Abstract
Antisense and RNA interference (RNAi)-mediated gene silencing systems are powerful reverse genetic methods for studying gene function. Most RNAi and antisense experiments used constitutive promoters to drive the expression of RNAi/antisense transgenes; however, several reports showed that constitutive promoters were not expressed in all cell types in cereal plants, suggesting that the constitutive promoter systems are not effective for silencing gene expression in certain tissues/organs. To develop an alternative method that complements the constitutive promoter systems, we constructed RNAi and/or antisense transgenes for four rice genes using a constitutive promoter or a cognate promoter of a selected rice target gene and generated many independent transgenic lines. Genetic, molecular, and phenotypic analyses of these RNAi/antisense transgenic rice plants, in comparison to previously-reported transgenic lines that silenced similar genes, revealed that expression of the cognate promoter-driven RNAi/antisense transgenes resulted in novel growth/developmental defects that were not observed in transgenic lines expressing constitutive promoter-driven gene-silencing transgenes of the same target genes. Our results strongly suggested that expression of RNAi/antisense transgenes by cognate promoters of target genes is a better gene-silencing approach to discovery gene function in rice.
Introduction
Plant genomic research has made remarkable progress in recent years. The genome sequence of a plant provides the foundation for detailed functional characterization of plant genes [1]. Rice was the first crop plant to have its complete genome sequenced [2]. Although 56,797 genes have been annotated from sequencing of the rice genome [3], [4], the functions of >60% of these predicted genes are unknown. Therefore, one of the most challenging goals of the rice functional genomics is to characterize the functions of these unknown rice genes.
Reverse genetics is a powerful tool for assessing gene function [5], and several reverse genetics approaches have been developed in recent years for functional genomic studies. Transfer DNA (T-DNA) insertional mutagenesis that creates loss of function mutations [6] is a very effective reverse genetics approach in studying gene functions. Although T-DNA insertional mutagen has been widely used, it has several disadvantages. One common drawback is complex organizations of many T-DNA inserts, resulting in an overall 40% to 50% failure rate in identifying the exact T-DNA insertional site [7]. Besides, T-DNA exhibits certain integration preference and may therefore not saturate the entire rice genome [8]. As a result, only 27,551 rice genes were found to be mutated by T-DNA insertions from collections of >400,000 independent rice T-DNA lines [8]. In addition, T-DNA insertion may lead to lethal phenotypes, preventing genetic studies of gene functions, or cause no observable phenotype due to functional redundancy of homologous genes.
Several alternative reverse genetic approaches to study gene function, such as RNA interference (RNAi) and antisense RNA technology could circumvent the limitations of T-DNA insertional mutagenesis. In RNAi technology, the introduction of double-stranded RNAs (dsRNAs) into cells inhibits the expression of the corresponding endogenous gene at transcriptional and post-transcriptional levels [9]. RNAi could silence the expression of an endogenous target gene without altering its gene structure or producing the permanent loss of gene function. The partial gene silencing-effect of the RNAi and antisense strategies could avoid potential lethality of a T-DNA insertional mutation. In addition, RNAi/antisense-initiated gene silencing could simultaneously inhibit the expression of several homologous genes, thus overcoming potential gene redundancy problems. These advantages have made the RNAi and antisense RNA strategies the method of choice for studying gene functions in plants in recent years.
The choice of promoter is a very important factor in RNAi and antisense RNA strategies. The most commonly used promoters in RNAi and antisense strategies are constitutive promoters, such as the 35S promoter from cauliflower mosaic virus (pCaMV35S) [10] and the promoter from the maize Ubiquitin-1 gene (pUbi1) [11]. Without species restriction, constitutive promoters drive high expression in virtually all tissues/organs of transgenic plants independently of tissue/organ-specific regulators or developmental/environmental signals. However, the constitutive promoter-driven expression of an RNAi/antisense-transgene could cause pleiotropic phenotypes or embryo lethality by silencing the expression of the target gene and its homologs, thus making it extremely difficult to study the functions of the target gene or to define a causal relationship between a silenced gene and the observed phenotypic alterations. On the other hand, recent studies revealed that constitutive promoters are not active in all cell types, especially in cereal crops [12], [13]. Therefore, gene functions cannot be fully defined, as the expression pattern of an RNAi/antisense transgene might not completely overlap with that of its target gene.
Regulated promoters such as organ/tissue- or developmental stage-specific promoters [14], [15] and physically/chemically-inducible promoters [16], [17], [18], [19], [20] have been used in the past to better control the expression of an RNAi/antisense transgene avoiding the adverse effects of constitutive promoters. However, these promoters have their own limitations as an RNAi/antisense-transgene driven by a regulated promoter will only be expressed in certain tissues/organs, at specific developmental stages, or in response to a unique chemical/physical signal but has no effect on the target gene in other relevant tissues/organs at certain important developmental stages [21].
By contrast, a cognate promoter of a target gene should drive the expression of a gene-knockdown RNAi/antisense-transgene in the native expression domains of the targeted endogenous gene, which could overcome many of the known limitations of constitutive/regulated promoters in driving the expression of gene-silencing transgenes to define the biological functions of their corresponding endogenous genes.
In this study, we investigated the effectiveness of constitutive/cognate promoter-driven RNAi/antisense-transgene in causing growth/developmental phenotype in transgenic rice plants. Four rice genes, Pyruvate Dehydrogenase Kinase 1 and 2 (OsPDK1 and OsPDK2), Silencing Information Regulator 2 (OsSRT1), and Metallothionein2b (OsMT2b), were selected for our studies. The physiological functions of these four genes were previously studied by gene silencing using constitutive promoter-driven RNAi/antisense transgenes [22], [23], [24], however, our studies using the cognate promoter-driven RNAi/antisense transgenes revealed additional functions of these genes in regulating rice growth/development. Our investigation with the two OsPDK genes also showed that the cognate promoter approach could specifically reduce the transcript level of one member gene without affecting the expression of other members of a gene family.
