Abstract
Induced pluripotent stem cells (iPSCs) have radically advanced the field of regenerative medicine by making possible the production of patient-specific pluripotent stem cells from adult individuals. By developing iPSCs to treat HIV, there is the potential for generating a continuous supply of therapeutic cells for transplantation into HIV-infected patients. In this study, we have used human hematopoietic stem cells (HSCs) to generate anti-HIV gene expressing iPSCs for HIV gene therapy. HSCs were dedifferentiated into continuously growing iPSC lines with four reprogramming factors and a combination anti-HIV lentiviral vector containing a CCR5 short hairpin RNA (shRNA) and a human/rhesus chimeric TRIM5α gene. Upon directed differentiation of the anti-HIV iPSCs toward the hematopoietic lineage, a robust quantity of colony-forming CD133+ HSCs were obtained. These cells were further differentiated into functional end-stage macrophages which displayed a normal phenotypic profile. Upon viral challenge, the anti-HIV iPSC-derived macrophages exhibited strong protection from HIV-1 infection. Here, we demonstrate the ability of iPSCs to develop into HIV-1 resistant immune cells and highlight the potential use of iPSCs for HIV gene and cellular therapies.
Introduction
HIV gene and cellular therapies hold enormous potential for not only treating HIV-infected individuals but also providing a functional cure. As HIV continues to be a global health problem, novel therapies need to be developed. Current antiretroviral drugs provide control of HIV replication, however, after prolonged use, these drugs can become toxic, escape mutants can arise, and they do not provide a cure.1,2,3,4 Advantages of HIV gene therapy using hematopoietic stem cells (HSCs) include the possibility of a one-time treatment, controlled or constitutive anti-HIV gene expression, and long-term viral inhibition upon HSC transplantation.5 Such HSCs have the capacity for self-renewal and the proliferation potential to differentiate into HIV-resistant target cells including CD4 T cells, macrophages, and dendritic cells.6 By engineering a sufficient quantity of HSCs to express anti-HIV genes, these cells may completely reconstitute the immune system with HIV-resistant immune cells after ablation of the host HSCs.
Clinical trials using retroviral and lentiviral vectors transferring anti-HIV genes into HSCs have demonstrated the feasibility of this approach.7,8,9,10,11 These strategies involve apheresis of the patient's mobilized peripheral stem cells followed by ex vivo gene transduction and transplantation back into the patient. Numerous anti-HIV genes have been employed in HIV gene therapy studies including siRNAs, RNA decoys, ribozymes, antisense molecules, and anti-HIV proteins.12,13,14,15,16,17,18,19,20 Pre-entry/preintegration inhibition of HIV infection is an ideal method to confer viral resistance. Therapies aimed at blocking HIV integration including CCR5 inhibitors (siRNAs, zinc-finger nucleases, ribozymes, and intrabodies), HIV fusion inhibitors, and TRIM5α prevent the generation of HIV provirus and the further establishment of viral reservoirs which are reasons for the failure to cure infected individuals.10,12,14,16,17,21,22,23
Recently, long-term control of HIV replication was observed in an infected individual who received an allogeneic bone marrow transplant from a donor who is homozygous for the CCR5 Δ32-bp deletion.24 The recipient has remained free from HIV replication for over 3 years post-transplantation. The results provided from this study, highlight the importance for developing anti-HIV gene and cellular therapies capable of generating HIV-resistant immune system cells in a similar way.
With current advancements in induced pluripotent stem cell (iPSC) technology, it is now possible to produce patient-specific pluripotent stem cells from adult individuals.25,26 Somatic cells, whether fibroblasts, T cells, amnion derived stem cells, HSCs, or many others, can be used as “starter cells” and be induced to pluripotency by the introduction of specific reprogramming factors including octamer-binding transcription factor 4 (OCT4), sex determining region Y-box 2 (SOX2), Kruppel-like factor 4 (KLF4), and cytoplasmic Myc (c-MYC).25,26,27,28,29 These iPSCs are similar to human embryonic stem cells (hESCs) in their capacity to differentiate into cells of all three germ layers, the ectoderm, mesoderm, and endoderm.26 Various cell types have been derived from iPSCs including cardiomyocytes, mesenchymal stem cells, hepatic endoderm, and hematopoietic cells.30,31,32,33,34 iPSCs, like hESCs, are also capable of indefinite growth and self-renewal with detected telomere elongation.35 They have enormous potential as a source of autologous cells for experimental and therapeutic use. If engineered to express anti-HIV genes, iPSCs have the potential to generate a continuous supply of anti-HIV HSCs. Thus, cells differentiated from iPSCs not only provide a source for cellular therapies but also hold the potential to correct genetic defects and inhibit infectious diseases such as HIV.
In our current studies, we have generated anti-HIV gene expressing iPSCs from cord blood (CB) CD34+ “starter cells”. We have used a combination of anti-HIV genes including a CCR5 short hairpin RNA (shRNA) and a chimeric human/rhesus TRIM5α molecule based on our previous work which demonstrated potent preintegration inhibition of HIV-1 infection.12 The iPSC lines were continually growing and did not contain any HSCs. Upon directed differentiation, the anti-HIV iPSCs generated a robust quantity of colony-forming hematopoietic progenitors which subsequently developed into phenotypically and functionally normal macrophages which were resistant to HIV-1 infection. The results presented here are the first to generate HIV-1 resistant immune cells from iPSCs and highlight the potential use of iPSC technology for HIV gene and cellular therapies.
Results
Generation of anti-HIV iPSCs
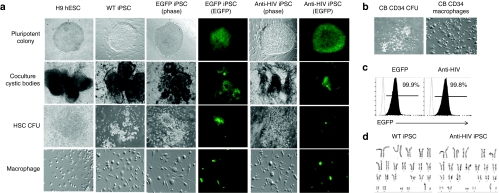
A third generation self-inactivating lentiviral vector CCLc-MNDU3-x-PGK-EGFP, was used to construct the combination anti-HIV vector. This vector contains a human/rhesus macaque TRIM5α isoform under the control of the modified retroviral MNDU3 promoter directly followed by a CCR5 shRNA under the control of a U6 pol-III promoter. The chimeric TRIM5α molecule was generated by replacing an 11-amino acid patch in the human isoform (#330–#340) with a 13-amino acid patch from the rhesus macaque isoform (#332–#344). By replacing amino acids in the human isoform with those essential for HIV restriction in the rhesus macaque isoform, immunogenicity of the chimeric protein is likely to be avoided compared to using the wild-type rhesus macaque TRIM5α. Another self-inactivating lentiviral vector, CCLc-TRE-Tight-PGK-rtTA (TRE-Tight, tetracycline-responsive promoter element; PGK, phosphoglycerate kinase promoter; rtTA, reverse tetracycline transcriptional activator) which is doxycycline-inducible, was used to construct the iPSC reprogramming vectors OCT4, SOX2, KLF4, and c-MYC. These four reprogramming vectors alone (wild-type (WT)), or together with the enhanced green fluorescent protein-alone (EGFP) or combination anti-HIV vector (anti-HIV), were transduced into CB CD34+ cells at a ratio of 1:1:1:1:1. Transduced CD34+ HSCs were plated on mouse embryonic fibroblasts with media containing doxycycline to induce expression of the four iPSC reprogramming factors. After 9 days of culture, “embryonic-like” colonies resembling hESCs had formed in the WT, EGFP, and anti-HIV iPSC cultures. These iPSC colonies were tightly packed and circular with defined edges similar to the control H9 hESCs (Figure 1a). In the cultures transduced with the four reprogramming factors in addition to either the EGFP-alone control vector or the anti-HIV vector, iPSC colonies were generated which expressed EGFP demonstrating successful transduction with either the EGFP-alone or anti-HIV vector (Figure 1). The transduction and expression of the anti-HIV genes did not adversely affect either iPSC generation or the morphology of the anti-HIV iPSC colonies as the colonies formed at the same time (9 days) as the control cells and also displayed similar morphology. The newly generated iPSC lines have retained their pluripotent and “embryonic-like” morphology beyond 21 passages as observed by visualization. The EGFP and anti-HIV iPSCs have remained EGFP positive beyond 21 passages indicating a lack of gene silencing of the EGFP transgenes (Figure 1c). Expression of the anti-HIV transgenes was also detected in the anti-HIV iPSCs beyond passage 21 as determined using a QuantiMir quantitative reverse transcriptase–PCR detection kit (data not shown). Levels of expression were comparable to those obtained in our previous experiments after transduction of CB CD34+ cells utilizing a similar combination anti-HIV vector.12
Figure 1.
