Abstract
Lentiviral vectors are now in clinical trials for a variety of inherited and acquired disorders. A challenge for moving any viral vector into the clinic is the ability to screen the vector product for the presence of replication-competent virus. Assay development for replication-competent lentivirus (RCL) is particularly challenging because recombination of vector packaging plasmids and cellular DNA leading to RCL has not been reported with the current viral vector systems. Therefore, the genomic structure of a RCL remains theoretical. In this report, we describe a highly sensitive RCL assay suitable for screening vector product and have screened large-scale vector supernatant, cells used in vector production, and cells transduced with clinical grade vector. We discuss the limitations and challenges of the current assay, and suggest modifications that may improve the suitability of this assay for screening US Food and Drug Administration (US FDA)-licensed products.
Introduction
The clinical application of lentiviral vectors requires a sensitive and reliable test for detecting replication-competent lentivirus (RCL) to ensure patients are not inadvertently exposed to replicating virus.1,2,3,4,5,6 Many investigators are seeking to generate clinical lentiviral vectors through the transient transfection method using plasmids expressing the transgene vector and the viral genes required for virion formation. To expand the range of cells susceptible to vector transduction, the HIV-1 envelope glycoprotein (which restricts infection to CD4+ cells) is replaced with an alternative envelope, most commonly the vesicular stomatitis virus G glycoprotein (VSV-G).7 Many clinical investigators are developing “third-generation” lentiviral vectors that have been modified to remove accessory proteins from the vector and packaging plasmids.8,9 As an added safety precaution, Rev responsive elements may be retained in the vector, thus requiring Rev expression during vector production. The most likely source of RCL is recombination between transfer vector and packaging construct sequences used in vector production. In addition, the possibility of recombination between packaging plasmids and human endogenous retroviral sequences must also be considered.10 If vector is generated using transient transfection methods, detecting RCL can be further complicated by contamination of vector supernatants with packaging plasmid DNA that contain the same viral sequences likely to be present in a RCL.11,12
A limited number of RCL assays have been described in the literature. A PCR assay to detect tat sequences from a packaging construct in cultures infected with lentiviral vectors has been described for bovine Jembrana disease viral vectors.13 As tat has been removed from most “third-generation” HIV-1-based lentiviral packaging constructs, this assay has limited applicability for many clinical applications. Syncytia formation assays for testing vector-transfected cells, producer cells, or transduced cells have been developed using cell lines permissive for HIV-1 infection.14 However, these assays detect a fully competent, env-containing lentivirus and may not detect the type of RCL generated with current HIV-1 vectors. Marker rescue assays involving mobilization of an integrated marker provirus following infection of an indicator cell line with RCL have also been used for RCL but whether an unusual recombinant virus will rescue the marker vector is unknown.1,4 Assays based on sensitive measures of reverse transcriptase activity do provide broad base screening for retroviruses but are associated with a significant rate of false positive as background activity varies with the cell type and media used for cell growth.15,16
In developing detection methods for RCL, the C8166 human T cell line has been shown to be highly infectable with HIV-1 and lentiviral vectors pseudotyped with the VSV-G envelope.17,18,19 As important, this cell line is able to amplify HIV-1 at high titer. Previously, we compared detection of virus using a commercially available p24 enzyme-linked immunosorbent assay (ELISA) method (sensitivity of 3 pg/ml–36,000 viral particles), a real-time PCR assay for VSV-G env DNA (sensitivity of ~5–50 copies/0.1 µg of genomic DNA), and a PCR assay (psi-gag PCR) that detects early recombination between the vector plasmid and the gag/pol plasmid (sensitivity 10 copies/0.1 µg of genomic DNA). These detection assays are not sensitive enough to detect small numbers of virus particles but are equally sensitive when limiting number of viruses are first amplified to high titer on cell lines such as the C8166 T cell line.18
As the true nature of a RCL remains theoretical, we chose two methods of virus detection to improve the chance of detecting an unusual recombinant rather than rely on a single read-out assay. Our assay currently uses the p24 ELISA to detect HIV-1 capsid protein along with PCR for psi-gag recombination. The latter was chosen based on our prior observations of rare recombinations occurring in vector preparation.18 We have utilized this assay to screen large-scale vector preparations for RCL and the experience and refinements in the assay are presented in this report.
Results
Assay design
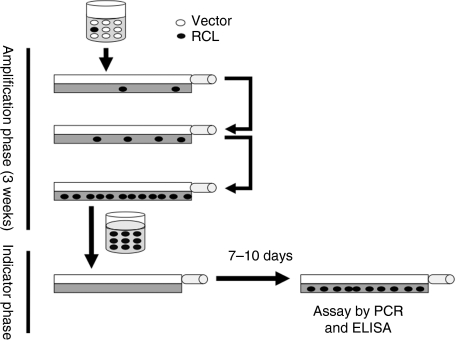
We developed a RCL assay that is modeled after those currently accepted by the US Food and Drug Administration (US FDA) for detecting replication-competent murine retroviruses (RCR).20 For RCL detection, the test article is first amplified in the C8166 T cell line that is permissive for viral infection.18 The virus is allowed to replicate during the 3-week culture period (amplification phase) so virus below the limits of detection at the start of the assay will be amplified and readily detected after 3 weeks (Figure 1). Our preliminary experience with the assay led us to add an indicator phase rather than test material at the end of the amplification phase for two reasons. First, when testing concentrated vector material (which has a high p24 content), we occasionally observed low levels of p24 in the amplification phase even after 3 weeks of culture. Therefore, the possibility of a slow-growing RCL cannot be excluded. Secondly, when evaluating cultures for viral DNA, we previously reported that there can be rare recombinations between vector and packaging plasmids.18 Although the recombinations do not reconstitute a RCL, they may be present (at very low levels) in the amplification cells leading to a false positive assay. The indicator phase ensures that the assay detects a true RCL by requiring passage from amplification phase cells to naive C8166 cells.
Figure 1.
Schematic representation of the replication-competent lentivirus (RCL) assay. Vector product is used to transduce C8166 cells. Small amount of virus which may be present in vector preparations will propagate in the C8166 culture over the 3 weeks of the amplification phase typically yielding virus concentration in excess of X ng/ml of p24. Cell-free media is harvested at the end of the amplification phase and used to transduce naive C8166 cells, which are propagated in the indicator phase. At the end of the indicator phase, the media is evaluated for virus using the p24 enzyme-linked immunosorbent assay (ELISA) assay. Cells are evaluated for evidence of virus using PCR amplification with primers within the viral packaging sequence and the gag gene region (psi-gag PCR).
For a positive control, we have selected the attenuated HIV-1 virus R8.71.17 This lacks the HIV-1 vif, vpr, vpu, and nef accessory genes that are also deleted in most third-generation lentiviral packaging systems. It should be noted that this virus expresses the native HIV-1 envelope whereas all of the vector testing in this report express the VSV-G glycoprotein envelope. While a RCL that arises from vector products is likely to also express the VSV-G envelope, we believed that generating a VSV-G pseudotyped replication-competent HIV-1 virus with the wide host range conferred by the VSV-G envelope represented an unacceptable risk to laboratory personnel.
