Abstract
Recombinant adeno-associated viruses (rAAVs) that can cross the blood–brain-barrier and achieve efficient and stable transvascular gene transfer to the central nervous system (CNS) hold significant promise for treating CNS disorders. However, following intravascular delivery, these vectors also target liver, heart, skeletal muscle, and other tissues, which may cause untoward effects. To circumvent this, we used tissue-specific, endogenous microRNAs (miRNAs) to repress rAAV expression outside the CNS, by engineering perfectly complementary miRNA-binding sites into the rAAV9 genome. This approach allowed simultaneous multi-tissue regulation and CNS-directed stable transgene expression without detectably perturbing the endogenous miRNA pathway. Regulation of rAAV expression by miRNA was primarily via site-specific cleavage of the transgene mRNA, generating specific 5′ and 3′ mRNA fragments. Our findings promise to facilitate the development of miRNA-regulated rAAV for CNS-targeted gene delivery and other applications.
Introduction
Gene transfer mediated by recombinant adeno-associated virus (rAAV) holds promise for treatment of a large number of neurological disorders, but the blood–brain barrier blocks systemic vector delivery to the central nervous system (CNS). Nonetheless, previous proof-of-concept clinical studies using intracranial injection of AAV serotype 2-based vectors to the brain provided sustained gene expression and therapeutic effects.1,2,3,4 Although localized vector delivery may be effective in treating CNS disorders that map to well defined anatomic and functional regions of the brain, the process requires invasive neurosurgical procedures and, therefore, is costly and potentially risky. If AAV vectors could cross the blood–brain barrier and be specifically expressed in the CNS, intravascular delivery of rAAV would provide a more effective method for gene therapy of CNS diseases that affect large areas of the brain and spinal cord.
Recent advancements in the discovery, rational design, and directed evolution of AAV have created a collection of distinct recombinant AAVs (rAAVs) appropriate for different gene transfer applications.5 Among these vectors, rAAV6, rAAV8, and rAAV9 stand out for their ability to deliver genes transvascularly to target tissues.6,7,8,9 Intravascularly administered rAAV9 and some other rAAVs with capsid modifications can cross the blood–brain barrier, efficiently transferring genes to the CNS in mice, cats, and nonhuman primates.10,11,12,13,14 Nonetheless, several potential challenges remain to using rAAV9 to treat CNS disorders. The first challenge is to deliver rAAV specifically to the CNS. The viral capsid is the principal determinant for AAV tissue tropism,5 and the liver is the major target for AAV vectors. Systemic administration of some rAAV serotypes transduces the liver and other tissues, including the CNS, skeletal muscle, heart, pancreas, and antigen-presenting cells.6,7 Such off-target transduction raises the specter of overexpression of transgenes outside the CNS, potentially eliciting toxic responses. Thus, the second challenge is to confine transgene expression to the CNS when rAAV9 is delivered systemically. Historically, CNS-specific promoters have been used to limit transgene expression to the CNS. However, tissue-specific, strong CNS promoters are often too large to be packaged into the rAAV genome.
As an alternative, we used endogenous microRNAs (miRNAs) to suppress transgene expression outside the CNS. miRNAs are small, noncoding RNAs that regulate gene expression by post-transcriptional silencing. miRNAs silence genes by two mechanisms. When partially complementary to mRNA sequences, they typically reduce target mRNA stability and protein expression by two- to fourfold or less, a mode of regulation thought to tune mRNA expression.15 In contrast, when miRNAs are nearly perfectly complementary to their mRNA targets, they cleave the mRNA, triggering its wholesale destruction. miRNA-binding sites were first used in lentiviral vectors to suppress transgene expression in hematopoietic cells, thereby attenuating transgene immunogenicity.16 The strategy was subsequently used to regulate naked DNA-mediated gene transfer, detarget oncolytic viral therapeutics from noncancer tissues to reduce host toxicity, and, in another application, attenuate live, attenuated virus-based viral vaccines to reduce toxicity in vaccine recipients.16,17,18,19,20,21 The use of miRNA detargeting poses special challenges for systemic delivery of rAAV to transduce the CNS. Intravascularly delivered rAAV for CNS-targeted transduction requires suppressing high levels of rAAV expression in multiple peripheral tissues even when transgene transcripts are successfully expressed in the brain.
Here, we report the use of miRNAs to detarget rAAV9 expression both separately and concurrently in the liver, heart, and skeletal muscle, the three tissues that are most efficiently targeted by intravenously delivered rAAV9.7,8 Silencing of transgene expression in liver, heart, and muscle exploited the natural expression of the abundant (≥60,000 copies/cell) miRNAs, miR-122, which is expressed in hepatocytes, and miR-1, a miRNA found in the heart and skeletal muscle of virtually all animals.22,23 miR-122-binding sites have been successfully used to prevent hepatotoxicity of a transgene from an adenovirus vector.18 Perfectly complementary sites for miR-1, miR-122, or both were engineered into the 3′ untranslated region (UTR) of a nuclear-targeted, β-galactosidase (nLacZ) reporter transgene whose expression was driven by a cytomegalovirus-enhancer, chicken β-actin (CB) promoter. We present multiple independent lines of evidence to show that the miRNAs repress nLacZ expression by cleaving the transgene mRNA at exactly the same site as by all Argonaute-bound small RNAs in eukaryotic cells.24 When delivered systemically in vivo, the miRNA-detargeted rAAV9 vector successfully expressed the reporter transgene in the CNS, but not the liver or heart or skeletal muscle.
Results
miRNAs efficiently repress reporter gene expression in cultured cells
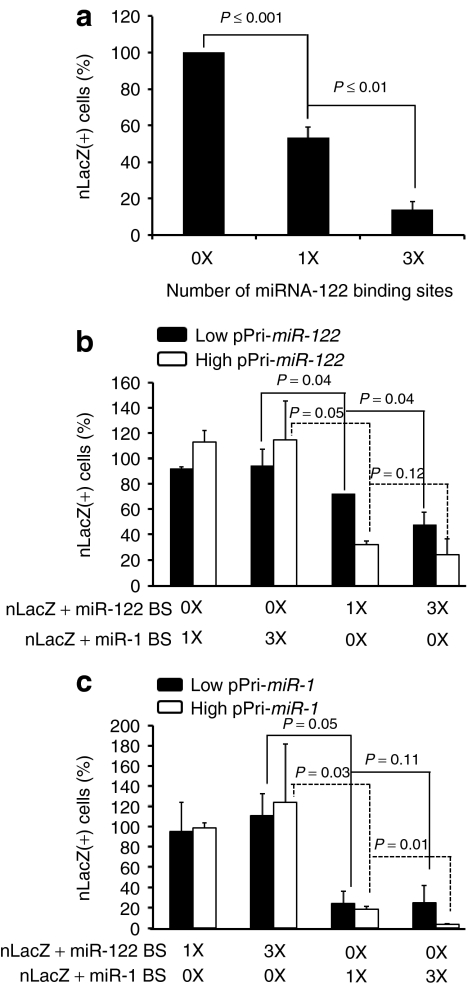
Endogenous miRNA-mediated post-transcriptional gene silencing has proven to be an effective and tissue-specific approach to regulate transgene or viral gene expression in vivo for plasmids, viral vectors, or live, attenuated viral vaccines.16,17,18,19,20,21 To evaluate this strategy for rAAV-mediated transduction, we introduced one or three tandem copies of a perfectly complementary binding site for miR-1 or miR-122 into the 3′ UTR of nLacZ in a rAAV plasmid vector. We transfected the constructs into HuH7 cells, a human hepatoma cell line expressing ~16,000 copies of miR-122 per cell,23 and measured the number of nLacZ-positive cells. The number of nLacZ-expressing HuH7 cells for the one-site plasmid was about half that of the no site control; three sites reduced the number of nLacZ-expressing cells more than sevenfold (Figure 1a).