Generation of anti-HIV induced pluripotent stem cells (iPSCs) and end-stage macrophages. (a) iPSCs were generated by transducing cord blood (CB) CD34+ human hematopoietic stem cells (HSCs) with four lentiviral vectors expressing the pluripotency factors octamer-binding transcription factor 4, sex determining region Y-box 2, Kruppel-like factor 4, and cytoplasmic Myc either alone [wild-type (WT)] or with an enhanced green fluorescent protein control vector (EGFP) or a combination anti-HIV vector (anti-HIV). iPSCs were further cocultured on OP9 stromal cells where cystic bodies developed. CD133+ HSCs were isolated from the cystic bodies and grown in semisolid methylcellulose media to form myeloid colony-forming units (CFUs). The CFUs were further cultured in media specific for macrophage development. EGFP and anti-HIV iPSCs and their differentiated progeny were visualized by both phase and EGFP fluorescence. H9 human embryonic stem cells (hESCs) and their differentiated progeny were used as controls. (b) CB CD34+ cells were used as a positive control and cultured in semisolid methylcellulose media and macrophage-specific media to derive myeloid CFUs and end-stage macrophages. (c) Representative fluorescence-activated cell sorting (FACS) plots displaying the EGFP percent of the EGFP and anti-HIV iPSCs at passage 21. WT iPSCs (EGFP negative) are displayed as the unshaded histogram. FACS analysis was performed in triplicate. (d) Representative karyotyping analyses of the WT and anti-HIV iPSCs derived from CB CD34+ HSCs. Karyotyping was performed in duplicate.
With the induction of pluripotency in CB CD34+ cells, it is possible that chromosomal and genetic abnormalities could arise. Therefore, karyotyping analyses were performed on the anti-HIV iPSCs. As displayed in Figure 1d, the anti-HIV iPSCs retained a normal chromosomal profile. Normal banding was observed and no translocations or chromosomal abnormalities could be detected. This was observed in comparison to WT iPSCs which also did not display any abnormalities.
Anti-HIV iPSCs express pluripotency markers
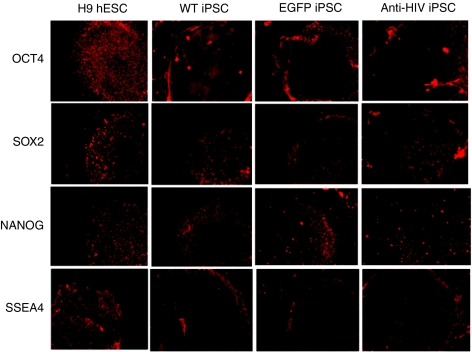
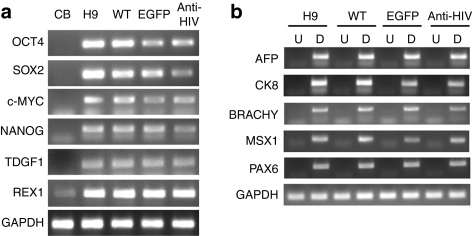
To determine whether the anti-HIV iPSCs were fully reprogrammed and expressed pluripotency markers, immunofluorescence was performed. iPSCs were stained with antibodies specific for various pluripotency markers including OCT4, SOX2, NANOG, and stage-specific embryonic antigen-4 (SSEA4). As displayed in Figure 2, all iPSC lines, WT, EGFP, and anti-HIV expressed all pluripotency markers analyzed as compared to control H9 hESCs. Reverse transcriptase-PCR (RT-PCR) was also performed to detect the gene expression of various pluripotency genes including OCT4, SOX2, c-MYC, NANOG, teratocarcinoma-derived growth factor 1 (TDGF1), and Zinc finger protein 42 (REX1). As displayed in Figure 3a, all iPSCs expressed the pluripotency genes as compared to control H9 hESCs. CB CD34+ cells, used as a negative control, expressed lower levels of the pluripotency markers as compared to the iPSCs or the H9 hESCs. GAPDH was used as an internal control.
Figure 2.
Expression of pluripotency markers by immunofluorescence. Induced pluripotent stem cells (iPSCs), wild-type (WT), enhanced green fluorescent protein (EGFP), and anti-HIV were stained with antibodies specific for the pluripotency markers octamer-binding transcription factor 4 (OCT4), sex determining region Y-box 2 (SOX2), NANOG, and stage-specific embryonic antigen-4 (SSEA4). Cells were visualized for fluorescence. H9 human embryonic stem cells (hESCs) were used as pluripotency positive controls. Pictures are representative of triplicate experiments.
Figure 3.
Expression of pluripotency and differentiation genes by reverse transcriptase-PCR. (a) Total RNA from undifferentiated induced pluripotent stem cells (iPSCs) was extracted and analyzed by reverse transcriptase-PCR for the expression of the pluripotency genes octamer-binding transcription factor 4 (OCT4), sex determining region Y-box 2 (SOX2), cytoplasmic Myc (c-MYC), NANOG, teratocarcinoma-derived growth factor 1 (TDGF1), and REX1. CB CD34+ human hematopoietic stem cells (HSCs) were used as a control to detect expression in the “starter cells.” H9 human embryonic stem cells (hESCs) were used as a pluripotency positive control. (b) Total RNA was extracted from differentiated (D) cells from the cystic bodies which formed in the iPSC/OP9 cocultures and analyzed by RT-PCR for the expression of genes from all three germ layers including α-fetoprotein (AFP), cytokeratin 8 (CK8), brachyury (BRACHY), Msh homeobox 1 (MSX1), and paired box gene 6 (PAX6). Undifferentiated (U) cells were used as negative controls for expression. Undifferentiated and differentiated H9 hESCs were used as negative and positive controls, respectively. GAPDH was used as an internal loading control. Experiments were performed in duplicate. CB, cord blood; EGFP, enhanced green fluorescent protein; WT, wild type.
Another characteristic of true pluripotent stem cells is their ability to differentiate into cells and tissues of all three germ layers, the ectoderm, mesoderm, and endoderm. To determine whether the anti-HIV iPSCs were capable of such differentiation, RT-PCR was performed to detect expression of specific genes from all three germ layers including α-fetoprotein (AFP), cytokeratin 8 (CK8), brachyury (BRACHY), Msh homeobox 1 (MSX1), and paired box gene 6 (PAX6). Cells were allowed to differentiate on mouse OP9 stromal cells followed by analysis by RT-PCR. As displayed in Figure 3b, upon differentiation (D), all iPSC lines expressed similar expression levels of each gene analyzed as compared to the positive control H9 hESC differentiated cells. Expression of the differentiated genes was not detected in undifferentiated (U) iPSCs or H9 hESCs. GAPDH was used as an internal control.
Directed differentiation of anti-HIV iPSCs to HSCs
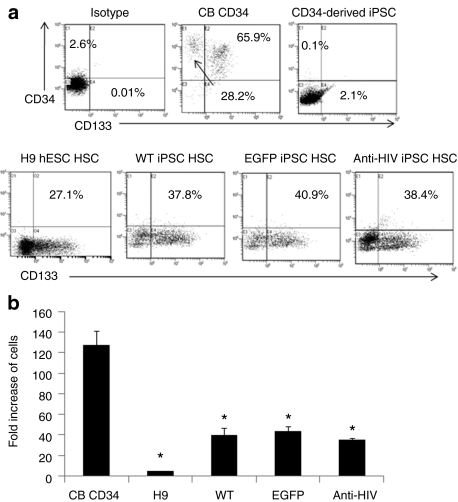
To directly differentiate the anti-HIV iPSCs into HSCs, cells were cocultured on mouse OP9 stromal cells with the addition of bone morphogenetic protein-4. This culture method has been previously used to promote hematopoietic differentiation of pluripotent stem cells.36,37 Upon coculturing the iPSCs on OP9 cells, dark cystic bodies formed which were similar to cystic bodies formed in H9 hESC/OP9 cocultures demonstrating differentiation of the iPSCs (Figure 1a). Cystic bodies formed in the EGFP and anti-HIV cocultures remained EGFP positive (Figure 1a). Single-cell suspensions of the cocultures were made and fluorescence-activated cell sorting (FACS) analysis was performed to determine the levels of HSC generation by staining for CD133. In previous experiments using this specific coculture method, we obtained a greater number of CD133+ HSCs than CD34+ HSCs (data not shown) and, therefore, used this hematopoietic stem and progenitor cell marker for HSC purification. As demonstrated by FACS analysis, the iPSC cocultures contained significant numbers of CD133+ HSCs, WT = 37.8%, EGFP = 40.9%, and anti-HIV = 38.4%. This was in comparison to H9 hESC cocultures which only contained 27.1% cells expressing CD133 (Figure 4a). To determine whether the iPSCs derived from CB CD34+ cells retained expression of CD34 or CD133 upon induction to pluripotency, FACS analysis was performed. As displayed in Figure 4a (top right panel), the CD34+ cell-derived iPSCs did not express CD34 or CD133 upon reprogramming. This was in comparison to CB CD34+ cells which expressed 94.1% CD34 and 65.9% CD133 upon immunomagnetic bead purification (Figure 4a, top middle panel).