Optimizing the concentration of vector product
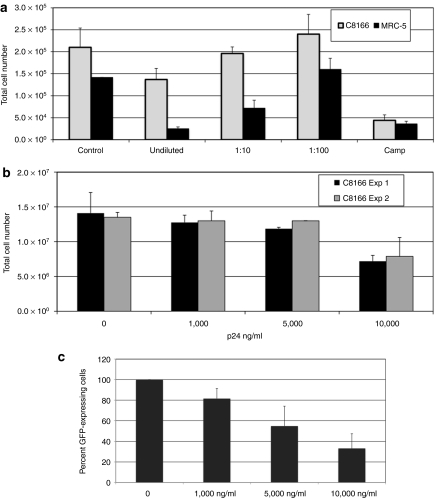
Although our initial test articles were unconcentrated vector material, most final products intended for clinical use will be concentrated to high titer. Upon testing concentrated vector, we noted significant inhibition of C8166 growth using material produced at our facility and in the laboratory of our collaborators and with vectors that expressed a variety of different transgenes (data not shown). To help define the relationship between inhibition and vector dose, CSCGW vector [an HIV-1-based vector expressing green fluorescent protein (GFP) and pseudotyped with the VSV-G envelope] was generated by transient transfection then concentrated 100-fold by ultracentrifugation. C8166 and MRC-5 cells were exposed to undiluted, 1:10, and 1:100 dilutions of vector and the effect on cell expansion was measured after 48 hours (Figure 2a). The addition of concentrated vector inhibited cell growth in a dose-dependent manner, with the greatest effect noted in MRC-5 cells. To help determine the optimal dose for use in the RCL assay, C8166 cells were exposed to CSCGW vector at defined amounts of p24 for 4 hours (the length of vector exposure in the RCL assay). As shown in Figure 2b, growth inhibition was noted with ~60% of the cells present after exposure to vector at the 10,000 ng/ml concentration. We have also observed growth inhibition using clinical grade vector produced at Indiana University (Indianapolis, IN) and at the City of Hope (data not shown). This data suggest that care must be taken when screening concentrated vectors. For example, MRC-5 cells are commonly used in the in vitro virus assay for adventitious agents, a certification assay that has been required by the US FDA before clinical use of gene therapy products. The data also suggests that at a vector concentration of 1,000 ng/ml the growth inhibition of C8166 cells is modest with ~90% of cells surviving.
Figure 2.
Effect of concentrated lentiviral vector on the growth and infectivity of C8166 cells. (a) The effect on concentrated vector was assessed in the nonadherent C8166 cell line and the adherent MRC-5 cell line. Cells were exposed to concentrated CSCGW vector for 4 hours and cell number counted after 48 hours. Concentrated vector (~5,000 ng/ml) was used undiluted or diluted 1:10 and 1:100. Camptothecin was used as a positive toxin control. The data represents the average of three cultures ± SD. (b) The effect of concentrated vector on the growth of C8166 cells was measured at three concentration, 1,000, 5,000, and 10,000 ng/ml of p24 using the CSCGW vector. Duplicate cultures were exposed to vector preparations on day 0 and the total number of cells was determined in each culture after 5 or 6 days of growth (experiment 1 or 2, respectively). Data represents the average of two cultures ± SD for two independent experiments. (c) The inhibitory effect of concentrated vector on C8166 cell infectivity was measured by introducing increasing amounts of a null vector (CSO) into a lentiviral vector preparation of the green fluorescent protein (GFP)-expressing CSCGW vector. The x-axis represents the amount of CSO vector as measured by p24 ELISA. The y-axis represents the GFP-expressing cells as a percentage of control (no CSO vector). The percentage of control cells expressing GFP ranged between 13.5 and 20.1%. The data represents the average of three independent experiment ± SD.
The concentration of vector is also important when optimizing the sensitivity of the RCL assay. For example, vector may be present in great excess relative to any RCL; the vector particles could block receptors and decrease the sensitivity of RCL detection. This was assessed by introducing a small amount of the GFP-expressing vector (CGCGW, sufficient to transduce 10–20% of C8166 cells) with varying concentrations of the CSO vector (a vector that lacks an expressed transgene). Infectivity was monitored by assessing the number of GFP-expressing cells after transfection. As shown in Figure 2c, there was a dose-dependent decrease in the number of GFP-expressing cells indicating that excess of CSO vector particles limited the ability of CSCGW vector to transduce the target cells. At 1,000 ng/ml, this inhibition is quite modest (~80%) and is not predicted to significantly decrease the sensitivity of the RCL assay. Therefore, for the RCL assay, we determine the p24 concentration on all vector supernatants and, if needed, dilute the material to 1,000 ng/ml and do not exceed a ratio of 1,000 ng of p24/5 × 106 C8166 cells.
Performance of assay controls
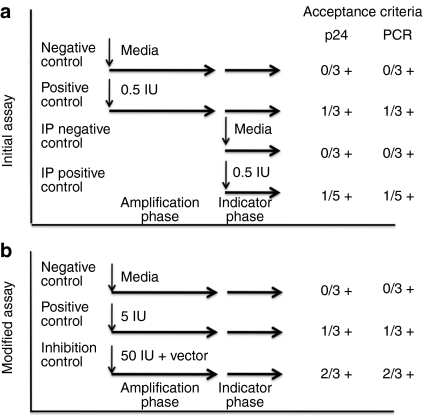
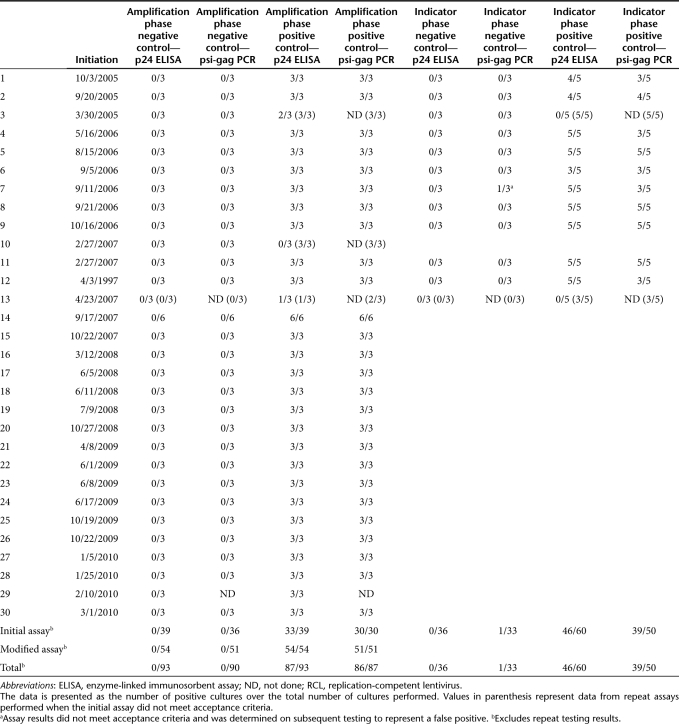
Critical to the usefulness of a clinical RCL detection assay is its ability to minimize false negatives and false positives. As shown in Figure 3a, our initial assay incorporated three positive and three negative control cultures at the start of the amplification phase and three negative and five positive cultures at the start of the indicator phase. All positive cultures were inoculated with the R8.71 attenuated HIV-1 virus at the TCID50 (~0.5 IU/culture). We chose five cultures for the indicator phase positive control since virus would only have 7 days to expand. For the assay to be acceptable, all negative control cultures must be negative for RCL and at least one of three amplification phase positive control cultures, and at least one of the five-indicator phase positive control cultures must be positive. For a culture to be positive, the amount of p24 must be above background for the ELISA, and a band of the expected size for the psi-gag PCR must be present on Southern blot, or both. Using this series of controls, we have performed 13 RCL assays under Good Manufacturing Practice conditions and the results are presented in Table 1 (assays 1–13).
