Abstract
Background:
Activating mutation of KRAS and BRAF are focused on as potential prognostic and predictive biomarkers in patients with colorectal cancer (CRC) treated with anti-EGFR therapies. This study investigated the clinicopathological features and prognostic impact of KRAS/BRAF mutation in advanced and recurrent CRC patients.
Method:
Patients with advanced and recurrent CRC treated with systemic chemotherapy (n=229) were analysed for KRAS/BRAF genotypes by cycleave PCR. Prognostic factors associated with survival were identified by univariate and multivariate analyses using the Cox proportional hazards model.
Results:
KRAS and BRAF mutations were present in 34.5% and 6.5% of patients, respectively. BRAF mutated tumours were more likely to develop on the right of the colon, and to be of the poorly differentiated adenocarcinoma or mucinous carcinoma, and peritoneal metastasis. The median overall survival (OS) for BRAF mutation-positive and KRAS 13 mutation-positive patients was 11.0 and 27.7 months, respectively, which was significantly worse than that for patients with wild-type (wt) KRAS and BRAF (40.6 months) (BRAF; HR=4.25, P<0.001, KRAS13; HR=2.03, P=0.024). After adjustment for significant features by multivariate Cox regression analysis, BRAF mutation was associated with poor OS (HR=4.23, P=0.019).
Conclusion:
Presence of mutated BRAF is one of the most powerful prognostic factors for advanced and recurrent CRC. The KRAS13 mutation showed a trend towards poor OS in patients with advanced and recurrent CRC.
Keywords: BRAF , KRAS , prognostic marker, colorectal cancer, chemotherapy
Although the epidermal growth factor receptor (EGFR) has important roles in cell differentiation and proliferation in normal cells, activation of EGFR signalling is frequently observed in colorectal cancer (CRC) cells, resulting in cell proliferation, migration and metastasis, evasion of apoptosis, or angiogenesis (Fang and Richardson, 2005). Indeed, ∼35% of CRC tissues carry a mutation in codons 12 or 13 of KRAS that leads to the constitutive activation of downstream pathways, including the Ras/Raf/MAP/MEK/ERK and/or PTEN/PI3K/Akt pathways (Kinzler and Vogelstein, 1999; Wan et al, 2004; Benvenuti et al, 2007; Di Nicolantonio et al, 2008; Souglakos et al, 2009). BRAF is a downstream molecule of KRAS. Although more than 40 somatic mutations in the BRAF kinase domain have been described, the most common mutation across various cancers is the classic GTG → GAG substitution at the position 1799 of exon 15, which results in the V600E amino acid change, and the subsequent constitutive activation of the EGFR signalling pathway. Recent studies from Western countries have suggested that BRAF mutations occur in 10–20% of patients with sporadic disease (Jass, 2007; Benvenuti et al, 2007; Di Nicolantonio et al, 2008; Souglakos et al, 2009; Fariña-Sarasqueta et al, 2010), whereas other reports have revealed that tumours harbouring BRAF mutations have different clinical and histopathological features compared with tumours that harbour KRAS mutations (Kim et al, 2006; Deng et al, 2008; Zlobec et al, 2010). However, the frequency and clinicopathological features of KRAS/BRAF mutation in Japanese CRC patients remain unknown.
Information on KRAS/BRAF genotype is extremely useful in systemic chemotherapy for advanced and recurrent CRC patients, not just for predicting the therapeutic efficiency of anti-EGFR therapy, but also for identifying patients with poor prognoses. Therefore, both KRAS and BRAF are currently being focused on as potential prognostic and predictive biomarkers in patients with metastatic disease treated with anti-EGFR therapies, such as panitumumab and cetuximab (Karapetis et al, 2008; Bokemeyer et al, 2009; Tol et al, 2009; Van Cutsem et al, 2009). A number of retrospective analyses have revealed that patients with KRAS mutations do not benefit from cetuximab treatment, suggesting that KRAS genotype is a useful predictive marker for cetuximab therapy in CRC (Karapetis et al, 2008; Bokemeyer et al, 2009; Van Cutsem et al, 2009). It has also been reported that wild-type (wt) BRAF is required for a successful response to panitumumab or cetuximab therapies in metastatic CRC (Di Nicolantonio et al, 2008; Laurent-Puig et al, 2009; Souglakos et al, 2009; De Roock et al, 2010). In contrast, the prognostic relevance of KRAS genotype in CRC has been controversial despite a number of multi-institutional investigations dating from the 1990s (Andreyev et al, 1998; French et al, 2008; Kakar et al, 2008; Ogino et al, 2009; Roth et al, 2010). Although few studies have investigated the impact of KRAS12 and KRAS13 mutations on CRC prognosis, a series of recent studies have highlighted the potential adverse prognostic impact of BRAF mutations, using both patients with stage II and III disease and patients across all disease stages (Ogino et al, 2009; Fariña-Sarasqueta et al, 2010). Although Tol et al (2009) analysed BRAF genotypes in 520 metastatic CRC patients, all the patients were treated with chemotherapy plus bevacizumab with or without cetuximab. Furthermore, BRAF genotypes were analysed in a large subgroup of 845 metastatic CRC treated with FOLFIRI and FOLFOX chemotherapy with or without cetuximab as the first-line treatment in the CRYSTAL and OPUS studies, respectively (Bokemeyer et al, 2010). Thus, although the prognostic value of BRAF has been analysed in CRC patients treated with specific chemotherapy regimens, it remains unclear what impact the KRAS12, KRAS13, and BRAF mutations have on clinical outcomes of all patients with advanced or recurrent CRC treated with systemic treatments.
We have previously introduced the cycleave PCR technique as applicable to the routine screening of KRAS/BRAF mutations in CRC from pathological specimens, such as surgical and biopsy specimens (Yokota et al, 2010). Cycleave PCR utilises chimeric DNA-RNA-DNA probes labelled with a fluorescent dye and quencher, and the accuracy of cycleave PCR in detecting KRAS/BRAF mutations has been confirmed by assessment of the concordance between cycleave PCR and reverse transcriptase PCR-coupled direct sequencing (Yatabe et al, 2006; Yokota et al, 2010).
The aim of this study was to evaluate the KRAS/BRAF genotypes of advanced and recurrent CRC patients and to assess the effects of these genotypes on clinical outcome. To this end, we analysed the frequencies of the KRAS12, KRAS13 and BRAF mutations, and correlated these results with the clinicopathological features of 229 Japanese CRC patients.