Results
The cognate promoter-driven expression of an RNAi-transgene revealed novel physiological functions of OsMT2b
Metallothioneins (MTs) are a family of low-molecular weight, cysteine rich intracellular proteins that are thought to play important roles in metal tolerance, detoxification, and homeostasis in plants via binding heavy metals [22], [25], [26]. The rice genome encodes 15 MT proteins that could be classified into four types [22]. OsMT2b, a type-2 MT, scavenges reactive oxygen species [22], [27]. Earlier studies using transgenic rice plants in which OsMT2b was silenced by an OsMT2b-RNAi transgene driven by the maize Ubi promoter showed that OsMT2b participates in epidermal cell death [28] and is involved in root development and seed embryo germination by modulating the endogenous cytokinin level [22].
To better understand the physiological functions of OsMT2b, we generated an OsMT2b RNAi transgene driven by the cognate promoter of the endogenous OsMT2b gene (Figure 1A) and transformed the resulting pOsMT2b::OsMT2b-RNAi transgene into wild-type rice plants. Ten independent transgenic lines were obtained and carefully analyzed, among which 6 transgenic lines exhibited phenotypic variations in the T0 generation. RNA blot analyses found that the expression of the endogenous OsMT2b gene was significantly reduced in two independent pOsMT2b::OsMT2b-RNAi transgenic lines exhibiting the growth defects (Figure 2A), while segregation analysis of T1 progeny of several T0 lines carrying single-copy transgene revealed a 3∶1 ratio for normal individuals vs. abnormal individuals. Analyses of the morphological/developmental defects of the 6 independent T0 transgenic lines and their offspring not only confirmed previously reported phenotypic alterations, including smaller mature embryos and reduced thickness of scutellum of embryos (Figure 2B), but also discovered novel growth phenotypes such as smaller spikelets, lower percentage of seed setting, and smaller seeds at the bottom of spikes (Figure 2C). Our study thus revealed a functional role of OsMT2b in spikelet/seed development, suggesting that the cognate promoter-driven gene silencing is a better strategy than the constitutive promoter-driven gene silencing to study gene functions in rice.
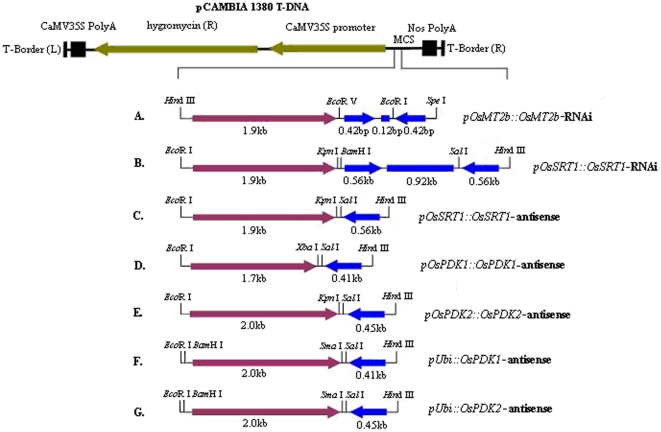
Figure 1. Schematic presentation of the constructed RNAi/antisense transgenes.
(A, B) Positions and orientations of independently amplified genomic/cDNA fragments for generating pOsMT2b::OsMT2b and pOsSRT1::OsSRT1 RNAi transgenes. (C–E) Schematic presentation of antisense transgenes of OsSRT1 (C), OsPDK1 (D) and OsPDK2 (E) driven by their cognate promoters. (F, G) Schematic presentation of the pUbi::OsPDK1 (F) and pUbi::OsPDK2 (G) antisense transgenes. Purple arrows represent promoters, blue right arrows indicate sense fragments, blue left arrows mean antisense fragments, and blue bars denote introns.
Figure 2. Phenotypic and RNA blot analyses of primary OsMT2b-RNAi transgenic lines.
A. RNA blot analysis of the endogenous OsMT2b transcript. Twenty µg of total RNAs isolated from two independent pOsMT2b::OsMT2b-RNAi transgenic lines (1 and 2) and the control line (Ctrl) were separated on denaturing agarose gel, stained with ethidium bromide, transferred to a nylon membrane, hybridized with α 32P-labeled OsMT2b cDNA fragment, and analyzed by autoradiography (upper panel). The lower panel shows the ethidium bromide-stained ribosomal RNAs used as a loading control. B. Comparison of the seed embryo between a representative OsMT2b-RNAi transgenic line (1) and the control (Ctrl). Scale Bar = 1 mm. C. Phenotypic comparison of panicles/spikelets between the representative pOsMT2b::OsMT2b-RNAi transgenic line (1) and the control (Ctrl).
Silencing of the rice OsSRT1 gene by cognate promoter-driven OsSRT1-RNAi or OsSRT1-antisense transgenes
To further confirm our discovery, we generated a cognate promoter-driven RNAi transgene for another rice gene, which encodes a protein homologous to the SILENT INFORMATION REGULATOR2 (SIR2), a highly conserved NAD+-dependent protein deacetylase [29], [30]. The rice genome encodes two SIR2-related proteins, named OsSRT1 and OsSRT2 [23], [31]. An earlier study showed that transgenic rice plants in which OsSIRT1 was silenced by an OsSRT1-RNAi transgene driven by the CaMV35S promoter exhibited brown dots on leaves, which became larger at later stages, leading to premature leaf senescence [23].
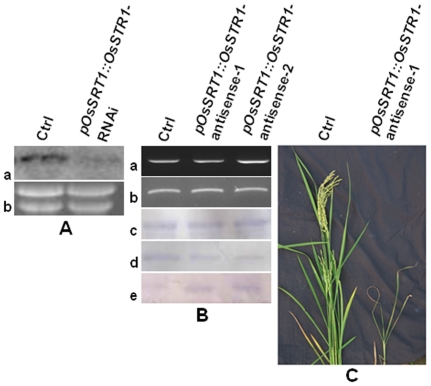
Despite numerous attempts, we were unable to generate a single pOsSRT1::OsSRT1-RNAi (Figure 1B) transgenic rice line from the OsSRT1-RNAi transgene-transformed calli. We suspected that the RNAi-mediated silencing of OsSRT1 in its native expression domains prevented transformed calli to regenerate. To test our hypothesis, we performed a Southern blot analysis with genomic DNAs isolated from antibiotic-resistant calli and found that these hygromycin-resistant calli carried the hygromycin-B-phosphotransferase gene, the antibiotic marker gene of the pOsSRT1::OsSRT1-RNAi plasmid and originated from different transformation events (data not shown). We also performed RNA blot analysis using total RNAs isolated from hygromycin-resistant and control calli and found that the OsSRT1 transcript level was significantly reduced in hygromycin-resistant calli (Figure 3A). Given the successful generation of multiple transgenic lines when an OsSRT1-RNAi transgene was driven by the CaMV35S promoter [23], our use of a cognate promoter-driven RNAi-transgene revealed a novel role of OsSRT1 in tissue regeneration.
