Abstract
Genetic material in plants is distributed into nucleus, plastids and mitochondria. Plastid has a central role of carrying out photosynthesis in plant cells. Plastid transformation is becoming more popular and an alternative to nuclear gene transformation because of various advantages like high protein levels, the feasibility of expressing multiple proteins from polycistronic mRNAs, and gene containment through the lack of pollen transmission. Recently, much progress in plastid engineering has been made. In addition to model plant tobacco, many transplastomic crop plants have been generated which possess higher resistance to biotic and abiotic stresses and molecular pharming. In this mini review, we will discuss the features of the plastid DNA and advantages of plastid transformation. We will also present some examples of transplastomic plants developed so far through plastid engineering, and the various applications of plastid transformation.
Keywords: Genetic engineering, genome, plastid transformation, plastome sequencing.
INTRODUCTION
Genetic material in plants is distributed into nucleus and the chloroplast and mitochondria in the cytoplasm. Each of these three compartments carries its own genome and expresses heritable traits [1, 2]. The chloroplast is one of organelles known as plastids in plant cells and eukaryotic algae [3]. According to Verhounig et al. [4], plastids and mitochondria are derived from formerly free-living bacteria and have largely prokaryotic gene expression machinery. The plastid (biosynthetic centre of the plant cell) carries out photosynthesis, in plant cells and eukaryotic algae, which provides the primary source of the world's food [3]. There are other important activities that occur in plastids. These include sequestration of carbon, production of starch, evolution of oxygen, synthesis of amino acids, fatty acids, and pigments, and key aspects of sulfur and nitrogen metabolism [5]. In spite of the prokaryotic past of the plastids, their gene expression has very different regulatory mechanisms from those operating in bacteria [6]. There are up to 300 plastids [7] in one plant cell. The plastid genome (plastome or plastid DNA, ptDNA), 1,000–10,000 copies per cell [8], contrasts strikingly with the nuclear DNA. In most species, plastids are usually strictly maternally inherited [9] in most (80%) angiosperm plant species [10, 11]. It is also not influenced by polyploidy, gene duplication and recombination that are widespread features of the nuclear genomes of plants [12, 13]. Therefore, ptDNA varies little among angiosperms in terms of size, structure and gene content [14]. Currently 170 chloroplast genomes from different species have been completely sequenced (NCBI Organelle Genome Resources; http://www.ncbi.nlm.nih.gov/genomes/). These also include many agriculturally important plant species like rice [15, 16] maize [17] sugarcane [18], wheat [19, 20], tomato [21], and mungbean [22] (Table 1). The plastid genome was determined to be circular double-stranded DNA through construction of complete genome maps. Its size ranges from 120.000 to 180.000 base pairs, depending on the species, that encode ~120 genes. Each plastid genome constitutes in almost all higher plant species of a large single copy (LSC), a small single copy (SSC), and duplication of a large (~25 kb) region (IRs) in an inverted orientation [2, 23].
Table 1.
A List of Sequenced Plastomes of Some Agriculturally Important Plants
| Plant species | Base pairs | Reference |
|---|---|---|
| Arabidopsis thaliana | 154,478 | [131] |
| Brassica napus | - | [132] |
| Citrus sinensis | 155,189 | [133] |
| Coffea arabica | - | [134] |
| Cucumis sativus | 155,293 | [135] |
| Daucus carota | 155,911 | [136] |
| Ficus sp. | - | [137] |
| Glycine max | 152,218 | [138] |
| Gossypium barbadense | 160,317 | [139] |
| Gossypium hirsutum | 160,301 | [140] |
| Helianthus annuus | 151,104 | [141] |
| Hordeum vulgare, Sorghum bicolor | - | [142] |
| Lycopersicon esculentum | 155,460 | [21] |
| Manihot esculenta | - | [143] |
| Morus indica | 156,599 | [144] |
| Musa acuminata | - | [145] |
| Nicotiana tabacum | 155,943 | [146] |
| Oryza sativa | 134,551 | [15] |
| Oryza nivara | 134,494 | [16] |
| Phaseolus vulgaris | 150,285 | [147] |
| Saccharum officinarum | 141,182 | [18] |
| Solanum tuberosu | 155,298 | [148] |
| Spinacia oleracea | 150,725 | [149] |
| Triticum aestivum | 134,545 | [19, 20] |
| Vitis vinifera | 160,928 | [150] |
| Zea mays | 22,784 | [17] |
| Jatropha curcas | 163,856 | [151] |
| Vigna radiata | 151,271 | [22] |
Genetic engineering has been experienced mostly in the nuclear genome [24, 25]. Inserting transgene(s) into the nuclear genome, however, has led to an increasing public concern of the possibility of escape of the transgene through pollen to wild or weedy relatives of the transgenic crops [26]. Scientists argued that since plastids are compared with prokaryotes, they can take up DNA as in bacterial transformation using naked DNA (http://www.studentsguide.in/ plant-biotechnology-genomics/transplastomic-plants-chloroplast-engineering/advantages-of-chloroplast-transformation. html). Therefore, during the past few years, researchers have begun to evaluate application of plastid engineering (transformation) in plant biotechnology as a viable alternative to conventional technologies for transformation of the nuclear DNA [27]. Recently, plastids have become attractive targets for genetic engineering efforts [28]. Plants with transformed plastid genomes are termed transplastomic [29].
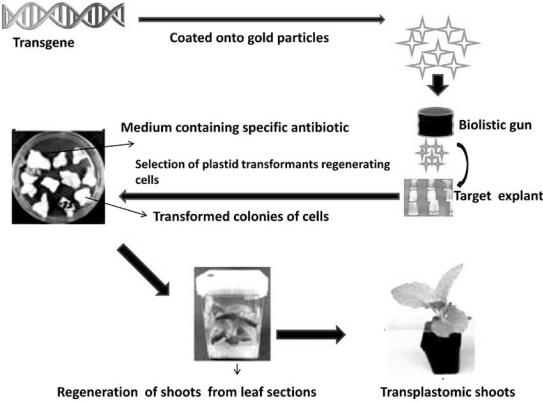
Diekmann et al. [30] believe that in order to develop plastid engineering, obtaining plastid genome sequences is crucial, and that efficient sequencing requires pure plastid DNA. The authors, therefore, developed a simple and inexpensive method to obtain plastid DNA from grass species by modifying and extending protocols optimized for the use in eudicots. Plastid engineering involves the targeting of foreign genes to the plastid's double-stranded circular DNA genome instead of chromosomal DNA [31], and as a consequence the production of the foreign protein of interest (Fig. 1). The advancement in the particle gun mediated transformation has enabled targeting the plastid genome for developing transgenic plants against many biotic (e.g. insects and pathogens) and abiotic stresses (e.g. drought and salinity), which reduce the plant productivity. Improving the quality of fruits has been another main target in plastid transformation [32].
Fig. (1).
Plant plastid engineering.
Plastid transformation was first achieved in a unicellular alga, Chlamydomonas reindhartii [33]. In 1990, Sváb et al. [34] reported the first successful chloroplast transformation of a higher plant (tobacco). This was followed by transformation of the plastid genome in tobacco by many researchers [35, 36]. Recently, tobacco plastid has been engineered to express the E7 HPV type 16 protein, which is an attractive candidate for anticancer vaccine development [37]. Similarly, a protocol for plastid transformation of an elite rapeseed cultivar (Brassica napus L.) has been developed [38]. More recently, a method for plastid transformation in eggplant (Solanum melongena L.) has been reported with pPRV111A plastid expression vector carrying the aadA gene encoding aminoglycoside 300-adenylyltransferase [26]. The authors believe that this may open up exciting possibilities to introduce and express novel genes in the engineered plants via plastid transformation for agronomic or pharmaceutical traits. Up to date, plastid transformation has been extended to many other higher plants, such as Arabidopsis thaliana [39], potato [40, 41], tomato [1, 42], Lesquerella fendleri, a kind of oilseed Brassicaceae [43], oilseed rape, [38, 44], petunia [45], lettuce [46], soybean [47], cotton [48], carrot [49], rice [50], poplar [51], tobacco, [28, 52, 53], mulberry , [54] and eggplant [26] (see review written by Wang et al. [3]).
Two interesting applications of plastid transformation were carried out by (i) [55] for the construction of a tobacco master line to improve Rubisco engineering in plastids, and (ii), [4] who explored the possibility of engineering riboswitches (natural RNA sensors that regulate gene expression in response to ligand binding) to function as translational regulators of gene and transgene expression in plastids.
The dominant trait that attracted the most attention for plastid transformation has been herbicide tolerance [28, 35, 56, 57]. Roudsari et al. [28] revealed the production of high level glyphosate tolerant plants (N. tabacum) through biolistic transformation of plastids by introduction of a mutated herbicide-tolerant gene coding for EPSP synthase. Plastid transformation is routine, however, only in tobacco and the efficiency of transformation is much higher in tobacco than in other plants [58]. Lee et al. [50] discussed the major obstacles to the extension of plastid transformation technology to other crop plants which includes regeneration via somatic embryogenesis.
ADVANTAGES OF PLASTID ENGINEERING
Production of transgenic plants, at laboratory level or commercially, has traditionally been mainly through expression of transgenes in the nucleus [24, 25]. Among the ecological concerns raised about genetically engineered organisms is that transgenes could move ("transgene flow", the process of transgene movement by recurrent hybridisation) via pollen from the crop and into relatives growing in natural or semi-natural communities [59]. Such concerns have led to a new field of transgene containment [60, 61].
Since plastids are inherited maternally in the majority of angiosperm species, they would therefore not be found in pollen grains of corps. Insertion of transgenes, therefore, into the plastid genome has the potential of preventing gene flow via pollen. Hence, genes expressed in the plastome will not be transferred through pollination to weedy or wild relatives of the transgenic crop. Bansal and Sharma, [62] believes that there is little risk of any transgene flow via pollen from transplastomic plants to the neighboring weedy or wild relatives since plastids are almost always maternally transferred to the next progeny. The authors suggested that plastid transformation could be a method of choice for generating improved transgenics in crops that grow along with their weedy or wild relatives in the same geographical region, such as rice, sorghum, cucurbits, solanaceous crops, Vigna and Cajanus species, and various Brassica crops. Therefore, the focus of many researchers has shifted to plastid engineering [26], rather than nuclear transformation. Singh et al. 2010 [26] reported that engineering of the plastid genome is gaining momentum as an attractive alternative to nuclear transformation.