Figure 3.
Controls for the replication-competent lentivirus (RCL) assay: inoculation and acceptance criteria. (a) The initial version of the RCL assay utilized three negative controls setup at the start of the amplification and three at the start of the indicator phase. All cultures are assayed for RCL at the end of the indicator phase. Two sets of positive controls were utilized, both inoculating cultures with R8.71 attenuated HIV-1 virus at the TCID50. Three cultures are set up at the start of the amplification phase and five cultures inoculated at the start of the indicator phase. Five cultures were chosen for the indicator phase positive control since virus expansion will only be occurring for 7 days and the level of virus after this time is expected to be low. However, both the amplification phase positive controls and the indicator phase positive controls must have at least one culture positive for RCL for the assay to be valid. The assay is considered positive is the p24, psi-gag PCR, or both are positive. The established acceptance required all negative cultures to be negative for RCL. (b) The RCL assay was modified to remove the indicator phase negative and positive controls. The inoculum at the start of the amplification phase was increased to 5 IU per positive culture. An inhibitor control was added to the assay in which the test articles (at a concentration ≤1,000 ng/ml of p24) are spiked with 50 IU. At least one of the amplification phase positive controls and two of the inhibitor controls must be positive for the assay to be valid. IP, intraperitoneal.
Table 1. Performance of the RCL assay positive and negative controls.
In terms of performance of the p24 ELISA assay there were no false positive assays detected in the negative control cultures. By ELISA, amplification phase positive control cultures were positive in 33 of 39 cultures (Table 1, column 5) and indicator phase positive control were positive in 46 of 60 cultures (Table 1, column 9).
For psi-gag PCR testing, all negative controls were negative except for one indicator phase control where a faint band was seen in one of two duplicate lanes that was interpreted upon further testing as a false positive. For amplification phase positive controls 30 of 30 cultures were positive (Table 1, column 6) whereas 39 of 50 were positive when inoculated at the start of the indicator phase (Table 1, column 10). These findings suggest that the p24 ELISA may be more sensitive than the psi-gag Southern when detecting low levels of virus (i.e., after only 7 days of expansion from the TCID50). Both ELISA and PCR performed well when higher concentrations of virus were being analyzed (after expansion in the amplification and indicator phase). Our data also suggests that our estimate of 0.5 IU/culture was close to the actual amount, with the actual inoculum being ~0.5–1 IU/culture.
As a quality control, we also measured p24 on the material collected at the end of the amplification phase in all 30 assays (data not shown). There was no instance where a positive p24 detected at the end of the amplification phase was negative after the indicator phase, providing assurance that the passage of cell-free media from the amplification phase cultures to the indicator phase does not decrease virus detection.
There were three assays that failed to meet the acceptability criteria. As discussed above, one negative control culture had a false positive psi-gag PCR assay. This analysis did not require retesting of vector product. Two assays failed because the positive controls failed to meet specifications, with one assay failing at the amplification phase and the other at the indicator phase. Since these failures called into question the sensitivity of the assay, the entire assays were repeated. On rerunning the assay, the positive controls met acceptance criteria and the test articles were found to be free of RCL.
Revising assay controls
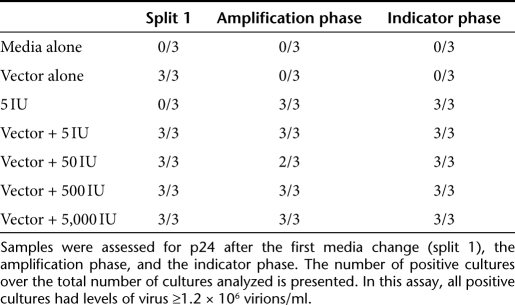
The failure of 2 of our first 13 assays to meet acceptance criteria because the positive control cultures were negative led us to re-evaluate the selection of 0.5 IU as the inoculum for positive cultures. At this level, we expect that only 50% of cultures would be positive but even using a careful methodology for preparing the positive controls, consistently inoculating this low level of virus into a culture is challenging. Coincident with this finding, we began an evaluation of assay sensitivity when the test article is a lentiviral vector designed to inhibit HIV-1 replication. We conducted a qualification protocol where the rHIV7-shI-TAR-CCR5RZ viral vector21,22 (generated at the City of Hope and diluted to 1,000 ng/ml) was spiked with the R8.71-attenuated HIV-1 virus at concentrations ranging from 5 to 5,000 IU. The rHIV7-shI-TAR-CCR5RZ vector contains three sequences targeted against HIV-1 with a U6 Pol III promoter-driven short hairpin RNA targeting the rev and tat mRNAs of HIV-1, a U6 transcribed nucleolar-localizing TAR RNA decoy, and a VA1-derived Pol III cassette that expresses an anti-CCR5 ribozyme. As shown in Table 2, the RCL assay was capable of detecting replicating virus, even at inoculums as low as 5 IU.
Table 2. Detection of attenuated HIV-1 contaminating the rHIV7-shI-TAR-CCR5RZ vector expressing sequences aimed at suppressing HIV-1 replication.
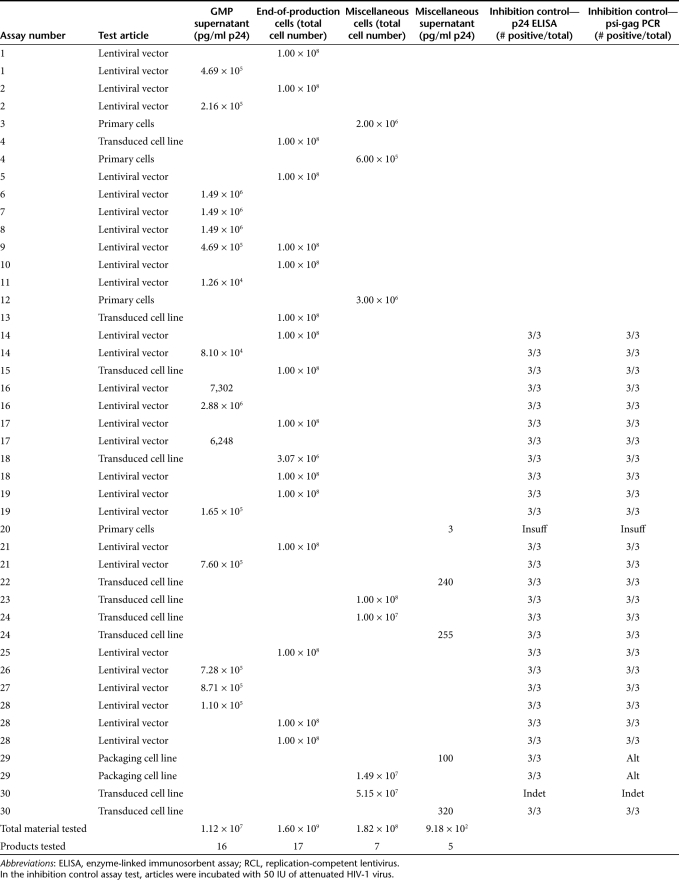
Based on our experience with the first 13 GMP RCL assays, and our finding with the above qualification protocol, we revised the controls in our RCL assay (see Figure 3b). Since the indicator phase negative and positive controls correlated with amplification phase controls, we no longer perform them and rely solely on the controls initiated at the start of the amplification phase. To decrease the likelihood that all positive controls would be negative, we inoculate the positive control cultures with 5 IU of virus. Furthermore, we have added an inhibition control where test articles (i.e., vector product or cells) are spiked with 50 IU of R8.71 virus. Three inhibition control cultures are inoculated and carried through the amplification and indicator phase. At least two of the three must be positive for the assay to be acceptable. We continue to utilize vector product at a maximum concentration of 1,000 ng/ml and do not exceed 5 × 106 C8166/1,000 ng of p24. To date, the modified RCL assay has proven reproducible and sensitive. We have not had to repeat an assay due to failure of positive controls nor have we seen significant inhibition by vector products (Table 3).