Figure 3. Phenotypic, molecular, and biochemical analyses of primary OsSRT1 antisense transgenic lines.
A. RNA blot analysis of the OsSRT1 transcript. Twenty µg of total RNAs isolated from calli derived from the control rice plant (Ctrl) and hygromycin-resistant calli transformed with the pOsSRT1::OsSRT1-RNAi transgene were separated on denaturing agarose gel, stained with ethidium bromide, transferred to a nylon membrane, hybridized with α-32P-labeled OsSRT1 cDNA fragment, and analyzed by autoradiography(a). The lower panel shows the ethidium bromide-stained ribosomal RNAs used as a loading control (b). B. The expression of the pOsSRT1::OsSRT1-antisense transgene had no effect on the OsSRT1 mRNA level but significantly reduced the OsSRT1 protein abundance. pOsSRT1::OsSRT1-antisense-1 and -2 are two independent OsSRT1-antisense transgenic lines. a) RT-PCR analysis of the transcript abundance of the endogenous OsSRT1 gene (see Materials and Methods for experimental details). b) β-actin was used as a loading control. c–e) Immunoblot analysis of the protein abundance of Tubulin (c), OsSRT1(d), and the level of H3K9 acetylation(e). Equal amounts of protein crude extracts were separated by SDS-polyacrylamide gel electrophoresis, transferred to nitrocellulose filters, and analyzed by immunoblotting with antibodies against Tubulin (for loading control), OsSRT1, and acetylated Lys-9 residue of histone 3 (H3K9). C. Phenotypic comparison between a representative pOsSRT1::OsSRT1 antisense transgenic line (1) and a wild-type control (Ctrl).
Because no transgenic plants were obtained with the pOsSRT1::OsSTR1-RNAi transgene, we created a pOsSRT1::OsSRT1 antisense transgene carrying the cognate promoter of the endogenous OsSRT1 gene (Figure 1C), as an antisense transgene is less effective in triggering gene silencing. A total of 12 independent transgenic lines were produced but none of them exhibited any observable growth alteration. However, at least 5 T0 lines segregated out T1 individuals displaying developmental defects with a 3∶1 ratio of normal plants vs. defective individuals (data not shown). Further genetic studies suggested that the defective T1 plants are likely homozygous for the pOsSRT1::OsSRT1-antisense transgene as they failed to segregate out normal plants in 4 subsequent generations. Two homozygous pOsSRT1::OsSRT1-antisense lines were selected to determine the gene silencing effect of the cognate-promoter-driven antisense transgene.
Although RT-PCR analysis detected no significant changes in the OsSRT1 transcript level (Figure 3B-a), our immunoblot experiment showed that the OsSRT1 protein abundance in the two pOsSRT1::OsSIRT1-antisense transgenic lines was significantly reduced (Figure 3B-d). Consistent with the known function of the yeast/mammalian SIR2 proteins in deacetylating the acetylated lysine-9 residue on histone 3 (H3K9) [23], an immunoblot assay using antibodies raised against the acetylated H3K9 revealed the increased H3K9 acetylation in the two selected transgenic lines (Figure 3B-e) , further supporting a reduction of OsSRT1 abundance in the two selected transgenic lines. These homozygous pOsSRT1::OsSRT1-antisense transgenic rice plants not only displayed brown spots on the leaves and early senescence symptom (Figure 3C), which are similar to what were previously observed on pCaMV35S::OsSRT1-RNAi transgenic plants [23], but also exhibited additional growth/developmental abnormalities, such as decreased tillering capacity and lower seed setting (Figure 3C and data not shown). Our studies using pOsSRT1::OsSRT1-RNAi/antisense transgenes therefore further supported our conclusion that expression of RNAi/antisense transgene using a cognate promoter of the target gene is a better silencing strategy in revealing its physiological functions in rice.
Direct comparison of the phenotypic differences of constitutive and cognate promoters in driving the expression of antisense transgenes in rice
To directly compare the differential effects of constitutive and cognate promoters on silencing rice genes, we created two antisense transgenes each for two highly-homologous rice genes encoding pyruvate dehydrogenase kinase 1 and 2 (OsPDK1 and OsPDK2), one using the maize Ubi promoter and the other with the cognate promoters of the OsPDK genes (Figure 1D–1G). An earlier study showed that silencing the OsPDK1 gene by a CaMV35S promoter-driven OsPDK1-RNAi transgene resulted in a weak dwarf phenotype in transgenic rice plants [24].