Patients and methods
Patients and tissues
Analysis of the genes encoding KRAS and BRAF was performed on surgically resected or biopsied specimens from CRC patients at our institution from 2002 to 2010. Hematoxylin and eosin (H and E)-stained slides were retrospectively collected and histologic subtypes were reviewed by an experienced gastrointestinal pathologist. Clinicopathological and survival analyses were subsequently performed on all patients with advanced and recurrent CRC who underwent systemic chemotherapy. Clinical data, including patient age at diagnosis, tumour location, and metastatic sites, were retrieved from patient records. Right-sided cancers included tumours from the caecum to transverse colon, left-sided included tumours from the splenic flexure to the rectosigmoid junction. Specimens used for KRAS/BRAF genotyping were either frozen or paraffin embedded tissues. For the KRAS/BRAF genotyping, appropriate approvals were obtained from the institutional review committee and written informed consent was obtained from all patients.
DNA extraction
DNA was extracted from surgical or biopsy specimens. Briefly, tumour cell-rich areas in H and E-stained sections were marked under a microscope, and tissues scratched from the same areas were sequentially deparaffinised and unstained. Recovered tissues were incubated in 1X PCR buffer containing 100 μg ml−1 proteinase K for 1 h at 54 °C. After heat inactivation at 95 °C for 3 min, samples were used directly as template DNA for PCR assay.
KRAS/BRAF genotyping by cycleave PCR
To detect point mutations at KRAS codons 12, 13 and 61, we used the cycleave PCR technique (Yatabe et al, 2006; Sakamoto et al, 2007; Yokota et al, 2010). Each chimeric DNA-RNA-DNA probe was labelled with a fluorescent dye and quencher at each end that targeted the G12D, G12V, G12R, G12C, G12S, or G12A mutations in codon 12, the G13D or G13C mutations in codon 13, or the G61H, G61L, G61E, or G61K mutations in codon 61 of KRAS. We also designed probes that targeted the V600E mutation in BRAF. The PCR reactions were performed using a cycleave PCR core kit (TAKARA, Co. Ltd, Ohtsu, Japan). Fluorescent signals were quantified using the Smart Cycler system (SC-100; Cepheid, Sunnyvale, CA, USA).
Statistical analysis
The χ2, Fischer's exact tests and Student's t-tests were used to analyse the relationship between variables using SYSTAT software (SYSTAT Software Inc., Richmond, CA, USA). The KRAS wt/BRAF wt (wild/wild), KRAS12 mutant (G12X), KRAS13 mutant (G13X), and BRAF mutant (V600E) groups were analysed separately. Overall survival (OS) was calculated from the starting date of the first-line chemotherapy until death from any cause, or censored at last follow-up visit. Survival data were analysed using the Kaplan–Meier product-limit method. Comparison of survival curves was carried out using the log-rank test. We first performed a univariate comparison of survival functions for factors that could potentially affect the survival time using the log-rank test, and then a multivariate analysis using the Cox proportional hazards model. P-values <0.05 were considered statistically significant, and all P-values represent two-sided significance tests.
Results
Frequency of KRAS and BRAF gene mutations in CRC patients
According to our previous investigation on the spectrum of KRAS genotypes in our database of CRC cases, the most frequent mutations at KRAS codon 12 were the G12D, G12V, G12R, G12C, G12S and G12A mutations, which accounted for more than 95% of the codon 12 mutations. Similarly, the G13D and G13C mutations at codon 13, and the G61H, G61L, G61E, and G61K mutations at codon 61 were also found to be the most common at each site (Yokota et al, 2010). All the KRAS mutations we located have been previously described as oncogenically active and were present in the COSMIC (catalogue of somatic mutations in cancer) database (Sanger Institute, Cambridge, UK). Therefore, a series of specific probes targeting the common mutations in KRAS codons 12, 13 and 61 were designed for subsequent analysis of KRAS mutation frequency in our population of CRC patients. Because the most common mutation in BRAF is a valine to glutamate transition at position 600 of the protein (V600E), we designed probes targeting the V600E mutation in BRAF.
We initially analysed the KRAS genotypes of 349 CRC patients at our institution for which pathological specimens were available by cycleave PCR. The KRAS mutations were present in 35.7% (n=126) of patients tested, including 24.4% (n=86) that exhibited codon 12 mutations and 11.3% (n=40) that exhibited codon 13 mutations. However, only 4.7% (n=15) of the patients tested were positive for the BRAF V600E mutation (n=319). None of the KRAS-mutated samples carried a concomitant BRAF mutation. Approximately 2–3% of the surgical specimens could not be evaluated by cycleave PCR, probably due to over-fixation by formalin, as we reported previously (Yokota et al, 2010).
For the subsequent clinicopathological and survival analysis, we picked out 229 patients with advanced and recurrent CRC for which we could access complete clinicopathological information. The KRAS mutations were present in 34.5% (n=79) of advanced and recurrent CRC patients, including 23.1% (n=53) with codon 12 mutations and 11.4% (n=26) with codon 13 mutations. The BRAF mutation was found in 6.6% (n=15) of this population (Table 1).
Table 1. Spectrum of KRAS/BRAF mutations in CRC.
|
KRAS
|
||||
|---|---|---|---|---|
| BRAF | Wild type | G12 | G13 | 61 |
| Wild type | 135 | 53 | 26 | 0 |
| V600E | 15 | 0 | 0 | 0 |
Abbreviation: CRC=colorectal cancer.
n=229.
Association of BRAF/KRAS mutations with clinicopathological features
We then correlated the KRAS and BRAF genotypes with clinicopathological features of CRC, including primary tumour location, histological findings, and sites of metastases. We categorised the population into four subtypes; those with wt KRAS and BRAF (wild/wild), KRAS12 mutations (G12X), KRAS13 mutations (G13X), and BRAF mutations (V600E).
For disease status, recurrent disease was more frequent in the KRAS12 and KRAS13 mutant groups than in the wild/wild group. There was no association between KRAS/BRAF genotype and age, gender or PS. Primary tumours were located at the rectum in almost half of the wild/wild and G12X populations. However, right-side tumour location was more frequent (60%) in patients with BRAF mutation in all subtypes (P=0.0391) (Table 2). Furthermore, 46.2% (12 out of 26) of the primary tumours with KRAS13 mutations were located on the right side whereas the frequencies of right-side location were 20.7% (28 out of 135) and 26.4% (14 out of 53), for the wild/wild and G12X groups, respectively (Table 2). The BRAF and KRAS13 mutations were present in 14.3% (9 out of 63) and 19.0% (17 out of 63) of right-sided CRC, respectively. These results suggested that the BRAF and KRAS codon 13 mutations were associated with a right-sided tumour location.