Table 3. Test articles evaluated for RCL.
Performance of the RCL assay in screening vector products
Table 3 summarizes the vector products tested and the results of the inhibition controls for materials tested using the modified RCL assay. All of the vector products have been produced using transient transfection methods, the majority using third-generation vector systems.8 With a total of 30 assays performed under GMP conditions, there has been no evidence of RCL detected in vector supernatants, end-of-production cells, engineered cell lines, and clinical samples transduced with lentiviral vectors. For materials intended for clinical use, we have analyzed 1.12 × 107 ng of p24, which is estimated to represent ~1.3 × 1014 virions. We have performed screening of end-of-production cells for 17 products, and screened an additional seven cell lines to be used in generating clinical trial material. This represents screening of ~1.8 × 109 cells generated in six different GMP facilities (~20% of the tested material was generated at Indiana University).
Discussion
When evaluating phase I trials using novel agents, safety not efficacy is the major factor used by the US FDA in determining whether these early phase trials may move forward. Inadvertent exposure of subjects to pathogens that contaminate gene therapy products is a top safety concern. In this article, we demonstrate consistent performance of a RCL detection assay. The testing of clinical grade material provides further evidence that the likelihood of RCL development using current lentiviral vector systems is low.
The initial retroviruses used for gene therapy were based on the murine leukemia viruses. RCR that arise during production of murine leukemia virus-based vectors are known to cause malignancy in mice and immunosuppressed nonhuman primates.23,24 Murine leukemia virus-based vectors are generally produced by packaging cell lines and RCR was frequently detected in early versions of these cell lines.25,26,27,28 RCR development has been decreased by minimizing homology between vector and packaging cell sequences and segregating packaging genes.29,30,31,32,33,34 Still, recombinations have been detected between packaging gene sequences and endogenous sequences within the packaging cells.35,36 The detailed evaluation of RCR that arise during the course of retroviral vector productions has facilitated the development of RCR assays and shaped US FDA recommendations for vector testing.37
Lentiviral vectors are replacing murine leukemia virus-based retroviral vector due to their improved ability to transduce nondividing cells, and their lower risk of insertional mutagenesis.11,38,39,40,41,42,43,44,45,46 Another advantage of the lentiviral system is an apparent lack of RCL generation during vector production. This may be due, in part, to the use of transient transfection methods for lentiviral production. The ability to segregate the vector components onto four plasmids, the use of self-inactivating long terminal repeat's, and retention of Rev dependence also contributes to the safety profile of lentiviral vectors. To date, no RCL has been reported using this method although vector material has not previously been screened as extensively as we described in this article.
A key component of any RCL assay is the positive control. While a VSV-G pseudotyped HIV-1 virus may better represent a RCL arising through recombination, that assumption remains theoretical and a VSV-G pseudotyped HIV-1 control presents significant risk to laboratory personnel. Most importantly, the positive control is used to establish that the sensitivity of the assay is consistent from assay to assay and the R8.71 HIV-1 is sufficient to provide this assurance. In terms of sensitivity, our assay appears to be able to detect R8.71 at 1 IU but with ~10% of cultures failing acceptance criteria in our initial RCL assay, we increased the positive control inoculum from the TCID50 (0.5 IU/culture) to 5 IU/culture. While it is possible that the original failure of virus detection in positive control cultures represents variability in the assay, the finding of consistent positives at the 5 IU suggest that the technical challenges of diluting virus to the TCID50 and the statistical probabilities of relying on three cultures at the TCID50 are the cause. Preparation of our positive control is done through a defined procedure under GMP protocol with extensive testing to establish the IU, a factor critical in setting the limits of detection of our assay (1–5 IU). We demonstrate that the vector decreases the sensitivity of RCL detection, but by diluting vector to 1,000 ng/ml and maintaining a high ratio of cells to vector (1,000 ng for 5 × 106 C8166 cells) this effect can be minimized. This was shown when comparing two VSV-G pseudotyped virions (Figure 2c) and when the R8.71 virus is spiked into vector known to inhibit HIV-1 replication (Table 2).
The assay described here was overdesigned by intent. Since lentiviral vectors are just entering the clinic, an assay, which could provide a high level of assurance that RCL could be detected, was required. To this aim, we relied on two methods of virus detection (p24 ELISA and psi-gag PCR) to provide redundant virus detection. This redundancy minimizes the possibility that a technical problem with a single assay (ELISA or PCR) would lead to a false positive or negative result. Two assays may also increase the chance of detecting a RCL that arose from vector sequences recombining with endogenous retroviral sequences. As the psi-gag PCR method involves detection of the PCR product by Southern blotting using a radiolabeled probe, we are now looking to replace this cumbersome assay with quantitative PCR for the VSV-G envelope DNA. We have previously shown the VSV-G quantitative PCR has similar sensitivity to psi-gag PCR in detecting RCL and are currently validating this alternative approach.18 We would continue to test for p24 that is expected to be positive, even if the RCL contained human endogenous retroviral or other viral envelope sequences.
The amount of vector used per cell (1,000 ng/ml p24 exposed to 5 × 106 C8166 cells or 1 × 106 cocultured with 5 × 106 C8166 cells) is approximately five times the ratio used in RCR assays. This excess of C8166 cells does suggest that the number of infections per cell is low, and may explain the ability of our assay to detect RCL in the presence of an anti-HIV-1 vector. Although our current assay has shown excellent sensitivity, it will be technically challenging to meet the current US FDA guidance requiring testing of 5% of the vector final product as manufacturing methodology allows further scale-up. Currently, the average clinical production scale is in the order of 20–50 l unconcentrated, which requires in excess of 1 × 1010 C8166 cells for a single RCL assay. To facilitate RCL testing of larger batches, we are now evaluating the possibility of decreasing the number of cells per 1,000 ng of p24. Purification of vector47,48,49,50,51,52 should also decrease toxicity and allow higher concentrations of vector to be used in the RCL assay. This will need to be assessed experimentally to validate that the increased concentration of test material does not decrease the sensitivity of the RCL assay. The ratio of C8166 to test material may also need to be adjusted based on the vector properties (i.e., a higher ratio when the test article is a vector that inhibits HIV-1). In addition, there was good correlation between the indicator phase and the amplification p24 data, and the possibility of eliminating the indicator phase or performing it only when the results of the amplification phase are equivocal, could be considered. The data presented here serves as a starting point for future RCL assay modifications as vector technology and manufacturing mature.