Figure 4.
Detection and proliferation of induced pluripotent stem cells (iPSCs) derived CD133+ human hematopoietic stem cells (HSCs). (a) iPSCs, wild-type (WT), enhanced green fluorescent protein (EGFP), and anti-HIV were differentiated toward the hematopoietic lineage by OP9 cocultures. On day 9, the cells were analyzed for their expression of CD133 by FACS. Isotype control stained coculture cells are displayed in the top-left panel. CB CD34+ HSCs were used as a positive control for CD34 and CD133 expression (top-middle panel). Undifferentiated iPSCs were used as a negative control for CD34 and CD133 expression (top-right panel). Representative fluorescence-activated cell sorting plots are displayed from duplicate experiments. (b) Total cell counts were performed from methylcellulose cultured colony-forming units and analyzed for fold expansion of the input CD133+ cells. CB CD34+ HSCs were used as a positive comparative control. Asterisks above the bars indicate statistically significant values as compared to cord blood (CB) HSCs. Experiments were performed in triplicate. hESCs; human embryonic stem cells.
To determine whether the CD133+ HSCs purified from the iPSC/OP9 cocultures were functional and could generate colony-forming units, the cells were cultured in Methocult methylcellulose media specific for the generation of myeloid colonies. The iPSC-derived CD133+ cells formed morphologically normal myeloid colony-forming units as compared to H9 hESC derived CD133+ HSCs and CB CD34+ HSCs (Figure 1a,b). EGFP and anti-HIV iPSC-derived colony-forming units were EGFP positive confirming continued expression of the EGFP and anti-HIV transgenes upon differentiation of the iPSCs into HSCs (Figure 1a). The iPSC-derived CD133+ HSCs were also evaluated to determine whether their proliferation potential was comparable to CB CD34+ cells. After 9 days of culture in Methocult media, total cell numbers were counted and compared to the initial cell input. As displayed in Figure 4b, iPSC-derived CD133+ HSCs, whether WT, EGFP, or anti-HIV, increased ~40-fold in total cell numbers. This was in comparison to H9 hESC derived CD133+ HSCs which only increased fivefold in cell numbers and CB CD34+ cells which increased 127-fold. The differences between iPSC-derived HSC cell counts were significant from CB derived HSC cell counts, however, when we compared anti-HIV HSC cell counts to WT and EGFP control HSC cell counts, there was no significant difference. These results demonstrate that the anti-HIV iPSCs were capable of directed differentiation toward the hematopoietic lineage in a robust quantity and that no significant differences were observed in the differentiation or growth characteristics possibly caused by the expression of the anti-HIV genes.
A major safety concern with utilizing cells for therapeutic purposes which were derived from pluripotent cells is the possible retention of pluripotency and the formation of teratomas. Therefore, to determine whether the CD133+ HSCs derived from the iPSCs retained any pluripotent capabilities, one to two million cells were injected into immunodeficient NOD/SCID−/−γ−/− knockout mice. After 2 months of daily monitoring, no teratomas were observed. This was in comparison to mice injected with a single-cell suspension of the WT or anti-HIV iPSCs (106 cells/mouse) which formed teratomas within 1 month of injection. The teratomas from the WT and anti-HIV iPSC injected mice contained tissues from all three germ layers, the ectoderm, mesoderm, and endoderm (data not shown).
Anti-HIV iPSC-derived phenotypically and functionally normal macrophages
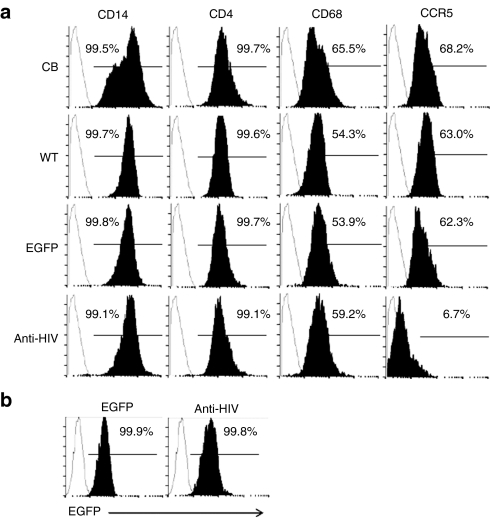
To evaluate the capacity of anti-HIV iPSC-derived HSCs to develop into phenotypically and functionally normal macrophages, colony-forming units were further cultured in a macrophage-specific differentiation media containing granulocyte-macrophage colony-stimulating factor and macrophage colony-stimulating factor. After 4 days, morphologically distinct macrophages had attached to the culture plates in the WT, EGFP, and anti-HIV iPSC-derived macrophage cultures (Figure 1a). This was similar to CB CD34+ cell-derived macrophage cultures which also displayed macrophage development in 4 days (Figure 1b). However, in the H9 hESC cultures, macrophages did not appear until day 10 (Figure 1a). To determine whether the macrophages derived from the anti-HIV iPSCs were phenotypically normal, FACS analysis was performed to detect expression of the normal macrophage markers, CD14, CD4, CD68 (lipopolysaccharide (LPS)-stimulated), and CCR5. Upon stimulation of macrophages with LPS, CD68 cell surface expression is upregulated.38 As displayed in Figure 5a, normal cell surface expression of CD14, CD4, and CD68 (LPS-stimulated) was detected in all of the iPSC-derived macrophage cultures and was similar to CB CD34+ cell-derived macrophages, CD14 >99%, CD4 >99%, and CD68 >53%. However, CCR5 cell surface expression was dramatically decreased (6.7% positive) in the anti-HIV iPSC-derived macrophages compared to CB (68.2% positive), WT iPSC (63.0% positive), and EGFP iPSC (62.3% positive) derived macrophages (Figure 5a). The decrease in CCR5 expression was due to the presence of the CCR5 shRNA anti-HIV gene and demonstrated the constitutive expression of this transgene upon differentiation from iPSC to end-stage macrophages. EGFP (99.9%) and anti-HIV (99.8%) iPSC-derived cells also remained EGFP positive throughout the differentiation process as determined by visualization (Figure 1a) and by FACS (Figure 5b).
Figure 5.
Phenotypic analysis of induced pluripotent stem cells (iPSCs) derived macrophages. (a) Macrophages derived from the wild-type (WT), enhanced green fluorescent protein (EGFP), and anti-HIV iPSCs were stained with antibodies specific for human CD14, CD4, CD68, and CCR5 and analyzed by fluorescence-activated cell sorting (FACS). Cord blood (CB) CD34+ HSC-derived macrophages were used as positive controls. Isotype controls are displayed as unshaded histograms. (b) EGFP and anti-HIV iPSC-derived macrophages were analyzed by FACS for EGFP expression. WT (EGFP−) cells were used as negative controls and are displayed as unshaded histograms.
