Abstract
Background
Intraoperative Gram stains have a reported low sensitivity but high specificity when used to help diagnose periprosthetic infections. In early 2008, we recognized an unexpectedly high frequency of apparent false-positive Gram stains from revision arthroplasties.
Questions/purposes
The purpose of this report is to describe the cause of these false-positive test results.
Methods
We calculated the sensitivity and specificity of all intraoperative Gram stains submitted from revision arthroplasty cases during a 3-month interval using microbiologic cultures of the same samples as the gold standard. Methods of specimen harvesting, handling, transport, distribution, specimen processing including tissue grinding/macerating, Gram staining, and interpretation were studied. After a test modification, results of specimens were prospectively collected for a second 3-month interval, and the sensitivity and specificity of intraoperative Gram stains were calculated.
Results
The retrospective review of 269 Gram stains submitted from revision arthroplasties indicated historic sensitivity and specificity values of 23% and 92%, respectively. Systematic analysis of all steps of the procedure identified Gram-stained but nonviable bacteria in commercial broth reagents used as diluents for maceration of periprosthetic membranes before Gram staining and culture. Polymerase chain reaction and sequencing showed mixed bacterial DNA. Evaluation of 390 specimens after initiating standardized Millipore filtering of diluent fluid revealed a reduced number of positive Gram stains, yielding 9% sensitivity and 99% specificity.
Conclusions
Clusters of false-positive Gram stains have been reported in other clinical conditions. They are apparently rare related to diagnosing periprosthetic infections but have severe consequences if used to guide treatment. Even occasional false-positive Gram stains should prompt review of laboratory methods. Our observations implicate dead bacteria in microbiologic reagents as potential sources of false-positive Gram stains.
Introduction
Many tests are used to help identify possible infection in patients with symptoms of a failed arthroplasty. Some tests such as culture of aspirated joint fluid can be performed preoperatively, but not all patients undergo preoperative aspiration, and sometimes surgeons do not become suspicious of an infection until the revision arthroplasty operation is underway. Tests with turnaround time rapid enough for intraoperative use include a frozen section of periprosthetic tissue, cell count of aspirated joint fluid, and Gram stain of aspirated fluid and periprosthetic tissue. The results of intraoperative Gram stains have a reported low sensitivity (ie, 0% [6] to 43% [2]) such that many authors do not recommend their routine use [7, 15]. However, positive Gram stain results are often believed to have high specificity (ie, 97% [17] to 100% [2, 3, 6, 12, 14, 17, 19]) and, if so, could have important implications. For example, a positive intraoperative Gram stain may influence a surgeon to convert an intended revision arthroplasty to the first stage of a two-stage operation for periprosthetic infection or may help guide antibiotic therapy before culture results become available. Positive Gram stain results could represent true-positives (clinically important viable organisms), dead bacteria secondary to antibiotic therapy, viable but nonculturable bacteria, or false-positive results resulting from contamination or misinterpretation. Correct interpretation of Gram stains is not always straightforward and to a large part influenced by experience. Clumping of dye, staining of cellular structures, or cellular debris other than bacteria can mimic Gram-positive bacteria. Conversely, a thick Gram smear with a strong background counterstain can cloak Gram-negative bacteria. The Gram stain has also been shown to have high intra- and interobserver variability as a result of an inherent degree of subjectivity [20].
At the beginning of 2008, we noticed an unexpectedly high number of positive intraoperative Gram stains of macerated tissue from hip and knee arthroplasty cases for which frozen sections lacked acute inflammation and subsequent cultures remained negative. Several such patients were discussed among orthopaedic surgeons, pathologists, and microbiologists and other clinical findings taken into account. Positive Gram stains in patients without any suspicion for infections prompted a retrospective review of 269 samples that had been obtained during revision arthroplasty operations during the preceding 3 months. We describe an investigation that determined the cause of false-positive intraoperative Gram stains and review published experience with Gram stains for diagnosing periprosthetic infections. To our knowledge, no report in the orthopaedic literature has been devoted to such an investigation.