Figure 1.
In vitro validation of artificial miRNA-binding sites for reporter silencing. Plasmids harboring the rAAVCBnLacZ genome with or without miR-1 or miR-122-binding sites were transfected into human hepatoma (HuH7) cells (a) which express miR-122 or cotransfected into 293 cells, together with a plasmid expressing either pri-miR-122 (b) or pri-miR-1 (c) at molar ratios of 1:3 (low) or 1:10 (high). 0X: no miRNA-binding site; 1X: one miRNA-binding site; 3X: three miRNA-binding sites. The cells were fixed and stained histochemically with X-gal 48 hours after transfection and blue cells counted. The percentage of nLacZ-positive cells in each transfection were compared to transfection of the control plasmid (prAAVCBnLacZ). CB, chicken β-actin; miR, microRNA; nLacZ, β-galactosidase reporter transgene; rAAV, recombinant adeno-associated viruses.
Next, we analyzed expression of the nLacZ constructs in human embryonic kidney 293 cells, which naturally express low levels of both miR-122 and miR-1, when miR-1 or miR-122 was introduced as a pri-miRNA from a second plasmid. We transfected 293 cells with the nLacZ reporter plasmids carrying 0, 1, or 3 miR-122 or miR-1-binding sites, together with a plasmid expressing either pri-miR-122 (Figure 1b) or pri-miR-1 (Figure 1c). To vary the concentration of the miRNA, we used either a low (1:3) or a high (1:10) molar ratio of the nLacZ-binding site plasmid to the miRNA expression plasmid. When miR-122 or miR-1 was introduced into the cells, nLacZ expression was repressed only when the nLacZ reporter mRNA contained the corresponding miRNA-binding sites; there was no reduction of nLacZ-positive cells when miR-1 was coexpressed with nLacZ containing miR-122-binding sites or when miR-122 was coexpressed with nLacZ containing miR-1-binding sites (Figure 1b,c).
Tissue-specific endogenous miRNAs can regulate expression of rAAV9 delivered systemically in adult mice
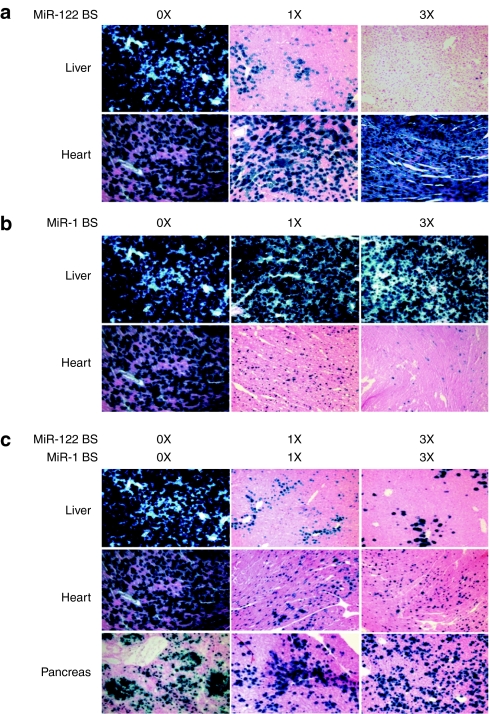
To evaluate miRNA regulation of systemically delivered AAV9CBnLacZ vectors in vivo, we produced AAV9CBnLacZ vectors carrying 0, 1, or 3 miRNA-binding sites perfectly complementary to either miR-122 or miR-1. The vectors were administered by tail vein injection to adult male C56BL/6 mice at a dose of 5 × 1013 genome copies per kg (GC/kg) body weight. Four weeks later, we examined the liver and heart of the transduced animals. LacZ staining revealed that the nLacZ transgene was silenced by the endogenous miRNAs in the cell type and organ in which they are predominantly expressed: the transgene was specifically silenced by miR-122 in the liver and by miR-1 in the heart (Figure 2a,b). While nLacZ positive cells were reduced in the livers of the animals treated with rAAV9CBnLacZ bearing one or three miR-122-binding sites, nLacZ expression levels in the hearts of the same animals were similar to those in the animals treated with AAV9CBnLacZ bearing no sites (Figure 2a). Similarly, nLacZ expression was not detected in the hearts of the animals that received AAV9CBnLacZ containing one or three miR-1-binding sites, but nLacZ expression in the livers of the same animals was not affected as compared to that in the control animal (Figure 2b). Our data suggest that the greater the number of sites for a miRNA in rAAV, the lower the nLacZ expression in the tissue where the corresponding miRNA is expressed (Figure 2a,b).
Figure 2.
In vivo evaluation of endogenous miRNA-mediated transgene silencing in rAAV9 transduction. (a–c) Adult male C58BL/6 mice were injected intravenously with 5 × 1013 genome copies per kg (GC/kg) each of rAAV9CBnLacZ (no binding site), (a) rAAVCB9nLacZ-miR-122BS (one miR-122-binding site) and rAAV9CBnLacZ-(miR-122BS)3 (three miR-122-binding sites), (b) rAAV9CBnLacZ-miR-1BS (one miR-1 binding site) and rAAV9CBnLacZ-(miR-1BS)3 (three miR-1-binding sites, and (c) rAAV9CBnLacZ-miR-1BS-miR-122BS (1X each binding site) and rAAV9CBnLacZ-(miR-1BS)3–(miR-122BS)3 (three miR-1 and three miR-122-binding sites). The animals were necropsied 4 weeks after vector administration, and appropriate tissues were harvested for cryosectioning and X-gal histochemical staining. miR, microRNA; nLacZ, β-galactosidase reporter transgene; rAAV, recombinant adeno-associated viruses.