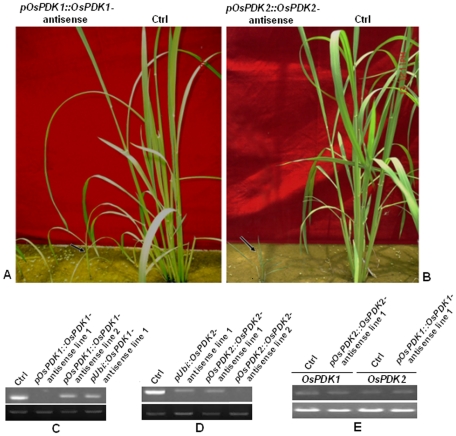
Transformation of pOsPDK1::OsPDK1 and pOsPDK2::OsPDK2 antisense transgenes resulted in generation of 16 and 13 independent transgenic lines of Zhonghua 11, respectively. Both transgenes caused two types of growth alterations.The first one is slightly-reduced plant height (∼10% reduction compared to the control), resembling that of the previously-reported pUbi::OsPDK1-RNAi transgenic lines [24]. The other type of growth defects included severe dwarfism (∼90% shorter than the control), single tillering, and semi-sterility (Figure 4A and 4B), which were not observed in p35S::OsPDK1-RNAi transgenic rice plants. RT-PCR analysis revealed a slight reduction of the OsPDK transcript abundance in weakly dwarfed transgenic plants but detected no OsPDK transcripts in severely dwarfed lines (Figure 4C and 4D). Interestingly, despite high sequence similarity between the two OsPDK genes, the antisense-triggered gene silencing was quite specific as the transcript levels of OsPDK1 and OsPDK2 were not obviously changed in OsPDK2-antisense and OsPDK1-antisense transgenic plants, respectively (Figure 4E). Consistently, the severely-dwarfed pOsPDK1::OsPDK1 and pOsPDK2::OsPDK2-antisense transgenic lines also exhibited unique phenotypes. The pOsPDK1::OsPDK1-antisense lines had longer life cycle than the control plants with pale yellow leaves, whereas pOsPDK2::OsPDK2-antisense dwarfs had shorter life cycle than the control with darker green leaves (Figure 4A and 4B), revealing different physiological functions for two highly homologous rice proteins.
Figure 4. Similar and different growth/developmental defects in pOsPDK::OsPDK-antisense transgenic plants.
A. Phenotypic comparison between a representative pOsPDK1::OsPDK1-antisense transgenic line (indicated by black arrow) and the wild-type control (Ctrl) of the same developmental age (booting stage). B. Phenotypic comparison between a representative pOsPDK2::OsPDK2-antisense transgenic line (indicated by black arrow) and the wild-type control (Ctrl) of the same developmental age (booting stage). C–E. RT-PCR analysis of the transcript abundance of the endogenous OsPDK1 and OsPDK2 genes in various pUbi/pOsPDK::OsPDK-antisense transgenic plants. Equal amounts of total RNAs isolated from the wild-type control (Ctrl) and selected transgenic plants were converted into 1st cDNAs. Half microliter of the resulting 1st-strand cDNAs was used as templates for PCR-amplification using gene-specific primers (see Materials and Methods for details) of the transcripts of the endogenous OsPDK1 (C plus lanes 1 and 2 in E) and OsPDK2 (D plus lanes 3 and 4 in E) genes. RT-PCR analysis of the rice β-actin gene (the lower strip in each panel) was used as a control.
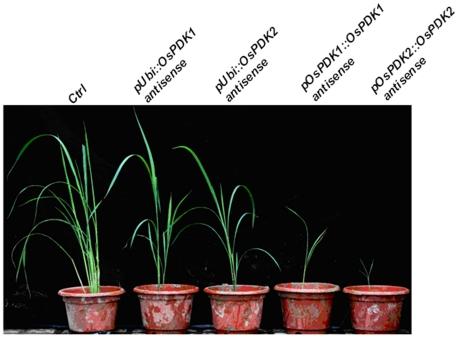
By contrast, expression of either OsPDK-antisense transgene driven by the constitutively-active Ubi promoter failed to cause extreme dwarfism but only resulted in the semidwarf phenotype (∼30% shorter than control plants) (Figure 5), which is slightly stronger than that caused by the expression of pUbi::OsPDK1-RNAi transgene [24]. Consistently, RT-PCR analysis revealed a slight reduction of OsPDK1 or OsPDK2 transcript level in the pUbi::OsPDK-antisense transgenic lines (Figure 4C and 4D). As expected from the failure of the pUbi::OsPDK-antisense transgenes to cause strong dwarfism, no obvious phenotypic difference was observed between pUbi:OsPDK1- and pUbi:OsPDK2-antisense transgenic plants. Taken together, our direct comparison study clearly demonstrated the superiority of the cognate promoter-driven transgenes in silencing the corresponding endogenous rice genes to reveal their physiological functions.
Figure 5. Phenotypic comparison between pOsPDK::OsPDK-antisense and pUbi::OsPDK-antisense transgenic plants.
Shown here from left to right are six-week old soil-grown rice plants of the wild-type control (Ctrl) and a representative transgenic line carrying an antisense transgene of pUbi::OsPDK1, pUbi::OsPDK2, pOsPDK1::OsPDK1 and pOsPDK2::OsPDK2.
Discussion
In this study, we investigated the differential effects of constitutive promoter-driven and cognate promoter-driven RNAi/antisense transgenes on gene silencing and causing growth/developmental defects in transgenic rice plants. By comparing the growth/developmental phenotypes of our transgenic plants with those of previously reported transgenic lines, we found that the expression of the cognate promoter-driven RNAi/antisense transgenes often gave rise to growth/developmental defects that were not observed on transgenic lines expressing constitutive promoter-driven RNAi/antisense transgenes of the same target genes. For example, some pOsPDK1::OsPDK1-antisense transgenic lines were severe dwarfs with yellow leaves, which were not observed in previously reported pUbi/p35S::OsPDK1- antisense transgenic rice plants [24]. On the other hand, the use of a cognate promoter-driven RNAi/antisense transgene could avoid potential lethal phenotype caused by expression of a constitutive promoter-driven RNAi/antisense transgene. For example, an earlier study reported that strong silencing of the OsSRT1 gene caused a postembryonic lethal phenotype in p35S::OsSRT1-RNAi transgenic plants [23], whereas no such a phenotype was observed in our pOsSRT1::OsSRT1-antisense transgenic lines. Our results demonstrated that gene silencing using cognate promoter-driven RNAi/antisense transgenes was a more effective and physiologically relevant approach than that driven by constitutive promoters to define the gene functions in rice. We have so far adopted this approach to investigate the physiological functions of more than 20 rice genes (unpublished data).