Patients and Methods
We performed two studies, one retrospective and one prospective. The goal of the first retrospective study was to test the suspicion of our orthopaedic surgeons that the specificity of the intraoperative Gram stain was lower than expected from the literature. After analysis and changes in procedures, called “intervention,” a second, prospective study was conducted to confirm that the intervention successfully increased specificity. We focused on the performance of Gram stains of macerated periprosthetic tissue; the “gold standard” against which the Gram stains were compared was the microbiologic culture of the same fluid or tissue sample. Sensitivity and specificity were determined using standard calculations [4], and all Gram stains were reviewed by a senior microbiologist (MO). Sensitivity and specificity calculations were then performed on 390 specimens obtained during the 3 months after technique modifications. We initially identified 14 of 269 specimens associated with a positive Gram stain for a sensitivity of 23% and specificity of 92% (Table 1). This sensitivity is comparable to that reported in the literature, but the specificity is lower than most reports (ie, 97% [17] to 100% [2, 3, 6, 12, 14, 17, 19]) (Table 2).
Table 1.
Two-by-two table for results of intraoperative Gram stains for diagnosis of joint infection before and after intervention*
| Reference method | Before intervention* | After intervention | ||||
|---|---|---|---|---|---|---|
| Gram stain | Culture results | Gram stain | Culture results | |||
| Positive | Negative | Positive | Negative | |||
| Culture-positive | 19 | 65 | 84 | 13 | 132 | 145 |
| Culture-negative | 14 | 171 | 185 | 3 | 242 | 245 |
| 33 | 236 | Sum 269 | 16 | 374 | Sum 390 | |
| Sensitivity 23% | Sensitivity 9% | |||||
| Specificity 92% | Specificity 99% | |||||
* The “intervention” comprised initiation of routine Millipore filtering of the diluent fluid used during maceration of periprosthetic tissue before Gram staining and culture.
Table 2.
Performance characteristics of intraoperative Gram Stains for the detection of periprosthetic infection as reported in the literature
| Author | Year | Reference method | Specimen* | Gram stain | Culture | Number of patients | Number of infections | Infected arthroplasties | |||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Sensitivity (%) | Specificity (%) | Sensitivity (%) | Specificity (%) | Hips | Knees | ||||||
| Chimento et al. [6] | 1996 | Culture and/or histopathology | Tissue | 0 | 0 | NG† | NG† | 169 | 32 | 11 | 21 |
| Barrack et al. [3] | 1997 | Culture and/or histopathology | Preoperative synovial fluid | 10 | 100 | 55 | 96 | 67 | 20 | – | 20 |
| Atkins et al. [1] | 1998 | Histopathology | Tissue | 12 | 98 | 65 | 99.6 | 297 | 41 | NG† | NG† |
| Kraemer et al. [14] | 1998 | Culture | Tissue | 23 | 100 | N/A | N/A | 55 | 3 | 3 | – |
| Della Valle et al. [7] | 1999 | Composite findings | Tissue | 14.7 | 98.8 | NG† | NG† | 413 | 68 | 260 | 153 |
| Spangehl et al. [19] | 1999 | Composite findings | Tissue | 19 | 98 | 96 | 97 | 178 | 35 | 35 | – |
| Banit et al. [2] | 2002 | Culture | Tissue | 43 | 100 | N/A | N/A | 121 | 21‡ | 11 | 9 |
| Ko et al. [12] | 2005 | Culture | Tissue | 0 | 0 | N/A | N/A | 40 | 9 | 8 | 1 |
| Parvizi et al. [17] | 2006 | Composite findings | Synovial fluid Tissue | 35 | 97 | 90 | 97 | 70 | 39 | – | 39 |
| 22 | 100 | 86 | 100 | ||||||||
| Morgan et al. [15] | 2009 | Composite findings | Synovial fluid and tissue | 27 | 99.9 | NG† | NG† | 921§ | 247 | – | 247 |
| Ghanem et al. [9] | 2009 | Composite findings | Tissue | 31 | 100 | 92.8 | NG† | 1004 | 321 | 150 | 171 |
| Oethinger et al. [current study] | 2010 | Culture | Synovial fluid and tissue | ||||||||
| Before intervention | 23 | 92 | N/A | N/A | 269|| | NG | NG† | NG† | |||
| After intervention | 9 | 99 | N/A | N/A | 390|| | NG† | NG† | ||||
* All specimens were obtained intraoperatively unless noted otherwise; †not given; ‡one case was a shoulder hemiarthroplasty; §number of arthroplasties, not patients; ||number of specimens, not patients; N/A = not applicable.