The current US FDA requirements to screen vector supernatant for RCL has proven to be a challenging endeavor. Concentrated vector pseudotyped with VSV-G can be toxic to a variety of cells, and the assay is difficult to perform even with modest scale clinical productions (10–50 l range). As production methodologies move to larger scale batches, what percentage of the final product is feasible and appropriate for RCL testing will require additional experience and assay development. The data presented in this paper does provide evidence that the incidence of RCL in lentiviral vector preparations is low and should be considered when assessing regulatory guidelines. This data may also be useful when assessing the need to test transduced cells for RCL, as currently recommended by the US FDA for transduced cells products incubated ex vivo for >4 days. While modifications are being tested to better adapt to the larger vector productions, additional work will be required as alternative pseudotypes to the VSV-G envelope and non-HIV-1-based vectors are developed for clinical trials.
Materials and Methods
Cell lines, and vector preparations. The C8166-45 (C8166; human T-lymphocyte) was obtained from the AIDS Research and Reference Reagent Program (Rockville, MD) and maintained in RPMI 1640. The HEK293T, HEK293, and MRC-5 cell lines were obtained from the American Type Culture Collection and maintained in Dulbecco's modified Eagle's medium. RPMI and Dulbecco's modified Eagle's medium (Invitrogen, Carlsbad, CA) are supplemented with 10% fetal calf serum (Hyclone, Logan, UT), 2 mmol/l -glutamine (Invitrogen), and 100 units/ml penicillin and 100 µg/ml streptomycin (Invitrogen). C8166 cells are maintained in cell banks and only those cells at ≤15 passages are utilized at the start of an assay.
Lentiviral vectors were produced by transient transfection of HEK293T cells (at a ratio of 5 × 106 cells/75 cm2) using calcium phosphate transfection (Profection kit; Promega, Madison, WI). Packaging plasmids were obtained from Cell Genesys (Foster City, CA) and include pMDLg (6.6 µg/75 cm2), pRSV/Rev (3.3 µg/75 cm2), and the VSV-G glycoprotein expression plasmid pMD.G (4.62 µg/75 cm2). Vector plasmids (18 µg/75 cm2) include the GFP-expressing vector pcDNA-HIV-CS-CGW (provided by Philip Zoltick, Children's Hospital, Philadelphia, PA) and the null vector CSO (provided by Donald Kohn, UCLA, Los Angeles, CA). After refeeding cells with fresh medium 16–18 hours post-transfection, vector-containing supernatants were harvested 48 hours after transfection, and filtered through a 0.45-µm filter. Nonclinical vector was concentrated 100-fold by ultracentrifugation (50,000g for 1 hour at 4 °C in a Beckman ultracentrifuge Optima XL-100k using a 45-Ti fixed-angle rotor). Clinical vector preparation were treated with Benzonase to remove residual plasmids as previously described53 and concentrated 100–200-fold by ultrafiltration using a MiniKros disposable hollow fiber tangential flow modules (Spectrum Laboratories, Rancho Dominguez, CA).
Growth inhibition assessment. To assess potential inhibition of vector on cell growth, serial dilutions of concentrated CSCGW vector were used to transduce 5 × 104 C8166 or MRC-5 cells. Vector was pelleted by ultracentrifugation (described above), and the pellet was resuspended in media with 10% fetal calf serum at 1/100th the original volume. Aliquots of the concentrated material were diluted 1:10 and 1:100 in serum-containing media and the vector material was placed directly onto pelleted C8166 or monolayers of MRC-5 where the media had been removed. After a 4-hour incubation in the presence of 8 µg/ml polybrene (Sigma, St Louis, MO), the vector was removed and fresh media was added to the culture (RMPI or Dulbecco's modified Eagle's medium based media for C8166 or MRC-5, respectively). Alternatively, a defined concentration of CSCGW vector (0, 1,000, 5,000, and 10,000 ng) was also used to evaluate C8166 cell growth, using the same procedure. Viable cell counts were assessed 2–6 days after vector exposure.
Inhibition of transduction by vector particles. The impact of competing vector particles on transduction efficiency was assessed by combining the GFP-expressing CSCGW with the null CSO vector. A limiting amount of CSCGW vector stock (previously determined to provide 15–25% GFP-expressing C8166 cells) was mixed with concentrated CSO vector (1,000, 5,000, and 10,000 ng/ml). The vector combination was used to transduce C8166 cells for 4 hours (8 µg/ml in polybrene) and the percentage of GFP-expressing cells was assessed after 72 hours using flow cytometry as previously described.12
Positive control preparation. The attenuated HIV-1 positive control is generated by transfecting HEK293T cells with the pR8.71 plasmid (provided from Cell Genesys, 112 µg plasmid/5 × 106 cells/) using the Profection kit (Promega). After a media change with RPMI at 24 hours, cell-free supernatant (0.45-µm filter) is collected 48 hours after transfection. Material is frozen in 0.5 ml aliquots and frozen at less than −70 °C. Potency is assessed by preparing 45 50-ml flat bottom culture tube with 1 × 106 cells/tube and incubating cells overnight. On day 0, six tubes are centrifuged and cells are resuspended in 1 ml of virus with polybrene at 8 µg/ml. The six dilutions tested range from 10−3 to 10−8 of the frozen viral stock. Three negative control tubes are also prepared. After 4-hour incubation, cells are pelleted, resuspended in RMPI, and transferred to 6-well plates. Cells are maintained in log-phase growth for at least 12 days, after which time cells are pelleted, and the media is filtered (0.45 µm) and assessed for p24 by ELISA and cultures read as positive or negative.
RCL assays. Before initiation of the assay, the p24 content of each test article is assessed to determine the number of C8166 cells required. For aqueous test article, 5 × 106 C8166 cells/ml of test material is required, and the maximal concentration of p24 used is 1,000 ng/ml. In this assay, all culture tubes/flasks are incubated at 37 °C, 5% CO2.
On day −1, triplicate negative, inhibition, and positive control cultures are prepared by adding 12 ml of RPMI and 5 × 106 C8166 cells into nine 50-ml culture tubes (flat-bottom 10 cm2). On day 0, three negative control cultures are prepared by centrifuging cells by replacing the media with 1 ml of RPMI and polybrene (8 µg/ml). Three inhibition controls are prepared in a similar manner except the cultures are inoculated with 1 ml of the test article, 50 IU of R8.71 virus, and 8 µg/ml polybrene. Three positive controls are prepared by inoculating cultures with 1 ml of RPMI, 5 IU of R8.71 virus, and 8 µg/ml polybrene.
Test article flasks are prepared on day −1 based on the volume of material to be tested. For clinical productions, multiple flasks are usually required. The amount of cell suspension (5 × 106/ml) added to each flask is equal to the volume of the test article (after dilution to 1,000 ng/ml). To accommodate the large volumes required for clinical production testing the following options are used: 12 ml/75-cm2 flask or 50 ml culture tube, 30 ml/175-cm2 flask, 60 ml/300 cm2 flask. On day 0, cells are transferred into a 50-ml conical tube, the media is removed after centrifugation, and the test article added to resuspend the cells and the mixture is returned to the flask with polybrene (8 µg/ml).
After 4-hour incubation, the negative, inhibition, and positive culture cells are centrifuged and the media is replaced with 12 ml of fresh RPMI medium and transferred to 75-cm2 flasks. For test articles, cultures from each tubes/flasks are centrifuged after 4-hour incubation and media is replaced with fresh RPMI medium (12–30 ml/75-cm2 flask, 30–60 ml/ 175-cm2 flask, 60–100 ml/300 cm2 flask) and returned to the original flask.