Ruf et al. [60] believe that plastid transformation is considered as a superb tool for ensuring transgene containment and improving the biosafety of transgenic plants. However, they pointed out that plastid transformation would only be effective as a biocontainment measure when applied on a landscape scale if it were combined additional mechanisms such as mitigating genes genetic use restriction technology, and/or male sterility [63]. In a recent study, it has been demonstrated that the use of plastid transformation would provide an imperfect biocontainment for GM oilseed rape (Brassica napus L.) in the United Kingdom [64]. In another study, Allainguillaume et al. [65] revealed that chloroplast transformation may slow transgene recruitment in two settings, but actually accelerate transgene spread in a third. Plastid transformation has become an attractive alternative to nuclear gene transformation due to several other advantages [3]. The high ploidy number of the plastid genome allows high levels (up to 1-40% of total protein) of protein expression or expression of the transgene [28]. Daniell et al. [66] and Hou et al. [44] reported that while nuclear transgenes typically result in 0.5 - 3% of total proteins, concentration of proteins expressed by plastid transgenes is much higher; up to 18%. The greater production of the expressed protein is possible because plastid transgenes are present as multiple copies per plant cell, and they are little affected by phenomena like pre- or post-transcriptional silencing. Other advantages of plastid engineering are the capacity to express multiple genes from polycistronic messenger RNA (mRNA) [31], and the absence of epigenetic effects and gene silencing [40].
Wang et al. [3] believe that transgene stacking in operons and a lack of epigenetic interference allowing stable transgene expression. Added to that, plastid transformation is more environmental friendly than transformation of the nuclear DNA for plant engineering because it eliminates the possibility of toxic transgenic pollen to nontarget insects [67]. Adverse effects of toxic proteins might be minimized by plastid compartmentalization but in case of nuclear transformation, toxic proteins accumulating within the cytosol might result in serious pleiotropic effects. Further, the expression of the transgene in case of plastid transformation is more uniform compared to that of trangenes inserted into the nuclear genome. Although there is a major drawback in the engineering of plastid gene expression, which is the lack of tissue-specific developmentally regulated control mechanisms [3], the many advantages of plastid engineering stated above attracted researchers to engineer the plastid genome to confer several useful agronomic traits, and hence the number of species whose plastome can be transformed continues to expand [68].
GENE DELIVERY INTO PLASTIDS
Gene delivery into the plastome was initially done by Agrobacterium mediated method [Block et al.1985] [69]. The discovery of biolistic DNA delivery led to plastid transformation via particle gun [Sanford 1990] [70]. In this method, the Escherichia coli plasmids contain a marker gene and the gene of interest is introduced into plastids. The foreign genes are inserted into plasmid DNA by homologous recombination via the flanking sequences at the insertion site [66]. Polyethylene glycol (PEG) mediated transformation of plastids was also utilized [71, 72]. PEG-mediated transformation of plastids requires enzymatically removing the cell wall to obtain protoplasts, then exposing the protoplasts to purified DNA in the presence of PEG. The protoplasts first shrink in the presence of PEG, then lyse due to disintegration of the cell membrane. Removing PEG before the membrane is irreversibly damaged reverses the process. Biolistic delivery is the routine system for most laboratories, as manipulation of leaves, cotyledons, or cultured cells in tissue culture is a simple practice than the alternative PEG treatment of protoplasts [58]. Recently, particle gun mediated plastid transformation has been demonstrated in rapeseed using cotyledons as explants [38].
The major difficulty in engineering plastid genome for production of transplastomic plants is in generating homoplasmic plants in which all the plastids are uniformly transformed, for that takes a long process of selection, thus hampering the production of genetically stable transplastomic plants (e.g. rice). This is due to the presence of about 10-100 plastids, each of which has up to 100 copies of the plastid genome, in one cell, that does not allow achieving homoplastomic state [73]. It was also stated that getting high level of protein expression, even though the gene copy number is high, is another problem. In 2005, however, Nguyen et al. [41] described the generation of homoplasmic plastid transformants of a commercial cultivar of potato (Solanum tuberosum L.) using two tobacco specific plastid transformation vectors, pZS197 (Prrn/aadA/psbA3′) and pMSK18 (trc/gfp/Prrn/aadA/psbA3′). Similarly, Liu et al. [74] were able to develop homoplasmic fertile plants of Brassica oleracea L. var. capitata L. (cabbage). Among other higher plants of which fertile homoplasmic plants with genetically modified plastid genomes have be been produced are Nicotiana tabacum (tobacco), Nicotiana plumbaginifolia (texmex tobacco), Solanum lycopersicum (tomato), Glycine max (soybean), Lesquerella fendleri (bladderpod), Gossypium hirsutum (cotton), Petunia hybrida (petunia), and Lactuca sativa (lettuce) [68]. The amino glycoside 3-adenylyltransferase (aadA) gene, which confers dual resistance to spectinomycin-streptomycin antibiotics, is still the selectable marker that is routinely used efficiently for plastid transformation [58, 75, 76]. Since the antibiotic resistant genes used in transformation are not desirable in the final products, different strategies have been developed to eliminate the necessity of using such selectable markers [56, 77].
IMPROVEMENT OF SOME AGRONOMIC TRAITS BY PLASTID ENGINEERING
Apel & Bock, [42] demonstrated the potential of plastids genome engineering for the nutritional enhancement of food crops when they enhanced carotenoid biosynthesis in transplastomic tomatoes by induced lycopene-to-provitamin A conversion. The transplastomic technology could also be useful for engineering agronomic traits including phytoremediation [78] reversible male sterility [79], and tolerance/resistance of stresses such as diseases, drought, insect pests, salinity and freezing that can severely limit plant growth and development [3].
Since plastids are transferred mostly through the “maternal inheritance” as identical copies, and hence a female plant transfers identical copies to all the seeds it produces without changes from one generation to the next, an important promise for applying plastid transformation for industry is the stable passing on to the next generation of the foreign DNA [80]. Therefore, the plastid genome has also been utilized for metabolic pathway engineering and in the field of molecular farming (the production of drugs and chemicals through engineered crops) [27, 68] for the expression and production of biomaterials and biopharmaceuticals in plants, human therapeutic proteins, and vaccines for use in humans or animals (reviewed in [26, 27, 50]. Singh et al. [26] believe that for such applications, plastid transformation technology offers solutions to the ecological and technical problems associated with conventional transgenic technologies such as outcrossing and transgene silencing.
Insect Resistance
Transformation of the nuclear genome of plants with genes (e.g. Bt genes) to confer insect resistance gives very low levels of expression unless extensive modifications are carried. Whereas, introduction of the same genes into the plastid genome results in high levels of toxin accumulation as the plastid genome is bacterial in origin [81]. Therefore, the insect resistance genes were investigated for high-level expression from the plastid genome [3]. Hence, when insect resistance genes are expressed into the plastid genome, leaves of these transplastomic plants proved highly toxic to herbivorous insect larvae.
One of the major advantages of introducing the Bt toxin into the plastid genome is the high levels of toxin accumulation (3% – 5% of total leaf protein as compared to > 0.2% of total soluble protein through nuclear genome transformation) [82]. Achievement of stable transformation of the plastid genome and transforming plastids in species other than tobacco (Nicotiana tabacum) are some of the hurdles for widespread adaptation of this technique [81]. Despite of overwhelming odds, many attempts have been made to produce transplastomic plants expressing Bt toxin for increased resistance against insect pests. [83] generated soybean plastid transformants expressing Bacillus thuringiensis Cry1Ab protoxin. Similarly, Chakrabarti et al. [84] reported the control of potato tuber moth (Phthorimaea operculella) by incorporating a truncated Bacillus thuringiensis cry9Aa2 gene in the plastid genome. The authors observed high-level expression (about 10% of total soluble protein) of the cry gene from the plastid genome which resulted in severe growth retardation.
Over-expression of the cry2Aa2 operon in plastids was proved effective in allowing a broad-spectrum of protection against a range of pests [81]. In cabbage, the cry1Ab gene was also successfully transferred into the plastid genome [85]. Expression of cry1Ab protein was detected in the range of 4.8–11.1% of total soluble protein in transgenic mature leaves of the two species. Insecticidal effects on Plutella xylostella were also demonstrated in cry1Ab transformed cabbage. In an attempt to increase insect resistance in transgenic rice plants, a synthetic truncated cry1Ac gene was linked to the rice rbcS promoter and its transit peptide sequence (tp) for plastid-targeted expression [86]. Use of the rbcS-tp sequence increased the cry1Ac transcript and protein levels by 25- and 100-fold, respectively, with the accumulated protein in plastids comprising up to 2% of the total soluble proteins. The high level of cry1Ac expression resulted in high levels of plant resistance to three common rice pests, rice leaf folder, rice green caterpillar, and rice skipper, as evidenced by insect feeding assays. It was concluded that targeting of cry1Ac protein to the plastid using the rbcS:tp system confers a high level of plant protection to insects. Several other cry proteins have also been expressed in plastids of tobacco [83, 84, 87, 88] and rice [50] (see Table 2).
Table 2.
A List of Some Transplastomic Plants that were Engineered for Various Agronomic Traits
| Plant species | Gene introduced | Reference |
|---|---|---|
| Nicotiana tabacum | rrn16 | [152] |
| Nicotiana tabacum | nptII | [153] |
| Nicotiana tabacum | uidA | [154] |
| Nicotiana tabacum | Human somatotropin (hST) | [155] |
| Nicotiana tabacum | cry | [88] |
| Nicotiana tabacum | cry9Aa2 | [84] |
| Nicotiana tabacum | Bar & aadA | [156] |
| Nicotiana tabacum | Cor 15a-FAD7 | [157] |
| Nicotiana tabacum | rbcL | [55] |
| Nicotiana tabacum | DXR | [158] |
| Nicotiana tabacum | aadA & gfp | [60] |
| Nicotiana tabacum | Delta(9) desaturase | [159] |
| Nicotiana tabacum | AsA2 | [160] |
| Nicotiana tabacum | PhaG & PhaC | [161] |
| Nicotiana tabacum | gfp | [4] |
| Nicotiana tabacum | A1AT | [162] |
| Arabidopsis thaliana | aadA | [39] |
| Solanum tuberosum | aadA & gfp | [40] |
| Oryza sativa | aadA & gfp | [50] |
| Solanum lycopersicon | aadA | [1] |
| Solanum lycopersicon | Lyc | [42] |
| Brassica napus | aadA & cry1Aa10 | [44] |
| Brassica napus | aadA | [38] |
| Lesquerella fendleri | aadA & gfp | [43] |
| Daucus carota | dehydrogenase (badh) | [48] |
| Gossypium hirsutum | aphA-6 | [49] |
| Glycine max | aadA | [47] |
| Petunia hybrida | aadA & gusA | [45] |
| Lactuca sativa | gfp | [46] |
| Brassica oleracea | gus & aadA | [163] |
| Lettuce | gfp | [164] |
| Populus alba | gfp | [51] |
| Brassica oleracea | aadA & uidA | [74] |
| Beta vulgaris | aadA & uidA | [165] |
| Crocus sativus | CstLcyB1& CstLcyB2a | [166] |
| Solanum melongena | aadA | [26] |
| Arabidopsis thaliana | pre-Tic40-His | [167] |
| Zea mays | ManA | [168] |
Disease Resistance
Plastid engineering offers a new and effective option in development of plant varieties which are resistant to various bacterial and fungal diseases. In tobacco, introduction of MSI-99 gene, an antimicrobial peptide, into plastids resulted in transplastomic plants resistant to fungal pathogen Colletotrichum destructive [89]. The plastids expressed MSI-99 at high levels and showed 88% (T1) and 96% (T2) inhibition of growth against Pseudomonas syringe, which is a major plant pathogen. In another study, Agrobacterium mediated transformation was used to develop tobacco plants carrying argK gene, which encodes ROCT [90]. Since OCT in plant cells is produced in the plastid, argK was fused to the plastid transit sequence of the pea rubisco small subunit (rbcS) gene for localized expression of the enzyme. The ROCT enzyme produced by the transgenic tobacco showed greater resistance (83-100%) to phaseolotoxin compared to the wild-type OCT (0-22%). When phaseolotoxin was applied exogenously to the leaves of plants, chlorosis was observed in 100% of wild-type tobacco, but not seen in the leaves of the transgenic tobacco plants carrying the argK gene from P. syringae pv. phaseolicola. Transgenic tobacco plants that constitutively expressed both entC and pmsB in the plastid have also been reported [91] where transformation was accomplished through biolistic methods. The transgenic tobacco plants expressing these bacterial genes showed accumulation of salicylic acid that were up to 1000 times higher than that observed in wild-type tobacco. When challenged with the fungus Oidium lycopersicon, the transgenic tobacco plants showed increased levels of resistance compared to the wild-type plants. It was revealed that the transgenic plants generated did not show any adverse effects due to the high level expression of salicylic acid. Thus gene transfer in plastids can provide a significant protection from various bacterial and fungal diseases.
