Abstract
DNA vaccination is an attractive approach to induce antigen-specific cytotoxic CD8+ T lymphocytes (CTLs), which can mediate protective antitumor immunity. The potency of DNA vaccines encoding weakly immunogenic tumor-associated antigens (TAAs) can be enhanced by codelivering gene-encoded adjuvants. Pattern recognition receptors (PRRs) that sense intracellular DNA could potentially be used to harness intrinsic immune-stimulating properties of plasmid DNA vaccines. Consequently, the cytosolic DNA sensor, DNA-dependent activator of interferon (IFN) regulatory factors (DAI), was used as a genetic adjuvant. In vivo electroporation (EP) of mice with a DAI-encoding plasmid (pDAI) promoted transcription of genes encoding type I IFNs, proinflammatory cytokines, and costimulatory molecules. Coimmunization with pDAI and antigen-encoding plasmids enhanced in vivo antigen-specific proliferation, and induction of effector and memory CTLs. Moreover, codelivery of pDAI effectively promoted CTL and CD4+ Th1 responses to the TAA survivin. The DAI-enhanced CTL induction required nuclear factor κB (NF-κB) activation and type I IFN signaling, but did not involve the IFN regulatory factor 3 (IRF3). Codelivery of pDAI also increased CTL responses to the melanoma-associated antigen tyrosinase-related protein-2 (TRP2), enhanced tumor rejection and conferred long-term protection against B16 melanoma challenge. This study constitutes “proof-of-principle” validating the use of intracellular PRRs as genetic adjuvants to enhance DNA vaccine potency.
Introduction
Cytotoxic CD8+ T lymphocytes (CTLs) are key effector cells of the immune system and critical components of protective immunity against infectious diseases and cancer. Activated CTLs have the potential to eradicate malignant cells with high specificity. Indeed, intratumoral CTL infiltration is often associated with favorable clinical outcomes such as decreased disease recurrence and improved survival in diverse malignancies.1,2,3 Adoptive transfer trials of tumor-specific CTLs that control disease progression in metastatic melanoma patients have provided direct evidence of the efficacy of CTLs.4 Accordingly, induction of effective and long-lasting T cell immunity represents a major goal of cancer vaccines that have shown promising results in the clinic, especially when used as adjuvant therapy to standard cancer treatments in patients with minimal residual disease.5
DNA vaccination is an attractive and safe approach to generate protective CTL responses against cancer. This approach has been successful in animal models, but has shown limited efficacy in clinical trials.6 One underlying reason is that most of the tumor-associated antigens (TAAs) recognized by T cells are normal nonmutated self-antigens and potentially self-reactive TAA-specific T cells are either eliminated or become regulatory T cells by mechanisms involving central and peripheral immune tolerance. Therefore, efficient delivery systems and potent adjuvants are needed for cancer DNA vaccines to overcome tumor-associated T cell tolerance. In vivo electroporation (EP) has emerged as a simple, efficient, and clinically applicable method for delivering DNA vaccines that greatly enhances plasmid uptake, antigen expression, and immune responses.7 Moreover, DNA EP activates innate immunity resulting in infiltration of immune cells and the production of proinflammatory molecules that contribute to the induction of the immune responses.8,9 The versatility of DNA vaccines facilitates codelivery of genes encoding immunomodulatory molecules, typically cytokines and chemokines, as genetic adjuvants. Moreover, the concerted action of several cytokines and costimulatory molecules clearly facilitates potent activation of the immune response.10 One means to this end is the use of adjuvants, which exploit the immune-stimulating effects of pathogens by activating pattern recognition receptors (PRRs). PRRs are a group of evolutionary conserved innate immune receptors that sense pathogen-associated molecular patterns and activate downstream master transcription factors initiating the production of an array of cytokines, chemokines, and type I interferons (IFNs) to promote activation and maturation of adaptive immune responses.11,12 A central feature of pattern recognition is the sensing of foreign nucleic acids. DNA-sensing PRRs include TLR9, located at endosomal compartments, and the recently described cytosolic sensors: DAI (also known as ZBP1 and DLM-1),13 absent in melanoma 2 (AIM2),14,15 and DNA-dependent RNA polymerase III.16,17 In vitro studies revealed that DAI, the first cytoplasmic DNA sensor described, recognizes double-stranded DNA and triggers the gene expression of type I IFNs, IFN-inducible chemokine Cxcl10, and proinflammatory cytokines, such as interleukin 6 (IL-6) and tumor necrosis factor-α (TNF-α) via two distinct signaling pathways. One involves DAI-mediated phosphorylation of the TANK-binding kinase-1 and subsequent activation of the transcription factor IFN regulatory factor 3 (IRF3).13,18 The other pathway requires phosphorylation of the receptor interacting protein-1 kinase, leading to phosphorylation of IκB-α, and subsequent activation of the transcription factor NF-κB.19
In the present study, we explored whether coexpressing a DNA-sensing PRR could further potentiate intrinsic adjuvant properties of bacterial plasmids used for DNA vaccines. Although DAI is dispensable for DNA vaccine immunogenicity,20 we hypothesized that in vivo overexpression of DAI would boost DNA-induced innate immune activation and ultimately enhance adaptive T cell immunity.
Results
DAI delivery promotes transcriptional upregulation of genes involved in innate and adaptive immunity in vivo
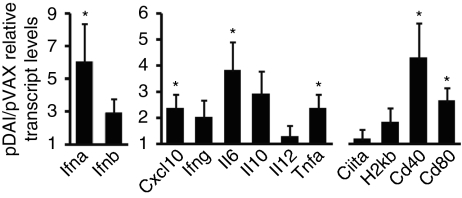
It has recently been established that intradermal (i.d.) DNA delivery followed by EP increases up to 200-fold the transcription of genes encoding proteins involved in immune regulation, such as chemokines, activation markers, and proinflammatory molecules.21 To evaluate whether in vivo overexpression of DAI would further stimulate innate immunity, transcript levels of innate immune mediators were measured after DNA vaccination. Mice were inoculated i.d. with either control (pVAX) or DAI-encoding (pDAI) plasmids, followed by EP. Total RNA isolated from skin biopsies taken 24 hours later was analyzed by reverse transcription real-time PCR. Gene expression was normalized to the L32 housekeeping gene. Although inoculation with pVAX or pDAI generated a similar local inflammatory response as determined by immunohistochemistry on electroporated skin sections (Supplementary Figure S1 and Supplementary Materials and Methods) and a similar upregulation of costimulatory molecules on dendritic cells isolated from the draining lymph node (Supplementary Figure S2), a significant increase (P < 0.05) was observed for genes encoding IFN-α, IL-6, TNF-α, and Cxcl10 in the pDAI-inoculated group relative to the control vector inoculated group (Figure 1). A trend toward increased IFN-β expression was also observed (P = 0.055). Interestingly, pDAI inoculation also led to significant upregulation of the genes encoding the costimulatory molecules CD40 and CD80 required for T cell activation and differentiation into effector T cells. Moreover, we found that i.d. EP with pDAI also upregulated transcription of genes involved in antigen presentation, T cell proliferation and maturation, growth factors, and antiviral responses as compared to control DNA using two real-time PCR arrays investigating the transcriptional regulation of >400 genes involved in the IFN and inflammatory responses (Supplementary Table S1). For instance, multiple genes encoding cytokines such as IL-5, -9, -13, -20, -21, -23, and -31 were upregulated. Interestingly, the most upregulated gene was Ly75 (also DEC-205 or CD205), a C-type lectin receptor expressed on skin-resident dendritic cells involved in directing captured antigens to antigen-processing compartments.22 Vaccines targeting antigen to DEC-205-expressing dendritic cells has been used as a strategy to enhance crosspresentation and T cell responses.23 A list of the 20 most strongly upregulated genes, in addition to the genes assayed by the real-time PCR analysis, is provided in Supplementary Table S1.
Figure 1.
DAI EP promotes transcription of type I IFNs, proinflammatory cytokines, chemokine, and costimulatory molecules. C57BL/6 mice were electroporated with pVAX or pDAI (n = 6 per group) and skin biopsies were taken 24 hours later for gene expression analysis. Transcript levels of target genes were determined by quantitative real-time PCR. Relative pDAI/pVAX transcript levels represent the ratio between the levels detected in pDAI- and pVAX-injected mice. Bars are the mean ± SEM. *P < 0.05. Data presented are pooled from two independent experiments. EP, electroporation.