To identify the potential cause(s) of the false-positive Gram stains, the procedure of intraoperative STAT Gram stains was dissected into various steps and each of them observed and analyzed for potential bacterial contamination. The processes of specimen collection in the operating room, transport to the microbiology laboratory, handling in the dedicated central processing and accessioning areas, stain reagent preparation, tissue handling to prepare the smears, and Gram staining were all observed. One aliquot of the macerated tissue was used for the Gram stain smear; other aliquots were plated onto sheep blood agar, chocolate agar, and McConkey agar and incubated aerobically for 3 days and plated onto CDC agar and incubated anaerobically for 5 days. Lastly, a small aliquot was added to thioglycollate enrichment broth, which was incubated for 7 days. During the investigation, sterile collection containers and sterile plastic bags for macerating the tissue specimens were rinsed with sterile saline and the fluid cultured. Gram stain reagents were exchanged, one by one, and different lots of brain heart infusion broth (Becton, Dickinson & Co, Sparks, MD) and thioglycollate broth (Becton, Dickinson & Co), both of which were sometimes added to the tissue specimens to facilitate the mixing process in a “Stomacher” (Seward Stomacher® Laboratory Biomaster; Seward Laboratory Systems Inc, Bohemia, NY), were tested. Twenty-milliliter samples of broth were centrifuged and the pellet cultured for up to 10 days. DNA was extracted (EasyMag®; Biomérieux, Durham, NC) from the very small pellet, and broad-range real-time polymerase chain reaction (PCR) for 16S rDNA using universal primers was used to identify the presence of bacteria as previously described [13]. If positive, the PCR was rerun in duplicate with one reaction containing SYBR green and the other reaction without SYBR green. Sanger sequencing was performed on amplicons generated in the latter reaction to determine the identity of bacteria.
In the second part of the study, we determined the effect of the intervention described by evaluating the number of “false-positive” Gram smears during the 3-month period after the intervention. For this analysis, all specimens that were interpreted as Gram stain “positive for bacteria” but failed to yield bacterial growth from the same sample were considered “false-positive.”
Results
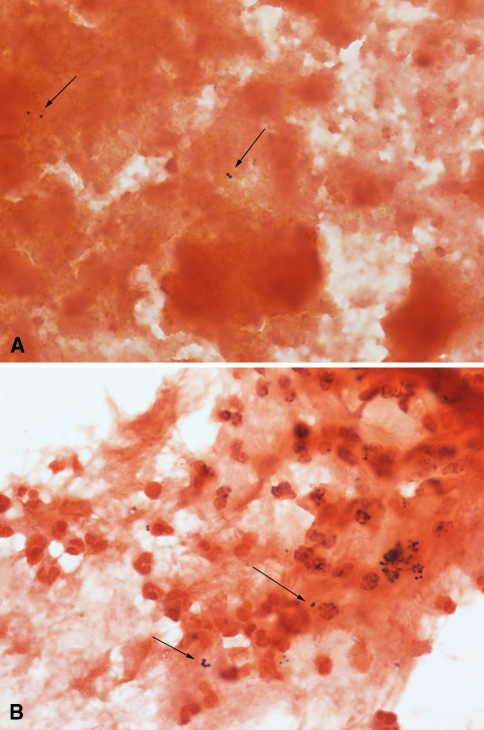
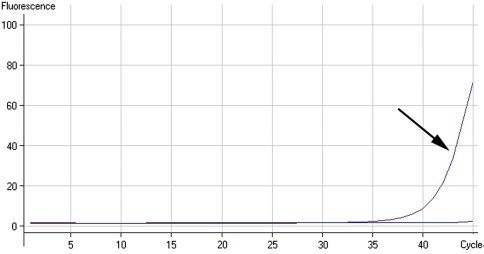
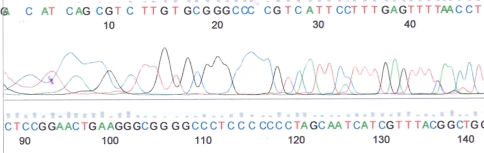
Review of the microscopic appearance of apparent false-positive specimens (Fig. 1A) confirmed the presence of bacteria morphologically indistinguishable from cases that were true-positives (Fig. 1B). No likely sources of contamination related to the way the samples were obtained in the operating room or transported to the microbiology laboratory were evident, and there was no contamination of the reagents used in the staining process itself. Changing Gram stain fluids or nutrient broths did not reduce the number of apparently false-positive Gram stains. Cultures of rinsing fluids from specimen containers were negative, and cultures of the brain heart infusion broth and thioglycollate broth remained sterile. However, Gram staining of the sediment of centrifuged sterile brain heart infusion broth and thioglycollate revealed a low number of predominantly Gram-positive bacteria that resembled the bacteria that had been identified in intraoperative Gram stains. Culture of the pellet yielded no growth, indicating these bacteria were either dead or “viable but not culturable.” PCR for 16S rDNA showed late amplification (Fig. 2), but sequencing showed the amplicons were derived from a mixture of bacteria (Fig. 3). The sequences aligned with different bacterial species, including but not limited to Ruminococcus gnavus, Clostridium spp, and Eubacterium contortum.