Table 2. Association of BRAF and KRAS mutational status with clinicopathological features in colorectal cancer.
| KRAS/BRAF status | Wild/wild |
KRAS mutant
|
BRAF mutant | ||||
|---|---|---|---|---|---|---|---|
| Clinicopathological features | n=135 | G12X n=53 | G13X n=26 | Total (G12X+G13X) n=79 | V600E n=15 | *P-value | Overall n=229 |
| Age at diagnosis (median) | 62 (27–83) | 62 (40–85) | 68 (41–79) | 63 (40–85) | 62 (30–80) | ||
| Gender | |||||||
| Female | 47 (34.8%) | 27 (50.9%) | 13 (50.0%) | 40 (50.6%) | 8 (53.3%) | 0.1082 | 95 |
| Male | 88 (65.2%) | 26 (49.1%) | 13 (50.0%) | 39 (49.4%) | 7 (46.7%) | 134 | |
| ECOG PS | |||||||
| 0–1 | 115 (85.2%) | 46 (86.8%) | 22 (84.6%) | 68 (86.1%) | 13 (86.7%) | 0.7898 | 196 |
| >2 | 9 (6.7%) | 4 (7.5%) | 3 (11.5%) | 7 (8.9%) | 2 (13.3%) | 18 | |
| Unknown | 11 (8.1%) | 3 (5.7%) | 1 (3.8%) | 4 (5.1%) | 0 (0.0%) | 15 | |
| Tumour location | |||||||
| Right sided | 28 (20.7%) | 14 (26.4%) | 12 (46.2%) | 26 (32.9%) | 9 (60.0%) | 0.0391 | 63 |
| Left sided | 41 (30.4%) | 13 (24.5%) | 3 (11.5%) | 16 (20.3%) | 3 (20.0%) | 60 | |
| Rectum | 64 (47.4%) | 25 (47.2%) | 11 (42.3%) | 36 (45.6%) | 3 (20.0%) | 103 | |
| Other | 2 (1.5%) | 1 (1.9%) | 0 (0.0%) | 1 (1.3%) | 0 (0.0%) | 3 | |
| Disease status | |||||||
| Advanced | 82 (60.7%) | 26 (49.1%) | 11 (42.3%) | 37 (46.8%) | 9 (60.0%) | 0.2269 | 128 |
| Recurrence | 53 (39.3%) | 27 (50.9 %) | 15 (57.7%) | 42 (53.2%) | 6 (40.0%) | 101 | |
| Histological subtype | |||||||
| Well | 28 (20.7%) | 8 (15.1%) | 7 (26.9%) | 15 (19.0%) | 1 (6.7%) | <0.0001 | 44 |
| Mod | 91 (67.4%) | 37 (69.8%) | 18 (69.2%) | 55 (69.6%) | 5 (33.3%) | 151 | |
| por/sig/muc | 10 (7.4%) | 5 (9.4%) | 1 (3.8%) | 6 (7.6%) | 9 (60.0%) | 25 | |
| Other | 1 (0.7%) | 0 (0.0%) | 0 (0.0%) | 0 (0%) | 0 (0.0%) | 1 | |
| Unknown | 5 (3.7%) | 3 (5.7%) | 0 (0.0%) | 3 (3.8%) | 0 (0.0%) | 8 | |
| Metastatic sites | |||||||
| Liver | 90 (66.7%) | 31 (58.5%) | 15 (57.7 %) | 46 (58.2%) | 10 (66.7%) | 0.6595 | 146 |
| Peritoneum | 30 (22.2%) | 11 (20.8%) | 4 (15.4%) | 15 (20.0%) | 9 (60.0%) | 0.0062 | 54 |
| Lung | 42 (31.1%) | 21 (39.6%) | 10 (38.5%) | 31 (39.2%) | 5 (33.3%) | 0.6867 | 78 |
| CNS | 1 (0.7%) | 0 (0.0%) | 1 (3.8%) | 1 (1.3%) | 0 (0.0%) | 0.3503 | 2 |
| Bone | 9 (6.7%) | 3 (5.7%) | 2 (7.7%) | 5 (6.3%) | 2 (13.3%) | 0.7736 | 16 |
| Number of metastatic sites | |||||||
| >2 | 64 (47.4%) | 23 (43.4%) | 14 (53.8%) | 37 (46.8%) | 10 (66.7%) | 0.4078 | 111 |
| <1 | 71 (52.6%) | 30 (56.6%) | 12 (46.2%) | 42 (53.2%) | 5 (33.3%) | 118 | |
| WBC | |||||||
| WBC>10000 | 9 (6.7%) | 4 (7.5%) | 2 (7.7%) | 6 (7.6%) | 0 (0.0%) | 0.7622 | 15 |
| WNL | 100 (74.1%) | 38 (71.7%) | 20 (76.9%) | 58 (73.4%) | 14 (93.3%) | 172 | |
| Unknown | 26 (19.3%) | 11 (20.8%) | 4 (15.4%) | 15 (20.2%) | 1 (6.7%) | 42 | |
| ALP | |||||||
| ALP>300 | 59 (43.7%) | 18 (34.0%) | 12 (46.2%) | 30 (38.0%) | 6 (40.0%) | 0.6635 | 95 |
| WNL | 49 (36.3%) | 24 (45.3%) | 10 (38.5%) | 34 (43.0%) | 8 (53.3%) | 91 | |
| Unknown | 27 (20.0%) | 11 (20.8%) | 4 (15.4%) | 15 (20.0%) | 1 (6.7%) | 43 | |
| First-line regimen | |||||||
| IRI-based | 24 (17.8%) | 6 (11.3%) | 2 (7.7%) | 8 (10.1%) | 1 (6.7%) | 0.4062 | 33 |
| OXA-based | 85 (63.0%) | 37 (69.8%) | 17 (65.4%) | 54 (68.4%) | 13 (86.7%) | 152 | |
| Others | 26 (19.3%) | 10 (18.9%) | 7 (26.9%) | 17 (21.5%) | 1 (6.7%) | 44 | |
| Anti-EGFR treatment | |||||||
| Yes | 86 (63.7%) | 1 (1.9%) | 1 (3.8%) | 2 (2.5%) | 5 (33.3%) | <0.0001 | 93 |
| No | 44 (32.6%) | 52 (98.1%) | 25 (96.2%) | 77 (97.5%) | 10 (66.7%) | 131 | |
| Unknown | 5 (3.7%) | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | 5 | |
Abbreviations: CNS=central nervous system; ECOG=Eastern Cooperative Oncology Group; EGFR=epidermal growth factor receptor; PS=performance status; well=well-differentiated adenocarcinoma; mod=moderately differentiated adenocarcinoma; por=poorly differentiated adenocarcinoma; muc=mucinous carcinoma; sig=signet-ring cell carcinoma; CNS=central nervous system; IRI=irinotecan; OXA=oxaliplatin, ALP=alkaline phosphatase; WNL=within normal range; WBC=white blood cells.