Amplification phase: Cells are maintained in log phase growth through the 3-week period by passing cells a minimum of five times during the 2-week period. For test articles in larger flasks, the initial splits are performed so that cells are placed in smaller size flasks with each subsequent split until the material is in a 75-cm2 flask. Split ratio should not exceed 1:10 for larger flasks (300 and 175-cm2 flasks) and 1:5 for 75-cm2 flasks. To meet acceptability criteria, cultures must contain sufficient cells for passage on days 3–4, 6–8, 9–11, 12–14, and 16–17 of the assay. After at least five splits (3 weeks of culture), cells are removed from the flask, centrifuged, and resuspended in 12 ml of fresh RPMI. At least 24 hours later, the media from each flask is collected and filter through a 0.45-µm filter. Filtered supernatant from each of the control flasks is used to inoculate the corresponding indicator phase culture. For a test article, the material from all flasks are pooled and used to inoculate two test article indicator phase flasks.
Indicator phase: In the last week of the amplification phase, naive C8166 cells are expanded for the indicator phase. The day before harvesting media from the indicator phase, nine control flasks are prepared plus an additional two flasks for each test article analyzed in the assay. Each control flask is to contain 1 × 106 C8166 cells in 4 ml of RPMI. After incubating overnight, cells are centrifuged and resuspended in 1 ml of filtered media from the corresponding amplification phase culture (with polybrene 8 µg/ml). After 4 hours, cells are centrifuged and resuspended in 4 ml of RPMI. After 24 hours, all contents in a culture tube are transferred to a 75-cm2 flask with 12 ml of fresh RPMI medium.
For the test article flasks, 12 ml of RPMI is added to duplicate 75-cm2 flasks along with 1 × 106 C8166 cells for each of the test article flasks from the amplification phase (e.g., if there are 25 amplification flasks for a test article 2.5 × 107 C8166 cells are added to each of 2 flasks). After incubating overnight, cells are centrifuged and resuspended in amplification phase media with polybrene (8 µg/ml). One milliliter of the pooled media for each of the amplification phase test article flasks is used for each duplicate flask. After 4 hours, cells are centrifuged and resuspended in 12 ml of RPMI. After 48 hours, all of the contents are transferred to a 175-cm2 flask with 30 ml of fresh RPMI medium.
Six days after the cells are inoculated, media is removed and fresh RPMI (12 ml/75 cm2, 30 ml/175 cm2) is added. At least 24 hours after media change, contents are transferred from flasks to a 15-ml conical tubes and centrifuged. Media is removed and individually filtered (0.45 µm) and analyzed for p24 content. Cells are then collected for DNA isolation.
Coculture assay: When setting up the inhibitor controls for the amplification phase, 5 × 106 test articles cells are incubated with 1 × 106 C8166 cells and 50 IU of R8.71 virus. For the test article cultures, the ratio is 1 test articles cell for every 5 C8166 cells in the amplification phase. The cultures are processed in a similar manner to aqueous test article except special attention is given to prevent overgrowth of the test article. Specifically, cultures are assessed 2 hours after passage and if there are significant adherent cells noted, the nonadherent cells are removed and replated into new flasks.
Interpretation of results: At the end of the indicator phase, the assay is acceptable if (i) all three negative controls are negative for p24 antigen and psi-gag sequences; (ii) two of three inhibition control flasks are positive for p24 antigen and psi-gag; and (iii) at least one of the positive controls flasks are positive for p24 antigen and psi-gag at the indicator phases. If the controls are acceptable and the test article flasks are negative for p24 antigen and psi-gag then the test article is reported as negative for RCL. If the test article is positive for both p24 and psi-gag, the sample is interpreted as RCL positive. If there is disparity in the p24 and psi-gag results of a test article, the indicator phase is repeated and extended for >14 days before samples are harvested.
Our assay also defines criteria when control samples do not meet the acceptability criteria. If one of the negative controls tested positive by p24 or psi-gag PCR (but not both), the samples are reassayed and if negative the negative control is considered acceptable. If the inhibition control is negative for p24 and psi-gag in ≥2 of the three cultures it is likely that an inhibitor of RCL infection or amplification is present. If the other controls are otherwise acceptable, the assay is considered acceptable but further analysis is performed to define the level of inhibition by spiking test article (1,000 ng/ml) with 100, 500, or 5,000 IU and assessing RCL detection by p24 ELISA after a 3-week amplification. The results are included in the assay findings and interpretation. If controls do not otherwise meet the acceptability criteria, then the assay is considered inconclusive and repeated.
Assessing p24 concentration. The p24 ELISA assay was performed using a commercially available kit (Alliance HIV-1 p24 ELISA kit; Perkin Elmer, Waltham, MA). The kit range is 12.5–100 pg/ml. All measurements were done in duplicate.
Psi-gag PCR assessment. Primers used in the psi-gag PCR are: GrecF1 (5-CAGGACTCGGCTTGCTGAA-3), and GrecR2 (5-TGTCTTATGTC CAGAATGCT-3). After PCR amplification, the product is transferred to a nitrocellulose membrane and probed with the P32 oligo probe GrecP (5-AAGATTTAAACACCATGCTA-3). In addition to test articles, two negative controls are used in the PCR assay; water as a no-template control and genomic DNA from untransduced C8166 cells. A PCR with 100 copies of pR8.71 plasmid in a background of C8166 genomic DNA serves as the positive control. Negative and positive control must demonstrate the expected result for the assay to be acceptable. The psi-gag methodology is described in detail in Sastry et al.18 and the assay has been modified to include a second PCR for human β-globin to validate that the test article DNA is of sufficient quantity and quality.
Acknowledgments
The authors would like to thank for assistance from Lakshmi Sastry, Scott Cross, Philip Zoltick, and Donald Kohn. This work was supported by a grant from the National Gene Vector Laboratory (U42RR11148) and the National Gene Vector Biorepository (P40 RR024928). T.H. received training grant support (T32 HL007910 Basic Science Studies on Gene Therapy of Blood Disease). K.C. was supported, in part, by the Indiana Genomic Initiative (INGEN) created through a grant from the Lilly Endowment, Inc. K.C. is a founder of Rimedion Inc. but there is no financial conflict of interest with the work described in this manuscript.