Drought and Salinity Tolerance
Transgenes that confer tolerance to abiotic stress may permanently transfer from transgenic crops to the nuclear genome of their weedy relatives which may result in drought tolerant superweeds [92] when the gene is inserted into the nuclear genome and the transgenic plant outcross with relative weeds. There is a great potential, therefore, for the genetic manipulation of key enzymes involved in stress metabolism in plants within plastids. Because plastid genomes of major crops including cotton and soybean have been successfully transformed, this offers an exciting new approach to create transgenic plants with abiotic stress tolerance [93]. Therefore, the authors believe that there appears to be tremendous potential for increasing tolerance in plants to a number of stresses by expression of appropriate genes within plastids due to the maternal inheritance of transgenes that confer tolerance to abiotic stress. Plastid engineering had been successfully applied for the development of plants with tolerance to salt, drought [94] and low temperature [reviewed in 3]. Djilianov et al. [95] demonstrated that enhanced tolerance to abiotic stresses has been achieved when the gene was directed to plastid genome.
Among many strategies used for development of abiotic stress tolerance in plants, the over-expression of compatible osmolytes like glycinebetaine was found to be successful [24]. Initial attempts for producing transplastomic plants through the introduction of CMO (Choline monooxygenase) and betaine aldehyde dehydrogenase (BADH) pathway were made in tobacco. Tobacco plants were transformed with cDNA for BADH from spinach (Spinacia oleracea) and sugar beet (Beta vulgaris) under the control of CaMV 35 S promoter. The BADH was produced in plastids of tobacco. Betaine aldehyde was converted to betaine by BADH, thus conferring resistance to betaine aldehyde. In another attempt, cDNA for choline monooxygenase from Spinacia oleracea was introduced into tobacco and the enzyme thus synthesized was transported to its functional place i.e., plastids. But the leaves of tobacco accumulated betaine at a very low concentration i.e., 10-100 folds lower [96]. The reason for insufficient synthesis of betaine most probably was the absence of engineered BADH activity in plastids. Therefore, both CMO and BADH need to be present in the plastids for efficient synthesis of betaine in transgenic plants which do not accumulate glycinebetaine. In carrot (Daucus carota), homoplasmic transgenic plants exhibiting high levels of salt tolerance were regenerated from bombarded cell cultures via somatic embryogenesis [48]. BADH enzyme activity was enhanced 8-fold in transgenic carrot cell cultures, grew 7-fold more, and accumulated 50- to 54-fold more betaine than untransformed cells grown in liquid medium containing 100 mM NaCl. Transgenic carrot plants expressing BADH grew in the presence of high concentrations of NaCl (up to 400 mM), the highest level of salt tolerance reported so far among genetically modified crop plants. Further, a gene for CMO, cloned from spinach (Spinacia oleracea) was introduced into rice through Agrobacterium mediated transformation. The level of glycinebetaine in rice was low to the expectations. The author has given several reasons for the low productivity of rice and low glycinebetaine accumulation. Firstly, the position of spinach CMO and endogenous BADH might be different and secondly the catalytic activity of spinach CMO in rice plants might be lower than it was in spinach [97]. Transplastomic plants constitutively expressing BvCMO under the control of the ribosomal RNA operon promoter and a synthetic T7 gene G10 leader were able to accumulate glycinebetaine in leaves, roots and seeds, and exhibited improved tolerance to toxic level of choline and to salt/drought stress when compared to wild type plants. Transplastomic plants showed higher net photosynthetic rate and apparent quantum yield of photosynthesis in the presence of 150 mM NaCl [98]. Thus, it can be concluded that glycinebetaine has a role as compatible solute and its engineering into non-accumulations will be a success only if both CMO and BADH pathways are introduced and if the localization of both CMO and BADH is in plastids. Very recently, George et al. [99] demonstrated how a chloroplast-localized and auxin-induced glutathione S-transferase from phreatophyte Prosopis juliflora conferred drought tolerance on tobacco. For more examples of conferring tolerance to abiotic stress to plants via plastid engineering, see review by [3].
PRODUCTION OF BIOPHARMACEUTICALS AND VACCINES IN PLANTS
Mulesky et al. [100] pointed out two reasons that make using crops to produce drugs interesting for industry. These are (i) crops can be employed more efficiently in this process than animals or bacteria, with a larger output achieved with fewer resources, and (ii) the oral delivery of the drugs produced to people and animals is easier. A third reason is the high-level production of antigens for use as vaccines and their tests for immunological efficacy in animal studies [3]. The hyper-expression of vaccine antigens or therapeutic proteins in transgenic chloroplasts (leaves) or chromoplasts (fruits/roots) and antibiotic-free selection systems available in plastid transformation systems made possible the oral delivery of vaccine antigens against cholera, tetanus, anthrax, plague, and canine parvovirus, [101-104] and reviews of [102, 105, 3 and 106] explained why plastid engineering can be regarded as an attractive strategy and environmentally friendly approach for the production of vaccines, therapeutic proteins, and biomaterials, and provided some examples. Kumar & Daniell [107] described various techniques for creating plastid transgenic plants and their biochemical and molecular characterization. They also provided suitable examples for application of chloroplast genetic engineering in human medicine.
Wang et al. [3] also discussed applying plastid transformation for metabolic pathway engineering in plants, the production of biopharmaceuticals, and marker gene excision system and how plastid transformation can be applied to study RNA editing. Similarly, Hefferon [67], considers plastid engineering as a valuable tool that gives enormous promise for the production of biopharmaceuticals and vaccines, because higher level of the protein expressed by the transgene inserted into the plastid genome can be achieved. Many vaccine antigens, which played a key role for the prevention of infectious diseases, and biopharmaceutical proteins, have been expressed at high levels via the chloroplast genome [108] and they proved to be functional using in vitro assays in cell cultures. [109] stated that production of therapeutic proteins in plastids eliminates the expensive fermentation technology, and that the oral delivery of plastid-derived therapeutic proteins eliminates cold storage, cold transportation, expensive purification steps, and delivery via sterile needles, and hence decrease their cost. The main goal for applications of plastid transformation by the biotech industry is molecular pharming, and food production is considered as only a secondary target [80].
To create an edible vaccine, selected desired genes should be introduced into plants and then inducing these altered plants to manufacture the encoded proteins. Like conventional subunit vaccines, edible vaccines are composed of antigenic proteins and are devoid of pathogenic genes. Plastids of green plants as bioreactors for the production of vaccines and biopharmaceuticals are of great potential as indicated from a number of published studies [110-114]. The significance of using plants as production platforms for pharmaceuticals is due to the low production and delivery costs, easy scale-up and high safety standards regarding less risk of product contamination with human pathogen [27]. Keeping in view the high efficiency of plastids to express foreign genes, it is meaningful to explore this property of plastids for the production of proteinaceous pharmaceuticals, such as antigens, antibodies and antimicrobials. The candidate subunit vaccine against Clostridium tetani, causing tetanus was the first plastid-produced antigen that proved to be immunologically active in experimental animals [111]. In this initial attempt, fragment C of the tetanus toxin (TetC), a non-toxic protein fragment, was expressed from the tobacco plastid genome which resulted in high levels of antigen protein expression (30% of the plant’s total soluble protein (TSP)). Anthrax is an acute infectious disease caused by the spore-forming bacterium Bacillus anthracis. Significant development has been achieved towards the production of plastid-based vaccine for this infectious disease. Expression (14% of the plants TSP) of the pagA gene encoding the protective antigen (PA) from the tobacco plastid genome gave rise to stable antigen protein [112, 113]. The plastid-derived PA was equally effective in cytotoxicity assays as the bacterial protein produced in B. anthracis. The potential of plastid transformation as an alternative tool to produce high levels of HIV-1 Nef and p24 antigens in plant cells have been also demonstrated [115]. Different constructs were designed to express the p27 Nef protein either alone or as p24-Nef or Nef-p24 fusion proteins. All constructs were utilized to transform tobacco (cv. Petite Havana) plastids and the transplastomic lines. Analysis of p24-Nef and Nefp24 fusion proteins showed that both can be expressed to relatively high levels in plastids. As the best results in terms of protein expression levels were obtained with the p24-Nef fusion protein, the correspondent gene was cloned in a new expression vector. This construct was introduced into the tobacco and tomato plastid genomes. Transplastomic tobacco and tomato plants were analyzed and protein accumulation was found to be close to 40% of the leaf’s total protein. Transcript and protein accumulation were analyzed in different ripening stage of tomato fruit and green tomatoes accumulated the fusion protein to 2.5% of the TSP [115]. Recently, a strategy for plastid production of antibiotics against pneumonia Streptococcus pneumonia has been outlined [116]. The authors describe it as a new technique for high level expression (to up to 30% of the plant’s TSP) of antmicrobial proteins that are toxic to E. coli. It was also shown that the plastid-produced antibiotics efficiently kill pathogenic strains of Streptococcus pneumoniae, the causative agent of pneumonia, thus providing a promising strategy for the production of next-generation antibiotics in plants. In 2007, Chebolu and Daniell [117] achieved a stable expression of Gal/GalNAc lectin of Entamoeba histolytica in transgenic chloroplasts and immunogenicity in mice towards vaccine development for amoebiasis. Shao et al. [118] reported the expression of the structural protein E2 of classical swine fever virus (CSFV), which has been shown to carry critical epitopes CFSV E2 gene in tobacco chloroplasts. In another study on tobacco, plastid transformation of the high-biomass tobacco variety `Maryland Mammoth` has been assessed by McCabe et al. [119] as a production platform for the human immunodeficiency virus type 1 (HIV-1) p24 antigen. Similarly, Meyers et al. 2008 [120] revealed the usefulness of plastid signal peptides in enhancing the production of recombinant proteins meant for use as vaccines. Transgenic plastids were also proved efficient for high-yield production of the vaccinia virus envelope protein A27L in plant cellsdagger by Rigano et al. [121], who revealed that chloroplasts are an attractive production vehicle for the expression of OPV subunit vaccines. Very recently, Youm et al. [122] were able to produce the human beta-site APP cleaving enzyme (BACE) via transformation of tobacco plastids. The authors argued that the successful production of plastid-based BACE protein has the potential for developing a plant-based vaccine against Alzheimer disease. Because recombinant extra domain A from fibronectin (EDA) could be used as an adjuvant for vaccine development, [104] aimed to express EDA from the tobacco plastome as a promising strategy in molecular farming. Tobacco plastids transformation was also evaluated by Lentz et al. [123] for the production of a highly immunogenic epitope containing amino acid residues 135-160 of the structural protein VP1 of the foot and mouth disease virus (FMDV). The authors concluded that this technology allows the production of large quantities of immunogenic proteins.