Antisense RNA, with the formation of antisense/target dsRNA, is a gene silencing mechanism resulting in mRNA degradation or affecting mRNA translation [32], [33]. It has been reported that the binding position of the antisense RNA may determine gene-silencing mechanisms [34], [35]. Antisense RNAs binding to the 3′ untranslated region (3′-UTR) represses translation [32], whereas antisense RNAs pairing with the 5′ UTR of the target gene could cause mRNA degradation. The full-length of OsSRT1 (NM_001058878) cDNA is 1891 bp, and the predicted antisense transcript of the OsSRT1-antisense transgene would hybridize to the region near the 3′-end, between nucleotides 1206 and 1770, of the endogenous OsSRT1 transcript. In pOsSRT1::OsSRT1-antisense transgenic plants, the transcript level of the endogenous OsSRT1 gene was not obviously changed but the OsSRT1 protein level and its predicted histone deacetylase activity were significantly reduced. The same antisense-transgene construction strategy was used for 8 additional rice genes, and similar effects on the abundance of the endogenous target genes and their protein products were observed (data not shown). The full-length of OsPDK1 (NM_001056731.1) and OsPDK2 (NM_001066936.1) cDNAs are 1535 bp and1480 bp, respectively. The cDNA fragments used for making antisense transgenes of OsPDK1 and OsPDK2 were derived from the regions spanning 434–845 bp and 153–594 bp near the 5′ ends of OsPDK1 and OsPDK2 cDNAs, respectively. In pOsPDK1::OsPDK1 and pOsPDK2::OsPDK2-antisense transgenic progeny, the mRNA levels of the corresponding target genes decreased significantly. Our studies thus further supported an earlier hypothesis that antisense RNA directed against the 5′ UTR often results in degradation of the target mRNA whereas antisense RNA targeted near the 3′ UTR causes translational inhibition.
Consistent with earlier discoveries that the gene-silencing efficiency of antisense transgene is lower than that of RNAi-transgene, growth/developmental defects were only observed in the homozygous progeny of OsSRT1/OsPDK1/OsPDK2-antisense transgenic lines. However, such a lower gene-silencing efficiency could be useful to avoid lethal phenotypes of RNAi-induced strong gene silencing. For example, despite numerous attempts and successful generation of antibiotic-resistant calli with an pOsSRT1::OsSRT1-RNAi transgene, no single pOsSRT1::OsSRT1-RNAi transgenic plants was regenerated; however, we were quite successful in generating pOsSRT1::OsSRT1-antisense transgenic lines with reduced transcript level of the endogenous OsSRT1 gene. We suggest that the antisense-mediated gene-silencing technology might be more appropriate than the RNAi technology to study rice genes that play roles in early stage of plant growth/development.
Gene redundancy is a major obstacle in functional genomic studies. 53% and 68% of the non-transposable element-related genes in rice and Arabidopsis are grouped into paralogous gene families, respectively [36]. Although family members show high sequence homology at the nucleic acid level, they often have different expression patterns and biological functions. Gene-silencing using a constitutive promoter-drive RNAi/antisense transgene could simultaneously knockdown the intended target gene and its potential homologs [37], making is extremely difficult in assigning a given biological function to a member of that gene family. For example, a recent report showed that four members of OsAGO1 gene family, OsAGO1a, OsAGO1b, OsAGO1c, and OsAGO1d, are highly similar in sequence with each other [38], and their transcript levels were all significantly reduced by the expression of a constitutive promoter-driven OsAGO1-RNAi transgene. In this work, we studied two members of the OsPDK gene family, OsPDK1 and OsPDK2, which share 82% similarity at the nucleotide level. OsPDK1 is expressed in mature leaves, while OsPDK2 is mainly expressed in actively-growing tissues. As expected, no phenotypic difference was observed in pUbi::OsPDK1/pUbi::OsPDK2-antisense transgenic lines, making it difficult to define the physiological function for each OsPDK gene. However, transgenic plants in which the OsPDK1/OsPDK2-antisense transgene was driven by the corresponding cognate promoter displayed different phenotypes. The pOsPDK1::OsPDK1-antisense transgene caused yellowish leaf color and longer life cycle, whereas the expression of the pOsPDK2::OsPDK2-antisense transgene resulted in darker green leaf color and a shortened life cycle with precocious flowering. Our results thus suggested that the expression of an antisense transgene by the cognate promoter of its target gene might be a better strategy to study the physiological functions of gene families.
Materials and Methods
Plant and Other Experimental Materials
Rice (Oryza sativa L. ssp. Japonica) cv. Zhonghua 11 was used for all experiments. Transgenic plants were grown in a greenhouse with normal daylight illumination. Escherichia coli DH10B and Agrobacterium tumefaciens strain EHA105 were used for cloning and transformation experiments. pCAMBIA1380 was used as the binary vector for Agrobacterium-mediated transformation [39].
Plasmid Construction
Two RNAi transgenes (OsSRT1 and OsMT2b) and three antisense transgenes (OsSRT1, OsPDK1 and OsPDK2) were constructed (Text S1). These 5 transgenes were driven by the cognate promoters of the corresponding target genes. To directly investigate the differential effect of cognate promoters and constitutive promoters on gene silencing, OsPDK1 and OsPDK2 antisense transgenes driven by the maize pUbi promoter were also constructed. Primers were designed based on published cDNA sequences of OsSRT1, OsMT2b, OsPDK1 and OsPDK2 (Table 1) and were used to amplify gene-specific cDNA fragments from total RNAs isolated from Zhonghua 11. The published genome sequences were also used to locate the 2.0-kb genomic fragment immediately upstream of the annotated ATG start codon for each gene (Table 2), which were amplified by PCR using the primer pairs listed in Table 1 and used as cognate promoters for RNAi/antisense transgene construction. The intron fragments of RNAi transgenes were directly amplified the genomic DNA of Zhonghua 11 (Figure 1A and 1B). Each of the constructed transgenes was fully sequenced to ensure no PCR error before being transformed into Agrobacterial cells.