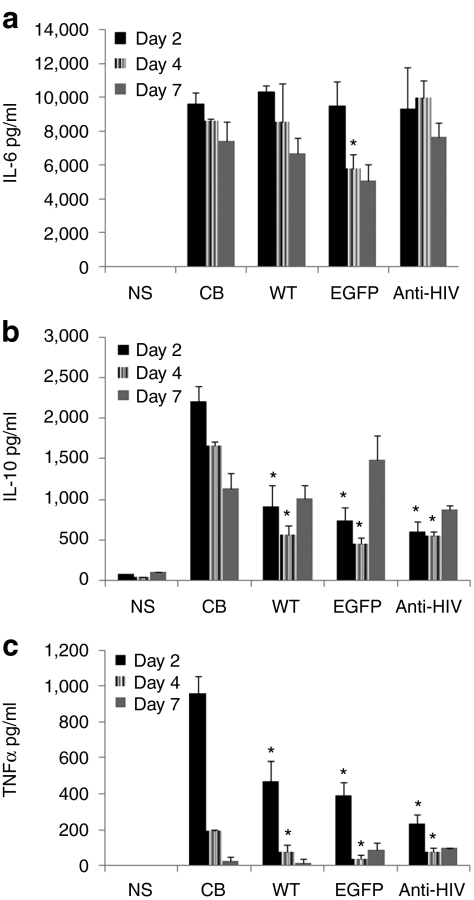
To determine whether the anti-HIV iPSC-derived macrophages were functionally normal, the levels of secretion of specific cytokines were measured. The iPSC and CB CD34+ HSC-derived macrophages were stimulated with LPS and the levels of interleukin-6 (IL-6), IL-10, and tumor necrosis factor-α (TNFα) were analyzed by a cytometric bead array. As displayed in Figure 6, the iPSC-derived macrophages were functional and secreted substantial amounts of IL-6 (Figure 6a), IL-10 (Figure 6b), and TNFα (Figure 6c) upon stimulation compared to nonstimulated (NS) macrophages. The secretion of IL-2, IL-4, and interferon-γ were not detected and were used as negative controls (data not shown). Cytokine secretion from the iPSC-derived macrophages were directly compared to CB derived macrophages. The levels of IL-6 secretion from the iPSC-derived macrophages were similar to CB derived macrophages and the differences, overall, were not statistically significant. The difference in levels of IL-10 and TNFα were statistically different on days 2 and 4 poststimulation from iPSC-derived macrophages compared to CB derived macrophages. However, when we compared the levels of cytokine secretion among the three iPSC lines, WT, EGFP, and anti-HIV, no significant differences were observed. The results obtained from these experiments demonstrated that the anti-HIV iPSC-derived macrophages are phenotypically normal and are functional upon stimulation with antigen.
Figure 6.
Cytokine secretion of induced pluripotent stem cells (iPSCs)-derived macrophages. Macrophages derived from the wild-type (WT), enhanced green fluorescent protein (EGFP), and anti-HIV iPSCs were stimulated with lipopolysaccharide. On days 2, 4, and 7, cell culture supernatants were analyzed for levels of secretion of (a) interleukin-6 (IL-6), (b) IL-10, and (c) tumor necrosis factor-α (TNFα). Cord blood (CB) CD34+ hematopoietic stem cells (HSC) derived macrophages were used as positive comparative controls. Asterisks above the bars indicate statistically significant values as compared to CB HSCs. Experiments were performed in triplicate. NS, nonstimulated.
Inhibition of HIV-1 infection by anti-HIV iPSC-derived macrophages
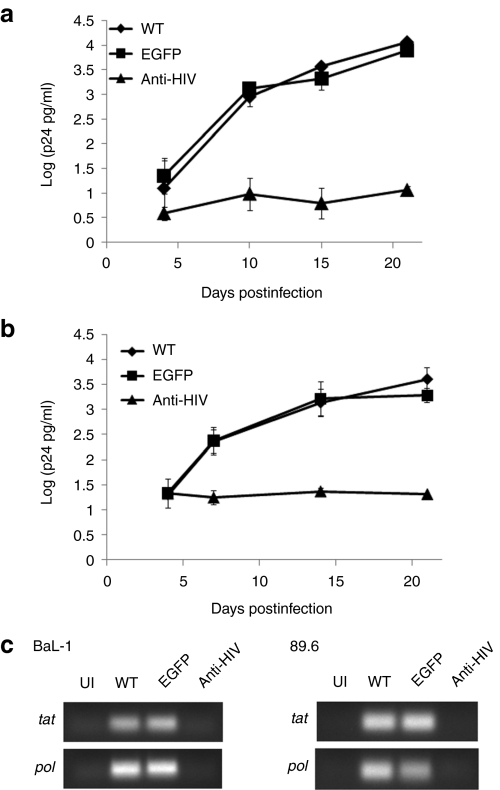
The ultimate goal of our study was to derive end-stage macrophages from iPSCs which were resistant to HIV infection. Therefore, to determine whether the anti-HIV iPSC derived macrophages were capable of blocking HIV-1 infection, the cells were challenged with either an R5-tropic BaL-1 or a dual-tropic 89.6 strain of HIV-1 at a multiplicity of infection of 0.05. On various days postinfection, culture supernatants were sampled and analyzed by p24 antigen enzyme-linked immunosorbent assay. As displayed in Figure 7a,b, potent inhibition of HIV-1 infection was observed. By day 15 postinfection, over a 2-log reduction in p24 antigen was detected in the anti-HIV macrophage cultures as compared to the WT and EGFP macrophage cultures. To confirm the results observed with the p24 antigen enzyme-linked immunosorbent assay, RT-PCR was performed to detect the expression of spliced (tat) and unspliced (pol) HIV transcripts in the challenged macrophages on day 21 postinfection. As displayed in Figure 7c, both tat and pol transcripts were detected in the WT and EGFP BaL-1 and 89.6 challenged macrophages. This was in comparison to the anti-HIV macrophages which displayed similar background levels as uninfected (UI) cells. These results demonstrate that HIV-resistant macrophages can be developed from anti-HIV gene expressing iPSCs.
Figure 7.
HIV-1 challenge of induced pluripotent stem cells (iPSCs) derived macrophages. Macrophages derived from the wild type (WT) (diamonds), enhanced green fluorescent protein (EGFP) (squares), and anti-HIV (triangles) iPSCs were challenged with (a) an R5-tropic BaL-1 or (b) a dual-tropic 89.6 strain of HIV-1 at an multiplicity of infection of 0.05. On various days postinfection, cell culture supernatants were sampled and analyzed for p24 antigen by enzyme-linked immunosorbent assay. Experiments were performed in triplicate. (c) On day 21 postinfection, total RNA from uninfected (UI) and HIV-1 challenged macrophages were analyzed by reverse transcriptase-PCR for the expression of spliced (tat) and unspliced (pol) HIV-1 transcripts.
Discussion
As a first step toward utilizing iPSCs for HIV gene and cellular therapies, for the first time, we have developed HIV-1 resistant macrophages from HSC-derived iPSCs which we could demonstrate to be both functional and phenotypically normal. Anti-HIV gene expressing iPSCs were also capable of generating a robust quantity of anti-HIV HSCs which were further differentiated into macrophages which inhibited HIV-1 infection. Current HIV stem cell gene therapy protocols rely on the apheresis of a patient's mobilized peripheral blood stem cells, ex vivo manipulation of these cells, and transplantation of the gene transduced cells back into the patient.7,8,9,10,11 However, a major disadvantage with this strategy is that the quantity of cells obtained for ex vivo manipulation is limited. By introducing anti-HIV genes into iPSCs, it is possible to generate a continuous supply and a large quantity of patient-specific anti-HIV HSCs for use as a cellular therapy. Additionally, this would allow for the production of a larger quantity of anti-HIV HSCs from banked stocks if subsequent transplantations were required.
Numerous “starter cells” have been used to generate iPSC lines including fibroblasts, lymphocytes, and HSCs.26,27,29 Our initial hypothesis for utilizing HSCs as “starter cells” to derive the anti-HIV iPSCs stemmed from our final goal of deriving anti-HIV HSCs from the reprogrammed cells. We hypothesized that CB CD34+ cells had already undergone normal hematopoietic differentiation and, therefore, iPSCs derived from these cells would be more efficient in generating functional HSCs upon directed differentiation. This hypothesis was recently confirmed by another laboratory demonstrating that derivation of a desired cell type (HSCs) from iPSCs is more efficiently achieved when using iPSCs generated from that same “starter cell” due to epigenetic memory.39 Indeed, our iPSCs derived from CB CD34+ cells were highly effective at generating hematopoietic colony-forming progenitors and macrophages upon directed differentiation.