Codelivery of DAI promotes in vivo proliferation and induction of CTLs
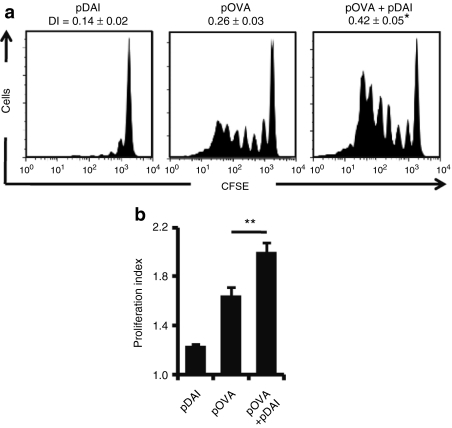
Given the observed upregulation of proinflammatory and costimulatory transcripts in the skin, we studied whether pDAI coimmunization enhanced adaptive immunity. To test this, mice were covaccinated with pDAI and an ovalbumin (OVA)-encoding plasmid (pOVA), and compared with mice vaccinated with pOVA alone. To correct for the immunostimulatory effects exerted by plasmid DNA, the total quantity of DNA was adjusted by addition of a non-coding pVAX plasmid, so that the amount of DNA was the same in every animal. CFSE-labeled splenocytes from OT-I mice, whose CD8+ T cells carry a transgenic TCR specific for the OVA(257–264) peptide, were adoptively transferred into the vaccinated mice. After 4 days, lymph nodes from recipient mice were isolated and the antigen-specific proliferation of OT-I CD8+ T cells was analyzed by flow cytometry. Both division (Figure 2a) and proliferation (Figure 2b) indexes were calculated. These analyses show that in the pOVA+pDAI immunized mice, compared to the pOVA immunized mice, OVA-specific CD8+ T cells divided significantly more (division indexes: 0.42 versus 0.26, P = 0.02; 0.14 is the basal level) and that dividing cells underwent more cycles of proliferation (proliferation indexes: 2.0 versus 1.6, P = 0.007; 1.23 is the basal level), indicating that codelivery of pDAI promoted a more efficient antigen presentation in vivo.
Figure 2.
Codelivery of pDAI enhances antigen-specific proliferation of CD8+ T cells. C57BL/6 mice were electroporated with pDAI, pOVA, or pOVA+pDAI (n = 6 per group). (a,b) In vivo antigen proliferation of adoptively transferred OT-I CD8+ T cells was evaluated 4 days later. Representative histograms showing proliferation profiles of OT-I CFSE+CD8+Vα2TCR+ populations for each group are displayed including the (a) duplication index (DI) and the (b) proliferation index (PI) calculated for each group (mean ± SEM). *P = 0.02, **P = 0.007. Data shown are from one representative of two independent experiments.
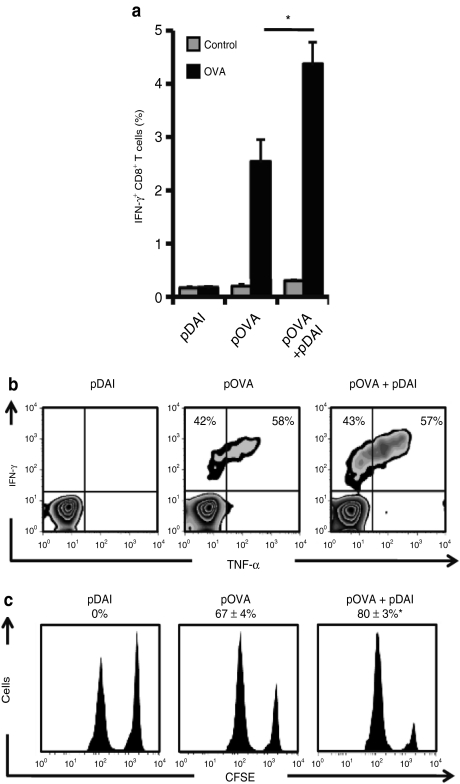
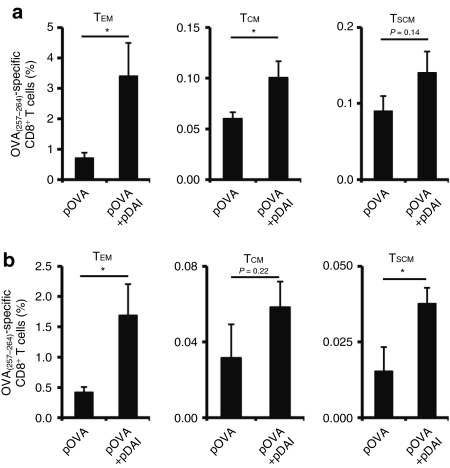
The ability of DAI to induce functional OVA-specific CD8+ T cells in peripheral blood following two immunizations was determined 13 days postimmunization by intracellular cytokine staining and flow cytometry analysis. Codelivery of pDAI increased the frequency of IFN-γ producing CD8+ T cells from 2.5% observed in pOVA immunized mice to 4.4% (P = 0.01) (Figure 3a). Functional OVA-specific IFN-γ producing CD8+ T cells could also produce TNF-α (Figure 3b). Using a H-2Kb:OVA(257–264) pentamer (Supplementary Figure S3a), we found that the OVA-specific CD8+ T cells displayed a similar cytotoxic effector phenotype (CD25low, CD69low, CD44high, CD62Llow; Supplementary Figure S3b) regardless of pDAI administration. To evaluate the cytotoxic potential of the antigen-specific CD8+ T cells, spleen cells from naive mice were labeled with 0.2 or 2 µmol/l of CFSE and pulsed with control or OVA(257–264) peptides, respectively, and adoptively transferred to immunized mice. After 6 hours, lymph nodes from pDAI, pOVA, and pOVA+pDAI vaccinated mice were removed and the killing of the OVA(257–264)-pulsed population (CFSEhigh) relative to the control population (CFSElow) was evaluated by flow cytometry. Vaccination with pOVA alone resulted in killing of 67% of the OVA(257–264)-pulsed target cells. Coadministration of pDAI significantly (P = 0.017) increased the killing of the target population to 80% (Figure 3c), consistent with the observed increase in CTL frequency. No difference in antibody titers (Supplementary Figure S4 and Supplementary Materials and Methods) or immunoglobulin subclasses (data not shown) were observed between mice vaccinated with pOVA alone and pDAI+pOVA despite the induction of IFN-α (Figure 1) and cytokines such as IL-5 (Supplementary Table S1). In addition, the induction of different OVA(257–264)-specific CD8+ memory T cell subsets was determined by CD44 and CD62L staining (Supplementary Figure S3c) 5 weeks after the last vaccination. Both effector memory (TEM; CD44high CD62Llow) and central memory (TCM; CD44high CD62Lhigh) CD8+ T cells were significantly increased (P = 0.024 and P = 0.031, respectively) in the spleen of mice coimmunized with pOVA+pDAI, whereas a similar trend was observed for CD8+ memory stem cells (TSCM; CD44low CD62Lhigh) (Figure 4a). This analysis also showed that TEM and TSCM subsets were significantly increased in the blood (Figure 4b; P = 0.025 and P = 0.030, respectively). Altogether, this data shows that coadministration of pDAI effectively improves T cell immunity by enhancing in vivo antigen-specific proliferation of CD8+ T cells and the induction and persistence of effector and memory CTLs.
Figure 3.
Codelivery of pDAI promotes antigen-specific cytotoxic T cell responses. C57BL/6 mice were electroporated with pDAI, pOVA, or pOVA+pDAI twice at 2-week interval. (a, b) CD8+ T cell responses were analyzed in blood collected 13 days after the last vaccination (n = 6). (a) Frequency of IFN-γ producing CD8+ T cells (over the gated CD8+ T cell population) after in vitro stimulation with trp2(180–188) (control) or OVA(257–264) (OVA) peptides (a). Bars are the mean ± SEM. *P = 0.01. (b) Representative histograms showing IFN-γ and TNF-α expression on gated CD8+ population. The relative proportion of IFN-γ+TNF-α– and IFN-γ+TNF-α+ CD8+ T cells is indicated. (c) In vivo cytotoxic killing of OVA(257–264)-pulsed target cells (CFSEhigh) relative to control trp2(180–188)-pulsed cells (CFSElow). A representative histogram per group is shown (n = 8) with the percentage of specific killing (mean ± SEM).*P = 0.017. Data shown are from one representative of at least two independent experiments. IFN, interferon; OVA, ovalbumin; TNF-α, tumor necrosis factor-α.
Figure 4.
Codelivery of pDAI promotes memory CTL responses. C57BL/6 mice were electroporated with pDAI, pOVA, or pOVA+pDAI twice at 2-weeks interval (n = 6) and lymphocytes from (a) spleen and (b) blood were analyzed 5 weeks later by immunofluorescence staining and flow cytometry. Frequency of OVA(257–264)-specific CD8+ T cells (over the gated CD8+ T cell population) showing one of the following memory subset phenotypes: effector memory (TEM; CD44highCD62Llow); central memory (TCM; CD44highCD62Lhigh); memory stem cells (TSCM; CD44lowCD62Lhigh). Bars are the mean ± SEM. *P < 0.05. CTL, cytotoxic CD8+ T lymphocytes.