Table 1. Sequences of primers.
| Names of primers | abbreviation | sequence (5′to 3′) | Description |
| OsMT2b promoter f | P-MT-F | aaaaaagcttgagatgctaatcaagtctctctg | Hind III |
| OsMT2b promoter r | P-MT-R | aaaagatatcagatgttgttgctgattgagctc | EcoR V |
| OsSRT1 promoter f | P-SRT-F | aaaagaattcgtgcttgtgtgtcattctaccc | EcoR I |
| OsSRT1 promoter r | P-SRT-R | aaaaggtaccggacatggtggttcagttgaaccc | Kpn I |
| OsPDK1 promoter f | P-PDK1-F | aaaagaattcgtagtgtcaggctgtcagcaac | EcoR I |
| OsPDK1 promoter r | P-PDK1-R | aaaatctagaccctaccgacaacagcaccac | Xba I |
| OsPDK2 promoter f | P-PDK2-F | aaaagaattccgctgtactatgagtcgtacc | EcoR I |
| OsPDK2 promoter r | P-PDK2-R | aaaaggtaccatcatgtagcgcaggctcac | Kpn I |
| Ubi promoter f | P-Ubi-F | aaaaggatccagtgcagcgtgacccggtc | BamH I |
| Ubi promoter r | P-Ubi-R | aaaacccgggcagaagtaacaccaaacaacagg | Sma I |
| OsMT2b RNAi 1 | R-MT-1 | aaaagaattcgctgctccatccaacaagg | EcoR I |
| OsMT2b RNAi 2 | R-MT-2 | aaaagatatcgaagcctggcacgcatgagg | EcoR V |
| OsMT2b RNAi 3 | R-MT-3 | aaaaactagtgaagcctggcacgcatgagg | Spe I |
| OsSRT1 RNAi 1 | R-SRT-1 | aaaagtcgacggctgttcgagctcttccattg | Sal I |
| OsSRT1 RNAi 2 | R-SRT-2 | aaaaggatccataccatcaagccccacaaccag | BamH I |
| OsSRT1 RNAi 3 | R-SRT-3 | aaaaaagcttcataccatcaagccccacaaccag | Hind III |
| OsPDK1 sense f | S-PDK1-F. | aaaagtcgactgggtctccatatatgttcac | Sal I |
| OsPDK1 sense r | S-PDK1-R | aaaaaagcttggactcattccgcgacttac | Hind III |
| OsPDK2 sense f | S-PDK2-R | aaaagtcgacgccaggctctgggtcag | Sal I |
| OsPDK1 sense r | S-PDK2-R | aaaaaagcttcgggtcgcgccccacg | Hind III |
Table 2. Promoter locations.
| Promoters | BAC clones | Locations in the BAC | Gene names | Locations in the rice genome |
| OsMT2b | AC079356 | 90035∼91973 | Os05g0111300 | −49∼-1987 |
| OsSRT1 | AL663014 | 143857∼141928 | Os04g0271000 | −49∼-1978 |
| OsPDK1 | AC082644 | 112200∼113877 | Os03g0370000 | −24∼-1701 |
| OsPDK2 | AP003749 | 87838∼85738 | Os07g0637300 | +94∼-2006 |
Note: “+” means upstream of ATG and “−” means downstream of ATG.
Plant transformation
To investigate the effectiveness of generated RNAi/antisense transgenes in silencing their target genes, these transgenes was then transformed into the A. tumefaciens strain EHA105, which were used to transform rice calli generated from mature dry seeds of Zhonghua11 following a previously described protocol [39]. Tranformed calli were allowed to generate T0 plants. After further analyses, they were transferred into soil to produce T1 seeds for the generation of T1 transgenic lines.
RNA preparation
Total RNAs were extracted using the Trizol method (Invitrogen) according to the manufacturer's protocols. Briefly, 0.1 g plant tissues from leaves and spikelets of different developmental stages of control/transgenic rice plants were ground in liquid N2 to fine powder, dissolved in the Trizol reagent, incubated at 15–30°C for 5 min, mixed with chloroform (0.2 mL/1 mL Trizol reagent), and centrifuged 12,000× g at 2–8°C for 15 min. The resulting supernatants were mixed with isopropanol (0.5 mL/1 mL Trizol reagent), incubated at 15–30°C for 10 min, and centrifuged at 12,000× g for 10 min at 2–8°C to collect RNA pellets. After twice washing with 75% ethanol, the resulting RNA pellets were dried and resuspended in water or an appropriate buffer.
Reverse transcriptase-PCR analysis
First strand cDNAs were synthesized at 42°C for 1 h in a 20 µL reaction that contains 2.0 µg of total RNAs, 4.0 µL of 5× reaction buffer, 1.0 µL of oligo d(T)15 (50 mmol/L), 2.0 µL dNTP mix (10 mM each), 1.0 µL Ribonuclease Inhibitor (40 U, TAKARA, Japan), 1 µL AMV reverse transcriptase (5 U, TAKARA, Japan). 0.5 µL of the reaction product was used for subsequent PCR amplification of gene-specific cDNA fragments in a 50 mL reaction containing 40 µL of RNase-free H2O, 5 µL of 10× PCR buffer, 1 µL dNTP mix (10 mM each), 1 µL of forward primer (10 µmol/L), 1 µL of reverse primer (10 µmol/L), and 0.4 µL of DNA polymerase (2.5 U/µL). The gene-specific primer pairs used for the RT-PCR reactions were: gaagaagaagatgtcttgctg and acagtagcagcatccatacg for OsMT2b; gtgcttgtgtgtcattctaccc and ggacatggtggttcagttgaaccc for OsSRT1; tgggtctccatatatgttcac and ggactcattccgcgacttac for OsPDK1; gccaggctctgggtcag and cgggtcgcgccccacg for OsPDK2.
RNA blot analysis
For RNA blot hybridization, equal amounts (∼20–30 µg) of total RNAs were separated on 1.2% denaturing agarose gels containing 12.5% formaldehyde and transferred on to a Hybond-N nylon membrane (Amersham Biosciences). The hybridization probes were amplified by gene-specific primers used for RT-PCR analysis and were labelled using an [α-32P]-dCTP random prime-labelling system. Hybridization was performed at 42°C following a previously described procedure [40]. After hybridization, the membrane was washed twice with 2× SSC containing 0.1% SDS (w/v) and twice with 0.1× SSC containing 0.1% SDS (w/v) at 50°C, and the hybridization signals were visualized by Molecular Imager PharosFX Plus System (Bio-Rad).