Next, we tested whether transgene silencing could be achieved simultaneously in multiple tissues. We inserted different numbers of both miR-122- and miR-1-binding sites in the 3′ UTR of the rAAV9CBnLacZ genome and examined their expression in rAAV9 transduced mice. Histochemical staining of tissue sections showed that nLacZ expression was suppressed in both heart and liver for rAAV9CBnLac containing one or three copies each of the miR-1- and miR-122-binding sites, but nLacZ was readily detectable in pancreas, where expression of both miR-122 and miR-1 is low25 (Figure 2c). Quantitative, β-galactosidase assays of homogenized liver tissue similarly showed that nLacZ expression was significantly lower when the transgene contained the miRNA-binding sites (one miR-122-binding site: 7.8 ± 7.4%, P value = 0.005; three miR-122-binding sites: 1.6 ± 1.0%, P value = 0.005; one miR-1- plus one miR-122-binding site: 8.6 ± 5.7%, P value = 0.005; three miR-1- plus three miR-122-binding sites: 3.1 ± 1.2%, P value = 0.005; three miR-1-binding sites: 105.7 ± 11.6%) (Supplementary Figure S1).
miRNA repression of rAAV expression does not perturb endogenous miRNA pathways
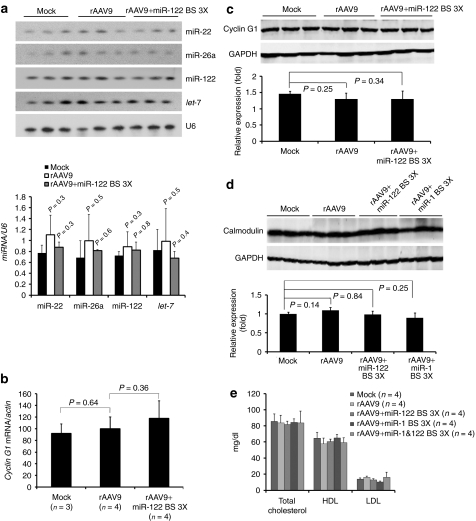
Highly expressed transgenes bearing miRNA-complementary sites have been reported to promote degradation of the corresponding miRNA.26 We examined the levels of miR-122, miR-22, miR-26a, and let-7 in rAAV transduced liver. We detected no difference in abundance of the four miRNAs among the three study groups (Figure 3a). Moreover, our preliminary data from high throughput sequencing analyses of small RNA from the livers of one animal each from the three study groups show no change in miRNA levels (data not shown).
Figure 3.
Analysis of expression levels of cognate miRNA, mRNA, and protein of endogenous miRNA target genes in mice transduced with rAAV9CBnLacZ with or without miRNA-binding sites. Total cellular RNA or protein was prepared from (a–c) liver or (d) heart. (a) Northern blot detection of miRNAs. U6 small nuclear RNA provides a loading control. (b) Quantitative reverse-transcription PCR measuring cyclin G1 mRNA. The data are presented as relative cyclin G1 mRNA levels normalized to β-actin. (c,d) Western blot analyses of protein levels of endogenous targets of miR-122 and miR-1. Total cellular protein prepared from (c) liver or (d) heart was analyzed for cyclin G1 and calmodulin. (e) Serum cholesterol levels. Serum samples from mice that received rAAV9 with or without miRNA-binding sites were collected after 4 weeks and measured for total cholesterol, high-density lipoprotein (HDL) and low-density lipoprotein (LDL). miR, microRNA; nLacZ, β-galactosidase reporter transgene; rAAV, recombinant adeno-associated viruses.
Introducing miRNA-complementary sites into a highly expressed transgene might also divert the corresponding miRNA to the transgene mRNA from its natural targets, derepressing them. We tested whether the miRNA-binding sites in the transgene transcripts would deregulate the expression of the known endogenous target mRNAs of miR-122 or miR-1. We analyzed the expression of cyclin G1, a miR-122 target in liver (Figure 3b,c) and calmodulin, a miR-1 target in heart (Figure 3d). We detected no significant alteration in cyclin G1 or calmodulin expression. miR-122 regulates cholesterol biosynthesis in the liver, and agents that block miR-122 function produce readily detectable changes in serum cholesterol levels.27 We could detect no change in total cholesterol, high-density lipoprotein, or low-density lipoprotein levels in mice 4 weeks after transduction with either control rAAV9 or rAAV9 expressing a transgene bearing miR-122-binding sites (Figure 3e). We conclude that miRNA-mediated detargeting of rAAV expression has no detectable effect on endogenous miRNA expression or function.
Endogenous miRNAs silence rAAV transduction by site-specific cleavage of transgene mRNA
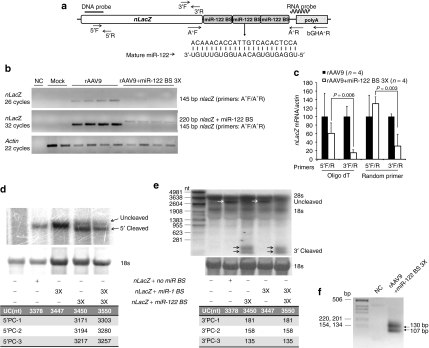
To determine how miRNAs suppress expression of transgenes delivered by rAAV in vivo, we characterized the transgene mRNA in liver by conventional PCR (Figure 4b), quantitative reverse-transcription PCR (qRT-PCR) (Figure 4c), Northern hybridization (Figure 4d,e), and rapid amplification of 5′ complimentary DNA (cDNA) ends (5′ RACE; Figure 4f). When we used primers that amplify the region between the 3′ end of nLacZ (A+F primer) and the 5′ end of the poly(A) signal (A+R primer), an amplicon that spans the miRNA-biding sites, we detected a 145 basepair (bp) product after 26 cycles of amplification for the samples that received control rAAV. An additional six cycles of amplification were required to detect a weak 220 bp band for the samples transduced by rAAV containing three miR-122-binding sites. These data are consistent with low levels of intact nLacZ mRNA (Figure 4a,b).
Figure 4.
Molecular characterization of transgene mRNAs with or without miRNA-binding sites. (a) Locations of the probes and primers, the sequences of mature miR-122 and its perfectly complementary binding site in the transgene mRNA are presented. (b) Total cellular RNA from liver was analyzed either by conventional reverse-transcription PCR (RT-PCR) by using primers that span a region between the 3′ end of nLacZ and the 5′ end of poly(A) signal (c) or by quantitative RT-PCR; data are presented as relative nLacZ mRNA levels normalized to β-actin. (d) For the northern blot analysis of nLacZ mRNA, 18S RNA served as a loading control, and the blots were hybridized with either a transgene DNA (e) or RNA probe. (f) In addition, poly(A) bearing mRNA from the liver of an animal received rAAV containing three miR-1- and three miR-122-binding sites was analyzed by 5′ RACE; the PCR product was resolved on an ethidium bromide-stained agarose gel. miR, microRNA; nLacZ, β-galactosidase reporter transgene; rAAV, recombinant adeno-associated viruses.
To quantitatively assess the extent of the miRNA-directed repression of the transgene transcripts, we performed qRT-PCR using either oligo(dT) or random hexamer primers for reverse-transcription and PCR primer pairs that span either a 5′ (nLacZ5′F/5′R), or 3′ (nLacZ 3′F/3′R) region of the nLacZ coding sequence (Figure 4a). We examined the levels of nLacZ mRNA with intact 5′ and 3′ ends in total liver RNA extracted from four animals that received the control rAAV9CBnLacZ and four that received rAAV9CBnLacZ containing three miR-122-binding sites in the 3′ UTR. We observed reductions ranging from 3 ± 1 (random hexamer) to 7 ± 1 (oligo[dT])-fold in nLacZ mRNA with an intact 3′ end in the animals that had received rAAV9 containing miR-122-binding sites, relative to the control. In contrast, we detected little or no decrease in nLacZ mRNA with an intact 5′ end for the same samples using the 5′F/5′R primer pair (Figure 4c). Our results suggest that the primary mode of turnover of the mRNA that has been cleaved by a miRNA is 3′-to-5′ exonucleolytic degradation.