Fig. 1A–B.
(A) A Gram stain of a clinically false-positive specimen that was interpreted as positive for Gram-positive cocci (Stain, Gram; original magnification, ×1000) is shown. (B) A Gram stain of a specimen that grew out Staphylococcus aureus for comparison is shown. There was no obvious difference that helped the observer decide whether the bacteria visualized in the specimen were true- or false-positives (Stain, Gram; original magnification, ×1000).
Fig. 2.
Polymerase chain reaction for 16S rDNA extracted from pellet of sediment in the centrifuged thioglycollate broth shows late amplification of bacterial DNA when compared with the negative control.
Fig. 3.
This figure shows a small segment of the entire Sanger sequencing demonstrating that the amplicons were derived from a mixture of different bacteria.
After the source of contamination was recognized, sterile saline was substituted for the brain heart infusion and thioglycollate broths that had been added to help with the maceration of tissue before Gram staining and plating. The saline was first sterile-filtered using a 0.22-μm filter (Millipore, Billerica, MA) and then autoclaved.
After implementation of these changes, sensitivity and specificity were calculated for the 390 orthopaedic tissue samples submitted during the next 3 months. The frequency of positive Gram stains decreased, resulting in a decrease in sensitivity from 23% to 9% but an increase in specificity from 92% to 99% (Table 1).
Discussion
Misinterpreted as aseptic loosening, a periprosthetic infection can severely compromise the results of revision total joint arthroplasty and is associated with high cost and patient morbidity. Although many infections are correctly diagnosed preoperatively by clinical features in combination with other diagnostic tests, surgeons still sometimes make intraoperative observations that increase their suspicion of a periprosthetic infection. Unfortunately, few tests are currently available that offer a rapid enough turnaround time to be of practical clinical use during the revision operation. The intraoperative Gram stain is one of these tests, and the charge for it is low. However, discrepancies between the results of intraoperative Gram stains and other tests performed at our institution prompted us to systemically evaluate each procedure associated with this test to help identify an important potential source of false-positive results.
This study has several limitations, among them the lack of a “gold standard” for diagnosing infection. It is recognized that some patients eventually show clinical features suggestive of infection but are culture-negative. Indeed, studies that used composite findings as the standard to diagnose infection have suggested that between 4% [19] and 45% [3] of apparent infections have at least one negative culture result. It is therefore possible that some of our cases represent true infections with positive Gram stains but negative cultures. However, we believe this is only a remote possibility, because we used the results of all available clinical and laboratory information to define the absence of infection in the cases we believe represent false-positive Gram stains.