In this study, we have demonstrated successful generation of anti-HIV iPSCs using CB CD34+ HSCs transduced with four reprogramming factors and a combination anti-HIV lentiviral vector containing two preintegration anti-HIV genes, a human/rhesus chimeric TRIM5α and a CCR5 shRNA. We used this combination anti-HIV vector based on our previous studies which demonstrated potent preintegration inhibition of HIV-1 infection.12 The expression of the anti-HIV transgenes did not show any adverse effect on iPSC generation or on the morphology of the iPSCs as displayed in Figure 1a. The anti-HIV iPSCs had similar morphology as compared to H9 hESCs displaying defined round colonies and tight edges. Also, expression of the anti-HIV genes in the iPSCs also did not have an effect on the expression of pluripotency markers as measured by immunofluorescence and RT-PCR. Upon reprogramming of the CB CD34+ cells, the anti-HIV iPSCs displayed normal expression of the various pluripotency markers analyzed. Another major concern with the transduction of therapeutic genes in pluripotent stem cells is the possibility for gene silencing, mutagenesis, and toxicity associated with the expression of foreign genes. As we observed in our experiments, no gene silencing had occurred with the EGFP or anti-HIV transgenes introduced by lentiviral vectors. This was confirmed by FACS analysis for EGFP expression in the anti-HIV iPSCs beyond passage 21 (Figure 1c). The cells also remained EGFP positive throughout their directed differentiation into end-stage macrophages (Figure 5b). Expression of the anti-HIV genes was also detected in the anti-HIV iPSCs and their differentiation macrophage progeny (data not shown). The anti-HIV iPSCs displayed similar growth kinetics compared to control iPSCs as determined by visualization after every passage. These data confirmed that the anti-HIV genes introduced into CB CD34+ cells at the same time as the reprogramming factors was feasible, did not produce toxicity, and did not have a negative effect on development, growth kinetics, or directed differentiation into HSCs and macrophages.
For use as a regenerative cellular therapy, anti-HIV HSCs differentiated from the anti-HIV iPSCs need to harness the capacity for cell proliferation and differentiation into phenotypically and functionally normal immune cells. As we have observed in our experiments, the anti-HIV HSCs derived from the anti-HIV iPSCs were capable of generating a robust quantity of CD133+ HSCs and hematopoietic progenitors as compared to H9 hESC derived CD133+ cells. Upon further differentiation, the anti-HIV iPSC-derived HSCs were capable of developing into normal macrophages at a rate similar to CB CD34+ cells. Anti-HIV iPSC-derived macrophages were phenotypically normal and expressed cell surface markers similar to CB CD34+ cell-derived macrophages. Additionally, they also secreted normal macrophage cytokines upon stimulation with LPS.
In a recent phase I clinical trial which used a triple combination anti-HIV lentiviral vector in patients with AIDS-related lymphoma, results obtained included an ex vivo transduction efficiency of between 5 and 23% and an in vivo gene marking of 0.02–0.32%.10 Anti-HIV gene expressing iPSCs have the potential to improve upon current HIV gene therapy protocols by providing HSCs which all contain anti-HIV activity thus alleviating the potential problem of low transduction efficiencies. Also, upon transplantation back into the patient, a higher percent of gene marking may be accomplished.
For HIV gene therapy to succeed, other aspects of HIV pathogenesis need to be considered including bystander cell death. Previous work has demonstrated that the HIV proteins negative regulatory factor, viral protein R, and envelope protein are involved in CD4+ and non-CD4+ cell death and that the numbers of CD4+ T cell depletion far exceeds the levels of infected T cells.40,41,42 By transplanting a mixed population of protected and unprotected cells back into patients, there is the risk of providing unprotected immune cells to become new targets for HIV to infect which could contribute to continued immune system failure. However, as mentioned above, by utilizing clonal anti-HIV HSCs generated from iPSCs, a population of cells which all contain anti-HIV activity could diminish this problem. These anti-HIV iPSC-derived HSCs have the potential to bridge the gap between current HIV gene therapy protocols and the successful suppression of HIV replication in the HIV-infected patient who received an allogeneic bone marrow transplant from a donor homozygous for the CCR5 Δ32-bp deletion.24 Another advantage in utilizing anti-HIV iPSCs for HIV gene therapy is that each iPSC line can be fully characterized for its safety and analysis of the integration site of the therapeutic transgenes. For future use of iPSCs for cellular therapies, the integration site of the therapeutic vector can be defined and the iPS lines with “safe harbor” sites can be selected, expanded, and further characterized for their anti-HIV efficacy. As demonstrated in Figure 7, potent inhibition (>2-logs) of HIV-1 infection was observed in the anti-HIV iPSC-derived macrophages. The strong inhibition of infection observed was due to the expression of the anti-HIV genes in every macrophage derived from the anti-HIV iPSCs.
By reprogramming CB CD34+ HSCs to pluripotency, we were able to efficiently generate anti-HIV iPSCs and successfully differentiate these cells in vitro into a robust quantity of functional HSCs with further differentiation to functional, HIV-resistant macrophages. Our results emphasize the advantages of utilizing anti-HIV iPSC-derived HSCs as therapeutic cells for HIV gene and cellular therapies.
Materials and Methods
Production of lentiviral vectors. Human OCT4, SOX2, KLF4, and c-MYC complementary DNA (cDNA) were cloned into the self-inactivating CCLc-TRE-Tight-PGK-rtTA doxycycline-inducible lentiviral vector as separate constructs (TRE-Tight = tetracycline-responsive promoter element, PGK = phosphoglycerate kinase promoter, rtTA = reverse tetracycline transcriptional activator). To generate the combination anti-HIV lentiviral vector, a self-inactivating lentiviral vector, CCLc-MNDU3-x-PGK-EGFP, was used. Briefly, human TRIM5α cDNA was cloned into pCR2.1 (Invitrogen, Carlsbad, CA). Site directed mutagenesis was performed to generate the chimeric human/rhesus macaque TRIM5α gene (HRH), as described previously,43 and subsequently inserted under the control of the MNDU3 promoter in the CCLc-MNDU4-x-PGK-EGFP lentiviral vector. The U6-CCR5 shRNA expression cassette was PCR amplified using a human genomic U6 promoter sequence and a long DNA oligo corresponding to the CCR5 shRNA sequence, as described previously.12 The U6-CCR5 shRNA expression cassette was then cloned downstream of the MNDU3-HRH expression cassette. To package the lentiviral vectors, HEK-293 T cells were transfected by lipofection with 25 µg of the lentiviral transfer vectors [OCT4, SOX2, KLF4, c-MYC, EGFP-alone (control vector), or the combination anti-HIV], 25 µg of pCMV-Δ8.9 (packaging plasmid expressing gag and pol), and 5 µg of pMDG-VSVG (vesicular stomatitis virus glycoprotein-pseudotyping envelope). Two days post-transfection, vector supernatants were collected, concentrated by ultrafiltration, and stored at −80 °C.
Isolation and transduction of human HSCs. Isolation of umbilical CB mononuclear cells was performed by Ficoll gradient centrifugation followed by CD34+ cell selection using a direct CD34+ microbead isolation kit (Miltenyi Biotech, Auburn, CA). Cells were purified through two LS separation columns (routine purity was >92%). CD34+ cells were cultured for 2 days in Iscove's modified Dulbecco's medium supplemented with 10% fetal bovine serum, and 50 ng/ml of (Flt3), (FMS-like tyrosine kinase 3), stem cell factor (SCF) and thrombopoietin (TPO). CD34+ cells were transduced with all four of the individual doxycycline-inducible lentivectors expressing human OCT4, SOX2, KLF4, and c-MYC alone or in addition to either the EGFP-alone (control vector) or the anti-HIV combination vector at a 1:1:1:1:1 ratio at an multiplicity of infection of 16.
Generation and culturing of iPSCs. On day 1 post-transduction, CD34+ HSCs were transferred onto irradiated mouse embryonic fibroblast feeder layers (Globalstem, Rockville, MD), with the culture medium switched to complete knockout Dulbecco's modified Eagle's medium-F12 which included 20% knockout serum replacement, 1 mmol/l -glutamine, 1% nonessential amino acids, 0.1 mmol/l β-mercaptoethanol, 20 ng/ml human basic fibroblast growth factor, and 2 µg/ml doxycycline (to induce the expression of the reprogramming factors). Fresh media changes were performed daily. By day 9, clearly recognizable, tightly packed colonies with morphologies similar to hESCs appeared. The newly generated iPSC colonies were mechanically lifted, passaged onto fresh mouse embryonic fibroblasts, and cultured in knockout complete Dulbecco's modified Eagle's medium-F12/20% knockout serum replacement/10 ng/ml human basic fibroblast growth factor without doxycycline. H9 hESCs were cultured in knockout complete Dulbecco's modified Eagle's medium-F12/20% knockout serum replacement/10 ng/ml human basic fibroblast growth factor. Periodical passaging of the iPSCs and H9 hESCs were performed to maintain their undifferentiated and pluripotent state.