DAI efficiently induces CTL immunity to survivin TAA and promotes tumor protection
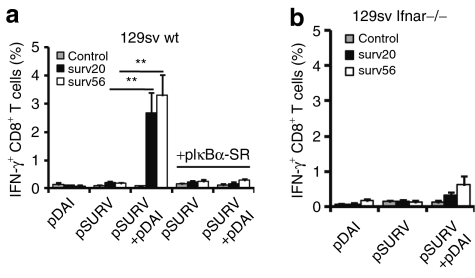
We next studied whether DAI coadministration would enhance the immunogenicity of a weakly immunogenic TAA. Survivin is an intracellular inhibitor of apoptosis that is strongly upregulated in many types of cancer cells and associated with enhanced tumor cell viability.24 While we were unable to detect responses to survivin in 129Sv mice vaccinated with a survivin-encoding plasmid (pSURV) alone, coadministration of pDAI overcame tolerance to the self-epitopes surv(20–28) and surv(56–64) and raised the frequency of IFN-γ-producing CD8+ T cells in peripheral blood from nondetectable (<0.2%) to 2.7% (P = 0.007) and 3.3% (P = 0.002) for surv(20–28) and surv(56–64), respectively (Figure 5a). Given that most of the transcripts upregulated by DAI overexpression observed in Figure 1 are potential targets for nuclear factor κB (NF-κB) (IFN-α, IL-6, TNF-α, Cxcl10, CD40, CD80),25 we asked whether activation of this pathway was responsible for the adjuvant effect in vivo. A plasmid encoding a nonphosphorylable, degradation-resistant mutant of the NF-κB inhibitor, IκBα super-repressor (pIκBα-SR),26 which blocks NF-κB-dependent transcription, was codelivered. Interestingly, coadministration of pIκBα-SR completely ablated the survivin-specific response observed in pDAI-vaccinated mice (Figure 5a), indicating that NF-κB activity is essential for the adjuvant effect of pDAI.
Figure 5.
Codelivery of pDAI overcomes CTL tolerance to survivin TAA via a mechanism requiring NF-κB activation and type I IFN production. Mice were electroporated twice at 2-week intervals with pDAI, pSURV, or pSURV+pDAI (n = 6). Where indicated, pIkBα-SR was also administrated. (a, b) Peripheral lymphocytes were obtained from (a) 129Sv WT or (b) Ifnar−/− mice 13 days after the last vaccination. The frequency of IFN-γ producing CD8+ T cells (over the gated CD8+ T cell population) after in vitro stimulation with trp2(180–188) (control), surv(20–28) (surv20), or surv(56–64) (surv56) peptides is shown. Bars are the mean ± SEM. **P = 0.002. Data shown are from one representative of at least two independent experiments. CTL, cytotoxic CD8+ T lymphocytes; IFN, interferon; NF-κB, nuclear factor κB; TAA, tumor-associated antigens; WT, wild type.
A hallmark of DAI signaling is the release of type I IFNs,13,18 which are essential for optimal clonal expansion and enhanced effector function of CTLs.27,28 Therefore, we investigated whether type I IFN production would contribute to the adjuvant effect of DAI. The induction of survivin-specific IFN-γ-producing CTLs observed in Sv129 mice vaccinated with pSURV+pDAI was drastically reduced in IFN-α/β receptor-deficient mice (Figure 5b). Codelivered pDAI could only generate an insignificant increase in the frequency of IFN-γ-producing CD8+ T cells. This indicates that type I IFN production was largely responsible for the adjuvant effect of DAI, although other factors may also contribute.
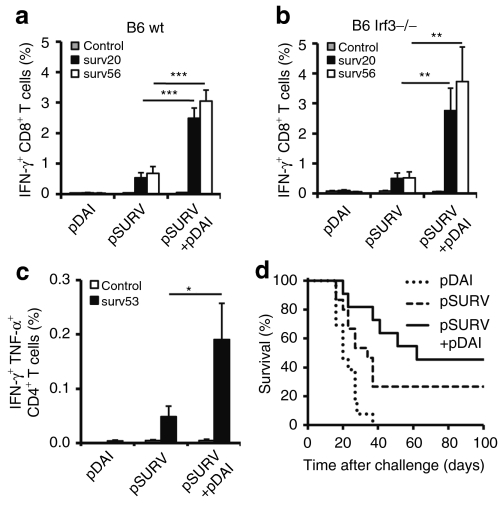
DAI-induced type I IFN-secretion depends, at least partially, on IRF3-activation in vitro.13 Therefore, we studied the effect of pDAI coadministration on survivin-specific CTL responses in IRF3-deficient mice. Coimmunization with pDAI+pSURV in both C57BL/6 wild type and Irf3−/− mice increased the frequency of antigen-specific IFN-γ-secreting CD8+ T cells more than fivefold compared to pSURV alone, from 0.45 to 2.4% (P < 0.0001), or from 0.5 to 2.8–3.7% (P < 0.005), respectively (Figure 6a,b). These results indicate that IRF3 is not required for DAI-enhanced CTL induction. Interestingly, survivin-specific CTLs could simultaneously produce TNF-α and a small proportion also produced IL-2 (Supplementary Figure S5). We further tested whether pDAI coimmunization would promote CD4+ Th1 responses to a major histocompatibility complex class II-restricted survivin epitope, surv(53–67). Indeed, pDAI increased the frequency of CD4+ T cells producing simultaneously IFN-γ and TNF-α after surv(53–67) peptide stimulation from 0.049 to 0.19% (Figure 6c, P < 0.05). In order to assess its relative potency as a genetic adjuvant, pDAI was directly compared to the granulocyte-macrophage colony-stimulating factor (GM-CSF)-encoding plasmid (pGM-CSF), a widely used genetic adjuvant.29 In contrast to pDAI, coimmunization with pSURV+pGM-CSF did not boost either CD8+ or CD4+ T cell responses to survivin (Supplementary Figure S6), indicating that DNA EP itself is a potent approach to elicit immune responses and probably masked the adjuvant effect of pGM-CSF observed by others including our group. Furthermore and considering the plethora of induced cytokines including IL-10, the potential induction of immunosuppressive cells such as CD25highFoxP3+ CD4+ regulatory T cells, and Gr1+CD11b+ myeloid-derived suppressor cells was investigated in the inguinal lymph nodes and spleens from vaccinated mice 13 days postimmunization. Neither the vaccination with the TAA survivin nor the coimmunization with pDAI altered the frequency of regulatory T cells (Supplementary Figure S7). A small but significant (P < 0.05) decrease in myeloid-derived suppressor cells was detected in the spleen of mice coimmunized with pDAI+pSURV as compared to pSURV alone. The enhanced CTL and CD4+ Th1 responses observed suggested that pDAI coadministration might enhance protection against tumor formation elicited by pSURV DNA vaccination. Indeed, administration of pDAI+pSURV to C57BL/6 mice demonstrated a trend (P = 0.079) toward improved rejection of a subcutaneous tumor challenge of the syngeneic B16 melanoma when compared to mice immunized with pSURV alone (Figure 6d).
Figure 6.
Codelivery of pDAI enhances CTL and CD4+ Th1 responses to survivin TAA and promotes tumor protection. Mice were electroporated twice at 2-week intervals with pDAI, pSURV, or pSURV+pDAI. (a, b) Peripheral lymphocytes were obtained from (a) C57BL/6 WT or (b) Irf3−/− mice (n = 8) 13 days after the last vaccination. The frequency of IFN-γ producing CD8+ T cells (over the gated CD8+ T cell population) after in vitro stimulation with trp2(180–188) (control), surv(20–28) (surv20), or surv(56–64) (surv56) peptides is shown. Bars are the mean ± SEM. ***P < 0.0001, **P = 0.005. Data shown are from one representative of at least two independent experiments. (c) Peripheral lymphocytes were obtained 13 days after the last vaccination (n = 7). The frequency of peripheral CD4+ T cells producing simultaneously IFN-γ and TNF-α (over the gated CD4+ T cell population) after in vitro stimulation with ova(323–339) (control) or surv(53–67) (surv53) peptides is shown. Bars are the mean ± SEM. *P < 0.05. (d) Survival of C57BL/6 WT mice challenged with B16 melanoma cells 2 weeks after the second vaccination (pDAI, n = 5; pSURV, n = 15; pSURV+pDAI, n = 11). P = 0.079 for the comparison between pSURV and pSURV+pDAI groups. Data presented were pooled from two independent experiments. CTL, cytotoxic CD8+ T lymphocytes; IFN, interferon; TAA, tumor-associated antigens; WT, wild type.