To further characterize the fate of the transgene mRNA targeted by miR-1 or miR-122, we performed Northern blot analyses. A transgene probe binding to the 5′ end of nLacZ mRNA detected a ~3.4 kb RNA in an animal injected with control rAAV9CbnLacZ, the expected size of the of the full-length nLacZ transcript; a slightly larger band was detected in the liver sample from a mouse treated with rAAV9CBnLacZ bearing three miR-1-binding sites (Figure 4a,d). In contrast to the single transcript detected for the rAAV9 expressing nLacZ bearing three miR-1-binding sites, two RNAs of different sizes were detected for the rAAV expressing nLacZ bearing three miR-122 sites (Figure 4d). The lengths of these transcripts suggest that the longer transcript likely represents the full-length mRNA, whereas the shorter, more abundant transcript corresponds to 5′ fragments of nLacZ RNA cleaved by miR-122 at the corresponding miR-122-binding sites in the 3′ UTR (Figure 4d).
To confirm this finding, we repeated the Northern analysis using an RNA probe spanning a portion of 3′ UTR of the transgene mRNA. In addition to detecting full-length nLacZ transcripts in the samples transduced by rAAV9 lacking miRNA-binding sites, two closely migrating species smaller than the 281 nucleotide RNA marker were detected. The size of these fragments is consistent with miRNA-directed 3′ cleavage products of the nLacZ mRNA (Figure 4e). These two 3′ cleavage products were also detected by gel electrophoresis of the product from the 5′ RACE experiment described below (Figure 4f).
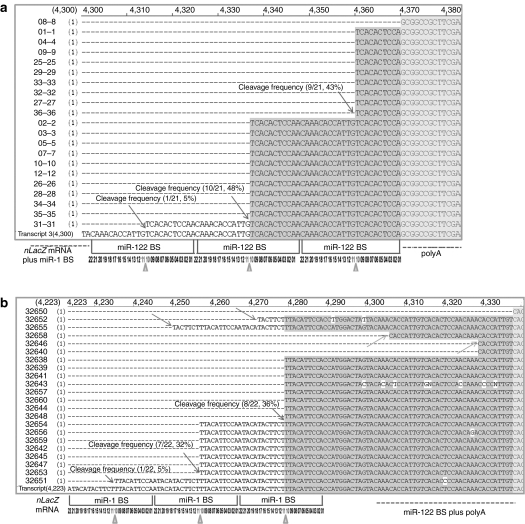
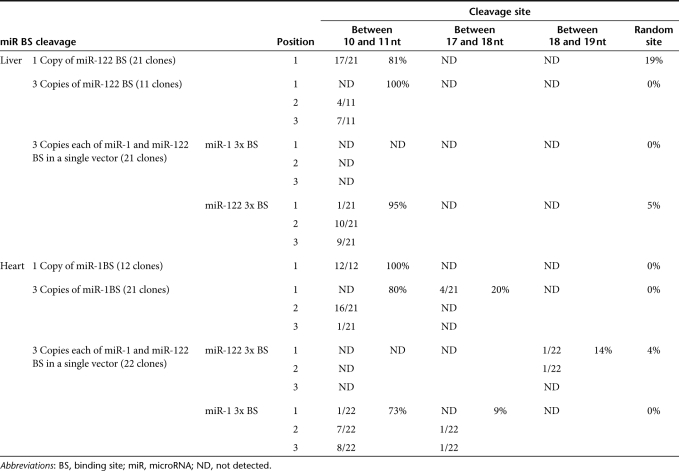
To determine whether such target cleavage occurs in vivo when the nLacZ transcript contained miR-1 or miR-122-binding sites, we performed rapid amplification of 5′ cDNA ends (5′ RACE). Figure 5 presents the sequences of 21 clones recovered using 5′ RACE from liver RNA (Figure 5a) and 22 clones isolated from heart RNA (Figure 5b) from the animals injected with rAAV9 in which the nLacZ 3′ UTR contained three miR-1 and three miR-122-binding sites. In liver, we detected the sequence signatures for miR-122-directed cleavage of the transgene mRNA at each miR-122-binding site: 5% for the first binding site, 48% for the second binding site, and 43% for the third binding site. All 5′ ends mapped to the phosphate that lies between the target nucleotides that pair with positions 10 and 11 of the sequence perfectly complementary to miR-122, the precise site cleaved by small RNAs bound to Argonaute proteins in all eukaryotes24 (Figure 5a). Similar results were obtained in the heart for the miR-1 sites (Figure 5b).
Figure 5.
Alignment of sequences spanning the miRNA-binding sites and poly(A) signal regions recovered by 5′ RACE. Poly(A)-containing mRNA was isolated from the (a) liver and (b) heart of an animal injected with rAAV9CBnLacZ-(miR-1BS)3–(miR-122BS)3. Twenty-one liver-derived and 22 heart-derived clones were sequenced. The putative cleavage sites in each clone are identified by arrows; the frequencies of miRNA-directed, site-specific cleavage for each miRNA-binding site are reported; triangles point to the positions of the expected miRNA-directed cleavage sites (a,b). miRNA, microRNA, nLacZ, β-galactosidase reporter transgene; rAAV, recombinant adeno-associated viruses.
Table 1 presents an expanded 5′ RACE analysis for additional vector groups. We note that none of the 5′ RACE products sequenced corresponded to miR-1-directed site-specific cleavage in liver or miR-122-directed site-specific cleavage in heart (Table 1). Although no cleavage was detected within miR-1-binding sites in the liver, some clones from heart were cleaved within the miR-122-binding sites, but not at the hallmark position for miRNA-directed cleavage.
Table 1. Summary of microRNA-guided transgene mRNA cleavage in mouse liver and heart.