Karyotyping. To determine the chromosomal and genetic stability of the HSC-derived iPSCs, karyotyping was performed. The iPSCs were treated with colcemid, a mitotic inhibitor, for 30 minutes at 37 °C to arrest the cells in metaphase followed by treatment with cell stripper for 10 minutes at 37 °C. The cell suspension was then treated with potassium chloride hypotonic solution and subsequent 3:1 methanol:acetic acid fixative solutions. Karyotyping slides were made and Giemsa banded. Karyotyping was performed on an Olympus Bx41 microscope (Olympus, Center Valley, PA)with a DP20 camera. Analysis was performed with an Applied Imaging System.
Immunofluorescence. To determine whether the derived iPSCs displayed normal pluripotency and self-renewal markers, immunofluorescence was performed. iPSCs and hESCs were plated in 12-well plates, cultured for 2 days, then rinsed with phosphate-buffered saline (PBS) and fixed with 2% paraformaldehyde in PBS for 10 minutes at room temperature. Cells were washed with PBS and blocked/permeabilized with 0.3% Triton-X100 and 3% normal goat serum. Cells were incubated overnight with primary antibodies specific for OCT4 (mouse antihuman; R&D Systems, Minneapolis, MN), SOX2 (mouse antihuman; R&D Systems), NANOG (mouse antihuman; BD Biosciences, San Jose, CA), or SSEA4 (mouse antihuman; R&D Systems) at 4 °C. Cells were washed three times with PBS and incubated with an Alexa 594 conjugated goat anti-mouse secondary antibody (Invitrogen) for 1 hour at room temperature. After washing with PBS, 50 µl of 4′,6-diamidino-2-phenylindole was added and the cells were incubated for 10 minutes at room temperature. Cells were imaged using an inverted Nikon fluorescent microscope. Experiments were performed in triplicate.
RT-PCR and quantitative RT-PCR. Total RNA was extracted from the undifferentiated iPSCs and hESCs, OP9 coculture differentiated iPSCs and hESCs, and the iPSC-derived macrophages using RNA-Stat-60 (Tel-Test, Friendswood, TX) according to the manufacturer's protocol. cDNA was synthesized using an Applied Biosystems Taqman Reverse Transcription Reagents kit (Applied Biosystems, Carlsbad, CA). To determine the expression of the undifferentiated and differentiated genes, RT-PCR were performed using 200 ng of cDNA for each reaction with primer sets specific for each gene of interest: OCT4(f) 5′-AAACCCTGGCACAAACTCC-3′ OCT4(r) 5′-GACCAGTGTCCTTTCCTCTG-3′ SOX2(f) 5′-CACATGTC CCAGCACTACC-3′ SOX2(r) 5′-CCATGCTGTTTCTTACTCTCCTC-3′ NANOG(f) 5′-CAGCCCCGATTCTTCCACCAGTCCC-3′ NANOG(r) 5′-CGGAAGATTCCCAGTCGGGTTCACC-3′ cMYC(f) 5′-GCGTCCTG GGAAGGGAGATCCGGAGC-3′ cMYC(r) 5-TTGAGGGGCATCGTC GCGGGAGGCTG-3′ TDGF1(f) 5′-CTGCTGCCTGAATGGGGGAAC CTGC-3′ TDGF1(r) 5′-GCCACGAGGTGCTCATCCATCACAAGG-3′ REX1(f) 5′-CAGATCCTAAACAGCTCGCAGAAT-3′ REX1(r) 5′-GCG TACGCAAATTAAAGTCCAGA-3′ AFP(f) 5′-GAATGCTGCAAACTG ACCACGCTGGAAC-3′ AFP(r) 5′-TGGCATTCAAGAGGGTTTTCAGT CTGGA-3′ CK8(f) 5′-CCTGGAAGGGCTGACCGACGAGATCAA-3′ CK8(r) 5′-CTTCCCAGCCAGGCTCTGCAGCTCC-3′ BRACHY(f) 5′-GC CCTCTCCCTCCCCTCCACGCACAG-3′ BRACHY(r) 5′-CGGCGCCG TTGCTCACAGACCACAGG-3′ MSX1(f) 5′-CGAGAGGACCCCGTGG ATGCAGAG-3′ MSX1(r) 5′-GGCGGCCATCTTCAGCTTCTCCAG-3′ PAX6(f) 5′-ACCCATTATCCAGATGTGTTTGCCCGAG-3′ PAX6(r) 5′-AT GGTGAAGCTGGGCATAGGCGGCAG-3′ and GAPDH(f) 5′-ACAGTC AGCCGCATCTTC-3′ GAPDH(r) 5′-CTCCGACCTTCACCTTCC-3′. PCR were analyzed by UV light on a 0.8% agarose gel containing ethidium bromide. Experiments were performed in duplicate.
To detect the expression of the anti-HIV genes in the anti-HIV iPSCs and macrophages, quantitative RT-PCR was performed. TRIM5α was detected using the SYBR Green PCR Master Mix Kit (Applied Biosystems) with a primer pair specific to the chimeric TRIM5α gene and not the native human TRIM5α gene: (f) 5′-CTGGGTTGATGTGACAGTGG-3′ and (r) 5′-CGTGAGTGACGGAAACGTAA-3′. To detect the expression of the CCR5 shRNA, a QuantiMir RT kit (System Biosciences, Mountain View, CA) was used according to the manufacturer's protocol. Total RNA was used from the RNA-STAT-60 extraction from the iPSCs and macrophages. The forward primer used for detection of the CCR5 shRNA was 5′-GT AGATGTCAGTCATGCTC-3′. These experiments were performed in triplicate.
Directed differentiation of iPS cells. For directed differentiation of iPSCs to the hematopoietic lineage, hESCs and iPSCs were cocultured on OP9 mouse stromal cells. One day before coculture, 1.2 × 106 OP9 cells were plated onto gelatinized 10 cm dishes. Cells were added to OP9 cultures at a density of 1.6 × 106 cells/dish in 20 ml of differentiation medium: α-MEM supplemented with 10% fetal bovine serum and 50 ng/ml bone morphogenetic protein-4. Half media changes were performed every 3 days. Cells from the cystic bodies which formed in the cocultures were harvested at day 9 and single-cell suspensions were prepared by treatment of the cocultures with Collagenase IV (BD Biosciences) (1 mg/ml in α MEM) for 10 minutes at 37 °C followed by treatment with 0.05% trypsin-EDTA for 3 minutes at 37 °C. Cells were washed twice with PBS, filtered through a cell strainer, and used for FACS analysis and HSC cell purification.
Single-cell suspensions from the hESC and iPSC OP9 cocultures were labeled with CD133 magnetic antibodies using a Direct CD133 microbead isolation kit (Miltenyi Biotech) and processed through two LS separation column. CD133+ cells (2 × 105–3× 105 cells/well) and CB CD34+ cells were cultured, separately, in 35 mm dishes with semisolid Methocult media containing a cocktail of cytokines specific for myeloid differentiation (Stem Cell Technologies, Vancouver, Canada). To derive macrophages, myelomonocytic colonies were harvested from the Methocult cultures and plated in complete Dulbecco's modified Eagle's medium containing 10% fetal bovine serum and 10 ng/ml each of granulocyte-macrophage colony-stimulating factor and macrophage colony-stimulating factor. The macrophages were then used for phenotypic, functional, and HIV-1 challenge experiments.
Teratoma assay. To determine whether the iPSCs were pluripotent and that their differentiated CD133+ HSCs lost their pluripotent capacity, teratoma assays were performed in NOD/SCID−/−γ−/− knockout mice. The iPSCs were dissociated into single-cell suspensions and were injected (106 cells/mouse) into the flank of adult mice. One to two million iPSC-derived CD133+ HSCs were also injected into another cohort of mice. Mice were monitored daily for the formation of teratomas (>2 months). Upon observance of a teratoma, the mice were killed and the tissue was sectioned and analyzed for cells from the endoderm, mesoderm, and ectoderm.