Immunoblot Analysis
Tissues were collected from the transgenic and wild type plants, and total proteins were extracted as described [41]. The protein extracts (100 µg per lane) were separated by SDS-polyacrylamide gel electrophoresis and transferred to Pure Nitrocellulose Blotting Membrane (Pall Corporation) using the wet transfer apparatus. The membranes were incubated in blocking buffer (5% [w/v] skimmed milk powder, 0.05% [v/v] Tween 20, 20 mM Tris-HCl, and 500 mM NaCl, pH 7.5) for 1 h, washed 5 times (5 min each) with TBST (0.05% [v/v] Tween 20, 20 mM Tris-HCl, and 500 mM NaCl, pH 7.5), and incubated with the primary antiserum (1∶500 dilution) for 2 h at room temperature. After 5 rinses (5 min each) with TBST, the membranes were incubated with the secondary antibody (alkaline phosphatase-conjugated goat anti-rabbit IgG [ALP], 1∶10000 dilution; Kirkegaard and Perry Laboratories) for 1.5 h at room temperature, washed 5 times (5 min each) with TBST, and subsequently incubated in the substrate buffer (0.33 mg/mL nitroblue tetrazolium [Sigma-Aldrich], 0.165 mg/mL BCIP [Bio-Basic], 0.1 M Tris, 0.1 M NaCl, and 5 mM MgCl2, pH 9.5) for several minutes in the dark, and the chemiluminescent signals were subsequently detected by autoradiography film.
Supporting Information
Construction of RNAi/antisense transgenes.
(DOC)
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported by the National Basic Research Program of China (2005CB120802, http://www.973.gov.cn/English/Index.aspx), the National High Technology Research and Development Program of China (2007AA10A144, http://www.most.gov.cn/eng/programmes1/200610/t20061009_36225.htm), the National Natural Science Foundation of China (30470983, http://www.973.gov.cn/English/Index.aspx) and Genetically Modified Organisms breeding Major Projects (2008ZX08001-004, http://www.most.gov.cn/index.htm). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.The Arabidopsis Genome Initiative. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- 2.Frantz J, Pinnock D, Klassen S, Bugbee B. Rice: Characterizing the environmental response of a gibberellic acid-deficient rice for use as a model crop. Agron J. 2004;96:1172–1181. [Google Scholar]
- 3. Rice Genome Annotation [ http://rice.plantbiology.msu.edu/riceInfo/info.shtml#Genes]
- 4.International Rice Genome Sequencing Project. The map-based sequence of the rice genome. Nature. 2005;436:793–800. doi: 10.1038/nature03895. [DOI] [PubMed] [Google Scholar]
- 5.Pastuglia M, Azimzadeh J, Camilleri C, Bouchez D. Forward and reverse genetics in Arabidopsis: isolation of cytoskeletal mutants. Cell Biol Intl. 2003;27:249–250. doi: 10.1016/s1065-6995(02)00306-2. [DOI] [PubMed] [Google Scholar]
- 6.Bouchez D, Höfte H. Functional genomics in plants. Plant Physiol. 1998;118:725–732. doi: 10.1104/pp.118.3.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Upadhyaya NM. Rice Functional Genomics—Challenges, Progress and Prospects. New York: Springer; 2007. 500 [Google Scholar]
- 8.An G. T-DNA Tagging Lines. In: Hirono H, Sano Y, Hirai A, Sasaki T, editors. Biotechnology in Agriculture and Forestry: Rice Biology in the Genomics Era. Vol. 62. New York: Springer; 2008. pp. 95–106. [Google Scholar]
- 9.Bernstein E, Caudy A, Hammond S, Hannon G. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- 10.Odell JT, Nagy F, Chua N-H. Identification of DNA sequences required for activity of the cauliflower mosaic virus 35S promoter. Nature. 1985;313:810–812. doi: 10.1038/313810a0. [DOI] [PubMed] [Google Scholar]
- 11.Christensen A, Sharrock R, Quail P. Maize polyubiquitin genes: structure, thermal perturbation of expression and transcript splicing, and promoter activity following transfer to protoplasts by electroporation. Plant Mol Biol. 1992;18:675–689. doi: 10.1007/BF00020010. [DOI] [PubMed] [Google Scholar]
- 12.Suwannaketchanatit C, Chaisuk P, Piluek J, Peyachoknakul S. Evaluation of constitutive promoters for gene expression in Dendrobium protocorms and flowers. Kasetsart J (Nat Sci) 2006;40:934–943. [Google Scholar]
- 13.Rooke L, Byrne D, Salgueiro S. Marker gene expression driven by the maize ubiquitin promoter in transgenic wheat. Ann Appl Biol. 2000;136:167–172. [Google Scholar]
- 14.Nakatsuka T, Pitaksutheepong C, Yamamura S, Nishihara M. Induction of differential flower pigmentation patterns by RNAi using promoters with distinct tissue-specific activity. Plant Biotechnol Rep. 2007;1:251–257. [Google Scholar]
- 15.Hirsche J, Engelke T, Völler D, Götz M, Roitsch T. Interspecies compatibility of the anther specific cell wall invertase promoters from Arabidopsis and tobacco for generating male sterile plants. Theor Appl Genet. 2009;118:235–345. doi: 10.1007/s00122-008-0892-2. [DOI] [PubMed] [Google Scholar]
- 16.Masclaux FG, Charpenteau M, Takahashi T, Pont-Lezica R, Galaud J-P. Gene silencing using a heat-inducible RNAi system in Arabidopsis. Biochem Biophys Res Commun. 2004;321:364–369. doi: 10.1016/j.bbrc.2004.06.154. [DOI] [PubMed] [Google Scholar]
- 17.Gatz C, Frohberg C, Wendenburg R. Stringent repression and homogeneous derepression by tetracycline of a modified CaMV 35S promoter in intact transgenic tobacco plants. Plant J. 1992;2:397–404. doi: 10.1111/j.1365-313x.1992.00397.x. [DOI] [PubMed] [Google Scholar]
- 18.Mett VL, Lochhead LP, Reynolds PH. Copper-controllable gene expression system for whole plants. Proc Natl Acad Sci U S A. 1993;90:4567–4571. doi: 10.1073/pnas.90.10.4567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ait-Ali T, Rands C, Harberd NP. Flexible control of plant architecture and yield via switchable expression of Arabidopsis gai. Plant Biotechnol J. 2003;1:337–343. doi: 10.1046/j.1467-7652.2003.00030.x. [DOI] [PubMed] [Google Scholar]