Patients with both wild-type KRAS and wild-type BRAF were designated as wild/wild. All patients with KRAS mutations (n=79) either in codon 12 (G12X) or in codon 13 (G13X) are shown as total (G12X+G13X).
*P-values calculated between wild-type KRAS and BRAF (wild/wild), KRAS12 mutant (G12X), KRAS13 mutant (G13X), and BRAF mutant (V600E) groups.
Analysis with respect to histology showed that the frequencies of poorly differentiated adenocarcinoma (por), mucinous carcinoma (muc) and signet-ring cell carcinoma (sig) were <10.9% in patients with wt BRAF, which supported previous reports that such histologies are rare in CRC (Ogino et al, 2006; Catalano et al, 2009). However, 60.0% (9 out of 15) of CRC cases with BRAF mutation were of the por or muc subtypes, although no signet-ring cell carcinomas were observed. The BRAF mutations were present in 36.0% (9 out of 25) of patients with por/muc histology. Furthermore, 60.0% (9 out of 15) of CRCs with BRAF mutation metastasised to the peritoneum, compared with ∼15% of CRCs with other subtypes (P=0.0062) (Table 2). However, Fisher's exact test indicated no statistically significant correlation between tumour histology and peritoneal metastasis in BRAF mutant patients. No other significant differences or trends in metastatic patterns with respect to KRAS/BRAF genotypes were observed.
Details of the first line chemotherapy regimens used are shown in Table 2. In all, 66.4% of patients were treated with oxaliplatin-based regimens, 14.4% with irinotecan-based regimens, and 19.2% with fluoropyrimidine-based chemotherapy without oxaliplatin or irinotecan. There were no significant differences in treatment regimens between KRAS/BRAF genotypes. A total of 86 (63.7%) patients with wild/wild tumours and five (33.3%) patients with BRAF mutation-positive tumours received anti-EGFR therapy, whereas few patients with KRAS12 or KRAS13 mutations received anti-EGFR therapy (1.9% and 3.8%, respectively).
Survival
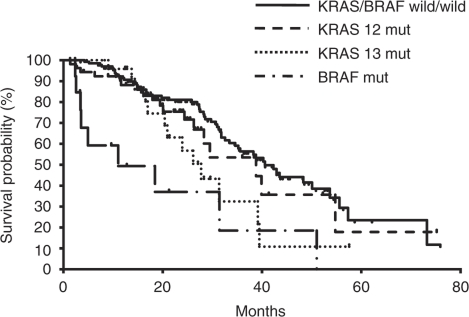
The median OS for BRAF mutation-positive patients was 11.0 months, which was significantly worse than for patients with wt KRAS and BRAF (40.6 months) (HR=4.25, 95% CI 2.08–8.67, P<0.001; Figure 1). The median OS for all KRAS mutation-positive patients, including those with KRAS12 or KRAS13 mutations, was not statistically different to that of wt KRAS and BRAF patients (HR=1.51, 95% CI 0.97–2.36, P=0.071). However, if OS for KRAS13 mutation-positive patients was analysed separately from KRAS12 mutation-positive patients, then the median OS for KRAS13 mutation-positive patients was significantly worse than that for wt KRAS and BRAF patients (27.7 months vs 40.6 months, HR=2.03, 95% CI 1.10–3.74, P=0.024; Figure 1). In contrast, the median OS for KRAS12 mutation-positive patients was 38.8 months, similar to that for wt KRAS and BRAF patients (HR=1.28, 95% CI 0.74–2.19, P=0.376; Figure 1). Univariate analysis showed that two other variables were also significantly associated with poor survival, PS ECOG⩾2 and gender (Table 3). KRAS13 mutation was not statistically associated with poor survival by univariate analysis. This was because we compared OS for KRAS13 mutation-positive patients with that for wt KRAS13 patients, which included KRAS12 and BRAF mutation-positive patients as well as wt KRAS and BRAF patients. The por/sig/muc histology and lung metastasis showed a trend towards poor OS (P=0.066 and P=0.061, respectively).
Figure 1.
Kaplan–Meier plot showing overall survival in metastatic and recurrent colon cancer patients according to KRAS and BRAF V600E mutational status (n=229). mut, mutated.
Table 3. Factors associated with overall survival in univariate and multivariate analyses.
|
Univariate analysis
|
Multivariate analysis
|
|||
|---|---|---|---|---|
| Variable | Hazard ratio (95% CI) | P-value | Hazard ratio (95% CI) | P-value |
| Age >65 | 0.74 (0.48–1.13) | 0.157 | 0.55 (0.34–0.90) | 0.018 |
| Female | 1.59 (1.06–2.37) | 0.025 | 1.35 (0.85–2.12) | 0.201 |
| PS (ECOG)⩾2 | 6.14 (3.15–12.0) | <0.001 | 7.66 (3.68–16.0) | <0.001 |
| BRAF mutant | 3.78 (1.89–7.54) | <0.001 | 4.23 (1.76–10.2) | 0.001 |
| KRAS 12 mutant | 1.03 (0.62–1.74) | 0.897 | 1.57 (0.88–2.81) | 0.128 |
| KRAS 13 mutant | 1.67 (0.93–3.02) | 0.086 | 1.51 (0.76–2.98) | 0.239 |
| Pathology, por/sig/muc | 1.74 (0.96–3.14) | 0.066 | 2.38 (1.16–4.90) | 0.018 |
| Number of metastasis ⩾2 | 0.93 (0.63–1.40) | 0.738 | 1.12 (0.61–2.05) | 0.714 |
| Liver metastasis | 1.36 (0.88–2.11) | 0.162 | 1.72 (1.02–2.90) | 0.042 |
| Lung metastasis | 0.66 (0.42–1.02) | 0.061 | 0.59 (0.32–1.11) | 0.100 |
| Peritoneal metastasis | 1.21 (0.76–1.93) | 0.417 | 1.56 (0.85–2.88) | 0.154 |
| WBC ⩾10000 | 1.27 (0.51–3.15) | 0.605 | — | — |
| ALP ⩾300 | 1.21 (0.78–1.88) | 0.395 | — | — |
| Anti-EGFR treatment | 0.80 (0.53–1.20) | 0.277 | — | — |
Abbreviations: ALP=alkaline phosphatase; PS=performance status; ECOG=Eastern Cooperative Oncology Group; EGFR=epidermal growth factor receptor; por=poorly differentiated adenocarcinoma; muc=mucinous carcinoma; sig=signet-ring cell carcinoma; CI=confidence interval; WBC=white blood cells.