In spite of this huge success in applying plastid engineering for molecular pharming, Ho & Cummins [73] referred to several risks of plastid engineering in producing GM pharmaceuticals that are associated with its advantages. For more details on the use of plastid engineering for biotechnology applications, see review by Verma & Daniell [5]. Other valuable reviews are available on (a) generation of plants with transgenic plastids, summary of our current understanding of the transformation process and highlights on selected applications of transplastomic technologies in basic and applied research [124], (b) Progress in expressing proteins that are biomedically relevant, in engineering metabolic pathways, and in manipulating photosynthesis and agronomic traits and the problems of implementing the technology in crops [125], (c) plastid transformation in higher plants [56], (d) the characteristics, applications of chloroplast genetic engineering and its promising prospects, [126], (e) Engineering the chloroplast genome [127] (f) exciting developments in this field and offers directions for further research and development, [128] (g) the expression of resistance traits, the production of biopharmaceuticals and metabolic pathway engineering in plants [27], (h) chloroplasts as bioreactors, and whether we can replace plants with plants [129], (i) how plastid transformation played an important role in understanding the RNA editing [3] and (j) comparison of opportunities and challenges between nuclear and plastid genetic engineering of plants [130].
CONCLUSIONS
Up to date, many transgenes have been successfully introduced into the plastid genome of model plant tobacco and many other important crop plants for various agronomic traits. Initially, this technology was limited to model plant species, but now it has been extended to some other important crops. Still there are many agronomically important cereals crops in which plastid engineering has not yet been standardized. Plastid transformation has been proved to result in high levels of transgene expression. It has also provided a baseline for production of proteinaceous pharmaceuticals, such as antigens, antibodies and antimicrobials in a cost effective manner. Bock [27] believes that a great progress has been achieved over years in investigating the mechanisms that govern transgene expression from the plastid genome and in using this technology for biotechnological applications. We agree with the author that "the routine use of plastid engineering in biotechnology is still a long way off, but would surely benefit the humanity in the near future". There is no doubt that plastid engineering holds a great potential in plant biotechnology; but like every new technology, there are some challenges which need to be addressed before its widespread adoption. Among other important factors to be solved are the protein purification and expression level control.
REFERENCES
- 1.Ruf S, Hermann M, Berger IJ, Carrer H, Bock R. Stable genetic transformation of tomato plastids and expression of a foreign protein in fruit. Nat. Biotechnol. 2001;19:870–875. doi: 10.1038/nbt0901-870. [DOI] [PubMed] [Google Scholar]
- 2.Siguira M. The chloroplast genome. Plant Mol. Biol. 1992;19:149–168. doi: 10.1007/BF00015612. [DOI] [PubMed] [Google Scholar]
- 3.Wang HH, Yin WB, Hu ZM. Advances in chloroplast engineering. J. Genet. Genomics. 2009;36:387–98. doi: 10.1016/S1673-8527(08)60128-9. [DOI] [PubMed] [Google Scholar]
- 4.Verhounig A, Karcher D, Bock R. Inducible gene expression from the plastid genome by a synthetic riboswitch. Proc. Natl. Acad. Sci. USA. 2010;107:6204–6209. doi: 10.1073/pnas.0914423107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Verma D, Daniell H. Chloroplast vector systems for biotechnology applications. Plant Physiol. 2007;145:1129–1143. doi: 10.1104/pp.107.106690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barkan A, Goldschmidt-Clermont M. Participation of nuclear genes in chloroplast gene expression. Biochimie. 2000;82:559–572. doi: 10.1016/s0300-9084(00)00602-7. [DOI] [PubMed] [Google Scholar]
- 7.Boffey SA, Leech RM. Chloroplast DNA levels and the control of chloroplast division in light-grown wheat leaves. Plant Physiol. 1982;69:1387–1391. doi: 10.1104/pp.69.6.1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bendich AJ. Why do chloroplasts and mitochondria contain so many copies of their genome? BioEssays. 1987;6:279–282. doi: 10.1002/bies.950060608. [DOI] [PubMed] [Google Scholar]
- 9.Corriveau JL, Coleman AW. Rapid screening method to detect potential biparental inheritance of plastid DNA and results for over 200 angiosperm species. Am. J. Bot. 1988;75:1443–1458. [Google Scholar]
- 10.Matsushima R, Hu Y, Toyoda K, Sodmergen Sakamoto W. The model plant Medicago truncatula exhibits biparental plastid inheritance. Plant Cell. Physiol. 2008;49:81–91. doi: 10.1093/pcp/pcm170. [DOI] [PubMed] [Google Scholar]
- 11.Hagemann R. The sexual inheritance of plant organelles. Mol. Biol. Biotechnol. Plant Organel. 2004:93–113. [Google Scholar]
- 12.Tanksley SD, Pichersky E. Organisation and evolution of sequences in the plant nuclear genome. In: Leslie DG, Subodh KJ, editors. Plant evolutionary biology. London - New York: Chapman and Hall; 1988. [Google Scholar]
- 13.Harris SA, Ingram R. Chloroplast DNA and biosystematics - the effects of intraspecific diversity and plastid transmission. Taxon. 1991;40:393–412. [Google Scholar]
- 14.Curtis SE, Clegg MT. Molecular evolution of chloroplast DNA sequences. Mol. Biol. Evol. 1984;1:291–301. doi: 10.1093/oxfordjournals.molbev.a040319. [DOI] [PubMed] [Google Scholar]
- 15.Hiratsuka J, Shimada H, Whittier R, Ishibashi T, Sakamoto M, Mori M, Kondo C, Honji Y, Sun CR, Meng BY, Li YQ, Kanno A, Nishizawa Y, Hirai A, Shinozaki K, Sugiura M. The complete sequence of the rice (Oryza sativa) chloroplast genome, Intermolecular recombination between distinct tRNA genes accounts for a major plastid DNA inversion during the evolution of the cereals. Mol. Gen. Genet. 1989;217:185–194. doi: 10.1007/BF02464880. [DOI] [PubMed] [Google Scholar]
- 16.Masood MS, Nishikawa T, Fukuoka S, Njenga PK, Tsudzuki T, Kadowaki K. The complete nucleotide sequence of wild rice (Oryza nivara) chloroplast genome, first genome wide comparative sequence analysis of wild and cultivated rice. Gene. 2004;340:133–139. doi: 10.1016/j.gene.2004.06.008. [DOI] [PubMed] [Google Scholar]
- 17.Maier RM, Neckermann K, Igloi GL, Kössel H. Complete Sequence of the Maize Chloroplast Genome, Gene Content, Hot-spots of Divergence and Fine Tuning of Genetic Information by Transcript Editing. J. Mol. Biol. 1995;251:614–628. doi: 10.1006/jmbi.1995.0460. [DOI] [PubMed] [Google Scholar]
- 18.Asano T, Tsudzuki T, Takahashi S, Shimada H, Kadowaki K. Complete nucleotide sequence of the sugarcane (Saccharum officinarum) chloroplast genome, a comparative analysis of four monocot chloroplast genomes. DNA Res. 2004;11:93–99. doi: 10.1093/dnares/11.2.93. [DOI] [PubMed] [Google Scholar]
- 19.Ogihara Y, Isono K, Kojima T, Endo A, Hanaoka M, Shiina T, Terachi T, Utsugi S, Murata M, Mori N, Takumi S, Ikeo K, Gojobori T, Murai R, Murai K, Matsuoka Y, Ohnishi Y, Tajiri H, Tsunewaki K. Chinese spring wheat (Triticum aestivum L.) chloroplast genome, complete sequence and contig clones. Plant Mol. Biol. Rep. 2000;18:243–253. [Google Scholar]
- 20.Ogihara Y, Isono K, Kojima T, Endo A, Hanaoka M, Shiina T, Terachi T, Utsugi S, Murata M, Mori N, Takumi S, Ikeo K, Gojobori T, Murai R, Murai K, Matsuoka Y, Ohnishi Y, Tajiri H, Tsunewaki K. Structural features of a wheat plastome as revealed by complete sequencing of chloroplast DNA. Mol. Genet. Genom. 2002;266:740–746. doi: 10.1007/s00438-001-0606-9. [DOI] [PubMed] [Google Scholar]
- 21.Kahlau S, Aspinall S, Gray JC, Bock R. Sequence of the tomato chloroplast DNA and evolutionary comparison of solanaceous plastid genomes. J. Mol. Evol. 2006;63:194–207. doi: 10.1007/s00239-005-0254-5. [DOI] [PubMed] [Google Scholar]
- 22.Tangphatsornruang S, Sangsrakru D, Chanprasert J, Uthaipai-sanwong P, Yoocha T, Jomchai N, Tragoonrung S. The chloroplast genome sequence of mungbean (vigna radiata) determined by high-throughput pyrosequencing: structural organization and phylogenetic relationships. DNA Res. 2010;17:11–22. doi: 10.1093/dnares/dsp025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wakasugi T, Tsudzuki T, Sugiura M. The genomics of land plant plastids: gene content and alteration of genomic information by RNA editing. Photosynth. Res. 2001;70:107–118. doi: 10.1023/A:1013892009589. [DOI] [PubMed] [Google Scholar]
- 24.Gosal SS, Wani SH, Kang MS. Biotechnology and drought tolerance. J. Crop Improv. 2009;23:19–54. [Google Scholar]
- 25.Wani SH, Gosal SS. Introduction of OsglyII gene into Indica rice through particle bombardment for increased salinity tolerance. Biol. Plant. 2010 in press. [Google Scholar]
- 26.Singh AK, Verma SS, Bansal KC. Plastid transformation in eggplant (Solanum melongena L.) Transgenic Res. 2010;19:113–119. doi: 10.1007/s11248-009-9290-z. [DOI] [PubMed] [Google Scholar]
- 27.Bock R. Plastid biotechnology: Prospects for herbicide and insect resistance, metabolic engineering and molecular farming. Curr. Opin. Biotechnol. 2007;18:100–106. doi: 10.1016/j.copbio.2006.12.001. [DOI] [PubMed] [Google Scholar]
- 28.Roudsari MF, Salmanian AH, Mousavi A, Sohi HH, Jafari M. Regeneration of glyphosate-tolerant Nicotiana tabacum after plastid transformation with a mutated variant of bacterial aroA gene. Iran. J. Biotechnol. 2009;7:247–253. [Google Scholar]
- 29.Maliga P. Towards plastid transformation in higher plants. Trends Biotech. 1993;11:101–107. [Google Scholar]
- 30.Diekmann K, Hodkinson TR, Fricke E, Barth S. An optimized chloroplast DNA extraction protocol for grasses (Poaceae) proves suitable for whole plastid genome sequencing and SNP detection. PLoS ONE. 2008;3:e2813. doi: 10.1371/journal.pone.0002813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maliga P. Plastid engineering bears fruit. Nat. Biotechnol. 2001;19:826–827. doi: 10.1038/nbt0901-826. [DOI] [PubMed] [Google Scholar]