- 20.Zuo J, Chua NH. Chemical-inducible systems for regulated expression of plant genes. Curr Opin Biotechnol. 2000;11:146–151. doi: 10.1016/s0958-1669(00)00073-2. [DOI] [PubMed] [Google Scholar]
- 21.Vasil IK. Molecular Improvement of Cereal Crops. Dordrecht: Kluwer Academic Publishers; 1999. 500 [Google Scholar]
- 22.Yuan J, Chen D, Ren Y, Zhang X, Zhao J. Characteristic and expression analysis of a metallothionein gene, OsMT2b, down-regulated by cytokinin suggests functions in root development and seed embryo germination of rice. Plant Physiol. 2007;146:1637–1650. doi: 10.1104/pp.107.110304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang L, Sun Q, Qin F, Li C, Zhao Y, et al. Down-regulation of a SILENT INFORMATION REGULATOR2-related histone deacetylase gene, OsSRT1, induces DNA fragmentation and cell death in rice. Plant Physiol. 2007;144:1508–1519. doi: 10.1104/pp.107.099473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jan A, Nakamura H, Handa H, Ichikawa H. Gibberellin regulates mitochondrial pyruvate dehydrogenase activity in rice. Plant Cell Physiol. 2006;47:244–253. doi: 10.1093/pcp/pci241. [DOI] [PubMed] [Google Scholar]
- 25.Cherian MG, Jayasuryab A, Bay B-H. Metallothioneins in human tumors and potential roles in carcinogenesis. Mutat Res/Fundam Mol Mech Mutage. 2003;533:201–209. doi: 10.1016/j.mrfmmm.2003.07.013. [DOI] [PubMed] [Google Scholar]
- 26.Leszczyszyn OI, Schmid R, Blindauer CA. Toward a property/function relationship for metallothioneins: Histidine coordination and unusual cluster composition in a zinc-metallothionein from plants. Proteins. 2007;68:922–935. doi: 10.1002/prot.21463. [DOI] [PubMed] [Google Scholar]
- 27.Wong HL, Sakamoto T, Kawasaki T, Umemura K, Shimamoto K. Down-regulation of metallothionein, a reactive oxygen scavenger, by the small GTPase OsRac1 in rice. Plant Physiol. 2004;135:1447–1456. doi: 10.1104/pp.103.036384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Steffens B, Sauter M. Epidermal cell death in rice is confined to cells with a distinct molecular identity and is mediated by ethylene and H2O2 through an autoamplified signal pathway. Plant Cell. 2009;21:184–196. doi: 10.1105/tpc.108.061887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smith JS, Brachmann CB, Celic I, Kenna MA, Muhammadi S, et al. A phylogenetically conserved NAD+-dependent protein deacetylase activity in the Sir2 protein family. Proc Natl Acad Sci U S A. 2000;97:6658–6663. doi: 10.1073/pnas.97.12.6658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brachmann CB, Sherman JM, Devine SE, Cameron EE, Pillus L, et al. The SIR2 gene family, conserved from bacteria to humans, functions in silencing, cell cycle progression, and chromosome stability. Genes Dev. 1995;9:2888–2902. doi: 10.1101/gad.9.23.2888. [DOI] [PubMed] [Google Scholar]
- 31.Fu W, Wu K, Duan J. Sequence and expression analysis of histone deacetylases in rice. Biochem Biophys Res Commun. 2007;356:843–850. doi: 10.1016/j.bbrc.2007.03.010. [DOI] [PubMed] [Google Scholar]
- 32.Good L. Translation repression by antisense sequences. Cell Mol Life Sci. 2003;60:854–861. doi: 10.1007/s00018-003-3045-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Praveena S, Mishra AK, Dasgupta A. Antisense suppression of replicase gene expression recovers tomato plants from leaf curl virus infection. Plant Sci. 2005;168:1011–1014. [Google Scholar]
- 34.Coopera SR, Taylora JK, Miragliaa LJ, Dean NM. Pharmacology of antisense oligonucleotide inhibitors of orotein expression. Pharmacol Ther. 1999;82:427–435. doi: 10.1016/s0163-7258(99)00002-9. [DOI] [PubMed] [Google Scholar]
- 35.van der Krol AR, Mur LA, de Lange P, Mol JNM, Stuitje AR. Inhibition of flower pigmentation by antisense CHS genes: promoter and minimal sequence requirements for the antisense effect. Plant Mol Biol. 1990;14:457–466. doi: 10.1007/BF00027492. [DOI] [PubMed] [Google Scholar]
- 36.Lin H, Ouyang S, Egan A, Nobuta K, Haas BJ, et al. Characterization of paralogous protein families in rice. BMC Plant Biol. 2008;8:18. doi: 10.1186/1471-2229-8-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Elomaa P, Helariutta Y, Kotilainen M, Teeri TH. Transformation of antisense constructs of the chalcone synthase gene superfamily into Gerbera hybrida: differential effect on the expression of family members. Molecular Breed. 1996;2:41–50. [Google Scholar]
- 38.Wu L, Zhang Q, Zhou H, Ni F, Wu X, et al. Rice microRNA effector complexes and targets. Plant Cell. 2009;21:3421–3435. doi: 10.1105/tpc.109.070938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hiei Y, Ohta S, Komari T, Kumashiro T. Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J. 1994;6:271–282. doi: 10.1046/j.1365-313x.1994.6020271.x. [DOI] [PubMed] [Google Scholar]
- 40.Yadetie F, Sandvik AK, Bergum H, Norsett K, Laegreid1 A. Miniaturized fluorescent RNA dot blot method for rapid quantitation of gene expression. BMC Biotechnol. 2004;4:12. doi: 10.1186/1472-6750-4-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yu S-M, Kuo Y-H, Sheu G, Sheu Y-J, Liu L-F. Metabolic derepression of alpha-amylase gene expression in suspension-cultured cells of rice. J Biol Chem. 1991;266:21131–21137. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Construction of RNAi/antisense transgenes.
(DOC)