To correct for significant prognostic factors, a Cox proportional hazards model that included age, gender, PS, KRAS status, BRAF status, pathological finding, number of metastasis and metastatic sites, was used. As two variables, WBC and ALP, had missing data, they were not included in the multivariate analysis. BRAF mutation and PS ECOG⩾2 were confirmed as poor prognostic factors. Specifically, the relative risk of death for patients with BRAF mutation was 4.23 (95% CI 1.76–10.2) compared with patients with wt BRAF tumours (P=0.001) (Table 3). Multivariate analysis also found that por/sig/muc histology, age>65, and liver metastasis were negative independent prognostic factors. However, KRAS13 mutation was not found to be an independent prognostic factor.
Discussion
In this study, we examined the incidence of KRAS and BRAF mutations in advanced and recurrent CRC patients, and clarified the relationship between KRAS/BRAF genotypes and clinicopathological features, including survival. Up to now, estimates of KRAS gene mutation frequency in metastatic CRCs have been based on selective clinical studies or drug admission trials with variable inclusion criteria. To our knowledge, the present report is the first to provide data on the frequency and type of KRAS/BRAF mutations from a large Japanese population of advanced and recurrent CRC patients tested in a routine setting.
Our results showed that KRAS mutation was observed in around 35% of CRC cases, which included 25% of patients with mutations at codon 12 and 10% of patients with mutations at codon 13. This observation agreed well with previous studies on selected cohorts that reported frequencies in the range of 30–42% (Table 1). The cycleave PCR technique was simultaneously applied to the detection of BRAF mutation, thought to be an adverse prognostic marker as well as a predictive marker for anti-EGFR therapy. Our analysis demonstrated that the BRAF V600E mutation was observed in ∼5% of CRC patients, which appeared to be lower than that previously reported from Western countries. None of the CRC patients in our study carried both KRAS and BRAF mutations, supporting the hypothesis that KRAS and BRAF mutations occur in a mutually exclusive manner (Rajagopalan et al, 2002; Frattini et al, 2004; Ahlquist et al, 2008). One possible explanation for the comparatively low frequency of BRAF mutation might be the different ethnic group. Indeed, several studies have reported that the mutation rates of DNA mismatch repair (MMR) genes, such as hMSH2 and hMLH1, in hereditary non-polyposis colorectal cancer, is variable between countries. Therefore, geographical variation may account for differences in the mutation spectrum of BRAF, as observed for MMR genes (Wei et al, 2003; Lee et al, 2005; Goldberg et al, 2008).
We also investigated the clinicopathological characteristics of CRC patients with respect to KRAS12, KRAS13 and BRAF mutations. In accordance with previous reports (Kim et al, 2006; Deng et al, 2008; Zlobec et al, 2010), BRAF mutation occurred more frequently in right-sided tumour locations. We also found that 60.0% of the BRAF mutation-positive specimens were of the poorly differentiated adenocarcinoma or mucinous carcinoma subtypes. It was recently reported that mucinous histology predicts a poor response to oxaliplatin- and/or irinotecan-based chemotherapies and is correlated with poor OS (Catalano et al, 2009). As BRAF mutation was more frequent in mucinous groups than non-mucinous carcinoma, as demonstrated by the present study and others (Ogino et al, 2006), the poor prognosis associated with mucinous histology may be at least partially explained by BRAF gene mutation. These specific clinicopathological features support the hypothesis that the BRAF mutation-mediated carcinogenesis in CRC is initiated by altered BRAF function as an early step in the serrated pathway (Bennecke et al, 2010), leading to activation of RAF-MEK-ERK-MAP signalling.
In contrast to BRAF mutation, no significant differences in clinicopathological parameters were observed according to KRAS genotype. However, our analysis did suggest that KRAS13 mutations were also associated with right-sided tumour location. This result raises the possibility that KRAS13 may have a distinct phenotype from that of other KRAS genotypes.
Using a representative cohort of 229 sporadic CRCs, we identified the BRAF V600E mutation as an independent prognostic factor for survival in patients with advanced and recurrent CRC. The presence of the BRAF mutation is associated with a significantly higher risk of dying of cancer-related causes, independently of other factors such as age, gender, PS, KRAS status, pathological finding, number of metastasis and metastatic sites, in agreement with other recent studies (Ogino et al, 2009; Tol et al, 2009; Bokemeyer et al, 2010; Fariña-Sarasqueta et al, 2010). For example, analysis of stage II and stage III CRC patients (Fariña-Sarasqueta et al, 2010) was consistent with the finding that 44% of our population included recurrent disease. The BRAF mutation was correlated with survival in a heterogeneous group of CRC patients that included all disease stages (Ogino et al, 2009). Furthermore, a positive correlation between BRAF mutation and shorter survival was demonstrated in a homogeneous group of metastatic CRC patients treated with a specific chemotherapy regimen with or without cetuximab (Tol et al, 2009; Bokemeyer et al, 2010). However, our study focused on the advanced and recurrent group who received systemic chemotherapy, including fluoropyrimidines, in combination with oxaliplatin, irinotecan, bevacizumab and anti-EGFR antibody in several lines. Even though all of the patients in our study received systemic chemotherapy, a positive correlation between BRAF mutation and shorter survival was still demonstrated, independent of treatment arm.