Several previous studies have reported low sensitivity but relatively high specificity of intraoperative Gram stains. For example, Atkins and coworkers prospectively evaluated several different tests to diagnose infection in patients undergoing revision arthroplasty [1]. Using histology as the gold standard of diagnosis, intraoperative Gram staining had a sensitivity of only 12% and a specificity of 98.8%. Surgeon confidence in negative Gram staining results was low so patient management was not altered in any negative Gram stain case in which clinical findings or the results of frozen section were suggestive of infection. A review of 169 Gram stain results obtained during 194 revision arthroplasty operations, 32 of which were infected, reported no positive Gram stain results [6], again emphasizing the low value of a negative stain. Similarly, in a study of tests to diagnose periprosthetic knee infections, Barrack et al. reported only 10% sensitivity but 100% specificity of Gram stains of aspirated joint fluid [3]. Gram stain results were also collected by Ko and coworkers during a retrospective study primarily designed to test the interpretation of frozen sections [12]. In that study, all Gram stains were reported as negative, including nine cases believed to be infected. Ghanem et al. [9] found that incorporating the neutrophil cell count with Gram stain result could improve specificity but still suggested the “Gram stain continues to have little value in ruling out periprosthetic infection.” The reported sensitivity and specificity of Gram stains reported in the literature are summarized (Table 2).
Taken together, these studies indicate that Gram stains related to revision arthroplasty have low sensitivity. Several studies have noted a few false-positive results [1, 17, 19], but in general, the reported series have described relatively high specificity. A low specificity of the Gram stain should be suspected when cultures are negative despite positive Gram stains and histopathology is negative for acute inflammation. Sometimes a uniform morphology of bacteria between different patients and an abnormal Gram stain reaction such as partial staining should prompt an investigation. Occasional episodes of “pseudoepidemics” resulting from false-positive Gram stains have been reported related to other clinical settings. For example, in 1973, Musher and Schell reported four cases of apparent false-positive Gram stains of cerebrospinal fluid that had been identified at a Veterans Administration Hospital [16]. Evaluation of staining processes showed bacteria and bacteria-like particles could be identified in glass specimen tubes and that the problem could be prevented by more thorough cleaning. In a comment reflecting practice in 1973, it was noted that glass microscope slides were free of bacteria “after they were washed in alcohol and dried with gauze or a shirttail.” Several years later, Ericsson and coworkers reported an “outbreak” of false-positive Gram stains of cerebrospinal fluid and eventually discovered flaming an alcohol-rinsed microscope slide tended to cause dead bacteria that were contaminating the alcohol to clump and adhere to the glass [8]. Some experienced microscopists recognized the clumps as contaminants, but others did not. In 1979, Hoke and coworkers recognized an increased number of false-positive Gram stains and discovered that stainable but nonviable Gram-negative bacteria were present in two of seven lots of commercially obtained transport media [10]. More recently, Southern and Colvin reported “an epidemic of pseudomeningitis” related to the identification of Gram-positive bacteria in cerebrospinal fluid smears for which the corresponding cultures were negative [18]. Subsequent investigation identified contamination from two different sources: funnels related to a cytocentrifuge machine used for Gram staining and the reagent storage wells used in an automated Gram stain instrument. Other studies have also demonstrated limitations in Gram stain results for specimens from different sites [5, 11].
As noted, the low sensitivity of intraoperative Gram stains has led most authors to discourage the use of the test at revision arthroplasty, but positive findings are generally believed to have relatively high specificity, so one might argue that Gram stains could be of value in cases of gross purulence to help identify an organism to guide early antibiotic treatment. Several cases of apparent false-positive results prompted this study, and the results document nonviable bacteria in commercially prepared enrichment broths. These bacteria or bacterial remnants stained mostly Gram-positive but were not culturable with routine media under both aerobic and anaerobic conditions. It should be noted that manufacturers of media guarantee their products to be sterile, but the package inserts note the possible presence of nonviable micro-organisms in the media. These reagents are routinely used in diagnostic bacteriology as enrichment broths that become turbid when viable bacteria proliferate to approximately 107 to 108 CFU/mL. A Gram stain may then be performed that usually shows the “correct” morphology of the numerous viable bacteria that had multiplied in the broth.