REFERENCES
- Segall HI, Yoo E., and, Sutton RE. Characterization and detection of artificial replication-competent lentivirus of altered host range. Mol Ther. 2003;8:118–129. doi: 10.1016/s1525-0016(03)00134-5. [DOI] [PubMed] [Google Scholar]
- Delenda C, Audit M., and, Danos O. Biosafety issues in lentivector production. Curr Top Microbiol Immunol. 2002;261:123–141. doi: 10.1007/978-3-642-56114-6_6. [DOI] [PubMed] [Google Scholar]
- Mautino MR, Ramsey WJ, Reiser J., and, Morgan RA. Modified human immunodeficiency virus-based lentiviral vectors display decreased sensitivity to trans-dominant Rev. Hum Gene Ther. 2000;11:895–908. doi: 10.1089/10430340050015509. [DOI] [PubMed] [Google Scholar]
- Srinivasakumar N., and, Schuening FG. A lentivirus packaging system based on alternative RNA transport mechanisms to express helper and gene transfer vector RNAs and its use to study the requirement of accessory proteins for particle formation and gene delivery. J Virol. 1999;73:9589–9598. doi: 10.1128/jvi.73.11.9589-9598.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans JT., and, Garcia JV. Lentivirus vector mobilization and spread by human immunodeficiency virus. Hum Gene Ther. 2000;11:2331–2339. doi: 10.1089/104303400750038444. [DOI] [PubMed] [Google Scholar]
- Wu X, Wakefield JK, Liu H, Xiao H, Kralovics R, Prchal JT, et al. Development of a novel trans-lentiviral vector that affords predictable safety. Mol Ther. 2000;2:47–55. doi: 10.1006/mthe.2000.0095. [DOI] [PubMed] [Google Scholar]
- Cronin J, Zhang XY., and, Reiser J. Altering the tropism of lentiviral vectors through pseudotyping. Curr Gene Ther. 2005;5:387–398. doi: 10.2174/1566523054546224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dull T, Zufferey R, Kelly M, Mandel RJ, Nguyen M, Trono D, et al. A third-generation lentivirus vector with a conditional packaging system. J Virol. 1998;72:8463–8471. doi: 10.1128/jvi.72.11.8463-8471.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zufferey R, Dull T, Mandel RJ, Bukovsky A, Quiroz D, Naldini L, et al. Self-inactivating lentivirus vector for safe and efficient in vivo gene delivery. J Virol. 1998;72:9873–9880. doi: 10.1128/jvi.72.12.9873-9880.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urnovitz HB., and, Murphy WH. Human endogenous retroviruses: nature, occurrence, and clinical implications in human disease. Clin Microbiol Rev. 1996;9:72–99. doi: 10.1128/cmr.9.1.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naldini L, Blömer U, Gallay P, Ory D, Mulligan R, Gage FH, et al. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science. 1996;272:263–267. doi: 10.1126/science.272.5259.263. [DOI] [PubMed] [Google Scholar]
- Sastry L, Johnson T, Hobson MJ, Smucker B., and, Cornetta K. Titering lentiviral vectors: comparison of DNA, RNA and marker expression methods. Gene Ther. 2002;9:1155–1162. doi: 10.1038/sj.gt.3301731. [DOI] [PubMed] [Google Scholar]
- Metharom P, Takyar S, Xia HH, Ellem KA, Macmillan J, Shepherd RW, et al. Novel bovine lentiviral vectors based on Jembrana disease virus. J Gene Med. 2000;2:176–185. doi: 10.1002/(SICI)1521-2254(200005/06)2:3<176::AID-JGM106>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- Chang LJ, Urlacher V, Iwakuma T, Cui Y., and, Zucali J. Efficacy and safety analyses of a recombinant human immunodeficiency virus type 1 derived vector system. Gene Ther. 1999;6:715–728. doi: 10.1038/sj.gt.3300895. [DOI] [PubMed] [Google Scholar]
- Sastry L, Xu Y, Duffy L, Koop S, Jasti A, Roehl H, et al. Product-enhanced reverse transcriptase assay for replication-competent retrovirus and lentivirus detection. Hum Gene Ther. 2005;16:1227–1236. doi: 10.1089/hum.2005.16.1227. [DOI] [PubMed] [Google Scholar]
- Miskin J, Chipchase D, Rohll J, Beard G, Wardell T, Angell D, et al. A replication competent lentivirus (RCL) assay for equine infectious anaemia virus (EIAV)-based lentiviral vectors. Gene Ther. 2006;13:196–205. doi: 10.1038/sj.gt.3302666. [DOI] [PubMed] [Google Scholar]
- Escarpe P, Zayek N, Chin P, Borellini F, Zufferey R, Veres G, et al. Development of a sensitive assay for detection of replication-competent recombinant lentivirus in large-scale HIV-based vector preparations. Mol Ther. 2003;8:332–341. doi: 10.1016/s1525-0016(03)00167-9. [DOI] [PubMed] [Google Scholar]
- Sastry L, Xu Y, Johnson T, Desai K, Rissing D, Marsh J, et al. Certification assays for HIV-1-based vectors: frequent passage of gag sequences without evidence of replication-competent viruses. Mol Ther. 2003;8:830–839. doi: 10.1016/j.ymthe.2003.08.003. [DOI] [PubMed] [Google Scholar]
- Manilla P, Rebello T, Afable C, Lu X, Slepushkin V, Humeau LM, et al. Regulatory considerations for novel gene therapy products: a review of the process leading to the first clinical lentiviral vector. Hum Gene Ther. 2005;16:17–25. doi: 10.1089/hum.2005.16.17. [DOI] [PubMed] [Google Scholar]
- FDA . U.S. Food and Drug Administration; 2006. Guidance for Industry – Supplemental Guidance on Testing for Replication Competent Retrovirus in Retroviral Vector Based Gene Therapy Products and During Follow-up of Patients in Clinical Trials Using Retroviral Vectors. [DOI] [PubMed] [Google Scholar]
- Li MJ, Kim J, Li S, Zaia J, Yee JK, Anderson J, et al. Long-term inhibition of HIV-1 infection in primary hematopoietic cells by lentiviral vector delivery of a triple combination of anti-HIV shRNA, anti-CCR5 ribozyme, and a nucleolar-localizing TAR decoy. Mol Ther. 2005;12:900–909. doi: 10.1016/j.ymthe.2005.07.524. [DOI] [PubMed] [Google Scholar]
- DiGiusto DL, Krishnan A, Li L, Li H, Li S, Rao A, et al. RNA-based gene therapy for HIV with lentiviral vector-modified CD34(+) cells in patients undergoing transplantation for AIDS-related lymphoma. Sci Transl Med. 2010;2:36ra43. doi: 10.1126/scitranslmed.3000931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornetta K, Nguyen N, Morgan RA, Muenchau DD, Hartley JW, Blaese RM, et al. Infection of human cells with murine amphotropic replication-competent retroviruses. Hum Gene Ther. 1993;4:579–588. doi: 10.1089/hum.1993.4.5-579. [DOI] [PubMed] [Google Scholar]
- Donahue RE, Kessler SW, Bodine D, McDonagh K, Dunbar C, Goodman S, et al. Helper virus induced T cell lymphoma in nonhuman primates after retroviral mediated gene transfer. J Exp Med. 1992;176:1125–1135. doi: 10.1084/jem.176.4.1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muenchau DD, Freeman SM, Cornetta K, Zwiebel JA., and, Anderson WF. Analysis of retroviral packaging lines for generation of replication-competent virus. Virology. 1990;176:262–265. doi: 10.1016/0042-6822(90)90251-l. [DOI] [PubMed] [Google Scholar]