DAI enhances TRP2-specific CTL responses and confers long-term tumor protection
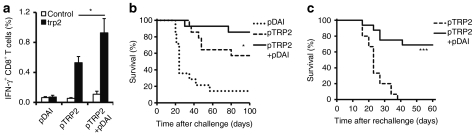
In order to confirm the ability of DAI to enhance antitumor CTL responses, we combined pDAI with a tyrosinase-related protein-2 (TRP2) encoding DNA vaccine (pTRP2). TRP2 is a highly expressed glycoprotein in human melanomas and a clinically relevant model antigen used for CTL-mediated targeting of mouse B16 melanoma.30 An increased frequency of trp2(180–188)-specific IFN-γ-producing CD8+ T cells was observed in mice covaccinated with pTRP2 and pDAI (pTRP2+pDAI) as compared to mice vaccinated with pTRP2 only (0.75 and 0.43% respectively, P = 0.043) (Figure 7a). We also tested whether pDAI covaccination would increase the rejection of B16 melanoma challenge and found that a higher proportion of the covaccinated mice rejected a B16 melanoma challenge compared to mice vaccinated with TRP2 alone (Figure 7b; P = 0.044). Given the enhancement of antigen-specific memory CD8+ T cell responses observed in Figure 4, mice that survived the B16 challenge from both pTRP2 and pTRP2+pDAI vaccinated groups were rechallenged 3 months after the initial challenge with a higher dose of B16 melanoma cells, without any additional immunization. Remarkably, almost 70% of the pTRP2+pDAI covaccinated mice rejected the melanoma cells whereas all mice vaccinated with pTRP2 alone succumbed to the second challenge (P < 0.0001) (Figure 7c). These results argue that DAI not only promotes the induction of functional effector cells, but also enhances immunological memory.
Figure 7.
Codelivery of pDAI enhances TRP2-specific CTL responses and confers long-term protection to B16 melanoma challenge. C57BL/6 mice were electroporated twice at 2-week intervals with pDAI, pTRP2, or pTRP2+pDAI. (a) Peripheral lymphocytes were obtained 13 days after the last vaccination. The frequency of IFN-γ producing CD8+ T cells (over the gated CD8+ T cell population) after in vitro stimulation with surv(20–28) (control) or trp2(180–188) (trp2) peptides is shown (n = 6). Bars are the mean ± SEM. *P = 0.043. Data presented are from one representative of two independent experiments. (b) Survival of C57BL/6 WT mice challenged with B16 melanoma cells two weeks after the second vaccination (n = 14). *P = 0.044 for the comparison between pTRP2 and pTRP2+pDAI groups. Data presented were pooled from two independent experiments. (c) Long-term protection of C57BL/6 WT mice that had rejected the initial tumor challenge and were rechallenged with a higher dose of B16 melanoma cells in the opposite flank 3 months after initial challenge (pTRP2, n = 15; pTRP2+pDAI, n = 16). Data presented were pooled from two independent experiments. ***P < 0.0001. CTL, cytotoxic CD8+ T lymphocytes; IFN, interferon; TRP2, tyrosinase-related protein-2; TAA, tumor-associated antigens; WT, wild type.
Discussion
In the present study, we describe for the first time the use of an intracellular PRR as a genetic adjuvant that enhances the immunogenicity of DNA vaccines. DNA vaccines encode single (or few) antigens, which makes them highly specific but also inherently less immunogenic than whole-cell or multicomponent vaccines developed in the past. Furthermore, DNA vaccines against cancer need to overcome the tumor-associated immune tolerance, which is generally characterized by low frequencies of TAA-specific T cell precursors or the presence of TAA-specific T cells with intermediate-low TCR affinity. Therefore, adjuvants that enhance the immunogenicity of the encoded antigen and efficiently induce TAA-specific effector T cells are essential components of antitumor DNA vaccines. Genes encoding single cytokines and chemokines have been extensively used as genetic adjuvants. Notably genes encoding IL-2, IL-12, IL-15, and GM-CSF have successfully adjuvanted DNA vaccines in mice and nonhuman primates31,32,33 as well as in humans.34 However, some of these molecules, e.g., IL-235 and GM-CSF,36,37 are important in maintaining immune tolerance to self-antigens. Moreover, recent results from clinical studies have shown that IL-2 or GM-CSF as adjuvant for cancer vaccines or immunotherapies can promote the induction and recruitment of immunosuppressive T regulatory cells38,39 and myeloid-derived suppressor cells.40 The homeostatic roles of certain cytokines and sometimes the contradictory effects observed in clinical trials have raised some concerns about the use of single cytokines as adjuvants for cancer vaccines. Strategies involving the concerted action of several cytokines and costimulatory molecules can result in a potent activation of the immune responses, in particular against weakly immunogenic antigens.10 The production of several immunostimulatory molecules can be achieved by stimulation of innate immune PRRs, as occurs during natural infection. Consequently, strategies that boost innate immune PRR signaling by coexpressing intracellular adaptor molecules or downstream transcription factors as genetic adjuvants, have been shown to enhance the potency of DNA vaccines.41,42 However, such strategies have not been combined with antitumor DNA vaccines. The strategy developed in this study consists of delivering both the DNA-encoded intracellular PRR (DAI) and its activating ligand (plasmid DNA) to stimulate downstream transcription factors and initiate the production of several proinflammatory and costimulatory molecules, as well as type I IFNs, which ultimately promote adaptive T cell responses. The coadministration of pDAI was indeed comparatively more potent than the plasmid encoding GM-CSF in enhancing survivin-specific T cell responses in the experimental setup described here (Supplementary Figure S6). Moreover, pDAI did not induce immunosuppressive cell populations, as it has been reported for other adjuvants,38,39,40 and rather showed decreased levels of myeloid-derived suppressor cells (Supplementary Figure S7).
In vitro studies have demonstrated that DAI signaling can activate both NF-κB and IRF3 to produce type I IFNs and other proinflammatory cytokines.13,18 Nevertheless, little is known about DAI signaling in vivo. Our data show that i.d. EP with pDAI induces transcriptional upregulation of molecules that are known downstream targets of NF-κB (Figure 1), indicating that this key mediator of the innate immunity was activated in mouse skin. NF-κB represents a master transcription factor for signaling through TLRs43 and intracellular DNA-sensing PRRs.13,15,16,19 Consistent with such a crucial role in DNA-induced innate immune activation, NF-κB was essential for DAI-promoted CTL induction (Figure 4a) and, quite unexpectedly, this effect was not dependent on IRF3 (Figure 5b). Based on these observations, the critical role of type I IFNs for DAI-enhanced CTL induction (Figure 4b) and that IRFs are the main transcription factors driving type I IFN expression, we speculate that NF-κB activation is required for the initial release of type I IFNs and the resulting signaling events then activate IRFs, other than IRF3, to ensure robust induction of innate immune responses. Type I IFN production has been described to occur in two waves (reviewed in ref. 44). In the first wave, PRR signaling results in activation of NF-κB, IRF3, or both, and subsequent release of IFN-β. In the second wave, IFN-β transmits the danger signal to neighboring cells through binding to the type I IFN receptor. This turns on a signaling cascade that promotes gene expression of IFN-inducible genes with antiviral activity as well as IRF7. Possibly, IRF7-signaling contributes to ensure type-I IFN responses in the absence of IRF3-signaling,13 thus sufficing the production of large amounts of type I IFNs.
The effects of type I IFNs include the release of cytokines and chemokines that modulate the function of dendritic cells resulting in, among other things, increased major histocompatibility complex class I crosspresentation, as well as improved development of effector and memory CTLs.45,46 Our results support the notion that, in addition to antigen and coreceptor mediated stimulation, a third cytokine signal is important for effective CTL induction.27,28 Among the cytokines that can provide the third signal, IFN-α but not IL-12 transcripts were upregulated after pDAI EP (Figure 1). Also, type I IFN signaling was essential for DAI-mediated CTL induction (Figure 4b). There is abundant evidence supporting the importance of type I IFNs in the generation of long-lasting antitumor immunity. Type I IFNs enhance tumor protection by increasing induction, proliferation, effector function, resistance to apoptosis, and long-term effector memory phenotype of CTLs.47 On the other hand, defective type I IFN signaling and downstream activation of T cells is a common immune dysfunction in patients with different types of cancer.48 Here, we demonstrate that pDAI coimmunization promotes the induction and persistence of memory CD8+ T cell of effector, central, and stem cell phenotypes (Figure 4). Accordingly, the combination of DAI- and TRP2-encoding plasmids elicited long-term protection against B16 melanoma (Figure 6c).
In summary, our studies show that in vivo overexpression of DAI boosts DNA-sensing innate immune activation and thereby generates a proinflammatory microenvironment essential for effective CTL induction and long-lasting antitumor immunity. Thus, this study validates the use of intracellular innate PRRs as genetic adjuvants that harness intrinsic innate immune-stimulating properties of plasmid DNA vaccines to enhance the immunogenicity of weakly immunogenic antigens. Hence, our findings are expected to improve the design of DNA vaccines for diseases where efficient cellular immunity is desired to confer protection.