We assume that the degree of contamination of enrichment broths varies between different lots, which would explain the surge of apparently false-positive Gram stains at the beginning of 2008 in our laboratory. Routine filtration through a 0.22-μm bacterial filter has been added to autoclaving during reagent preparation, and the specificity of Gram stains from revision arthroplasty specimens has improved from 92% to 99%. Hence, our report documents it is necessary for any additives to be not only sterile, but also free of dead, stainable bacteria and bacterial remnants. Although we do not recommend the use of intraoperative Gram stains at revision arthroplasty, if surgeons continue to request the test, then we recommend surgeons and microbiologists assess the specificity of the intraoperative Gram stain periodically and make sure that it does not fall below 97%.
Our study and a body of literature from decades ago give clues as to what can go wrong and cause false-positive Gram stains potentially harming the patient. Nevertheless, with a sensitivity of only 9% and occasional false-positives, we do not recommend the use of routine Gram stains during revision arthroplasty for guidance of clinical decisions. Infection is best diagnosed using a combination of clinical and laboratory tests, some of them obtained preoperatively, but when a surgeon’s suspicion of infection is suddenly increased during the course of a revision of a presumed aseptic implant, a frozen section of the periprosthetic tissue usually provides higher sensitivity and specificity than a Gram stain. A cell count of fluid aspirated intraoperatively also has a high predictive value for diagnosing infection as long as the patient does not have an underlying noninfectious inflammatory arthropathy [9].
Acknowledgments
We thank Geraldine Hall, PhD, and Gary Procop, MD, MS, for their guidance during the course of this study and formulation of the manuscript.
Footnotes
Each author certifies that he or she has no commercial associations (eg, consultancies, stock ownership, equity interest, patent/licensing arrangements, etc) that might pose a conflict of interest in connection with the submitted article.
References
- 1.Atkins BL, Athanasou N, Deeks JJ, Crook DWM, Simpson H, Peto TEA, McLardy-Smith P, Berendt AR. Prospective evaluation of criteria for microbiological diagnosis of prosthetic-joint infection at revision arthroplasty. J Clin Microbiol. 1998;36:2932–2939. doi: 10.1128/jcm.36.10.2932-2939.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Banit DM, Kaufer H, Hartford JM. Intraoperative frozen section analysis in revision total joint arthroplasty. Clin Orthop Relat Res. 2002;401:230–238. doi: 10.1097/00003086-200208000-00026. [DOI] [PubMed] [Google Scholar]
- 3.Barrack RL, Jennings RW, Wolfe MW, Bertot AJ. The value of preoperative aspiration before total knee revision. Clin Orthop Relat Res. 1997;345:8–16. doi: 10.1097/00003086-199712000-00003. [DOI] [PubMed] [Google Scholar]
- 4.Bauer TW, Parvizi J, Kobayashi N, Krebs V. Diagnosis of periprosthetic infection. J Bone Joint Surg Am. 2006;88:869–882. doi: 10.2106/JBJS.E.01149. [DOI] [PubMed] [Google Scholar]
- 5.Blot F, Raynard B, Chachaty E, Tancrede C, Antoun S, Nitenberg G. Value of Gram stain examination of lower respiratory tract secretions for early diagnosis of nosocomial pneumonia. Am J Respir Crit Care Med. 2000;160:1731–1737. doi: 10.1164/ajrccm.162.5.9908088. [DOI] [PubMed] [Google Scholar]
