CEA scFv/IL-2 fusion protein. Anticancer Res. 2001;21 3B:1673–1680. [PubMed] [Google Scholar]
- Kuroki M, Arakawa F, Haruno M, Murakami M, Wakisaka M, Higuchi H, et al. Biochemical characterization of 25 distinct carcinoembryonic antigen (CEA) epitopes recognized by 57 monoclonal antibodies and categorized into seven groups in terms of domain structure of the CEA molecule. Hybridoma. 1992;11:391–407. doi: 10.1089/hyb.1992.11.391. [DOI] [PubMed] [Google Scholar]