The prognostic value of KRAS mutations in CRC remains controversial, even though KRAS mutations have been associated with a poor response to anti-EGFR antibody therapy in metastatic CRC (Karapetis et al, 2008; Bokemeyer et al, 2009; Van Cutsem et al, 2009). Despite a number of studies investigating a prognostic role for KRAS mutations, no definitive conclusions can be drawn (Castagnola and Giaretti, 2005). This may be due to differences between the studies in terms of study size, patient selection, tumour sampling, use of archival versus fresh/frozen material, or laboratory methods and data analyses. More importantly, few studies have differentiated KRAS mutations at codon 12 from those at codon 13 with respect to clinicopathological features and survival (Bazan et al, 2002). Our analysis revealed that mutation at KRAS12 had no effect on patient OS. In contrast, our Kaplan–Meier curves clearly demonstrated that OS for patients with KRAS13 mutations were significantly worse than for those who had wt KRAS and BRAF. It has been reported that stage III patients with KRAS mutations displayed significantly worse disease-free survival, as compared with those with wt KRAS (Fariña-Sarasqueta et al, 2010). This finding may be partially explained by the impact of KRAS13 mutations on prognosis. As both univariate and multivariate analysis failed to confirm KRAS13 mutation as an independent prognostic factor, the prognostic value of mutations at KRAS13 remains unclear in advanced and recurrent CRC. In non-small-cell lung cancer there are differences in transforming potential and EGFR tyrosine kinase inhibitor sensitivity associated with EGFR somatic mutations L858R and deletion mutant Del (746-750) (Carey et al, 2006). Therefore, it remains a possibility that the different KRAS mutations at codons 12 and 13 may have different biological consequences that could influence the prognosis for CRC.
With respect to technical issue on KRAS and BRAF genotyping, we evaluated the prognostic value of the mutations frequently found in KRAS and BRAF using specific PCR probes. In contrast, direct sequencing is able to detect all possible KRAS and BRAF mutations including some more rare mutations. In fact, it is reported that KRAS codon 146 mutation, which was identified by direct sequencing, was associated with resistance to cetuximab plus irinotecan therapy although this is a minor oncogenic KRAS mutation (Loupakis et al, 2009). Therefore, direct sequencing may be able to obtain further insights into predictive and prognostic impact of these mutations.
Our study found that the median OS of patients with wt BRAF was generally longer than that observed in other reports. It could be argued that the selection of patients with good prognosis could bias the results in this study. Indeed, more than half of our study population was screened for KRAS/BRAF genotype to determine the use of anti-EGFR antibody, and 42% of the patients were treated with cetuximab combined therapy mostly as a second- or third-line chemotherapy. Although treatment selection may be a major reason for the longer survival observed in the present study as compared with previous studies involving metastatic CRC patients, univariate analysis revealed no significant differences in survival between patients with and without anti-EGFR therapy (38.8 months vs 32.6 months, P=0.277) (Table 3). Furthermore, almost all recurrent and advanced CRC patients are routinely screened for KRAS/BRAF genotype at the initiation of the first line chemotherapy in our institution since the use of cetuximab was approved for the treatment of CRC patients in Japan.
Another key point of discussion is the potential treatment bias in this retrospective analysis. The focus of the present study is the patient group with advanced and recurrent CRC who received systemic chemotherapy. However, we need to take the difference in the specific treatment regimen among four genotypes into consideration. In particular, 63.7% (86 out of 135) of wt KRAS and BRAF patients have received anti-EGFR therapy whereas 33.3% (6 out of 15) and 2.5% (2 out of 79) of patients with BRAF and KRAS12/13 mutations have received anti-EGFR therapy, respectively. Therefore, the prognostic advantage of wt KRAS and BRAF patients over BRAF or KRAS13 mutation might be partially explained by the presence of anti-EGFR therapy. Nevertheless, it is noteworthy that the prognosis of wt KRAS and BRAF patients was similar to that of the patients with KRAS12 mutation despite the frequent use of anti-EGFR therapy.
In conclusion, our retrospective analysis demonstrated that BRAF mutation was an independent prognostic factor in advanced and recurrent CRC. Although the presence of KRAS12 mutation had no apparent effect on OS in advanced and recurrent disease, the prognostic value of KRAS13 mutation remains uncertain. Our results are useful not only for predicting the efficacy of anti-EGFR therapy, but also for identifying patients with shorter OS in response to systemic chemotherapy, regardless of the use of anti-EGFR therapy. The exact effects of KRAS12 and KRAS13 mutations on survival require further study. The application of novel strategies targeting BRAF kinase is warranted for the treatment of CRC patients with BRAF mutation.
Acknowledgments
We thank Motoko Nimura and Akiko Yoshinari for their excellent technical assistance with the molecular genetic experiments.
References
- Ahlquist T, Bottillo I, Danielsen SA, Meling GI, Rognum TO, Lind GE, Dallapiccola B, Lothe RA (2008) RAS signaling in colorectal carcinomas through alteration of RAS, RAF, NF1, and/or RASSF1A. Neoplasia 10: 680–686, 2 p following 686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreyev HJ, Norman AR, Cunningham D, Oates JR, Clarke PA (1998) Kirsten ras mutations in patients with colorectal cancer: the multicenter ‘RASCAL’ study. J Natl Cancer Inst 90: 675–684 [DOI] [PubMed] [Google Scholar]