- 6.Chimento GF, Finger S, Barrack RL. Gram stain detection of infection during revision arthroplasty. J Bone Joint Surg Br. 1996;78:838–839. [PubMed] [Google Scholar]
- 7.Della Valle C, Scher DM, Kim YH, Oxley CM, Desai P, Zuckerman JD, DeCesare PE. The role of intraoperative Gram stain in revision total joint arthroplasty. J Arthroplasty. 1999;14:500–504. doi: 10.1016/S0883-5403(99)90108-0. [DOI] [PubMed] [Google Scholar]
- 8.Ericsson CD, Carmichael M, Pickering LK, Mussett R, Kohl S. Erroneous diagnosis of meningitis due to false-positive Gram stains. South Med J. 1978;71:1524–1525. doi: 10.1097/00007611-197812000-00023. [DOI] [PubMed] [Google Scholar]
- 9.Ghanem E, Ketonis C, Restrepo C, Joshi A, Barrack R, Parvizi J. Periprosthetic infection: where do we stand with regard to Gram stain? Acta Orthop. 2009;80:37–40. doi: 10.1080/17453670902804943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hoke CH, Batt JM, Mirrett S, Cox RL, Reller B. False-positive Gram-stained smears. JAMA. 1979;241:478–479. doi: 10.1001/jama.241.5.478. [DOI] [PubMed] [Google Scholar]
- 11.Keighley MRB, McLeish AR, Bishop HM, Burdon DW, Quoraishi AH, Oates GD, Dorricott NJ, Alexander-Williams J. Identification of the presence and type of biliary microflora by immediate Gram stains. Surgery. 1977;81:469–472. [PubMed] [Google Scholar]
- 12.Ko PS, Ip D, Chow KP, Cheung F, Lee OB, Lam JJ. The role of intraoperative frozen section in decision making in revision hip and knee arthroplasty in a local community hospital. J Arthroplasty. 2005;20:189–195. doi: 10.1016/j.arth.2004.06.034. [DOI] [PubMed] [Google Scholar]
- 13.Kobayashi N, Procop GW, Krebs V, Kobayashi H, Bauer TW. Molecular identification of bacteria from aseptically loose implants. Clin Orthop Relat Res. 2008;466:1716–1725. doi: 10.1007/s11999-008-0263-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kraemer WJ, Saplys R, Waddell JP, Morton J. Bone scan, gallium scan and hip aspiration in the diagnosis of infected total hip arthroplasty. J Arthroplasty. 1993;8:611–615. doi: 10.1016/0883-5403(93)90008-R. [DOI] [PubMed] [Google Scholar]
- 15.Morgan PM, Sharkey P, Ghanem E, Parvizi J, Clohisy JC, Burnett RSJ, Barrack RL. The value of intraoperative Gram stain in revision total knee arthroplasty. J Bone Joint Surg Am. 2009;91:2124–2129. doi: 10.2106/JBJS.H.00853. [DOI] [PubMed] [Google Scholar]
- 16.Muscher DM, Schell RF. False-positive Gram stains of cerebrospinal fluid. Ann Intern Med. 1973;79:603–604. doi: 10.7326/0003-4819-79-4-603. [DOI] [PubMed] [Google Scholar]
- 17.Parvizi J, Ghanem E, Menashe S, Barack BL, Bauer TW. Periprosthetic infection: what are the diagnostic challenges? J Bone Joint Surg Am. 2006;88:138–147. doi: 10.2106/JBJS.F.00609. [DOI] [PubMed] [Google Scholar]
- 18.Southern PM, Jr, Colvin DD. Pseudomeningitis again. Association with cytocentrifuge funnel and Gram-stain reagent contamination. Arch Pathol Lab Med. 1996;120:456–458. [PubMed] [Google Scholar]
- 19.Spangehl MJ, Masterson E, Masri BA, O’Connell JX, Duncan CP. The role of intraoperative Gram stain in the diagnosis of infection during total hip arthroplasty. J Arthroplasty. 1999;14:952–956. doi: 10.1016/S0883-5403(99)90009-8. [DOI] [PubMed] [Google Scholar]
- 20.Willcox JR, Adler MW, Belsey EM. Observer variation in the interpretation of Gram-stained urethral smears: implications for the diagnosis of non-specific urethritis. Br J Vener Dis. 1981;57:134–136. doi: 10.1136/sti.57.2.134. [DOI] [PMC free article] [PubMed] [Google Scholar]