- Bodine DM, McDonagh KT, Brandt SJ, Ney PA, Agricola B, Byrne E, et al. Development of a high-titer retrovirus producer cell line capable of gene transfer into rhesus monkey hematopoietic stem cells. Proc Natl Acad Sci USA. 1990;87:3738–3742. doi: 10.1073/pnas.87.10.3738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarpa M, Cournoyer D, Muzny DM, Moore KA, Belmont JW., and, Caskey CT. Characterization of recombinant helper retroviruses from Moloney-based vectors in ecotropic and amphotropic packaging cell lines. Virology. 1991;180:849–852. doi: 10.1016/0042-6822(91)90105-k. [DOI] [PubMed] [Google Scholar]
- Otto E, Jones-Trower A, Vanin EF, Stambaugh K, Mueller SN, Anderson WF, et al. Characterization of a replication-competent retrovirus resulting from recombination of packaging and vector sequences. Hum Gene Ther. 1994;5:567–575. doi: 10.1089/hum.1994.5.5-567. [DOI] [PubMed] [Google Scholar]
- Bosselman RA, Hsu RY, Bruszewski J, Hu S, Martin F., and, Nicolson M. Replication-defective chimeric helper proviruses and factors affecting generation of competent virus: expression of Moloney murine leukemia virus structural genes via the metallothionein promoter. Mol Cell Biol. 1987;7:1797–1806. doi: 10.1128/mcb.7.5.1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markowitz D, Goff S., and, Bank A. A safe packaging line for gene transfer: separating viral genes on two different plasmids. J Virol. 1988;62:1120–1124. doi: 10.1128/jvi.62.4.1120-1124.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markowitz D, Goff S., and, Bank A. Construction and use of a safe and efficient amphotropic packaging cell line. Virology. 1988;167:400–406. [PubMed] [Google Scholar]
- Danos O., and, Mulligan RC. Safe and efficient generation of recombinant retroviruses with amphotropic and ecotropic host ranges. Proc Natl Acad Sci USA. 1988;85:6460–6464. doi: 10.1073/pnas.85.17.6460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller AD., and, Rosman GJ. Improved retroviral vectors for gene transfer and expression. BioTechniques. 1989;7:980–2, 984. [PMC free article] [PubMed] [Google Scholar]
- Miller AD, Garcia JV, von Suhr N, Lynch CM, Wilson C., and, Eiden MV. Construction and properties of retrovirus packaging cells based on gibbon ape leukemia virus. J Virol. 1991;65:2220–2224. doi: 10.1128/jvi.65.5.2220-2224.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong H, Starkey W., and, Vile RG. A replication-competent retrovirus arising from a split-function packaging cell line was generated by recombination events between the vector, one of the packaging constructs, and endogenous retroviral sequences. J Virol. 1998;72:2663–2670. doi: 10.1128/jvi.72.4.2663-2670.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrett E, Miller AR, Goldman JM, Apperley JF., and, Melo JV. Characterization of recombination events leading to the production of an ecotropic replication-competent retrovirus in a GP+envAM12-derived producer cell line. Virology. 2000;266:170–179. doi: 10.1006/viro.1999.0052. [DOI] [PubMed] [Google Scholar]
- Cornetta, K, Wilson, CA. Dropulic BaCB. Concepts in Genetic Medicine. John Wiley & Sons; 2008. Safety of retroviral vectors: Regulatory and technical considerations; pp. 277–288. [Google Scholar]
- Amado RG., and, Chen IS. Lentiviral vectors–the promise of gene therapy within reach. Science. 1999;285:674–676. doi: 10.1126/science.285.5428.674. [DOI] [PubMed] [Google Scholar]
- Blömer U, Naldini L, Kafri T, Trono D, Verma IM., and, Gage FH. Highly efficient and sustained gene transfer in adult neurons with a lentivirus vector. J Virol. 1997;71:6641–6649. doi: 10.1128/jvi.71.9.6641-6649.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lever AM. Lentiviral vectors: progress and potential. Curr Opin Mol Ther. 2000;2:488–496. [PubMed] [Google Scholar]
- Podsakoff GM. Lentiviral vectors approach the clinic but fall back: National Institutes of Health Recombinant DNA Advisory Committee review of a first clinical protocol for use of a lentiviral vector. Mol Ther. 2001;4:282–283. doi: 10.1006/mthe.2001.0470. [DOI] [PubMed] [Google Scholar]
- Watson DJ, Kobinger GP, Passini MA, Wilson JM., and, Wolfe JH. Targeted transduction patterns in the mouse brain by lentivirus vectors pseudotyped with VSV, Ebola, Mokola, LCMV, or MuLV envelope proteins. Mol Ther. 2002;5 5 Pt 1:528–537. doi: 10.1006/mthe.2002.0584. [DOI] [PubMed] [Google Scholar]
- Li Z, Düllmann J, Schiedlmeier B, Schmidt M, von Kalle C, Meyer J, et al. Murine leukemia induced by retroviral gene marking. Science. 2002;296:497. doi: 10.1126/science.1068893. [DOI] [PubMed] [Google Scholar]
- Hacein-Bey-Abina S, Von Kalle C, Schmidt M, McCormack MP, Wulffraat N, Leboulch P, et al. LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1. Science. 2003;302:415–419. doi: 10.1126/science.1088547. [DOI] [PubMed] [Google Scholar]
- Ott MG, Schmidt M, Schwarzwaelder K, Stein S, Siler U, Koehl U, et al. Correction of X-linked chronic granulomatous disease by gene therapy, augmented by insertional activation of MDS1-EVI1, PRDM16 or SETBP1. Nat Med. 2006;12:401–409. doi: 10.1038/nm1393. [DOI] [PubMed] [Google Scholar]
- Nienhuis AW, Dunbar CE., and, Sorrentino BP. Genotoxicity of retroviral integration in hematopoietic cells. Mol Ther. 2006;13:1031–1049. doi: 10.1016/j.ymthe.2006.03.001. [DOI] [PubMed] [Google Scholar]
- Transfiguracion J, Jaalouk DE, Ghani K, Galipeau J., and, Kamen A. Size-exclusion chromatography purification of high-titer vesicular stomatitis virus G glycoprotein-pseudotyped retrovectors for cell and gene therapy applications. Hum Gene Ther. 2003;14:1139–1153. doi: 10.1089/104303403322167984. [DOI] [PubMed] [Google Scholar]
- Segura MM, Garnier A, Durocher Y, Coelho H., and, Kamen A. Production of lentiviral vectors by large-scale transient transfection of suspension cultures and affinity chromatography purification. Biotechnol Bioeng. 2007;98:789–799. doi: 10.1002/bit.21467. [DOI] [PubMed] [Google Scholar]
- Slepushkin, V, Chang, N, Cohen, R, Gan, Y, Jiang, B, Deausen, E, et al. Large-scale purification of a lentiviral vector by size exclusion chromatography or mustang Q ion exchange capsule. Bioprocessing J: 2003. pp. 89–95.
- Kutner RH, Zhang XY., and, Reiser J. Production, concentration and titration of pseudotyped HIV-1-based lentiviral vectors. Nat Protoc. 2009;4:495–505. doi: 10.1038/nprot.2009.22. [DOI] [PubMed] [Google Scholar]
- Scherr M, Battmer K, Eder M, Schüle S, Hohenberg H, Ganser A, et al. Efficient gene transfer into the CNS by lentiviral vectors purified by anion exchange chromatography. Gene Ther. 2002;9:1708–1714. doi: 10.1038/sj.gt.3301848. [DOI] [PubMed] [Google Scholar]
- Merten OW, Charrier S, Laroudie N, Fauchille S, Dugué C, Jenny C, et al. Hum Gene Ther. Nov 2010; 2010. Large scale manufacture and characterisation of a lentiviral vector produced for clinical ex vivo gene therapy application. [DOI] [PubMed] [Google Scholar]
- Sastry L, Xu Y, Cooper R, Pollok K., and, Cornetta K. Evaluation of plasmid DNA removal from lentiviral vectors by benzonase treatment. Hum Gene Ther. 2004;15:221–226. doi: 10.1089/104303404772680029. [DOI] [PubMed] [Google Scholar]