- Bazan V, Migliavacca M, Zanna I, Tubiolo C, Grassi N, Latteri MA, La Farina M, Albanese I, Dardanoni G, Salerno S, Tomasino RM, Labianca R, Gebbia N, Russo A (2002) Specific codon 13 K-ras mutations are predictive of clinical outcome in colorectal cancer patients, whereas codon 12 K-ras mutations are associated with mucinous histotype. Ann Oncol 13: 1438–1446 [DOI] [PubMed] [Google Scholar]
- Bennecke M, Kriegl L, Bajbouj M, Retzlaff K, Robine S, Jung A, Arkan MC, Kirchner T, Greten FR (2010) Ink4a/Arf and oncogene-induced senescence prevent tumor progression during alternative colorectal tumorigenesis. Cancer Cell 18: 135–146 [DOI] [PubMed] [Google Scholar]
- Benvenuti S, Sartore-Bianchi A, Di Nicolantonio F, Zanon C, Moroni M, Veronese S, Siena S, Bardelli A (2007) Oncogenic activation of the RAS/RAF signaling pathway impairs the response of metastatic colorectal cancers to anti-epidermal growth factor receptor antibody therapies. Cancer Res 67: 2643–2648 [DOI] [PubMed] [Google Scholar]
- Bokemeyer C, Bondarenko I, Makhson A, Hartmann JT, Aparicio J, de Braud F, Donea S, Ludwig H, Schuch G, Stroh C, Loos AH, Zubel A, Koralewski P (2009) Fluorouracil, leucovorin, and oxaliplatin with and without cetuximab in the first-line treatment of metastatic colorectal cancer. J Clin Oncol 27: 663–671 [DOI] [PubMed] [Google Scholar]
- Bokemeyer C, Köhne CH, Rougier P, Stroh C, Schlichting M, Van Cutsem E (2010) Cetuximab with chemotherapy (CT) as first-line treatment for metastatic colorectal cancer (mCRC): analysis of the CRYSTAL and OPUS studies according to KRAS and BRAF mutation status. ASCO Annual Meeting, Abstract No. 3506. J Clin Oncol 28: 15S [Google Scholar]
- Carey KD, Garton AJ, Romero MS, Kahler J, Thomson S, Ross S, Park F, Haley JD, Gibson N, Sliwkowski MX (2006) Kinetic analysis of epidermal growth factor receptor somatic mutant proteins shows increased sensitivity to the epidermal growth factor receptor tyrosine kinase inhibitor, erlotinib. Cancer Res 66: 8163–8171 [DOI] [PubMed] [Google Scholar]
- Castagnola P, Giaretti W (2005) Mutant KRAS, chromosomal instability and prognosis in colorectal cancer. Biochim Biophys Acta 1756: 115–125 [DOI] [PubMed] [Google Scholar]
- Catalano V, Loupakis F, Graziano F, Torresi U, Bisonni R, Mari D, Fornaro L, Baldelli AM, Giordani P, Rossi D, Alessandroni P, Giustini L, Silva RR, Falcone A, D'Emidio S, Fedeli SL (2009) Mucinous histology predicts for poor response rate and overall survival of patients with colorectal cancer and treated with first-line oxaliplatin- and/or irinotecan-based chemotherapy. Br J Cancer 100: 881–887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Roock W, Claes B, Bernasconi D, De Schutter J, Biesmans B, Fountzilas G, Kalogeras KT, Kotoula V, Papamichael D, Laurent-Puig P, Penault-Llorca F, Rougier P, Vincenzi B, Santini D, Tonini G, Cappuzzo F, Frattini M, Molinari F, Saletti P, De Dosso S, Martini M, Bardelli A, Siena S, Sartore-Bianchi A, Tabernero J, Macarulla T, Di Fiore F, Gangloff AO, Ciardiello F, Pfeiffer P, Qvortrup C, Hansen TP, Van Cutsem E, Piessevaux H, Lambrechts D, Delorenzi M, Tejpar S (2010) Effects of KRAS, BRAF, NRAS, and PIK3CA mutations on the efficacy of cetuximab plus chemotherapy in chemotherapy-refractory metastatic colorectal cancer: a retrospective consortium analysis. Lancet Oncol 11: 753–762 [DOI] [PubMed] [Google Scholar]
- Deng G, Kakar S, Tanaka H, Matsuzaki K, Miura S, Sleisenger MH, Kim YS (2008) Proximal and distal colorectal cancers show distinct gene-specific methylation profiles and clinical and molecular characteristics. Eur J Cancer 44: 1290–1301 [DOI] [PubMed] [Google Scholar]
- Di Nicolantonio F, Martini M, Molinari F, Sartore-Bianchi A, Arena S, Saletti P, De Dosso S, Mazzucchelli L, Frattini M, Siena S, Bardelli A (2008) Wild-type BRAF is required for response to panitumumab or cetuximab in metastatic colorectal cancer. J Clin Oncol 26: 5705–5712 [DOI] [PubMed] [Google Scholar]
- Fang JY, Richardson BC (2005) The MAPK signalling pathways and colorectal cancer. Lancet Oncol 6: 322–327 [DOI] [PubMed] [Google Scholar]
- Fariña-Sarasqueta A, van Lijnschoten G, Moerland E, Creemers GJ, Lemmens VE, Rutten HJ, van den Brule AJ (2010) The BRAF V600E mutation is an independent prognostic factor for survival in stage II and stage III colon cancer patients. Ann Oncol 21: 2396–2402 [DOI] [PubMed] [Google Scholar]
- Frattini M, Balestra D, Suardi S, Oggionni M, Alberici P, Radice P, Costa A, Daidone MG, Leo E, Pilotti S, Bertario L, Pierotti MA (2004) Different genetic features associated with colon and rectal carcinogenesis. Clin Cancer Res 10: 4015–4021 [DOI] [PubMed] [Google Scholar]
- French AJ, Sargent DJ, Burgart LJ, Foster NR, Kabat BF, Goldberg R, Shepherd L, Windschitl HE, Thibodeau SN (2008) Prognostic significance of defective mismatch repair and BRAF V600E in patients with colon cancer. Clin Cancer Res 14: 3408–3415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg Y, Porat RM, Kedar I, Shochat C, Sagi M, Eilat A, Mendelson S, Hamburger T, Nissan A, Hubert A, Kadouri L, Pikarski E, Lerer I, Abeliovich D, Bercovich D, Peretz T (2008) Mutation spectrum in HNPCC in the Israeli population. Fam Cancer 7: 309–317 [DOI] [PubMed] [Google Scholar]
- Jass JR (2007) Classification of colorectal cancer based on correlation of clinical, morphological and molecular features. Histopathology 50: 113–130 [DOI] [PubMed] [Google Scholar]