Materials and Methods
Mice and immunizations. C57BL/6, OT-I Rag-1−/−, Irf3−/−, and 129Sv and Ifnar−/− mice were kept according to the guidelines of the Regional Animal Ethics Committee. Mice anesthetized with isoflurane were injected i.d. with 40 µl of phosphate-buffered saline containing 20 µg of each plasmid at two different sites (20 µl each). pVAX was used to equalize DNA quantity within the same experiment. A parallel needle array electrode (two rows of four 2-mm pins, 1.5 × 4 mm gaps) was applied to deliver the electric pulses (two 1,125 V/cm, 0.05 ms pulses followed by eight 275 V/cm, 10 ms pulses)49 using the Derma Vax DNA Vaccine Skin Delivery System (Cyto Pulse Sciences, now Cellectics, Romainville, France). Mice were immunized two times with 2 weeks between immunizations. The following plasmids were used for immunizations: pOVA encoding membrane-bound OVA (kindly provided by Dr A. Lew, Walter and Eliza Hall Institute of Medical Research, Melbourne, Australia), pSURV encoding the human survivin gene has been described,50 pTRP2 encoding the human TRP2 (kindly provided by Dr T. Wölfel, Johannes Gutenberg University, Gutenberg, Germany), pIκBα-SR encoding the IκBα super-repressor (kindly provided by Dr R. Toftgård, Karolinska Institutet, Stockholm, Sweden) and pDAI. pDAI was produced by cloning the DAI coding sequence from mouse splenocytes into the pVAX vector (Invitrogen, Carlsbad, CA) using standard cloning procedures. Primers used for DAI cloning are listed in Supplementary Table S1. Plasmids were purified using the GigaPrep Endofree Kit (Qiagen, Hilden, Germany). Tumor challenge and rechallenge was performed by injecting subcutaneously 1 × 105 and 2 × 105 B16 cells, respectively.
Quantitative real-time reverse transcription-PCR. Total RNA was isolated from skin biopsies taken 24 hours after DNA EP and cDNA was prepared (iScript cDNA synthesis kit; Bio-Rad, Hercules, CA). Transcript levels were determined by quantitative real-time PCR (iQ SYBR Green Supermix; Bio-Rad; ABI7500, Applied Biosystems, Foster City, CA) using a two-step cycling program (1 minute at 95 °C, followed by 40 cycles of 15 seconds at 95 °C and 1 minute at 62–64 °C) and normalized to the L32 housekeeping gene. Primers are listed in Supplementary Table S2. Pooled cDNA from the pVAX and pDAI groups was added to the RT2 SYBR Green qPCR Master Mix (SABiosciences, Frederick, MD) and each sample was aliquoted on the Mouse IFN-α, β response, and the mouse inflammatory response and autoimmunity RT2Profiler PCR-arrays, respectively. All steps were done according to the manufacturer's protocol for the ABI Prism 7500 and 7900 HT Sequence Detection System. To analyze the PCR-array data, excel macros were downloaded from the manufacturer's website (http://www.sabiosciences.com/pcrarraydataanalysis.php). Data normalization was based on correcting all Ct values for the average Ct values of several constantly expressed housekeeping genes present on the array.
Antibodies and flow cytometry. Monoclonal antibodies anti-mouse CD8α (clone 53-6.7), IFN-γ (clone XMG1.29), TNF-α (clone MP6-XT22), Vα2 TCR (clone B20.1), IL-2 (clone JES6-5H4), CD11b (clone M1/70), CD11c (clone N418), CD25 (clone PC61), CD40 (clone 1C10), CD44 (clone IM7), CD62L (clone MEL-14), CD69 (clone H1.2F3), CD80 (clone 16-10A1), FOXP3 (MF23), major histocompatibility complex class II (clone M5/114.15.2), Gr1 (RB6-85C) (BD Biosciences, San Jose, CA) and H-2Kb/OVA(257–264) pentamer (Proimmune, Oxford, UK) were used for immunoflorescence staining and flow cytometry analysis. Nonspecific binding was blocked by adding unconjugated rat anti-mouse CD16/CD32 antibody (mouse BD Fc block, clone 2.4G2; BD Biosciences). Samples were analyzed on a FACSCalibur cytometer (BD Biosciences) and the data were processed using FlowJo version 6.4.7 (Tree Star, Ashland, OR).
Intracellular cytokine staining. Peripheral blood was collected 13 days after the last immunization and lymphocytes were stimulated with peptides (1 µg/ml) for 8 hours. GolgiPlug (BD Biosciences) was added after 2 hours. Intracellular staining was performed using Cytofix/Cytoperm Fixation/Permeabilization Solution set (BD Biosciences) according to the manufacturer's instructions.
In vivo antigen proliferation of OT-I CD8+ T cells. Spleen and lymph node cells from naive OT-I mice were isolated. Splenocytes were RBC depleted using PharmLyse buffer. Cells (108 cells/ml; phosphate-buffered saline 0.5% RPMI) were stained with 2 µmol/l CFSE (Sigma-Aldrich, St Louis, MO) for 5 minutes. A tenfold larger volume of 20% fetal bovine serum RPMI was added to stop CFSE staining. Cells were then washed twice, resuspended in phosphate-buffered saline at a final concentration of 107 cells/ml and 2 × 106 cells were intravenously transferred into vaccinated recipients. Inguinal lymph nodes were sampled after 4 days and analyzed by flow cytometry.
In vivo cytotoxicity assay. Target spleen cells were labeled with 0.2 and 2 µmol/l of CFSE, and pulsed with control or OVA peptide, respectively. Then, 107 cells from each population were mixed and injected intravenously into vaccinated mice. Lymph nodes were removed 6 hours later and analyzed by flow cytometry.50 CFSE+ donor target splenocytes were differentiated from host cells and the percentage of specific killing was determined as follows: 100 – [(% of OVA peptide-pulsed targets/% of TRP2 peptide-pulsed targets in vaccinated recipients)/(% of OVA peptide-pulsed targets/% of TRP2 peptide-pulsed targets in control vaccinated recipients) × 100].
Statistical analysis. Statistical analysis was performed using the Graphpad Prism software (Graphpad Software, La Jolla, CA). Unpaired t tests were performed pair wise between relevant groups. No multiple comparisons were performed to control for type I errors. Statistical analyses of survival curves were performed using the one-tailed Mantel–Cox log-rank test.
SUPPLEMENTARY MATERIAL Figure S1. Intradermal DNA electroporation induces tissue inflammation. Mice were vaccinated and biopsies were collected as in Figure 1. H&E staining reveals massive infiltration of lymphocytes (black cells, black arrowheads) in DNA electroporated skin (left panel). Significant skin infiltration by CD207+ DCs (brown, blue arrowheads) is observed in pVAX and pDAI electroporated mice when compared to untreated controls (p=0.007 and 0.0004, respectively) as detected by anti-CD207 staining (right panel). Figure S2. DNA vaccine-induced upregulation of costimulatory molecules in dendritic cells from skin-draining lymph nodes. C57BL/6 mice (n=6) were vaccinated as in Figure 1 and the inguinal lymph nodes were taken 24 h later. The levels of costimulatory molecules CD40 and CD80 were analyzed in dendritic cells (CD11c+, MHC class IIhigh) by surface staining (black histograms) using naïve mice as controls (red histograms) and the mean fluorescence intensity (MFI) was calculated for each group. Bars are the mean±SEM. **indicates p=0.007; ***indicates p=0.0001. Figure S3. Phenotypic analysis of DNA vaccine-induced OVA-specific CTLs. C57BL/6 mice were vaccinated twice as in Figure 3 (n=6). (a) Detection of OVA(257-264)-specific CD8+ T cells (R1) was performed by pentamer staining either at two (b) or five (c) weeks after the last vaccination. (b) Surface staining of phenotypic markers on OVA(257-264)-specific CD8+ T cells was evaluated in pooled peripheral blood from mice immunized with pOVA (—, thin line profiles) or pOVA+pDAI (—, thick line profiles). The bulk CD8+ T cell population (▪, solid grey profiles) was used as control. (c) Analysis of OVA(257-264)-specific CD8+ T cells to identify the different memory sub-set phenotypes: effector memory (TEM; CD44highCD62Llow); central memory (TCM; CD44highCD62Lhigh); memory stem cells (TSCM; CD44lowCD62Lhigh). Figure S4. Detection of DNA vaccine-induced OVA-specific antibodies. C57BL/6 mice were vaccinated twice as in Figure 3 and sera collected 13 days after the last vaccination. OVA-specific antibodies were detected by indirect ELISA in sera from mice immunized with pDAI (♦, diamonds), pOVA (▪, squares) or pOVA+pDAI (▴, triangles) (mean±SEM; n=8). No statistically significant differences in antibody levels were observed between pOVA and pOVA+pDAI groups. Figure S5. DNA vaccine-induced survivin-specific CTLs produced IFN-γ, TNF-α, and IL-2 after peptide stimulation. C57BL/6 mice were vaccinated as in Figure 5 and blood collected 13 days after the last vaccination. Detection of survivin-specific CD8+ T cells was performed after in vitro stimulation with surv(56-64) peptide. Intracellular staining of IFN-γ simultaneously with TNF-a or IL-2 in gated CD8+ T cells from mice immunized with pDAI (left panels), pSURV (middle panels) or pSURV+pDAI (right panels). Dot plots from a representative mouse per group are displayed indicating the mean±SEM for each group (n=4). Similar results were obtained after stimulation with surv(20-28) peptides. Figure S6. Comparison of the adjuvant efficacy of pDAI and pGM-CSF to enhance survivin-specific CTL and Th1 responses. C57BL/6 mice were electroporated twice at two-week intervals with pDAI, pSURV, pSURV+pDAI or pSURV+pGM-CSF (n=7) and blood collected 13 days after the last vaccination. (a) The frequency of peripheral IFN-γ-producing CD8+ T cells (over the gated CD8+ T cell population) after in vitro stimulation with trp2(180-188) (Control), surv(20-28) (surv20) or surv(56-64) (surv56) peptides is shown. (b) The frequency of peripheral IFN-γ- and TNF-α-producing CD4+ T cells (over the gated CD4+ T cell population) after in vitro stimulation with ova(323-339) (Control) or surv(53-67) (surv53) peptides is shown. Bars are the mean±SEM. *indicates p=0.026; ***indicates p=0.0003. Figure S7. Analysis of immunosuppressive cell populations. C57BL/6 mice were vaccinated twice at two-week intervals with pDAI, pSURV or pSURV+pDAI (n=8). Two weeks later, T regulatory (Treg) and myeloid-derived suppressor cells (MDSC) were analyzed in spleen and inguinal lymph nodes by immunofluorescence staining and flow cytometry. The frequency of Treg (CD4+; CD25high; FOXP3+) over the total CD4+ T cell population (a) and MDSC (CD11b+; Gr1+) over the total cell population (b) were determined and the results are shown. Bars are the mean±SEM. *indicates p<0.05. Table S1. The 20 most strongly upregulated gene transcripts in mice electroporated with pDAI as compared to mice electroporated with pVAX control vector. Table S2. Primers used for cloning and quantitative real-time PCR analysis. Materials and Methods.