- 32.Wang S, Liu J, Feng Y, Niu X, Giovannoni J, Liu Y. Altered plastid levels and potential for improved fruit nutrient content by downregulation of the tomato DDB1-interacting protein Cul4. Plant J. 2008;55:89–103. doi: 10.1111/j.1365-313X.2008.03489.x. [DOI] [PubMed] [Google Scholar]
- 33.Boynton JE, Gillham NW, Harris EH, Hosler JP, Johnson AM, Jones AR, Randolph-Anderson BL, Robertson D, TM Klein TM, Shark KB. Plastid transformation in Chlamydomonas with high velocity microprojectiles. Sci. 1988;240:1534–15838. doi: 10.1126/science.2897716. [DOI] [PubMed] [Google Scholar]
- 34.Svab Z, Hajdukiewicz P, Maliga P. Stable transformation of plastids in higher plants. Proc. Natl. Acad. Sci. USA. 1990;87:8526–8530. doi: 10.1073/pnas.87.21.8526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kang TJ, Loc NH, Jang MO, Jang YS, Kim YS, Seo JE, Yang MS. Expression of the B subunit of E. coli heat-labile enterotoxin in the chloroplasts of plants and its characterization. Transgenic Res. 2003;12:683–691. doi: 10.1023/B:TRAG.0000005114.23991.bc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jabeen R, Khan MS, Zafar Y, Anjum T. Codon optimization of cry1Ab gene for hyper expression in plant organelles. Mol. Bio. Rep. 2010;37:1011–1017. doi: 10.1007/s11033-009-9802-1. [DOI] [PubMed] [Google Scholar]
- 37.Morgenfeld M, Segretin ME, Wirth S, Lentz E, Zelada A, Mentaberry A, Gissmann L, Almonacid FB. Potato virus X coat protein fusion to human papillomavirus 16 E7 oncoprotein enhance antigen stability and accumulation in tobacco chloroplast. Biotechnol. 2009;43:243–249. doi: 10.1007/s12033-009-9195-3. [DOI] [PubMed] [Google Scholar]
- 38.Cheng L, Li HP, Qu1 B, Huang T, Tu JX, Fu TD, Liao YC. Chloroplast transformation of rapeseed (Brassica napus L.) by particle bombardment of cotyledons. Plant. Cell. Rep. 2010;29:371–381. doi: 10.1007/s00299-010-0828-6. [DOI] [PubMed] [Google Scholar]
- 39.Sikdar SR, Serino G, Chaudhuri S, Maliga P. Plastid transformation in Arabidopsis thaliana. Plant Cell Rep. 1998;18:20–24. [Google Scholar]
- 40.Sidorov VA, Kasten D, Pang SZ, Hajdukiewicz PTJ, Staub JM, and Nehra NS. Stable plastid transformation in potato: Use of green fluorescent protein as a plastid marker. Plant J. 1999;19:209–216. doi: 10.1046/j.1365-313x.1999.00508.x. [DOI] [PubMed] [Google Scholar]
- 41.Nguyen TT, Nugent G, Cardi T, Dix PJ. Generation of homoplasmic plastid transformants of a commercial cultivar of potato (Solanum tuberosum L.) Plant. Sci. 2005;168:1495–1500. [Google Scholar]
- 42.Apel W, Bock R. Enhancement of carotenoid biosynthesis in transplastomic tomatoes by induced lycopene-to-provitamin A conversion. Plant Physiol. 2009;151:59–66. doi: 10.1104/pp.109.140533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Skarjinskaia M, Svab Z, Maliga P. Plastid transformation in Lesquerella Fendleri, an oilseed brassicacea. Transgenic Res. 2003;12:115–122. doi: 10.1023/a:1022110402302. [DOI] [PubMed] [Google Scholar]
- 44.Hou BK, Zhou YH, Wan LH, Zhang ZL, Shen GF, Chen ZH, Hu ZM. Plastid transformation in oilseed rape. Transgenic Res. 2003;12:111–114. doi: 10.1023/a:1022180315462. [DOI] [PubMed] [Google Scholar]
- 45.Zubko MK, Zubko EI, Zuilen KV, Meyer P, Day A. Stable transformation of petunia plastids. Transgenic Res. 2004;13:523–530. doi: 10.1007/s11248-004-2374-x. [DOI] [PubMed] [Google Scholar]
- 46.Lelivelt C, McCabe M, Newell C, DeSnoo C, Dun K, Birch-Machin I, Gray J, Mills K, Nugent J. Stable plastid transformation in lettuce (Lactuca sativa L.) Plant Mol. Biol. 2005;58:763–774. doi: 10.1007/s11103-005-7704-8. [DOI] [PubMed] [Google Scholar]
- 47.Dufourmantel N, Pelissier B, Garcon F, Peltier G, Ferullo JM, Tissot G. Generation of fertile transplastomic soybean. Plant Mol. Biol. 2004;55:479–489. doi: 10.1007/s11103-004-0192-4. [DOI] [PubMed] [Google Scholar]
- 48.Kumar S, Dhingra A, Daniell H. Plastid-expressed betaine aldehyde dehydrogenase gene in carrot cultured cells, roots, and leaves confers enhanced salt tolerance. Plant Physiol. 004a;136:2843–2854. doi: 10.1104/pp.104.045187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kumar S, Dhingra A, Daniell H. Stable transformation of the cotton plastid genome and maternal inheritance of transgenes. Plant Mol. Biol. 004b;56:203–216. doi: 10.1007/s11103-004-2907-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lee SM, Kang K, Chung H, Yoo SH, Xu XM, Lee SB, Cheong JJ, Daniell H, Kim M. Plastid transformation in the monocotyledonous cereal crop, rice (Oryza sativa) and transmission of transgenes to their progeny. Mol. Cells. 2006;21:401–410. [PMC free article] [PubMed] [Google Scholar]
- 51.Okumura S, Sawada M, Park Y, Hayashi T, Shimamura M, Takase H, Tomizawa KI. Transformation of poplar (Populus alba) plastids and expression of foreign proteins in tree plastids. Transgenic Res. 2006;15:637–646. doi: 10.1007/s11248-006-9009-3. [DOI] [PubMed] [Google Scholar]
- 52.Svab Z, Maliga P. High-frequency plastid transformation in tobacco by selection for a chimeric aadA gene. Proc. Natl. Acad. Sci. USA. 1993;90:913–917. doi: 10.1073/pnas.90.3.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ko SM, Yoo BH, Lim JM, Oh KH, Liu JI, Kim SW, Liu JR, Choi KS, Yoon ES. Production of Fibrinolytic Enzyme in Plastid-Transformed Tobacco Plants. Plant Mol. Biol. Rep. 2009;27:448–453. [Google Scholar]
- 54.Umate P. Mulberry improvements via plastid transformation and tissue culture engineering. Plant Signal. Behav. 2010;5 doi: 10.4161/psb.5.7.12035. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Whitney SM, Sharwood RE. Construction of a tobacco master line to improve Rubisco engineering in plastids. J. Exp. Bot. 2008;59:1909–1921. doi: 10.1093/jxb/erm311. [DOI] [PubMed] [Google Scholar]
- 56.Daniell H, Datta R, Varma S, Gray S, Lee SB. Containment of herbicide resistance through genetic engineering of the chloroplast genome. Nat. Biotechnol. 1998;16:345–348. doi: 10.1038/nbt0498-345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Haghani K, Salmanian AH, Ranjbar B, Zakikhan K, Khajeh K. Comparative studies of wild type E. coli 5-enolpyruvylshikimate 3-phosphate synthase with three glyphosateinsensitive mutated forms, activity, stability and structural characterization. Biochim. Biophys. Acta. 2008;1784:1167–1175. doi: 10.1016/j.bbapap.2007.07.021. [DOI] [PubMed] [Google Scholar]
- 58.Maliga P. Plastid transformation in higher plants. Annu. Rev. Plant Biol. 2004;55:289–313. doi: 10.1146/annurev.arplant.55.031903.141633. [DOI] [PubMed] [Google Scholar]
- 59.Wilkinson MJ, Sweet J, Poppy GM. Risk assessment of GM plants, avoiding gridlock? Trends Plant Sci. 2003;8:208–212. doi: 10.1016/S1360-1385(03)00057-8. [DOI] [PubMed] [Google Scholar]
- 60.Ruf S, Karcher D, Bock R. Determining the transgene containment level provided by plastid transformation. Proc. Natl. Acad. Sci. USA. 2007;104:6998–7002. doi: 10.1073/pnas.0700008104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Daniell H. Transgene containment by maternal inheritance, Effective or elusive? . Proc. Natl. Acad. Sci. USA. 2007;104:6879–6880. doi: 10.1073/pnas.0702219104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bansal KC, Sharma RK. Chloroplast transformation as a tool for prevention of gene flow from GM crops to weedy or wild relatives. Curr. Sci. 2003;84:12–86. [Google Scholar]
- 63.Wang T, Li Y, Shi Y, Reboud X, Darmency H, Gressel J. Low frequency transmission of a plastid-encoded trait in Setaria italica. Theor. Appl. Genet. 2004;108:315–320. doi: 10.1007/s00122-003-1424-8. [DOI] [PubMed] [Google Scholar]
- 64.Haider N, Allainguillaume J, Wilkinson MJ. The inefficiency of chloroplast transformation in preventing transgene flow, a case study of chloroplast capture. Curr. Genet. 2009;55:39–50. doi: 10.1007/s00294-009-0230-5. [DOI] [PubMed] [Google Scholar]
- 65.Allainguillaume J, Harwood T, Ford CS, Cuccato G, Norris C, Allender CJ, Welters R, King GJ, Wilkinson MJ. Rapeseed cytoplasm gives advantage in wild relatives and complicates genetically modified crop biocontainment. New Phytol. 2009;183:1201–211. doi: 10.1111/j.1469-8137.2009.02877.x. [DOI] [PubMed] [Google Scholar]
- 66.Daniell H, Khan MS, Allison L. Milestones in chloroplast genetic engineering an environmentally friendly era in biotechnology. Trends Plant Sci. 2002;7:84–91. doi: 10.1016/s1360-1385(01)02193-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hefferon KL. Chloroplast engineering and production of bio-pharmaceuticals. In: Hefferon KL, editor. Biopharmaceuticals in plants toward the next century of medicine. CRC Press; 2010. pp. 57–76. [Google Scholar]
- 68.Koop HU, Herz S, Golds TJ, Nickelson J. The genetic transformation of plastids. In: Bock R, editor. Cell and molecular biology of plastids, Topics Curr. Genet. Vol. 19. Berlin Heidelberg: Springer-Verlag; 2007. pp. 457–510. [Google Scholar]
- 69.Block MD, Schell J, Montagu MV. Plastid transformation by Agrobacterium tumefaciens. EMBO J. 1985;4:1367–1372. doi: 10.1002/j.1460-2075.1985.tb03788.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sanford JC. Biolistic plant transformation. Physiol. Plant. 1990;79:206–209. [Google Scholar]
- 71.Sporlein B, Streubel M, Dahlfeld G, Westhoff P, Koop HU. PEG-mediated plastid transformation, A new system for transient gene expression assays in plastids. Theor. Appl. Genet. 1991;82:717–722. doi: 10.1007/BF00227316. [DOI] [PubMed] [Google Scholar]
- 72.O’Neill C, Horvath GV, Horvath E, Dix PJ, Medgyesy P. Plastid transformation in plants, polyethylene glycol (PEG) treatment of protoplasts is an alternative to biolistic delivery systems. Plant J. 1993;3:729–38. [PubMed] [Google Scholar]
- 73.Ho MW, Cummins J. Molecular pharming by chloroplast transformation. Institute of Science in Society http://www.isis.org.uk/MPBCT.php . 2005.