Flow cytometry. To determine the levels of HSC generation in the hESC and iPSC OP9 cocultures, single-cell suspensions obtained from the OP9 cocultures were labeled with a PE-conjugated antibody specific for human CD133 (Miltenyi Biotech). Control CB CD34+ HSCs were labeled with the CD133-PE antibody and also with an APC-conjugated antibody specific for human CD34 (BD Biosciences). The CD34+ HSC-derived iPSCs were labeled with the CD133-PE and CD34-APC antibodies to determine if they retained expression of HSC markers upon induction of pluripotency. To determine whether the iPSC-derived macrophages expressed normal macrophage cell surface markers, iPSC and CB CD34+ derived macrophages were labeled with PE-conjugated antibodies specific for human CD14, CCR5, and CD4 (BD Biosciences). LPS-stimulated macrophages were labeled with a PE-conjugated antibody specific for human CD68 (BD Biosciences). FACS analyses were performed on a Beckman Coulter FC500 and analyzed on Expo32 software. Experiments were performed in triplicate.
Functional analyses of iPSC-derived macrophages. To determine whether the iPSC-derived macrophages were functionally normal, both iPSC and CB CD34+ cell-derived macrophages were stimulated with LPS (2 µg/ml). On various days poststimulation, culture supernatants were sampled and analyzed for IL-2, IL-4, IL-6, IL-10, TNFα, and interferon-γ cytokine secretion using a BD Cytokine Array detection kit (BD Biosciences) according to the manufacturer's protocol. Experiments were performed in triplicate.
HIV-1 challenge of iPSC-derived macrophages. To determine whether the expression of the anti-HIV genes in the anti-HIV iPSC-derived macrophages conferred resistance to HIV-1 infection, cells were challenged with an R5-tropic (BaL-1) or a dual-tropic (89.6) strain of HIV-1 at an multiplicity of infection of 0.05. On various days postinfection, challenge supernatants were sampled and analyzed with an HIV-1 p24 antigen enzyme-linked immunosorbent assay kit (Zeptometrix Corp., Buffalo, NY). Experiments were performed in triplicate. To confirm the infection of the iPSC-derived macrophages, RT-PCR was performed, as previously described, to detect the expression of spliced and unspliced HIV transcripts.44 On day 21 postinfection total RNA was extracted from the macrophages using RNA-Stat-60. cDNA was made using an Applied Biosystems Taqman Reverse Transcription Reagents kit using the primers SC52R (spliced) 5′-TAAGTCTCTCAAGCGGTGGTAGCTGAA-3′ or SCApolR (unspliced) 5′-CCTATTATGTTGACAGGTGTAGGTCCTA-3′. PCR was performed using 400 ng cDNA with primers specific for tat (forward) 5′-CCTGG AATCATCCAGGAAGTCAGCCTA-3′ (reverse) 5′-GGATCTGTCTCTG TCTCTCTCTCCA-3′ or pol (forward) 5′-AAGCTCTATTAGATACAGG AGCAGATGA-3′ (reverse) 5′-ACAGGTGTAGGTCCTACTAATACTG TA-3′. PCR were analyzed by UV light on a 1.0% agarose gel containing ethidium bromide.
Acknowledgments
This work was supported by the University of California-Davis Health System Stem Cell Program start-up funds from the Dean's office and by the James B. Pendleton Charitable Trust. Y.J. was supported by the UC Davis Howard Hughes Medical Institute Med into Grad program. G.M. was supported by a charitable gift from the Randall family and C.N. received a stipend from philanthropic donations to the UC Davis Stem Cell Program. J.A.N. is supported by the NIH and by the California Institute for Regenerative Medicine. We thank Frank Kwong (Sutter Memorial Hospital) for assistance with karyotyping and to the Sutter Cytogenetics staff for allowing the use of their karyotyping microscopes. The NIH AIDS Research and Reference Reagent Program provided the HIV-1 BaL-1 strain used in this work. We have filed a record of invention for the anti-HIV iPSCs.
REFERENCES
- Marcelin AG, Ceccherini-Silberstein F, Perno CF., and, Calvez V. Resistance to novel drug classes. Curr Opin HIV AIDS. 2009;4:531–537. doi: 10.1097/COH.0b013e328331d4b1. [DOI] [PubMed] [Google Scholar]
- Martinez-Picado J, DePasquale MP, Kartsonis N, Hanna GJ, Wong J, Finzi D, et al. Antiretroviral resistance during successful therapy of HIV type 1 infection. Proc Natl Acad Sci USA. 2000;97:10948–10953. doi: 10.1073/pnas.97.20.10948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winters MA, Baxter JD, Mayers DL, Wentworth DN, Hoover ML, Neaton JD, et al. Frequency of antiretroviral drug resistance mutations in HIV-1 strains from patients failing triple drug regimens. The Terry Beirn Community Programs for Clinical Research on AIDS. Antivir Ther (Lond) 2000;5:57–63. [PubMed] [Google Scholar]
- Lafeuillade A, Poggi C, Hittinger G., and, Chadapaud S. Phenotypic and genotypic resistance to nucleoside reverse transcriptase inhibitors in HIV-1 clinical isolates. HIV Med. 2001;2:231–235. doi: 10.1046/j.1468-1293.2001.00078.x. [DOI] [PubMed] [Google Scholar]
- Strayer DS, Akkina R, Bunnell BA, Dropulic B, Planelles V, Pomerantz RJ, et al. Current status of gene therapy strategies to treat HIV/AIDS. Mol Ther. 2005;11:823–842. doi: 10.1016/j.ymthe.2005.01.020. [DOI] [PubMed] [Google Scholar]
- Rossi JJ, June CH., and, Kohn DB. Genetic therapies against HIV. Nat Biotechnol. 2007;25:1444–1454. doi: 10.1038/nbt1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohn DB, Bauer G, Rice CR, Rothschild JC, Carbonaro DA, Valdez P, et al. A clinical trial of retroviral-mediated transfer of a rev-responsive element decoy gene into CD34(+) cells from the bone marrow of human immunodeficiency virus-1-infected children. Blood. 1999;94:368–371. [PubMed] [Google Scholar]
- Mitsuyasu RT, Merigan TC, Carr A, Zack JA, Winters MA, Workman C, et al. Phase 2 gene therapy trial of an anti-HIV ribozyme in autologous CD34+ cells. Nat Med. 2009;15:285–292. doi: 10.1038/nm.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humeau LM, Binder GK, Lu X, Slepushkin V, Merling R, Echeagaray P, et al. Efficient lentiviral vector-mediated control of HIV-1 replication in CD4 lymphocytes from diverse HIV+ infected patients grouped according to CD4 count and viral load. Mol Ther. 2004;9:902–913. doi: 10.1016/j.ymthe.2004.03.005. [DOI] [PubMed] [Google Scholar]
- DiGiusto DL, Krishnan A, Li L, Li H, Li S, Rao A, et al. RNA-based gene therapy for HIV with lentiviral vector-modified CD34(+) cells in patients undergoing transplantation for AIDS-related lymphoma. Sci Transl Med. 2010;2:36ra43. doi: 10.1126/scitranslmed.3000931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podsakoff GM, Engel BC, Carbonaro DA, Choi C, Smogorzewska EM, Bauer G, et al. Selective survival of peripheral blood lymphocytes in children with HIV-1 following delivery of an anti-HIV gene to bone marrow CD34(+) cells. Mol Ther. 2005;12:77–86. doi: 10.1016/j.ymthe.2005.02.024. [DOI] [PubMed] [Google Scholar]
- Anderson JS, Javien J, Nolta JA., and, Bauer G. Preintegration HIV-1 inhibition by a combination lentiviral vector containing a chimeric TRIM5 α protein, a CCR5 shRNA, and a TAR decoy. Mol Ther. 2009;17:2103–2114. doi: 10.1038/mt.2009.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee NS, Dohjima T, Bauer G, Li H, Li MJ, Ehsani A, et al. Expression of small interfering RNAs targeted against HIV-1 rev transcripts in human cells. Nat Biotechnol. 2002;20:500–505. doi: 10.1038/nbt0502-500. [DOI] [PubMed] [Google Scholar]
- Anderson J., and, Akkina R. Human immunodeficiency virus type 1 restriction by human-rhesus chimeric tripartite motif 5α (TRIM 5α) in CD34(+) cell-derived macrophages in vitro and in T cells in vivo in severe combined immunodeficient (SCID-hu) mice transplanted with human fetal tissue. Hum Gene Ther. 2008;19:217–228. doi: 10.1089/hum.2007.108. [DOI] [PubMed] [Google Scholar]