Acknowledgments
This work was supported by grants to R.K. from the Swedish Cancer Society, the Swedish Medical Research Council, the Cancer Society of Stockholm, the European Union (Grant “EUCAAD” and “DC-THERA”), the Karolinska Institutet, “ALF-Project” grants from the Stockholm City Council. AFGQ has received support from ICGEB (International Center of Genetic Engineering and Biotechnology, Trieste, Italy) grant CRP/CH102-01, Wellcome Trust award WT06491I/Z/01/Z and FONDAP grant 15010006. A.L. has been supported by a Fellowship for Postgraduate Studies “Presidente de la República” from CONICYT, Chile. D.M. was supported by a grant of the German Research Association (DFG). K.L. has been supported by a postdoctoral fellowship from the Swedish Society for Medical Research.
Supplementary Material
Intradermal DNA electroporation induces tissue inflammation. Mice were vaccinated and biopsies were collected as in Figure 1. H&E staining reveals massive infiltration of lymphocytes (black cells, black arrowheads) in DNA electroporated skin (left panel). Significant skin infiltration by CD207+ DCs (brown, blue arrowheads) is observed in pVAX and pDAI electroporated mice when compared to untreated controls (p=0.007 and 0.0004, respectively) as detected by anti-CD207 staining (right panel).
DNA vaccine-induced upregulation of costimulatory molecules in dendritic cells from skin-draining lymph nodes. C57BL/6 mice (n=6) were vaccinated as in Figure 1 and the inguinal lymph nodes were taken 24 h later. The levels of costimulatory molecules CD40 and CD80 were analyzed in dendritic cells (CD11c+, MHC class IIhigh) by surface staining (black histograms) using naïve mice as controls (red histograms) and the mean fluorescence intensity (MFI) was calculated for each group. Bars are the mean±SEM. **indicates p=0.007; ***indicates p=0.0001.
Phenotypic analysis of DNA vaccine-induced OVA-specific CTLs. C57BL/6 mice were vaccinated twice as in Figure 3 (n=6). (a) Detection of OVA(257-264)-specific CD8+ T cells (R1) was performed by pentamer staining either at two (b) or five (c) weeks after the last vaccination. (b) Surface staining of phenotypic markers on OVA(257-264)-specific CD8+ T cells was evaluated in pooled peripheral blood from mice immunized with pOVA (—, thin line profiles) or pOVA+pDAI (—, thick line profiles). The bulk CD8+ T cell population (▪, solid grey profiles) was used as control. (c) Analysis of OVA(257-264)-specific CD8+ T cells to identify the different memory sub-set phenotypes: effector memory (TEM; CD44highCD62Llow); central memory (TCM; CD44highCD62Lhigh); memory stem cells (TSCM; CD44lowCD62Lhigh).
Detection of DNA vaccine-induced OVA-specific antibodies. C57BL/6 mice were vaccinated twice as in Figure 3 and sera collected 13 days after the last vaccination. OVA-specific antibodies were detected by indirect ELISA in sera from mice immunized with pDAI (♦, diamonds), pOVA (▪, squares) or pOVA+pDAI (▴, triangles) (mean±SEM; n=8). No statistically significant differences in antibody levels were observed between pOVA and pOVA+pDAI groups.
DNA vaccine-induced survivin-specific CTLs produced IFN-γ, TNF-α, and IL-2 after peptide stimulation. C57BL/6 mice were vaccinated as in Figure 5 and blood collected 13 days after the last vaccination. Detection of survivin-specific CD8+ T cells was performed after in vitro stimulation with surv(56-64) peptide. Intracellular staining of IFN-γ simultaneously with TNF-a or IL-2 in gated CD8+ T cells from mice immunized with pDAI (left panels), pSURV (middle panels) or pSURV+pDAI (right panels). Dot plots from a representative mouse per group are displayed indicating the mean±SEM for each group (n=4). Similar results were obtained after stimulation with surv(20-28) peptides.
Comparison of the adjuvant efficacy of pDAI and pGM-CSF to enhance survivin-specific CTL and Th1 responses. C57BL/6 mice were electroporated twice at two-week intervals with pDAI, pSURV, pSURV+pDAI or pSURV+pGM-CSF (n=7) and blood collected 13 days after the last vaccination. (a) The frequency of peripheral IFN-γ-producing CD8+ T cells (over the gated CD8+ T cell population) after in vitro stimulation with trp2(180-188) (Control), surv(20-28) (surv20) or surv(56-64) (surv56) peptides is shown. (b) The frequency of peripheral IFN-γ- and TNF-α-producing CD4+ T cells (over the gated CD4+ T cell population) after in vitro stimulation with ova(323-339) (Control) or surv(53-67) (surv53) peptides is shown. Bars are the mean±SEM. *indicates p=0.026; ***indicates p=0.0003.
Analysis of immunosuppressive cell populations. C57BL/6 mice were vaccinated twice at two-week intervals with pDAI, pSURV or pSURV+pDAI (n=8). Two weeks later, T regulatory (Treg) and myeloid-derived suppressor cells (MDSC) were analyzed in spleen and inguinal lymph nodes by immunofluorescence staining and flow cytometry. The frequency of Treg (CD4+; CD25high; FOXP3+) over the total CD4+ T cell population (a) and MDSC (CD11b+; Gr1+) over the total cell population (b) were determined and the results are shown. Bars are the mean±SEM. *indicates p<0.05.
The 20 most strongly upregulated gene transcripts in mice electroporated with pDAI as compared to mice electroporated with pVAX control vector.
Primers used for cloning and quantitative real-time PCR analysis.