- 74.Liu CW, Lin CC, Chen J, Tseng MJ. Stable plastid transformation in cabbage (Brassica oleracea L. var. capitata L.) by particle bombardment. Plant Cell Rep. 2007;26:1733–1744. doi: 10.1007/s00299-007-0374-z. [DOI] [PubMed] [Google Scholar]
- 75.Ye GN, Hajdukiewicz P, Broyles D, Rodriguez D, Xu CW, Nehra N, Staub JM. Plastid-expressed 5-enolpyruvylshikimate 3-phosphate synthase genes provide high level glyphosate tolerance in tobacco. Plant J. 2001;25:261–270. doi: 10.1046/j.1365-313x.2001.00958.x. [DOI] [PubMed] [Google Scholar]
- 76.Ye GN, Colburn S, Xu C, Hajdukiewic P, Staub JM. Persistance of unselected DNA during a plastid transformation and segregation approach to herbicide resistance. Plant Physiol. 2003;133:402–410. doi: 10.1104/pp.103.021949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Maliga P. Progress towards commercialization of plastid transformation technology. Trends Biotechnol. 2003;21:20–28. doi: 10.1016/s0167-7799(02)00007-0. [DOI] [PubMed] [Google Scholar]
- 78.Ruiz ON, Hussein HS, Terry N, Daniell H. Phytoremediation of organomercurial compounds via chloroplast genetic engineering. Plant Physiol. 2003;132:1344. doi: 10.1104/pp.103.020958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ruiz ON, Daniell H. Engineering cytoplasmic male sterility via the chloroplast genome by expression of ketothiolase. Plant Physiol. 2005;138:1232–1246. doi: 10.1104/pp.104.057729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Rady A. The new weapons of genetic engineering. http://www.opednews.com/articles/The-new-weapons-of-genetic-by-Rady-Ananda-090315-278.html . 2009.
- 81.Gatehouse JA. Biotechnological prospects for engineering insect-resistant plants. Plant Physiol. 2008;146:881–887. doi: 10.1104/pp.107.111096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.McBride KE, Svab Z, Schael DJ, Hogan PS, Stalker KM, Maliga P. Amplification of a chimeric Bacillus gene in plastids leads to an extraordinary level of an insecticidal protein in tobacco. Biotechnology. 1995;13:362–365. doi: 10.1038/nbt0495-362. [DOI] [PubMed] [Google Scholar]
- 83.Dufourmantel N, Tissot G, Goutorbe F, Garçon F, Muhr C, Jansens S, Pelissier B, Peltier G, Dubald M. Generation and analysis of soybean plastid transformants expressing Bacillus thuringiensis Cry1Ab protoxin. Plant Mol. Biol. 2005;58:659–668. doi: 10.1007/s11103-005-7405-3. [DOI] [PubMed] [Google Scholar]
- 84.Chakrabarti S, Lutz K, Lertwiriyawong B, Svab Z, Maliga P. Expression of the cry9Aa2 B.t. gene in tobacco plastids confers resistance to potato tuber moth. Transgenic Res. 2006;15:481–488. doi: 10.1007/s11248-006-0018-z. [DOI] [PubMed] [Google Scholar]
- 85.Liu CW, Lin CC, Yiu JC, Chen JJW, Tseng MJ. Expression of a Bacillus thuringiensis toxin (cry1Ab) gene in cabbage (Brassica oleracea L. var. capitata L.) plastids confers high insecticidal efficacy against Plutella xylostella. Theor. Appl. Genet. 2008;117:75–88. doi: 10.1007/s00122-008-0754-y. [DOI] [PubMed] [Google Scholar]
- 86.Kim EH, Suh SC, Park BS, Shin KS, Kweon SJ, Han EJ, Park SH, Kim YS, Kim JK. Plastid-targeted expression of synthetic cry1Ac in transgenic rice as an alternative strategy for increased pest protection. Planta. 2009;230:397–405. doi: 10.1007/s00425-009-0955-x. [DOI] [PubMed] [Google Scholar]
- 87.Kota M, Daniell H, Varma S, Garczynski SF, Gould F, Moar WJ. Overexpression of the Bacillus thuringiensis (Bt) Cry2Aa2 protein in plastids confers resistance to plants against susceptible and Bt-resistant insects. Proc. Natl. Acad. Sci. USA. 1999;96:1840–1845. doi: 10.1073/pnas.96.5.1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.De Cosa B, Moar W, Lee SB, Miller M, Daniell H. Overexpression of the Bt cry2Aa2 operon in plastids leads to formation of insecticidal crystals. Nat. Biotechnol. 2001;19:71–74. doi: 10.1038/83559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.DeGray G, Rajasekaran K, Smith F, Sanford J, Daniell H. Expression of an antimicrobial peptide via the chloroplast genome to control phytopathogenic bacteria and fungi. Plant Physiol. 2001;127:852–862. [PMC free article] [PubMed] [Google Scholar]
- 90.Hatziloukas E, Panopoulos NJ. Origin, structure, and regulation of argK, encoding the phaseolotoxin-resistant ornithine carbamoyltransferase in Pseudomonas syringae pv. phaseolicola, and functional expression of argK in transgenic tobacco. J. Bacteriol. 1992;174:5895–5909. doi: 10.1128/jb.174.18.5895-5909.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Verberne C, Verpoorte R, Bol J F, Mercado-Blanco J, Linthorst HJM. Overproduction of salicylic acid in plants by bacterial transgenes enhances pathogen resistance. Nat. Biotechnol. 2000;18:779–783. doi: 10.1038/77347. [DOI] [PubMed] [Google Scholar]
- 92.Zhang JY, Zhang Y, Song YR. Chloroplast genetic engineering in higher plants. Acta Bot. Sinica. 2003;45:509–516. [Google Scholar]
- 93.Cherry JH, Nielsen BL. Metabolic engineering of chloroplasts for abiotic stress tolerance. Mol. Biol. Biotechnol. Plant Organel. 2007;3:513–525. [Google Scholar]
- 94.Lee SB, Kwon HB, Kwon SJ, Park SC, Jeong MJ, Han SE, Byun MO, Daniell H. Accumulation of trehalose within transgenic chloroplasts confers drought tolerance. Mol. Breed. 2004;11:1–13. [Google Scholar]
- 95.Djilianov D, Georgieva T, Moyankova D, Atanassov A, Shinozaki K, Smeeken SCM, Verma DPS, Murata N. Improved abiotic stress tolerance in plants by accumulation of osmoprotectants - gene transfer approach. Biotechnol. Biotechnol. 2005;19:63–71. [Google Scholar]
- 96.Nuccio ML, Russell BL, Nolte KD, Rathinasabapathi B, Gage DA, Hanson AD. The endogenous choline supply limits glycinebetaine synthesis in transgenic tobacco expressing choline monooxygenase. Plant J. 1998;16:487–496. doi: 10.1046/j.1365-313x.1998.00316.x. [DOI] [PubMed] [Google Scholar]
- 97.Shirasawa K, Takabe T, Takabe T, Kishitani S. Accumulation of glycinebetaine in rice plants that overexpress choline monooxygenase from spinach and evaluation of their tolerance to abiotic stress. Ann. Bot. 2006;98:565–571. doi: 10.1093/aob/mcl126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zhang J, Tan W, Yang XH, Zhang HX. Plastid- expressed choline monooxygenase gene improves salt and drought tolerance through accumulation of glycine betaine in tobacco. Plant Cell. Rep. 2008;27:1113–1124. doi: 10.1007/s00299-008-0549-2. [DOI] [PubMed] [Google Scholar]
- 99.George S, Venkataraman G, Parida A. A chloroplast-localized and auxin-induced glutathione S-transferase from phreatophyte Prosopis juliflora confer drought tolerance on tobacco. J. Plant Physiol. 2010;167:311–318. doi: 10.1016/j.jplph.2009.09.004. [DOI] [PubMed] [Google Scholar]
- 100.Mulesky M, Oishi KK, Williams D. Chloroplasts, transforming biopharmaceutical manufacturing. Biopharm. Internat. 2004 http://tinyurl.com/8em3je . [Google Scholar]
- 101.Daniell H, Chebolu S, Kumar S, Singleton M, Falconer R. Chloroplast-derived vaccine antigens and other therapeutic proteins. Vaccine. 005a;23:1779–1183. doi: 10.1016/j.vaccine.2004.11.004. [DOI] [PubMed] [Google Scholar]
- 102.Grevich JJ, Daniell H. Chloroplast genetic engineering, recent advances and future perspectives. Crit. Rev. Plant. Sci. 2005;24:83–107. [Google Scholar]
- 103.Daniell H, Wiebe PO, Millan AF. Antibiotic-free chloroplast genetic engineering - an environmentally friendly approach. Trends Plant Sci. 2001;6:237–239. doi: 10.1016/s1360-1385(01)01949-5. [DOI] [PubMed] [Google Scholar]
- 104.Farran I, McCarthy-Suárez I, Río-Manterola F, Mansilla C, Lasarte JJ, Mingo-Castel AM. The vaccine adjuvant extra domain A from fibronectin retains its proinflammatory properties when expressed in tobacco chloroplasts. Planta. 2010;231:977–990. doi: 10.1007/s00425-010-1102-4. [DOI] [PubMed] [Google Scholar]
- 105.Daniell H. Production of biopharmaceuticals and vaccines in plants via the chloroplast genome. Biotechnol. J. 2006;1:1071–1079. doi: 10.1002/biot.200600145. [DOI] [PubMed] [Google Scholar]
- 106.Chebolu S, Daniell H. Chloroplast-derived vaccine antigens and biopharmaceuticals, expression, folding, assembly and functionality. Curr. Top. Microbiol. Immunol. 2009;332:33–54. doi: 10.1007/978-3-540-70868-1_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kumar S, Daniell H. Engineering the chloroplast genome for hyperexpression of human therapeutic proteins and vaccine antigens. Method Mol. Biol. 2008;267:365–383. doi: 10.1385/1-59259-774-2:365. [DOI] [PubMed] [Google Scholar]
- 108.Li HY, Ramalingam S, Chye ML. Accumulation of recombinant SARS-CoV spike protein in plant cytosol and chloroplasts indicate potential for development of plant-derived oral vaccines. Exp. Biol. Med. 2006;23:1346–1352. doi: 10.1177/153537020623100808. [DOI] [PubMed] [Google Scholar]