Intravascularly delivered rAAV9 can be efficiently controlled by endogenous miRNAs
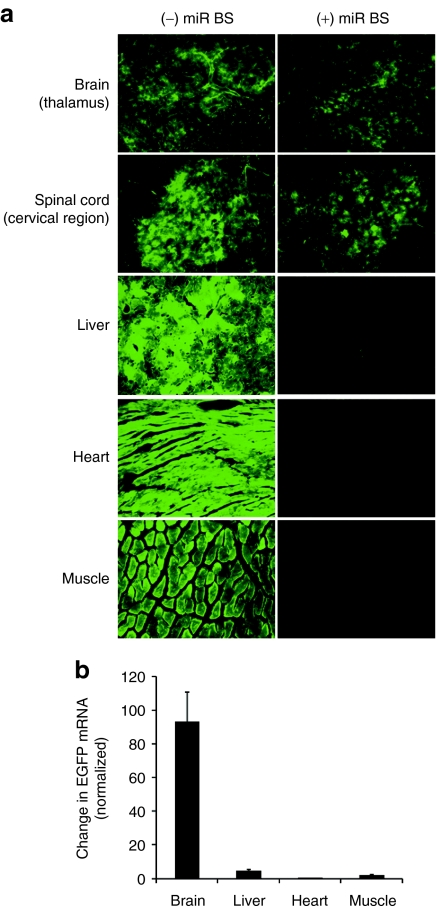
Recent studies suggest that high doses of self-complementary AAV9 (scAAV9), delivered intravascularly efficiently transduce the CNS in both mice and cats.11,12 However, intravenous administration of such vectors also transduces liver, heart, and pancreas. We added miRNA-1 and miRNA-122-binding sites into the scAAV9CB enhanced GFP (EGFP) vector genome and injected 10-week-old C57BL/6 male mice with 2 × 1014 GC/kg. After 3 weeks, we prepared 40 µm sections of brain and spinal cord and 8 µm sections of liver, heart, and skeletal muscle and examined EGFP protein expression. As reported previously,10,11,12,13 intravenously delivered scAAV9CBEGFP efficiently transduced the CNS; EGFP was readily detectable in the thalamus region of the brain and the cervical region of the spinal cord, but also in non-CNS tissues such as liver, heart, and muscle (Figure 6a). In contrast, transgene expression in those non-CNS tissues was reduced when miR-1 and miR-122-binding sites were included in the transgene; EGFP expression was unaltered in the CNS, where miR-1 and miR-122 are not present (Figure 6a). We also used qRT-PCR to measure the differential expression of the miRNA-repressed EGFP transgene in brain (41.2 ± 7.7%), liver (3.0 ± 0.5%), heart (0.4 ± 0.1%), and muscle (1.3 ± 0.4%), relative to the EGFP transgene lacking miRNA-binding sites (Figure 6b). To eliminate changes caused by variations in transduction efficiency between experiments, we normalized the data to the number of vector genomes detected in the experimental and control samples. Like the microscopic analyses of native EGFP expression, the qRT-PCR data show that the presence of miR-122- or miR-1-binding sites reduced transgene expression in liver (20-fold), heart (100-fold), and muscle (50-fold), but did not detectably alter transgene expression in brain.
Figure 6.
Endogenous miRNA-repressed, CNS-directed EGFP gene transfer by systemically delivered rAAV9. Ten-week-old male C57BL/6 mice were injected intravenously with scAAV9CBEGFP or scAAV9CBnLacZ-(miR-1BS)3–(miR-122BS)3 at a dose of 2 × 1014 genome copies per kg (GC/kg) body weight. The animals were necropsied 3 weeks later for whole body fixation by transcardiac perfusion. (a) Brain, spinal cord, liver, heart, and muscle were harvested for cryosectioning, immunofluorescent staining for EGFP (brain and cervical spinal cord), and fluorescence microscopy to detect EGFP. Total cellular DNA and RNA were extracted from brain, liver, heart and muscle to measure the amount of persistent vector genome by qPCR and EGFP mRNA by qRT-PCR. (b) For each tissue, the relative abundance of the EGFP mRNA containing miRNA-binding sites was compared to that of the EGFP mRNA lacking miRNA-binding sites. For each sample, mRNA abundance was normalized to the amount of vector genome detected in the tissue. EGFP, enhanced green fluorescent protein; miRNA, microRNA; nLacZ, β-galactosidase reporter transgene; qRT-PCR, quantitative reverse-transcription PCR; rAAV, recombinant adeno-associated viruses.
Discussion
Discovery that rAAV9 can efficiently cross the blood–brain barrier to deliver genes to the CNS represented a major advance toward the goal of developing gene therapy for CNS disorders.12,14 Here, we report that rAAV9 can be engineered so that endogenous miRNAs repress transgene expression outside the CNS. This strategy provides another step toward the development of systematically deliverable, CNS-targeted rAAV-based neurotherapeutics.
Recent advances in AAV vector development have substantially improved primate AAV vectors that achieve efficient and stable transvascular transduction in multiple tissues simultaneously.6,7 For example, systemic delivery of rAAV9 achieves extensive and robust transduction of astrocytes throughout the CNS of adult mice.11,12 This unique feature of rAAV9 promises to provide a protective therapy for the degenerating neurons associated with Parkinson's disease, Alzheimer's disease and amyotrophic lateral sclerosis, by expressing neurotrophic growth factors such as insulin-like growth factor, brain-derived neurotrophic factor or glial-derived neurotrophic factor in the transduced astrocytes.28 Yet, non-CNS expression derived from the peripheral tissues transduced by systemically delivered rAAV9 could lead to long-term circulation of super-physiological and potentially toxic levels of those growth factors.29,30,31 Thus, in some applications, CNS-directed gene transfer may also cause toxicity and immune responses if the transgene is expressed outside the CNS.
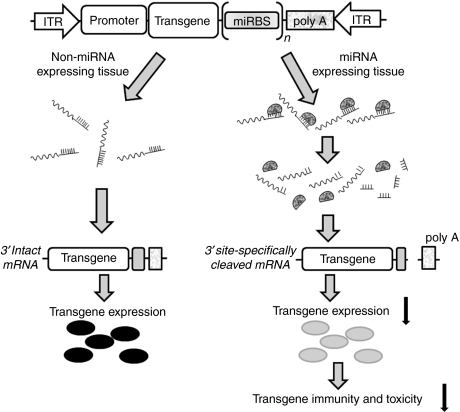
Achieving transgene expression in only the target tissues is vital for the clinical development of safe CNS gene delivery. Here, we have shown that endogenous miRNAs can be harnessed to restrict the tissue- and cell-type specificity of rAAV expression, as was initially shown for lentiviral vectors.16 Our data demonstrate that endogenous miRNAs can effectively repress transgene expression from rAAV. In both heart and liver, the miRNAs repressed transgene expression by directing endonucleolytic cleavage of the transgene mRNA (Figure 7). Importantly, miRNA regulation of rAAV expression did not perturb the expression or function of the corresponding endogenous miRNA, allowing transgene expression to be restricted to the CNS in mice. Our data suggest that a strategy that combines multiple binding sites for miRNAs expressed in the periphery but not the CNS may enable the development of safer, CNS-specific gene therapy vectors. In the future, validation of miRNA-mediated restriction of rAAV transgene expression in large animal models is required to pave the way for the development of intravascularly delivered rAAV-based gene therapy to treat disorders that affect the CNS.
Figure 7.
A molecular model for endogenous miRNA-regulated rAAV expression. miRNA, microRNA; rAAV, recombinant adeno-associated viruses.
Materials and Methods
Vector design, construction, and production. Perfectly complementary miRNA-binding sites were designed based on the annotated miR-1 and miR-122 sequences in miRBase32 and inserted into the BstBI restriction site in the 3′ UTR of the nLacZ expression cassette of the ubiquitously expressed pAAVCB nuclear-targeted β-galactosidase (nLacZ) plasmid using synthetic oligonucleotides (Figure 4a and Supplementary Table S1). This vector uses a hybrid cytomegalovirus enhancer/CB promoter cassette that is active in most cells and tissues. To express miR-122 and miR-1, pri-miR-122 and pri-miR-1 fragments were amplified by PCR from C57/B6 mouse genomic DNA (Supplementary Table S1) and inserted into the XbaI restriction site 3′ to a firefly luciferase cDNA in the pAAVCBFLuc plasmid. The identity of each pri-miRNA was verified by sequencing. AAV9 vectors used in this study were generated, purified, and titered as described.33
Cell culture and transfection. HEK-293 and HuH7 cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum and 100 mg/l of penicillin-streptomycin (Hyclone, South Logan, UT). Cells were maintained in a humidified incubator at 37 °C and 5% CO2. Plasmids were transiently transfected using Lipofectamine 2000 (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions.