REFERENCES
- Gao Q, Qiu SJ, Fan J, Zhou J, Wang XY, Xiao YS, et al. Intratumoral balance of regulatory and cytotoxic T cells is associated with prognosis of hepatocellular carcinoma after resection. J Clin Oncol. 2007;25:2586–2593. doi: 10.1200/JCO.2006.09.4565. [DOI] [PubMed] [Google Scholar]
- Nakano O, Sato M, Naito Y, Suzuki K, Orikasa S, Aizawa M, et al. Proliferative activity of intratumoral CD8(+) T-lymphocytes as a prognostic factor in human renal cell carcinoma: clinicopathologic demonstration of antitumor immunity. Cancer Res. 2001;61:5132–5136. [PubMed] [Google Scholar]
- Sato E, Olson SH, Ahn J, Bundy B, Nishikawa H, Qian F, et al. Intraepithelial CD8+ tumor-infiltrating lymphocytes and a high CD8+/regulatory T cell ratio are associated with favorable prognosis in ovarian cancer. Proc Natl Acad Sci USA. 2005;102:18538–18543. doi: 10.1073/pnas.0509182102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg SA., and, Dudley ME. Adoptive cell therapy for the treatment of patients with metastatic melanoma. Curr Opin Immunol. 2009;21:233–240. doi: 10.1016/j.coi.2009.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendandi M, Gocke CD, Kobrin CB, Benko FA, Sternas LA, Pennington R, et al. Complete molecular remissions induced by patient-specific vaccination plus granulocyte-monocyte colony-stimulating factor against lymphoma. Nat Med. 1999;5:1171–1177. doi: 10.1038/13928. [DOI] [PubMed] [Google Scholar]
- Rice J, Ottensmeier CH., and, Stevenson FK. DNA vaccines: precision tools for activating effective immunity against cancer. Nat Rev Cancer. 2008;8:108–120. doi: 10.1038/nrc2326. [DOI] [PubMed] [Google Scholar]
- Luxembourg A, Evans CF., and, Hannaman D. Electroporation-based DNA immunisation: translation to the clinic. Expert Opin Biol Ther. 2007;7:1647–1664. doi: 10.1517/14712598.7.11.1647. [DOI] [PubMed] [Google Scholar]
- Ahlén G, Söderholm J, Tjelle T, Kjeken R, Frelin L, Höglund U, et al. In vivo electroporation enhances the immunogenicity of hepatitis C virus nonstructural 3/4A DNA by increased local DNA uptake, protein expression, inflammation, and infiltration of CD3+ T cells. J Immunol. 2007;179:4741–4753. doi: 10.4049/jimmunol.179.7.4741. [DOI] [PubMed] [Google Scholar]
- Babiuk S, Baca-Estrada ME, Foldvari M, Middleton DM, Rabussay D, Widera G, et al. Increased gene expression and inflammatory cell infiltration caused by electroporation are both important for improving the efficacy of DNA vaccines. J Biotechnol. 2004;110:1–10. doi: 10.1016/j.jbiotec.2004.01.015. [DOI] [PubMed] [Google Scholar]
- Iwasaki A, Stiernholm BJ, Chan AK, Berinstein NL., and, Barber BH. Enhanced CTL responses mediated by plasmid DNA immunogens encoding costimulatory molecules and cytokines. J Immunol. 1997;158:4591–4601. [PubMed] [Google Scholar]
- Kumar H, Kawai T., and, Akira S. Pathogen recognition in the innate immune response. Biochem J. 2009;420:1–16. doi: 10.1042/BJ20090272. [DOI] [PubMed] [Google Scholar]
- Medzhitov R. Recognition of microorganisms and activation of the immune response. Nature. 2007;449:819–826. doi: 10.1038/nature06246. [DOI] [PubMed] [Google Scholar]
- Takaoka A, Wang Z, Choi MK, Yanai H, Negishi H, Ban T, et al. DAI (DLM-1/ZBP1) is a cytosolic DNA sensor and an activator of innate immune response. Nature. 2007;448:501–505. doi: 10.1038/nature06013. [DOI] [PubMed] [Google Scholar]
- Bürckstümmer T, Baumann C, Blüml S, Dixit E, Dürnberger G, Jahn H, et al. An orthogonal proteomic-genomic screen identifies AIM2 as a cytoplasmic DNA sensor for the inflammasome. Nat Immunol. 2009;10:266–272. doi: 10.1038/ni.1702. [DOI] [PubMed] [Google Scholar]
- Hornung V, Ablasser A, Charrel-Dennis M, Bauernfeind F, Horvath G, Caffrey DR, et al. AIM2 recognizes cytosolic dsDNA and forms a caspase-1-activating inflammasome with ASC. Nature. 2009;458:514–518. doi: 10.1038/nature07725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ablasser A, Bauernfeind F, Hartmann G, Latz E, Fitzgerald KA., and, Hornung V. RIG-I-dependent sensing of poly(dA:dT) through the induction of an RNA polymerase III-transcribed RNA intermediate. Nat Immunol. 2009;10:1065–1072. doi: 10.1038/ni.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu YH, Macmillan JB., and, Chen ZJ. RNA polymerase III detects cytosolic DNA and induces type I interferons through the RIG-I pathway. Cell. 2009;138:576–591. doi: 10.1016/j.cell.2009.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Choi MK, Ban T, Yanai H, Negishi H, Lu Y, et al. Regulation of innate immune responses by DAI (DLM-1/ZBP1) and other DNA-sensing molecules. Proc Natl Acad Sci USA. 2008;105:5477–5482. doi: 10.1073/pnas.0801295105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser WJ, Upton JW., and, Mocarski ES. Receptor-interacting protein homotypic interaction motif-dependent control of NF-kappa B activation via the DNA-dependent activator of IFN regulatory factors. J Immunol. 2008;181:6427–6434. doi: 10.4049/jimmunol.181.9.6427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii KJ, Kawagoe T, Koyama S, Matsui K, Kumar H, Kawai T, et al. TANK-binding kinase-1 delineates innate and adaptive immune responses to DNA vaccines. Nature. 2008;451:725–729. doi: 10.1038/nature06537. [DOI] [PubMed] [Google Scholar]
- Roos AK, Eriksson F, Timmons JA, Gerhardt J, Nyman U, Gudmundsdotter L, et al. Skin electroporation: effects on transgene expression, DNA persistence and local tissue environment. PLoS ONE. 2009;4:e7226. doi: 10.1371/journal.pone.0007226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W, Swiggard WJ, Heufler C, Peng M, Mirza A, Steinman RM, et al. The receptor DEC-205 expressed by dendritic cells and thymic epithelial cells is involved in antigen processing. Nature. 1995;375:151–155. doi: 10.1038/375151a0. [DOI] [PubMed] [Google Scholar]
- Bonifaz LC, Bonnyay DP, Charalambous A, Darguste DI, Fujii S, Soares H, et al. In vivo targeting of antigens to maturing dendritic cells via the DEC-205 receptor improves T cell vaccination. J Exp Med. 2004;199:815–824. doi: 10.1084/jem.20032220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambrosini G, Adida C., and, Altieri DC. A novel anti-apoptosis gene, survivin, expressed in cancer and lymphoma. Nat Med. 1997;3:917–921. doi: 10.1038/nm0897-917. [DOI] [PubMed] [Google Scholar]
- Liang Y, Zhou Y., and, Shen P. NF-kappaB and its regulation on the immune system. Cell Mol Immunol. 2004;1:343–350. [PubMed] [Google Scholar]
- van Hogerlinden M, Rozell BL, Ahrlund-Richter L., and, Toftgård R. Squamous cell carcinomas and increased apoptosis in skin with inhibited Rel/nuclear factor-kappaB signaling. Cancer Res. 1999;59:3299–3303. [PubMed] [Google Scholar]
- Curtsinger JM, Valenzuela JO, Agarwal P, Lins D., and, Mescher MF. Type I IFNs provide a third signal to CD8 T cells to stimulate clonal expansion and differentiation. J Immunol. 2005;174:4465–4469. doi: 10.4049/jimmunol.174.8.4465. [DOI] [PubMed] [Google Scholar]
- Kolumam GA, Thomas S, Thompson LJ, Sprent J., and, Murali-Krishna K. Type I interferons act directly on CD8 T cells to allow clonal expansion and memory formation in response to viral infection. J Exp Med. 2005;202:637–650. doi: 10.1084/jem.20050821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindencrona JA, Preiss S, Kammertoens T, Schüler T, Piechocki M, Wei WZ, et al. CD4+ T cell-mediated HER-2/neu-specific tumor rejection in the absence of B cells. Int J Cancer. 2004;109:259–264. doi: 10.1002/ijc.11654. [DOI] [PubMed] [Google Scholar]
- Bronte V, Apolloni E, Ronca R, Zamboni P, Overwijk WW, Surman DR, et al. Genetic vaccination with “self” tyrosinase-related protein 2 causes melanoma eradication but not vitiligo. Cancer Res. 2000;60:253–258. [PMC free article] [PubMed] [Google Scholar]
- Halwani R, Boyer JD, Yassine-Diab B, Haddad EK, Robinson TM, Kumar S, et al. Therapeutic vaccination with simian immunodeficiency virus (SIV)-DNA + IL-12 or IL-15 induces distinct CD8 memory subsets in SIV-infected macaques. J Immunol. 2008;180:7969–7979. doi: 10.4049/jimmunol.180.12.7969. [DOI] [PubMed] [Google Scholar]
- Lai L, Vödrös D, Kozlowski PA, Montefiori DC, Wilson RL, Akerstrom VL, et al. GM-CSF DNA: an adjuvant for higher avidity IgG, rectal IgA, and increased protection against the acute phase of a SHIV-89.6P challenge by a DNA/MVA immunodeficiency virus vaccine. Virology. 2007;369:153–167. doi: 10.1016/j.virol.2007.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barouch DH, Santra S, Schmitz JE, Kuroda MJ, Fu TM, Wagner W, et al. Control of viremia and prevention of clinical AIDS in rhesus monkeys by cytokine-augmented DNA vaccination. Science. 2000;290:486–492. doi: 10.1126/science.290.5491.486. [DOI] [PubMed] [Google Scholar]
- Cassaday RD, Sondel PM, King DM, Macklin MD, Gan J, Warner TF, et al. A phase I study of immunization using particle-mediated epidermal delivery of genes for gp100 and GM-CSF into uninvolved skin of melanoma patients. Clin Cancer Res. 2007;13 2 Pt 1:540–549. doi: 10.1158/1078-0432.CCR-06-2039. [DOI] [PubMed] [Google Scholar]