- 109.Singh ND, Ding Y, Daniell H. Chloroplast-derived vaccine antigens and biopharmaceuticals, protocols for expression, purification, or oral delivery and functional evaluation. Methods Mol. Biol. 2009;483:163–192. doi: 10.1007/978-1-59745-407-0_10. [DOI] [PubMed] [Google Scholar]
- 110.Fernandez-San Millan A, Mingo-Castel A, Miller M, Daniell H. A chloroplast transgenic approach to hyper-express and purify human serum albumin, a protein highly susceptible to proteolytic degradation. Plant. Biotechnol. J. 2003;1:71. doi: 10.1046/j.1467-7652.2003.00008.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Tregoning J, Maliga P, Dougan G, Nixon PJ. New advances in the production of edible plant vaccines, chloroplast expression of a tetanus vaccine antigen. Tet. C. Phytochemistry. 2004;65:989. doi: 10.1016/j.phytochem.2004.03.004. [DOI] [PubMed] [Google Scholar]
- 112.Watson J, Koya V, Leppla SH, Daniell H. Expression of Bacillus anthracis protective antigen in transgenic chloroplasts of tobacco, a non-food/feed crop. Vaccine. 2004;22:4374. doi: 10.1016/j.vaccine.2004.01.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Koya V, Moayeri M, Leppla SH, Daniell H. Plant-based vaccine, mice immunized with chloroplast-derived anthrax protective antigen survive anthrax lethal toxin challenge. Infect. Immun. 2005;73:8266–8274. doi: 10.1128/IAI.73.12.8266-8274.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Daniell H, Lee SB, Grevich J, Saski C, Quesada-Vargas T, Guda C, Tomkins J, Jansen RK. Complete chloroplast genome sequences of Solanum bulbocastanum, Solanum lycopersicum and comparative analyses with other Solanaceae genomes. Theor. Appl. Genet. 2006;112:1503–1518. doi: 10.1007/s00122-006-0254-x. [DOI] [PubMed] [Google Scholar]
- 115.Zhou F, Badillo-Corona JA, Karcher D, Gonzalez-Rabade N, Piepenburg K, Borchers MI, Maloney AP, Kavanagh TA, Gray JC, Bock R. High-level expression of HIV antigens from the tobacco and tomato plastid genomes. Plant Biotechnol. J. 2008;6:897–913. doi: 10.1111/j.1467-7652.2008.00356.x. [DOI] [PubMed] [Google Scholar]
- 116.Oey M, Lohse M, Scharff LB, Kreikemeyer B, Bock R. Plastid production of protein antibiotics against pneumonia via a new strategy for high-level expression of antimicrobial proteins. Proc. Natl. Acad. Sci. USA. 2009;106:6579–6584. doi: 10.1073/pnas.0813146106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Chebolu S, Daniell H. Stable expression of Gal/GalNAc lectin of Entamoeba histolytica in transgenic chloroplasts and immunogenicity in mice towards vaccine development for amoebiasis. Plant. Biotechnol. J. 2007;5:230–239. doi: 10.1111/j.1467-7652.2006.00234.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Shao HB, He DM, Qian KX, Shen GF, Su ZL. The expression of classical swine fever virus structural protein E2 gene in to-bacco chloroplasts for applying chloroplasts as bioreactors. C. R. Biologies. 2008;331:179–184. doi: 10.1016/j.crvi.2007.12.007. [DOI] [PubMed] [Google Scholar]
- 119.McCabe MS, Klaas M, Gonzalez-Rabade N, Poage M, Badillo-Corona JA, Zhou F, Karcher D, Bock R, Gray JC, Dix PJ. Plastid transformation of high-biomass tobacco variety Maryland Mammoth for production of human immunodeficiency virus type 1 (HIV-1) p24 antigen. Plant Biotechnol. J. 2008;6:914–929. doi: 10.1111/j.1467-7652.2008.00365.x. [DOI] [PubMed] [Google Scholar]
- 120.Meyers A, Chakauya E, Shephard E, Tanzer FL, Maclean J, Lynch A, Williamson AL, Rybicki EP. Expression of HIV-1 antigens in plants as potential subunit vaccines. BMC. Biotechnol. 2008;23:42–53. doi: 10.1186/1472-6750-8-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Rigano MM, Manna C, Giulini A, Pedrazzini E, Capobianchi M, Castilletti C, Di Caro A, Ippolito G, Beggio P, De Giuli Morghen C, Monti L, Vitale A, Cardi T. Transgenic chloroplasts are efficient sites for high-yield production of the vaccinia virus envelope protein A27L in plant cellsdagger. Plant Biotechnol. J. 2009;7:577–591. doi: 10.1111/j.1467-7652.2009.00425.x. [DOI] [PubMed] [Google Scholar]
- 122.Youm JW, Jeon JH, Kim H, Min SR, Kim MS, Joung H, Jeong WJ, Kim HS. High-level expression of a human beta-site APP cleaving enzyme in transgenic tobacco chloroplasts and its immunogenicity in mice. Transgenic Res. 2010 doi: 10.1007/s11248-010-9383-8. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Lentz EM, Segretin ME, Morgenfeld MM, Wirth SA, Dus Santos MJ, Mozgovoj MV, Wigdorovitz A, Bravo-Almonacid FF. High expression level of a foot and mouth disease virus epitope in tobacco transplastomic plants. Planta. 2010;231:387–395. doi: 10.1007/s00425-009-1058-4. [DOI] [PubMed] [Google Scholar]
- 124.Bock R. Transgenic plastids in basic research and plant biotechnology. J. Mol. Biol. 2001;312:425–438. doi: 10.1006/jmbi.2001.4960. [DOI] [PubMed] [Google Scholar]
- 125.Maliga P. Engineering the plastid genome of higher plants. Curr. Opin. Plant. Biol. 2002;5:164–172. doi: 10.1016/s1369-5266(02)00248-0. [DOI] [PubMed] [Google Scholar]
- 126.Su T, Zhan YG, Han M, Hao AP. Chloroplast genetic engineering, a new approach in plant biotechnology. Chin. J. Biotechnol. 2005;21:674–680. [PubMed] [Google Scholar]
- 127.Elizabeth PM. Engineering the chloroplast genome. Resonance. 2005;10:94–95. [Google Scholar]
- 128.Daniell H, Kumar S, Dufourmantel N. Breakthrough in chloroplast genetic engineering of agronomically important crops. Trends Biotechnol. 005b;23:238–245. doi: 10.1016/j.tibtech.2005.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Ruhlman T, Daniell H. Plastid pathways, Metabolic engineering via the chloroplast genome. In: Verpoorte R, et al., editors. Applications of Plant Metabolic Engineering. Berlin Heidelberg: Springer.Verlag; 2007. pp. 79–108. [Google Scholar]
- 130.Meyers B, Zaltsman A, Lacroix B, Kozlovsky SV, Krichevsky A. Nuclear and plastid genetic engineering of plants, Comparison of opportunities and challenges. Biotechnol. Adv. 2010 doi: 10.1016/j.biotechadv.2010.05.022. [DOI] [PubMed] [Google Scholar]
- 131.Sato S, Nakamura Y, Kaneko T, Asamizu E, Tabata S. Complete structure of the chloroplast genome of Arabidopsis thaliana. DNA Res. 1999;6:283–290. doi: 10.1093/dnares/6.5.283. [DOI] [PubMed] [Google Scholar]
- 132.Gao JG, Wang XM, Wang Q. The complete nucleotide sequence of Brassica napus chloroplast 16S rRNA gene. Acta Genet. Sinica. 1989;16:263–8. [PubMed] [Google Scholar]
- 133.Bausher MG, Singh ND, Lee SB, Jansen RK, Daniell H. The complete chloroplast genome sequence of Citrus sinensis L. Osbeck var Ridge Pineapple organization and phylogenetic relationships to other angiosperms. BMC Plant Biol. 2006;6:21–32. doi: 10.1186/1471-2229-6-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Samson N, Bausher MG, Lee SB, Jansen RK, Daniell H. The complete nucleotide sequence of the coffee (Coffea arabica L.) chloroplast genome organization and implications for biotechnology and phylogenetic relationships amongst angiosperms. Plant Biotechnol. J. 2007;5:339–353. doi: 10.1111/j.1467-7652.2007.00245.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Plader W, Yukawa Y, Sugiura M, Malepszy S. The complete structure of the cucumber (Cucumis sativus L.) chloroplast genome its composition and comparative analysis. Cell. Mol. Biol. Lett. 2007;12:584–594. doi: 10.2478/s11658-007-0029-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Ruhlman T, Lee SB, Jansen RK, Hostetler JB, Tallon LJ, Town CD, Daniell H. Complete plastid genome sequence of Daucus carota, Implications for biotechnology and phylogeny of angiosperms. BMC Genomics. 2006;7:222–232. doi: 10.1186/1471-2164-7-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Moore MJ, Soltis PS, Bell CD, Burleigh JG, Soltis DE. Phylogenetic analysis of 83 plastid genes further resolves the early diversification of eudicot. Proc. Natl. Acad. Sci. USA. 2010;107:4623–4628. doi: 10.1073/pnas.0907801107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Saski C, Lee SB, Daniell H, Wood TC, Tomkins J, Kim HG, Jansen RK. Complete chloroplast genome sequence of Glycine max and comparative analyses with other legume genome. Plant Mol. Biol. 2005;59:309–322. doi: 10.1007/s11103-005-8882-0. [DOI] [PubMed] [Google Scholar]
- 139.Ibrahim RIH, Azuma JI, Sakamoto M. Complete nucleotide sequence of the cotton (Gossypium barbadense L.) chloroplast genome with a comparative analysis of sequences among 9 dicot plants. Genes Genet. Syst. 2006;81:311–321. doi: 10.1266/ggs.81.311. [DOI] [PubMed] [Google Scholar]