Mouse studies. Male C57BL/6 mice (Charles River Laboratories, Wilmington, MA) were obtained and maintained and all animal procedures performed according to the guidelines of the Institutional Animal Care and Use Committee of the University of Massachusetts Medical School. To monitor lipid profiles of the study animals, serum samples were collected 4 weeks after rAAV9 injection and analyzed for total cholesterol, high-density lipoprotein and low-density lipoprotein on a COBAS C 111 analyzer (Roche Diagnostics, Lewes, UK). To evaluate endogenous miRNA-mediated, CNS-restricted EGFP gene transfer, 10-week-old male C57BL/6 mice were injected intravenously (tail vein) with AAV9CBnLacZ-[miR-122-binding site (BS)1], AAV9CBnLacZ-(miR-122BS)3, AAV9CBnLacZ-(miR-1BS)1, AAV9CBnLacZ-(miR-1BS)3, AAV9CBnLacZ-(miR-1BS)1-(miR-122BS)1, and AAV9CBnLacZ-(miR-1BS)3-(miR-122BS)3122BS)3, respectively, at 5 × 1013 GC/kg body weight) or scAAV9CBEGFP at 2 × 1014 GC/kg body weight). Animals receiving nLacZ vectors were necropsied 4 weeks later; 8 µm cryosections of liver, heart, and pancreas tissues were prepared for X-gal-histochemical staining. Animals that received EGFP vectors were necropsied 3 weeks later and fixed by transcardial perfusion with 4% (wt/vol) paraformaldehyde. Brain, spinal cord, liver, heart, and muscle were harvested for cryosectioning. Brain and cervical spinal cord tissue were stained as floating sections in a 12-well plate using rabbit anti-EGFP antibody (Invitrogen) diluted 1:500, followed by goat anti-rabbit secondary antibody (Invitrogen) diluted 1:400. Outside the CNS, EGFP expression was detected directly by fluorescence. EGFP and antibody fluorescence was recorded using a Nikon TE-2000S inverted microscope at ×10 magnification and an exposure time of 3 seconds for liver, heart, and muscle, and 5 seconds for thalamus (brain) and cervical spinal cord.
Vector genome quantification by qPCR. Genome DNA was extracted from the selected tissues using QIAamp DNA Mini Kit (Qiagen, West Sussex, UK), according to the manufacturer's instructions. Quantitative PCR were carried out in triplicate using 50 ng DNA and 0.3 µmol/l EGFP-specific primers (EGFP-F and EGFP-R) using GoTaq qPCR master mix (Promega, Madison, WI) in a StepOne Plus real-time PCR instrument (Applied Biosystems, Foster City, CA).
qRT-PCR analysis. RNA was extracted using Trizol (Invitrogen), according to the manufacturer's instructions. Total RNA (0.5–1.0 µg) was primed with random hexamers or oligo(dT) and reverse-transcribed with MultiScribe Reverse Transcriptase (Applied Biosystems). Quantitative PCR were performed in triplicate with 0.3 µmol/l gene-specific primer pairs (nLacZ5′F/5′R, nLacZ 3′F/3′R, cyclinG1F/R and EGFP-F/EGFP-R) using the GoTaq qPCR master mix in a StepOne Plus Real-time PCR device. The specificity of qRT-PCR products derived from the 5′ and 3′ ends of nLacZ mRNA was confirmed by gel electrophoresis.
Northern blot analysis. Total RNA was extracted from mouse liver and analyzed by Northern hybridization.34 To detect nLacZ mRNA, a 618 bp fragment of nLacZ cDNA was isolated by NcoI and PciI digestion of pAAVCBnLacZ and labeled with α-32P dCTP by random priming (Takara, Shiga, Japan). To detect 3′ fragments of the cleaved nLacZ mRNA, an 111 bp fragment of the poly(A) sequence in the vector genome was cloned into pCR4-TOPO (Invitrogen) for preparation of antisense RNA probe labeled with α-32P CTP during in vitro transcription using the Riboprobe System T7 kit (Promega). To detect miR-122, miR-26a, miR-22, and let-7 or U6 in total liver RNA, small RNAs were resolved by denaturing 15% polyacrylamide gels, transferred to Hybond N+ membrane (Amersham BioSciences, Pittsburgh, PA), and crosslinked with 254 nm light (Stratagene, La Jolla, CA). Synthetic oligonucleotides, 5′ end-labeled with γ-32P ATP using T4 polynucleotide kinase (New England Biolabs, Beverly, MA), were used as DNA probes (Supplementary Table S1) and hybridized in Church buffer (0.5 mol/l NaHPO4, pH 7.2, 1 mmol/l EDTA, 7% (w/v) sodium dodecyl sulphate) at 37 °C. Membranes were washed using 1× SSC (150 mM sodium chloride, 15 mM sodium citrate), 0.1% sodium dodecyl sulphate buffer, and then visualized using an FLA-5100 Imager (Fujifilm, Tokyo, Japan).
Western blot analysis. Proteins were extracted with radioimmunoprecipitation assay buffer [25 mmol/l Tris–HCl, pH 7.6, 150 mmol/l NaCl, 1% (vol/vol) NP-40, 1% (wt/vol) sodium deoxycholate, 0.1% (w/v) sodium dodecyl sulphate] containing a protease inhibitor mixture (Boston BP, Boston, MA). Protein concentration was determined using the Bradford method (Bio-Rad, Melville, NY). Protein samples, 50 µg each, were loaded onto 12% polyacrylamide gels, electrophoresed, and transferred to nitrocellulose membrane (Amersham BioSciences). Briefly, membranes were blocked with blocking buffer (LI-COR Biosciences, Lincoln, NE) at room temperature for 2 hours, followed by incubation with either anti-GAPDH (Millipore, Billerica, MA), anti-cyclin G1 (Santa Cruz Biotechnology, Santa Cruz, CA) or anti-calmodulin (Millipore) for 2 hours at room temperature. After three washes with PBS containing 0.1% (vol/vol) Tween-20, membranes were incubated with secondary antibodies conjugated to LI-COR IRDye for 1 hour at room temperature, and then antibodies detected using the Odyssey Imager (LI-COR).
β-Galactosidase assay. Proteins were extracted with radioimmunoprecipitation assay buffer and quantified as described above. Fifty micrograms of protein was used for each β-galactosidase assay using the Galacto-Star System (Applied Biosystems), according to the manufacturer's instructions.