- Kakar S, Deng G, Sahai V, Matsuzaki K, Tanaka H, Miura S, Kim YS, (2008) Clinicopathologic characteristics, CpG island methylator phenotype, and BRAF mutations in microsatellite-stable colorectal cancers without chromosomal instability. Arch Pathol Lab Med 132: 958–964 [DOI] [PubMed] [Google Scholar]
- Karapetis CS, Khambata-Ford S, Jonker DJ, O'Callaghan CJ, Tu D, Tebbutt NC, Simes RJ, Chalchal H, Shapiro JD, Robitaille S, Price TJ, Shepherd L, Au HJ, Langer C, Moore MJ, Zalcberg JR (2008) K-ras mutations and benefit from cetuximab in advanced colorectal cancer. N Engl J Med 359: 1757–1765 [DOI] [PubMed] [Google Scholar]
- Kim IJ, Kang HC, Jang SG, Kim K, Ahn SA, Yoon HJ, Yoon SN, Park JG (2006) Oligonucleotide microarray analysis of distinct gene expression patterns in colorectal cancer tissues harboring BRAF and K-ras mutations. Carcinogenesis 27: 392–404 [DOI] [PubMed] [Google Scholar]
- Kinzler KW, Vogelstein B (1999) Colorectal tumors. In: Kinzler KW, Vogelstein B, editors. The Genetic Basis of Human Cancer. McGraw-Hill: London, UK, pp 565–587 [Google Scholar]
- Laurent-Puig P, Cayre A, Manceau G, Buc E, Bachet JB, Lecomte T, Rougier P, Lievre A, Landi B, Boige V, Ducreux M, Ychou M, Bibeau F, Bouché O, Reid J, Stone S, Penault-Llorca F (2009) Analysis of PTEN, BRAF, and EGFR status in determining benefit from cetuximab therapy in wild-type KRAS metastatic colon cancer. J Clin Oncol 27: 5924–5930 [DOI] [PubMed] [Google Scholar]
- Lee SC, Guo JY, Lim R, Soo R, Koay E, Salto-Tellez M, Leong A, Goh BC (2005) Clinical and molecular characteristics of hereditary non-polyposis colorectal cancer families in Southeast Asia. Clin Genet 68: 137–145 [DOI] [PubMed] [Google Scholar]
- Loupakis F, Ruzzo A, Cremolini C, Vincenzi B, Salvatore L, Santini D, Masi G, Stasi I, Canestrari E, Rulli E, Floriani I, Bencardino K, Galluccio N, Catalano V, Tonini G, Magnani M, Fontanini G, Basolo F, Falcone A, Graziano F (2009) KRAS codon 61, 146 and BRAF mutations predict resistance to cetuximab plus irinotecan in KRAS codon 12 and 13 wild-type metastatic colorectal cancer. Br J Cancer 101: 715–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogino S, Brahmandam M, Cantor M, Clark JW, Ryan DP, Kulke MH, Enzinger PC, Wolpin BM, Loda M, Fuchs CS (2006) Distinct molecular features of colorectal carcinoma with signet ring cell component and colorectal carcinoma with mucinous component. Mod Pathol 19: 59–68 [DOI] [PubMed] [Google Scholar]
- Ogino S, Nosho K, Kirkner GJ, Kawasaki T, Meyerhardt JA, Loda M, Giovannucci EL, Fuchs CS (2009) CpG island methylator phenotype, microsatellite instability, BRAF mutation and clinical outcome in colon cancer. Gut 58: 90–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajagopalan H, Bardelli A, Lengauer C, Kinzler KW, Vogelstein B, Velculescu VE (2002) Tumorigenesis: RAF/RAS oncogenes and mismatch-repair status. Nature 418: 934. [DOI] [PubMed] [Google Scholar]
- Roth AD, Tejpar S, Delorenzi M, Yan P, Fiocca R, Klingbiel D, Dietrich D, Biesmans B, Bodoky G, Barone C, Aranda E, Nordlinger B, Cisar L, Labianca R, Cunningham D, Van Cutsem E, Bosman F (2010) Prognostic role of KRAS and BRAF in stage II and III resected colon cancer: results of the translational study on the PETACC-3, EORTC 40993, SAKK 60-00 trial. J Clin Oncol 28: 466–474 [DOI] [PubMed] [Google Scholar]
- Sakamoto H, Shimizu J, Horio Y, Ueda R, Takahashi T, Mitsudomi T, Yatabe Y (2007) Disproportionate representation of KRAS gene mutation in atypical adenomatous hyperplasia, but even distribution of EGFR gene mutation from preinvasive to invasive adenocarcinomas. J Pathol 212: 287–294 [DOI] [PubMed] [Google Scholar]
- Souglakos J, Philips J, Wang R, Marwah S, Silver M, Tzardi M, Silver J, Ogino S, Hooshmand S, Kwak E, Freed E, Meyerhardt JA, Saridaki Z, Georgoulias V, Finkelstein D, Fuchs CS, Kulke MH, Shivdasani RA (2009) Prognostic and predictive value of common mutations for treatment response and survival in patients with metastatic colorectal cancer. Br J Cancer 101: 465–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tol J, Nagtegaal ID, Punt CJ (2009) BRAF mutation in metastatic colorectal cancer. N Engl J Med 361: 98–99 [DOI] [PubMed] [Google Scholar]
- Van Cutsem E, Köhne CH, Hitre E, Zaluski J, Chang Chien CR, Makhson A, D'Haens G, Pintér T, Lim R, Bodoky G, Roh JK, Folprecht G, Ruff P, Stroh C, Tejpar S, Schlichting M, Nippgen J, Rougier P (2009) Cetuximab and chemotherapy as initial treatment for metastatic colorectal cancer. N Engl J Med 360: 1408–1417 [DOI] [PubMed] [Google Scholar]
- Wan PT, Garnett MJ, Roe SM, Lee S, Niculescu-Duvaz D, Good VM, Jones CM, Marshall CJ, Springer CJ, Barford D, Marais R, Cancer Genome Project (2004) Mechanism of activation of the RAF-ERK signaling pathway by oncogenic mutations of B-RAF. Cell 116: 855–867 [DOI] [PubMed] [Google Scholar]
- Wei SC, Yu CY, Tsai-Wu JJ, Su YN, Sheu JC, Wu CH, Wang CY, Wong JM (2003) Low mutation rate of hMSH2 and hMLH1 in Taiwanese hereditary non-polyposis colorectal cancer. Clin Genet 64: 243–251 [DOI] [PubMed] [Google Scholar]
- Yatabe Y, Hida T, Horio Y, Kosaka T, Takahashi T, Mitsudomi T (2006) A rapid, sensitive assay to detect EGFR mutation in small biopsy specimens from lung cancer. J Mol Diagn 8: 335–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokota T, Shibata N, Ura T, Takahari D, Shitara K, Muro K, Yatabe Y (2010) Cycleave polymerase chain reaction method is practically applicable for V-Ki-ras2 Kirsten rat sarcoma viral oncogene homolog (KRAS)/V-raf murine sarcoma viral oncogene homolog B1 (BRAF) genotyping in colorectal cancer. Transl Res 156: 98–105 [DOI] [PubMed] [Google Scholar]
- Zlobec I, Bihl MP, Schwarb H, Terracciano L, Lugli A (2010) Clinicopathological and protein characterization of BRAF- and K-RAS-mutated colorectal cancer and implications for prognosis. Int J Cancer 127: 367–380 [DOI] [PubMed] [Google Scholar]