- 140.Lee SB, Kaittanis C, Jansen RK, Hostetler JB, Tallon LJ, Town CD, Daniell H. The complete chloroplast genome sequence of Gossypium hirsutum, organization and phylogenetic relationships to other angiosperms. BMC Genomics. 2006;7:61–73. doi: 10.1186/1471-2164-7-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Timme RE, Kuehl JV, Boore JL, Jansen RK. A comparative analysis of the Lactuca and Helianthus (Asteraceae) plastid genomes, identification of divergent regions and categorization of shared repeats. Am. J. Bot. 2007;94:302–312. doi: 10.3732/ajb.94.3.302. [DOI] [PubMed] [Google Scholar]
- 142.Saski C, Lee SB, Fjellheim S, Guda C, Jansen RK, Luo H, Tomkins J, Rognli OA, Daniell H, Clarke JL. Complete chloroplast genome sequences of Hordeum vulgare, Sorghum bicolor and Agrostis stolonifera, and comparative analyses with other grass genomes. Theor. Appl. Genet. 2007;115:571–590. doi: 10.1007/s00122-007-0567-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Daniell H, Wurdack KJ, Kanagaraj A, Lee SB, Saski C, Jansen RK. The complete nucleotide sequence of the cassava (Manihot esculenta) chloroplast genome and the evolution of atpF in Malpighiales, RNA editing and multiple losses of a group II intron. Theor. Appl. Genet. 2008;116:723–737. doi: 10.1007/s00122-007-0706-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Ravi V, Khurana JP, Tyagi AK, Khurana P. The chloroplast genome of mulberry, complete nucleotide sequence, gene organization and comparative analysis. Tree Genet. Genom. 2006;3:49–59. [Google Scholar]
- 145.Jansen RK, Cai Z, Raubeson LA, Daniell H, DePamphilis CW, Leebens-Mack J, Müller KF, Guisinger-Bellian M, Haberle RC, Hansen AK, Chumley TW, Lee SB, Peery R, McNeal RJ, Kuehl JV, Boore JL. Analysis of 81 genes from 64 plastid genomes resolves relationships in angiosperms and identifies genome-scale evolutionary patterns. Proc. Natl. Acad. Sci. USA. 2007;104:19369–19374. doi: 10.1073/pnas.0709121104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Shinozaki K, Ohme M, Tanaka M, Wakasugi T, Hayashida N, Matsubayashi T, Zaita N, Chunwongse J, Obokata J, Yamaguchi-Shinozaki K, Ohto C, Torazawa K, Meng BY, Sugita M, Deno H, Kamogashira T, Yamada K, Kusuda J, Takaiwa F, Kato A, Tohdoh N, Shimada H, Sugiura M. The complete nucleotide sequence of the tobacco chloroplast genome, its gene organization and expression. EMBO J. 1986;5:2043–2049. doi: 10.1002/j.1460-2075.1986.tb04464.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Guo X, Castillo-Ramírez S, González V, Bustos P, Fernández-Vázquez JL, Santamaría RI, Arellano J, Cevallos MA, Dávila G. Rapid evolutionary change of common bean (Phaseolus vulgaris L) plastome, and the genomic diversification of legume chloroplasts. BMC Genomics. 2007;8:288–300. doi: 10.1186/1471-2164-8-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Chung HJ, Jung JD, Park HW, Kim JH, Cha HW, Min SR, Jeong WJ, Liu JR. The complete chloroplast genome sequences of Solanum tuberosum and comparative analysis with Solanaceae species identified the presence of a 241-bp deletion in cultivated potato chloroplast DNA sequence. Plant Cell Rep. 2006;25:1369–1379. doi: 10.1007/s00299-006-0196-4. [DOI] [PubMed] [Google Scholar]
- 149.Schmitz-Linneweber C, Maier RM, Alcaraz JP, Cottet A, Herrmann RG, Mache R. The plastid chromosome of spinach (Spinacia oleracea), complete nucleotide sequence and gene organization. Plant Mol. Biol. 2001;45:307–315. doi: 10.1023/a:1006478403810. [DOI] [PubMed] [Google Scholar]
- 150.Jansen RK, Kaittanis C, Saski C, Lee SB, Tomkins J, Al-verson AJ, Daniell H. Phylogenetic analyses of Vitis (Vitaceae) based on complete chloroplast genome sequences, effects of taxon sampling and phylogenetic methods on resolving relationships among rosids. BMC Evol. Biol. 2006;6:32–46. doi: 10.1186/1471-2148-6-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Asif MH, Mantri SS, Sharma A, Srivastava A, Trivedi I, Gupta P, Mohanty CS, Sawant SV, Tuli R. Complete sequence and organisation of the Jatropha curcas (Euphorbiaceae) chloroplast genome. Tree Genet. Genom. 2010 doi, 10.1007 / s 11295 -010-0303-0. [Google Scholar]
- 152.Golds T, Maliga P, Koop HU. Stable plastid transformation in PEG-treated protoplasts of Nicotiana tabacum. Nat. Biotechnol. 1993;11:95–97. [Google Scholar]
- 153.Carrer H, Hockenberry TN, Svab Z, Maliga P. Kanamycin resistance as a selectable marker for plastid transformation in tobacco. Mol. Gen. Genet. 1993;241:49–56. doi: 10.1007/BF00280200. [DOI] [PubMed] [Google Scholar]
- 154.Staub JM, Maliga P. Expression of a chimeric uidA gene indicates that polycistronic mRNAs are efficiently translated in tobacco plastids. Plant J. 1995;7:845–848. doi: 10.1046/j.1365-313x.1995.07050845.x. [DOI] [PubMed] [Google Scholar]
- 155.Staub JM, Garcia B, Graves J, Hajdukiewicz PTJ, Hunter P, Nehra N, Paradkar V, Schlittler M, Carroll JA, Spatola L, Ward D, Ye G, Russell DA. High-yield production of a human therapeutic protein in tobacco plastids. Nat. Biotech. 2000;18:333–338. doi: 10.1038/73796. [DOI] [PubMed] [Google Scholar]
- 156.Lutz KA, Bosacchi MH, Maliga P. Plastid marker-gene excision by transiently expressed CRE recombinase. Plant J. 2006;45:447–456. doi: 10.1111/j.1365-313X.2005.02608.x. [DOI] [PubMed] [Google Scholar]
- 157.Khodakovskaya M, McAvoy R, Peters J, Wu H, Li Y. Enhanced cold tolerance in transgenic tobacco expressing a plastid omega-3 fatty acid desaturase gene under the control of a cold-inducible promoter. Planta. 2006;223:1090–100. doi: 10.1007/s00425-005-0161-4. [DOI] [PubMed] [Google Scholar]
- 158.Hasunuma T, Takeno S, Hayashi S, Sendai M, Bamba T, Yoshimura S, Tomizawa K, Fukusaki E, Miyak C. Overexpression of 1-Deoxy-d-xylulose-5-phosphate reductoisomerase gene in plastid contributes to increment of isoprenoid production. J. Biosci. Bioeng. 2008;105:518–526. doi: 10.1263/jbb.105.518. [DOI] [PubMed] [Google Scholar]
- 159.Craig W, Lenzi P, Scotti N, De Palma M, Saggese P, Car-bone V, McGrath Curran N, Magee AM, Medgyesy P, Kavanagh TA, Dix PJ, Grillo S, Cardi T. Transplastomic tobacco plants expressing a fatty acid desaturase gene exhibit altered fatty acid profiles and improved cold tolerance. Transgenic Res. 2008;17:769–782. doi: 10.1007/s11248-008-9164-9. [DOI] [PubMed] [Google Scholar]
- 160.Barone P, Zhang XH, Widholm JM. Tobacco plastid transformation using the feedback insensitive anthranilate synthase [a]-subunit of tobacco (ASA2) as a new selectable marker. J. Exp. Bot. 2009;60:3195–3202. doi: 10.1093/jxb/erp160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Wang YH, Wu ZY, Zhang XH, Wang YQ, Huang CL, Jia JF. Transformation of phag and phac genes into tobacco plastid genome and genetic analysis. Acta. Agron. Sinica. 2009;35:1949–1957. [Google Scholar]
- 162.Nadai M, Bally J, Vitel M, Job C, Tissot G, Botterman J, Dubald M. High-level expression of active human alpha1-antitrypsin in transgenic tobacco plastids. Transgenic Res. 2009;18:173–183. doi: 10.1007/s11248-008-9209-0. [DOI] [PubMed] [Google Scholar]
- 163.Nugent GD, Coyne S, Nguyen TT, Kavanaghb TA, Dix PJ. Nuclear and plastid transformation of Brassica oleracea var. botrytis (cauliflower) using PEG-mediated uptake of DNA into protoplasts. Plant Sci. 2006;170:135–142. [Google Scholar]
- 164.Kanamoto H, Yamashita A, Asao H, Okumura S, Takase H, Hattori M, Yokota A, Tomizawa KI. Efficient and stable transformation of Lactuca sativa L.cv. cisco (lettuce) plastids . Transgenic Res. 2006;15:205–217. doi: 10.1007/s11248-005-3997-2. [DOI] [PubMed] [Google Scholar]
- 165.De Marchis F, Wang Y, Stevanato P, Arcioni S, Bellucci M. Genetic transformation of the sugar beet plastome. Transgenic Res. 2009;18:17–30. doi: 10.1007/s11248-008-9193-4. [DOI] [PubMed] [Google Scholar]
- 166.Ahrazem O, Rubio-Moraga A, Castillo López R, Gómez-Gómez L. The expression of a chromoplast-specific lycopene beta cyclase gene is involved in the high production of saffron's apocarotenoid precursors. J. Exp. Bot. 2010;61:105–119. doi: 10.1093/jxb/erp283. [DOI] [PubMed] [Google Scholar]
- 167.Singh ND, Li M, Lee SB, Danny Schnell D, Daniell H. Arabidopsis Tic40 expression in tobacco plastids results in massive proliferation of the inner envelope membrane and upregulation of associated proteins. Plant Cell. 2008;20:3405–3417. doi: 10.1105/tpc.108.063172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Ahmadabadi M, Ruf S, Bock R. A leaf-based regeneration and transformation system for maize (Zea mays L.) Transgenic Res. 2007;16:437–448. doi: 10.1007/s11248-006-9046-y. [DOI] [PubMed] [Google Scholar]