5′ RACE. 5′ RACE was performed as described.35 The 5′ RACE Outer Primer and the nLacZ gene-specific primer bGHpolyAR (Supplementary Table S1) were used for the first round of nested PCR. The 5′ RACE Inner Primer and the nLacZ gene-specific primer nLacZpolyR, which is located near the stop codon of nLacZ cDNA, were used for the second round of nested PCR (Supplementary Table S1). PCR products were TOPO-cloned into pCR-4.0 (Invitrogen) and sequenced.
Statistical analysis. All results are reported as mean ± SD and compared between groups using the two-tailed Student's t-test.
SUPPLEMENTARY MATERIAL Figure S1. Quantification of β-galactosidase activities in liver tissue from animals that received rAAVnLacZ vectors with and without miRNA-binding sites. Adult male C58BL/6 mice were injected intravenously with 5 × 1013 GC/kg each of rAAV9CBnLacZ (no miRNA binding site), rAAVCB9nLacZ-miR-122BS (one miR-122-binding site) or rAAV9CBnLacZ-(miR-122BS)3 (three miR-122-binding site), rAAV9CBnLacZ-miR-1BS-miR-122BS (one miR-1 and one miR-122-binding site), rAAV9CBnLacZ-(miR-1BS)3-(miR-122BS)3 (three miR-1 and three miR-122-binding sites) and rAAV9CBnLacZ-(miR-1BS)3 (three miR-1-binding sites). The animals were necropsied 4 weeks after vector administration, and liver tissues were harvested for total proteins and β-Galactosidase assays. Percentages report the β-Galactosidase activity of each vector group compared to the control group, rAAV9CBnLacZ. Table S1. Oligonucleotide primers and probes used in the study.
Acknowledgments
We greatly appreciate the contributions of the Vector Core of Gene Therapy Center and Analytic Cores of Diabetes Center of the University of Massachusetts Medical School. This work was funded by an internal grant to G.G. from the University of Massachusetts Medical School. The contributions of C.M. and T.R.F. to this work were supported with a PO1grant from the National Institute of Health (DK 58327). G.G., P.D.Z., C.C.M., and T.R.F. are members of UMASS DERC (DK 32520). This publication was made possible by grant 5P30DK 32520 from National Institute of Diabetes and Digestive and Kidney Diseases.
Supplementary Material
Quantification of β-galactosidase activities in liver tissue from animals that received rAAVnLacZ vectors with and without miRNA-binding sites. Adult male C58BL/6 mice were injected intravenously with 5 × 1013 GC/kg each of rAAV9CBnLacZ (no miRNA binding site), rAAVCB9nLacZ-miR-122BS (one miR-122-binding site) or rAAV9CBnLacZ-(miR-122BS)3 (three miR-122-binding site), rAAV9CBnLacZ-miR-1BS-miR-122BS (one miR-1 and one miR-122-binding site), rAAV9CBnLacZ-(miR-1BS)3-(miR-122BS)3 (three miR-1 and three miR-122-binding sites) and rAAV9CBnLacZ-(miR-1BS)3 (three miR-1-binding sites). The animals were necropsied 4 weeks after vector administration, and liver tissues were harvested for total proteins and β-Galactosidase assays. Percentages report the β-Galactosidase activity of each vector group compared to the control group, rAAV9CBnLacZ.
Oligonucleotide primers and probes used in the study.
REFERENCES
- Kaplitt MG, Feigin A, Tang C, Fitzsimons HL, Mattis P, Lawlor PA, et al. Safety and tolerability of gene therapy with an adeno-associated virus (AAV) borne GAD gene for Parkinson's disease: an open label, phase I trial. Lancet. 2007;369:2097–2105. doi: 10.1016/S0140-6736(07)60982-9. [DOI] [PubMed] [Google Scholar]
- Muramatsu S, Fujimoto K, Kato S, Mizukami H, Asari S, Ikeguchi K, et al. A phase I study of aromatic L-amino acid decarboxylase gene therapy for Parkinson's disease. Mol Ther. 2010;18:1731–1735. doi: 10.1038/mt.2010.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Federici T., and, Boulis NM. Invited review: festschrift edition of neurosurgery peripheral nervous system as a conduit for delivering therapies for diabetic neuropathy, amyotrophic lateral sclerosis, and nerve regeneration. Neurosurgery. 2009;65 4 Suppl:A87–A92. doi: 10.1227/01.NEU.0000335653.52938.F2. [DOI] [PubMed] [Google Scholar]
- Bjorklund T., and, Kordower JH. Gene therapy for Parkinson's disease. Mov Disord. 2010;25 Suppl 1:S161–S173. doi: 10.1002/mds.22785. [DOI] [PubMed] [Google Scholar]
- Vandenberghe LH, Wilson JM., and, Gao G. Tailoring the AAV vector capsid for gene therapy. Gene Ther. 2009;16:311–319. doi: 10.1038/gt.2008.170. [DOI] [PubMed] [Google Scholar]
- Wang Z, Zhu T, Qiao C, Zhou L, Wang B, Zhang J, et al. Adeno-associated virus serotype 8 efficiently delivers genes to muscle and heart. Nat Biotechnol. 2005;23:321–328. doi: 10.1038/nbt1073. [DOI] [PubMed] [Google Scholar]
- Zincarelli C, Soltys S, Rengo G., and, Rabinowitz JE. Analysis of AAV serotypes 1-9 mediated gene expression and tropism in mice after systemic injection. Mol Ther. 2008;16:1073–1080. doi: 10.1038/mt.2008.76. [DOI] [PubMed] [Google Scholar]
- Pacak CA, Mah CS, Thattaliyath BD, Conlon TJ, Lewis MA, Cloutier DE, et al. Recombinant adeno-associated virus serotype 9 leads to preferential cardiac transduction in vivo. Circ Res. 2006;99:e3–e9. doi: 10.1161/01.RES.0000237661.18885.f6. [DOI] [PubMed] [Google Scholar]
- Foust KD, Poirier A, Pacak CA, Mandel RJ., and, Flotte TR. Neonatal intraperitoneal or intravenous injections of recombinant adeno-associated virus type 8 transduce dorsal root ganglia and lower motor neurons. Hum Gene Ther. 2008;19:61–70. doi: 10.1089/hum.2007.093. [DOI] [PubMed] [Google Scholar]
- Chen YH, Chang M., and, Davidson BL. Molecular signatures of disease brain endothelia provide new sites for CNS-directed enzyme therapy. Nat Med. 2009;15:1215–1218. doi: 10.1038/nm.2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duque S, Joussemet B, Riviere C, Marais T, Dubreil L, Douar AM, et al. Intravenous administration of self-complementary AAV9 enables transgene delivery to adult motor neurons. Mol Ther. 2009;17:1187–1196. doi: 10.1038/mt.2009.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foust KD, Nurre E, Montgomery CL, Hernandez A, Chan CM., and, Kaspar BK. Intravascular AAV9 preferentially targets neonatal neurons and adult astrocytes. Nat Biotechnol. 2009;27:59–65. doi: 10.1038/nbt.1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray SJ, Blake BL, Criswell HE, Nicolson SC, Samulski RJ, McCown TJ, et al. Directed evolution of a novel adeno-associated virus (AAV) vector that crosses the seizure-compromised blood–brain barrier (BBB) Mol Ther. 2010;18:570–578. doi: 10.1038/mt.2009.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foust KD, Wang X, McGovern VL, Braun L, Bevan AK, Haidet AM, et al. Rescue of the spinal muscular atrophy phenotype in a mouse model by early postnatal delivery of SMN. Nat Biotechnol. 2010;28:271–274. doi: 10.1038/nbt.1610. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown BD, Venneri MA, Zingale A, Sergi Sergi L., and, Naldini L. Endogenous microRNA regulation suppresses transgene expression in hematopoietic lineages and enables stable gene transfer. Nat Med. 2006;12:585–591. doi: 10.1038/nm1398. [DOI] [PubMed] [Google Scholar]