- Bauer G, Selander D, Engel B, Carbonaro D, Csik S, Rawlings S, et al. Gene therapy for pediatric AIDS. Ann N Y Acad Sci. 2000;918:318–329. doi: 10.1111/j.1749-6632.2000.tb05501.x. [DOI] [PubMed] [Google Scholar]
- An DS, Donahue RE, Kamata M, Poon B, Metzger M, Mao SH, et al. Stable reduction of CCR5 by RNAi through hematopoietic stem cell transplant in non-human primates. Proc Natl Acad Sci USA. 2007;104:13110–13115. doi: 10.1073/pnas.0705474104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson J, Li MJ, Palmer B, Remling L, Li S, Yam P, et al. Safety and efficacy of a lentiviral vector containing three anti-HIV genes–CCR5 ribozyme, tat-rev siRNA, and TAR decoy–in SCID-hu mouse-derived T cells. Mol Ther. 2007;15:1182–1188. doi: 10.1038/sj.mt.6300157. [DOI] [PubMed] [Google Scholar]
- Kumar P, Ban HS, Kim SS, Wu H, Pearson T, Greiner DL, et al. T cell-specific siRNA delivery suppresses HIV-1 infection in humanized mice. Cell. 2008;134:577–586. doi: 10.1016/j.cell.2008.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi JJ. The application of ribozymes to HIV infection. Curr Opin Mol Ther. 1999;1:316–322. [PubMed] [Google Scholar]
- ter Brake O, Legrand N, von Eije KJ, Centlivre M, Spits H, Weijer K, et al. Evaluation of safety and efficacy of RNAi against HIV-1 in the human immune system (Rag-2(-/-)γc(-/-)) mouse model. Gene Ther. 2009;16:148–153. doi: 10.1038/gt.2008.124. [DOI] [PubMed] [Google Scholar]
- Holt N, Wang J, Kim K, Friedman G, Wang X, Taupin V, et al. Human hematopoietic stem/progenitor cells modified by zinc-finger nucleases targeted to CCR5 control HIV-1 in vivo. Nat Biotechnol. 2010;28:839–847. doi: 10.1038/nbt.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swan CH, Bühler B, Steinberger P, Tschan MP, Barbas CF., 3rd, and, Torbett BE. T-cell protection and enrichment through lentiviral CCR5 intrabody gene delivery. Gene Ther. 2006;13:1480–1492. doi: 10.1038/sj.gt.3302801. [DOI] [PubMed] [Google Scholar]
- Perez EE, Riley JL, Carroll RG, von Laer D., and, June CH. Suppression of HIV-1 infection in primary CD4 T cells transduced with a self-inactivating lentiviral vector encoding a membrane expressed gp41-derived fusion inhibitor. Clin Immunol. 2005;115:26–32. doi: 10.1016/j.clim.2005.02.019. [DOI] [PubMed] [Google Scholar]
- Hütter G, Nowak D, Mossner M, Ganepola S, Müssig A, Allers K, et al. Long-term control of HIV by CCR5 Δ32/Δ32 stem-cell transplantation. N Engl J Med. 2009;360:692–698. doi: 10.1056/NEJMoa0802905. [DOI] [PubMed] [Google Scholar]
- Takahashi K., and, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Brown ME, Rondon E, Rajesh D, Mack A, Lewis R, Feng X, et al. Derivation of induced pluripotent stem cells from human peripheral blood T lymphocytes. PLoS ONE. 2010;5:e11373. doi: 10.1371/journal.pone.0011373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagata S, Toyoda M, Yamaguchi S, Hirano K, Makino H, Nishino K, et al. Efficient reprogramming of human and mouse primary extra-embryonic cells to pluripotent stem cells. Genes Cells. 2009;14:1395–1404. doi: 10.1111/j.1365-2443.2009.01356.x. [DOI] [PubMed] [Google Scholar]
- Okabe M, Otsu M, Ahn DH, Kobayashi T, Morita Y, Wakiyama Y, et al. Definitive proof for direct reprogramming of hematopoietic cells to pluripotency. Blood. 2009;114:1764–1767. doi: 10.1182/blood-2009-02-203695. [DOI] [PubMed] [Google Scholar]
- Narazaki G, Uosaki H, Teranishi M, Okita K, Kim B, Matsuoka S, et al. Directed and systematic differentiation of cardiovascular cells from mouse induced pluripotent stem cells. Circulation. 2008;118:498–506. doi: 10.1161/CIRCULATIONAHA.108.769562. [DOI] [PubMed] [Google Scholar]
- Lian Q, Zhang Y, Zhang J, Zhang HK, Wu X, Zhang Y, et al. Functional mesenchymal stem cells derived from human induced pluripotent stem cells attenuate limb ischemia in mice. Circulation. 2010;121:1113–1123. doi: 10.1161/CIRCULATIONAHA.109.898312. [DOI] [PubMed] [Google Scholar]
- Sullivan GJ, Hay DC, Park IH, Fletcher J, Hannoun Z, Payne CM, et al. Generation of functional human hepatic endoderm from human induced pluripotent stem cells. Hepatology. 2010;51:329–335. doi: 10.1002/hep.23335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi KD, Yu J, Smuga-Otto K, Salvagiotto G, Rehrauer W, Vodyanik M, et al. Hematopoietic and endothelial differentiation of human induced pluripotent stem cells. Stem Cells. 2009;27:559–567. doi: 10.1634/stemcells.2008-0922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufman DS. Toward clinical therapies using hematopoietic cells derived from human pluripotent stem cells. Blood. 2009;114:3513–3523. doi: 10.1182/blood-2009-03-191304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suhr ST, Chang EA, Rodriguez RM, Wang K, Ross PJ, Beyhan Z, et al. Telomere dynamics in human cells reprogrammed to pluripotency. PLoS ONE. 2009;4:e8124. doi: 10.1371/journal.pone.0008124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vodyanik MA, Bork JA, Thomson JA., and, Slukvin II. Human embryonic stem cell-derived CD34+ cells: efficient production in the coculture with OP9 stromal cells and analysis of lymphohematopoietic potential. Blood. 2005;105:617–626. doi: 10.1182/blood-2004-04-1649. [DOI] [PubMed] [Google Scholar]
- Chadwick K, Wang L, Li L, Menendez P, Murdoch B, Rouleau A, et al. Cytokines and BMP-4 promote hematopoietic differentiation of human embryonic stem cells. Blood. 2003;102:906–915. doi: 10.1182/blood-2003-03-0832. [DOI] [PubMed] [Google Scholar]
- Ramprasad M, Terpstra V, Kondratenko N, Quehenberger O, Steinberg D. Cell surface expression of mouse macrosialin and human CD68 and their role as macrophage receptors for oxidized low density lipoprotein. Proc Natl Acad Sci. 1996;93:14833–14838. doi: 10.1073/pnas.93.25.14833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polo JM, Liu S, Figueroa ME, Kulalert W, Eminii S, Tan KY, et al. Cell type of origin influences the molecular and functional properties of mouse induced pluripotent stem cells. Nat Biotechnol. 2010;28:848–55. doi: 10.1038/nbt.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg H., and, Blumenthal R. Role of HIV Gp41 mediated fusion/hemifusion in bystander apoptosis. Cell Mol Life Sci. 2008;65:3134–3144. doi: 10.1007/s00018-008-8147-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seelamgari A, Maddukuri A, Berro R, de la Fuente C, Kehn K, Deng L, et al. Role of viral regulatory and accessory proteins in HIV-1 replication. Front Biosci. 2004;9:2388–2413. doi: 10.2741/1403. [DOI] [PubMed] [Google Scholar]
- Azad AA. Could Nef and Vpr proteins contribute to disease progression by promoting depletion of bystander cells and prolonged survival of HIV-infected cells. Biochem Biophys Res Commun. 2000;267:677–685. doi: 10.1006/bbrc.1999.1708. [DOI] [PubMed] [Google Scholar]
- Sawyer SL, Wu LI, Emerman M., and, Malik HS. Positive selection of primate TRIM5α identifies a critical species-specific retroviral restriction domain. Proc Natl Acad Sci USA. 2005;102:2832–2837. doi: 10.1073/pnas.0409853102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael NL, Mo T, Merzouki A, O'Shaughnessy M, Oster C, Burke DS, et al. Human immunodeficiency virus type 1 cellular RNA load and splicing patterns predict disease progression in a longitudinally studied cohort. J Virol. 1995;69:1868–1877. doi: 10.1128/jvi.69.3.1868-1877.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]