- Setoguchi R, Hori S, Takahashi T., and, Sakaguchi S. Homeostatic maintenance of natural Foxp3(+) CD25(+) CD4(+) regulatory T cells by interleukin (IL)-2 and induction of autoimmune disease by IL-2 neutralization. J Exp Med. 2005;201:723–735. doi: 10.1084/jem.20041982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enzler T, Gillessen S, Manis JP, Ferguson D, Fleming J, Alt FW, et al. Deficiencies of GM-CSF and interferon γ link inflammation and cancer. J Exp Med. 2003;197:1213–1219. doi: 10.1084/jem.20021258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinushi M, Nakazaki Y, Dougan M, Carrasco DR, Mihm M., and, Dranoff G. MFG-E8-mediated uptake of apoptotic cells by APCs links the pro- and antiinflammatory activities of GM-CSF. J Clin Invest. 2007;117:1902–1913. doi: 10.1172/JCI30966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemoine FM, Cherai M, Giverne C, Dimitri D, Rosenzwajg M, Trebeden-Negre H, et al. Massive expansion of regulatory T-cells following interleukin 2 treatment during a phase I-II dendritic cell-based immunotherapy of metastatic renal cancer. Int J Oncol. 2009;35:569–581. doi: 10.3892/ijo_00000368. [DOI] [PubMed] [Google Scholar]
- Wei S, Kryczek I, Edwards RP, Zou L, Szeliga W, Banerjee M, et al. Interleukin-2 administration alters the CD4+FOXP3+ T-cell pool and tumor trafficking in patients with ovarian carcinoma. Cancer Res. 2007;67:7487–7494. doi: 10.1158/0008-5472.CAN-07-0565. [DOI] [PubMed] [Google Scholar]
- Filipazzi P, Valenti R, Huber V, Pilla L, Canese P, Iero M, et al. Identification of a new subset of myeloid suppressor cells in peripheral blood of melanoma patients with modulation by a granulocyte-macrophage colony-stimulation factor-based antitumor vaccine. J Clin Oncol. 2007;25:2546–2553. doi: 10.1200/JCO.2006.08.5829. [DOI] [PubMed] [Google Scholar]
- Takeshita F, Tanaka T, Matsuda T, Tozuka M, Kobiyama K, Saha S, et al. Toll-like receptor adaptor molecules enhance DNA-raised adaptive immune responses against influenza and tumors through activation of innate immunity. J Virol. 2006;80:6218–6224. doi: 10.1128/JVI.00121-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki S, Amara RR, Yeow WS, Pitha PM., and, Robinson HL. Regulation of DNA-raised immune responses by cotransfected interferon regulatory factors. J Virol. 2002;76:6652–6659. doi: 10.1128/JVI.76.13.6652-6659.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai T., and, Akira S. Signaling to NF-kappaB by Toll-like receptors. Trends Mol Med. 2007;13:460–469. doi: 10.1016/j.molmed.2007.09.002. [DOI] [PubMed] [Google Scholar]
- Haller O, Kochs G., and, Weber F. The interferon response circuit: induction and suppression by pathogenic viruses. Virology. 2006;344:119–130. doi: 10.1016/j.virol.2005.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinman RM., and, Hemmi H. Dendritic cells: translating innate to adaptive immunity. Curr Top Microbiol Immunol. 2006;311:17–58. doi: 10.1007/3-540-32636-7_2. [DOI] [PubMed] [Google Scholar]
- Stetson DB., and, Medzhitov R. Type I interferons in host defense. Immunity. 2006;25:373–381. doi: 10.1016/j.immuni.2006.08.007. [DOI] [PubMed] [Google Scholar]
- Sikora AG, Jaffarzad N, Hailemichael Y, Gelbard A, Stonier SW, Schluns KS, et al. IFN-α enhances peptide vaccine-induced CD8+ T cell numbers, effector function, and antitumor activity. J Immunol. 2009;182:7398–7407. doi: 10.4049/jimmunol.0802982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Critchley-Thorne RJ, Simons DL, Yan N, Miyahira AK, Dirbas FM, Johnson DL, et al. Impaired interferon signaling is a common immune defect in human cancer. Proc Natl Acad Sci USA. 2009;106:9010–9015. doi: 10.1073/pnas.0901329106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roos AK, Eriksson F, Walters DC, Pisa P., and, King AD. Optimization of skin electroporation in mice to increase tolerability of DNA vaccine delivery to patients. Mol Ther. 2009;17:1637–1642. doi: 10.1038/mt.2009.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lladser A, Ljungberg K, Tufvesson H, Tazzari M, Roos AK, Quest AF, et al. Intradermal DNA electroporation induces survivin-specific CTLs, suppresses angiogenesis and confers protection against mouse melanoma. Cancer Immunol Immunother. 2010;59:81–92. doi: 10.1007/s00262-009-0725-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Intradermal DNA electroporation induces tissue inflammation. Mice were vaccinated and biopsies were collected as in Figure 1. H&E staining reveals massive infiltration of lymphocytes (black cells, black arrowheads) in DNA electroporated skin (left panel). Significant skin infiltration by CD207+ DCs (brown, blue arrowheads) is observed in pVAX and pDAI electroporated mice when compared to untreated controls (p=0.007 and 0.0004, respectively) as detected by anti-CD207 staining (right panel).
DNA vaccine-induced upregulation of costimulatory molecules in dendritic cells from skin-draining lymph nodes. C57BL/6 mice (n=6) were vaccinated as in Figure 1 and the inguinal lymph nodes were taken 24 h later. The levels of costimulatory molecules CD40 and CD80 were analyzed in dendritic cells (CD11c+, MHC class IIhigh) by surface staining (black histograms) using naïve mice as controls (red histograms) and the mean fluorescence intensity (MFI) was calculated for each group. Bars are the mean±SEM. **indicates p=0.007; ***indicates p=0.0001.
Phenotypic analysis of DNA vaccine-induced OVA-specific CTLs. C57BL/6 mice were vaccinated twice as in Figure 3 (n=6). (a) Detection of OVA(257-264)-specific CD8+ T cells (R1) was performed by pentamer staining either at two (b) or five (c) weeks after the last vaccination. (b) Surface staining of phenotypic markers on OVA(257-264)-specific CD8+ T cells was evaluated in pooled peripheral blood from mice immunized with pOVA (—, thin line profiles) or pOVA+pDAI (—, thick line profiles). The bulk CD8+ T cell population (▪, solid grey profiles) was used as control. (c) Analysis of OVA(257-264)-specific CD8+ T cells to identify the different memory sub-set phenotypes: effector memory (TEM; CD44highCD62Llow); central memory (TCM; CD44highCD62Lhigh); memory stem cells (TSCM; CD44lowCD62Lhigh).
Detection of DNA vaccine-induced OVA-specific antibodies. C57BL/6 mice were vaccinated twice as in Figure 3 and sera collected 13 days after the last vaccination. OVA-specific antibodies were detected by indirect ELISA in sera from mice immunized with pDAI (♦, diamonds), pOVA (▪, squares) or pOVA+pDAI (▴, triangles) (mean±SEM; n=8). No statistically significant differences in antibody levels were observed between pOVA and pOVA+pDAI groups.
DNA vaccine-induced survivin-specific CTLs produced IFN-γ, TNF-α, and IL-2 after peptide stimulation. C57BL/6 mice were vaccinated as in Figure 5 and blood collected 13 days after the last vaccination. Detection of survivin-specific CD8+ T cells was performed after in vitro stimulation with surv(56-64) peptide. Intracellular staining of IFN-γ simultaneously with TNF-a or IL-2 in gated CD8+ T cells from mice immunized with pDAI (left panels), pSURV (middle panels) or pSURV+pDAI (right panels). Dot plots from a representative mouse per group are displayed indicating the mean±SEM for each group (n=4). Similar results were obtained after stimulation with surv(20-28) peptides.
Comparison of the adjuvant efficacy of pDAI and pGM-CSF to enhance survivin-specific CTL and Th1 responses. C57BL/6 mice were electroporated twice at two-week intervals with pDAI, pSURV, pSURV+pDAI or pSURV+pGM-CSF (n=7) and blood collected 13 days after the last vaccination. (a) The frequency of peripheral IFN-γ-producing CD8+ T cells (over the gated CD8+ T cell population) after in vitro stimulation with trp2(180-188) (Control), surv(20-28) (surv20) or surv(56-64) (surv56) peptides is shown. (b) The frequency of peripheral IFN-γ- and TNF-α-producing CD4+ T cells (over the gated CD4+ T cell population) after in vitro stimulation with ova(323-339) (Control) or surv(53-67) (surv53) peptides is shown. Bars are the mean±SEM. *indicates p=0.026; ***indicates p=0.0003.
Analysis of immunosuppressive cell populations. C57BL/6 mice were vaccinated twice at two-week intervals with pDAI, pSURV or pSURV+pDAI (n=8). Two weeks later, T regulatory (Treg) and myeloid-derived suppressor cells (MDSC) were analyzed in spleen and inguinal lymph nodes by immunofluorescence staining and flow cytometry. The frequency of Treg (CD4+; CD25high; FOXP3+) over the total CD4+ T cell population (a) and MDSC (CD11b+; Gr1+) over the total cell population (b) were determined and the results are shown. Bars are the mean±SEM. *indicates p<0.05.
The 20 most strongly upregulated gene transcripts in mice electroporated with pDAI as compared to mice electroporated with pVAX control vector.
Primers used for cloning and quantitative real-time PCR analysis.