- Wolff LJ, Wolff JA., and, Sebestyén MG. Effect of tissue-specific promoters and microRNA recognition elements on stability of transgene expression after hydrodynamic naked plasmid DNA delivery. Hum Gene Ther. 2009;20:374–388. doi: 10.1089/hum.2008.088. [DOI] [PubMed] [Google Scholar]
- Suzuki T, Sakurai F, Nakamura S, Kouyama E, Kawabata K, Kondoh M, et al. miR-122a-regulated expression of a suicide gene prevents hepatotoxicity without altering antitumor effects in suicide gene therapy. Mol Ther. 2008;16:1719–1726. doi: 10.1038/mt.2008.159. [DOI] [PubMed] [Google Scholar]
- Edge RE, Falls TJ, Brown CW, Lichty BD, Atkins H., and, Bell JC. A let-7 MicroRNA-sensitive vesicular stomatitis virus demonstrates tumor-specific replication. Mol Ther. 2008;16:1437–1443. doi: 10.1038/mt.2008.130. [DOI] [PubMed] [Google Scholar]
- Barnes D, Kunitomi M, Vignuzzi M, Saksela K., and, Andino R. Harnessing endogenous miRNAs to control virus tissue tropism as a strategy for developing attenuated virus vaccines. Cell Host Microbe. 2008;4:239–248. doi: 10.1016/j.chom.2008.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown BD., and, Naldini L. Exploiting and antagonizing microRNA regulation for therapeutic and experimental applications. Nat Rev Genet. 2009;10:578–585. doi: 10.1038/nrg2628. [DOI] [PubMed] [Google Scholar]
- Lagos-Quintana M, Rauhut R, Yalcin A, Meyer J, Lendeckel W., and, Tuschl T. Identification of tissue-specific microRNAs from mouse. Curr Biol. 2002;12:735–739. doi: 10.1016/s0960-9822(02)00809-6. [DOI] [PubMed] [Google Scholar]
- Chang J, Nicolas E, Marks D, Sander C, Lerro A, Buendia MA, et al. miR-122, a mammalian liver-specific microRNA, is processed from hcr mRNA and may downregulate the high affinity cationic amino acid transporter CAT-1. RNA Biol. 2004;1:106–113. doi: 10.4161/rna.1.2.1066. [DOI] [PubMed] [Google Scholar]
- Elbashir SM, Lendeckel W., and, Tuschl T. RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev. 2001;15:188–200. doi: 10.1101/gad.862301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee EJ, Gusev Y, Jiang J, Nuovo GJ, Lerner MR, Frankel WL, et al. Expression profiling identifies microRNA signature in pancreatic cancer. Int J Cancer. 2007;120:1046–1054. doi: 10.1002/ijc.22394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ameres SL, Horwich MD, Hung JH, Xu J, Ghildiyal M, Weng Z, et al. Target RNA-directed trimming and tailing of small silencing RNAs. Science. 2010;328:1534–1539. doi: 10.1126/science.1187058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krützfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M, et al. Silencing of microRNAs in vivo with ‘antagomirs'. Nature. 2005;438:685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- Hester ME, Foust KD, Kaspar RW., and, Kaspar BK. AAV as a gene transfer vector for the treatment of neurological disorders: novel treatment thoughts for ALS. Curr Gene Ther. 2009;9:428–433. doi: 10.2174/156652309789753383. [DOI] [PubMed] [Google Scholar]
- Bensimon G, Lacomblez L., and, Meininger V. A controlled trial of riluzole in amyotrophic lateral sclerosis. ALS/Riluzole Study Group. N Engl J Med. 1994;330:585–591. doi: 10.1056/NEJM199403033300901. [DOI] [PubMed] [Google Scholar]
- Kordower JH, Palfi S, Chen EY, Ma SY, Sendera T, Cochran EJ, et al. Clinicopathological findings following intraventricular glial-derived neurotrophic factor treatment in a patient with Parkinson's disease. Ann Neurol. 1999;46:419–424. doi: 10.1002/1531-8249(199909)46:3<419::aid-ana21>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- Su X, Kells AP, Huang EJ, Lee HS, Hadaczek P, Beyer J, et al. Safety evaluation of AAV2-GDNF gene transfer into the dopaminergic nigrostriatal pathway in aged and parkinsonian rhesus monkeys. Hum Gene Ther. 2009;20:1627–1640. doi: 10.1089/hum.2009.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones S. miRBase: the microRNA sequence database. Methods Mol Biol. 2006;342:129–138. doi: 10.1385/1-59745-123-1:129. [DOI] [PubMed] [Google Scholar]
- Gao G, Alvira MR, Somanathan S, Lu Y, Vandenberghe LH, Rux JJ, et al. Adeno-associated viruses undergo substantial evolution in primates during natural infections. Proc Natl Acad Sci USA. 2003;100:6081–6086. doi: 10.1073/pnas.0937739100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu ZF, Qi WQ, Ouyang XZ, Yeung E., and, Chye ML. A proteinase inhibitor II of Solanum americanum is expressed in phloem. Plant Mol Biol. 2001;47:727–738. doi: 10.1023/a:1013623628857. [DOI] [PubMed] [Google Scholar]
- Guo HS, Xie Q, Fei JF., and, Chua NH. MicroRNA directs mRNA cleavage of the transcription factor NAC1 to downregulate auxin signals for arabidopsis lateral root development. Plant Cell. 2005;17:1376–1386. doi: 10.1105/tpc.105.030841. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Quantification of β-galactosidase activities in liver tissue from animals that received rAAVnLacZ vectors with and without miRNA-binding sites. Adult male C58BL/6 mice were injected intravenously with 5 × 1013 GC/kg each of rAAV9CBnLacZ (no miRNA binding site), rAAVCB9nLacZ-miR-122BS (one miR-122-binding site) or rAAV9CBnLacZ-(miR-122BS)3 (three miR-122-binding site), rAAV9CBnLacZ-miR-1BS-miR-122BS (one miR-1 and one miR-122-binding site), rAAV9CBnLacZ-(miR-1BS)3-(miR-122BS)3 (three miR-1 and three miR-122-binding sites) and rAAV9CBnLacZ-(miR-1BS)3 (three miR-1-binding sites). The animals were necropsied 4 weeks after vector administration, and liver tissues were harvested for total proteins and β-Galactosidase assays. Percentages report the β-Galactosidase activity of each vector group compared to the control group, rAAV9CBnLacZ.
Oligonucleotide primers and probes used in the study.