Some type 2 diabetes-associated SNPs identified by genome-wide association scans may act as cis-eQTL and increase disease susceptibility by modulating expression of nearby transcripts in human tissues.
Abstract
Context:
Genome-wide association scans (GWAS) have identified novel single nucleotide polymorphisms (SNPs) that increase T2D susceptibility and indicated the role of nearby genes in T2D pathogenesis.
Objective:
We hypothesized that T2D-associated SNPs act as cis-regulators of nearby genes in human tissues and that expression of these transcripts may correlate with metabolic traits, including insulin sensitivity (SI).
Design, Settings, and Patients:
Association of SNPs with the expression of their nearest transcripts was tested in adipose and muscle from 168 healthy individuals who spanned a broad range of SI and body mass index (BMI) and in transformed lymphocytes (TLs). We tested correlations between the expression of these transcripts in adipose and muscle with metabolic traits. Utilizing allelic expression imbalance (AEI) analysis we examined the presence of other cis-regulators for those transcripts in TLs.
Results:
SNP rs9472138 was significantly (P = 0.037) associated with the expression of VEGFA in TLs while rs6698181 was detected as a cis-regulator for the PKN2 in muscle (P = 0.00027) and adipose (P = 0.018). Significant association was also observed for rs17036101 (P = 0.001) with expression of SYN2 in adipose of Caucasians. Among 19 GWAS-implicated transcripts, expression of VEGFA in adipose was correlated with BMI (r = −0.305) and SI (r = 0.230). Although only a minority of the T2D-associated SNPs were validated as cis-eQTLs for nearby transcripts, AEI analysis indicated presence of other cis-regulatory polymorphisms in 54% of these transcripts.
Conclusions:
Our study suggests that a small subset of GWAS-identified SNPs may increase T2D susceptibility by modulating expression of nearby transcripts in adipose or muscle.
Type 2 diabetes mellitus (T2D) is one of the most highly prevalent metabolic disorders for which the role of genetic susceptibility loci is well accepted (1). Until recently, the progress in unraveling the role of genes in the complex pathophysiology of T2D has been very limited. Publication of results from large genome-wide association scans (GWAS) for T2D in Caucasians ignited rapid progress through the identification of novel common single nucleotide polymorphisms (SNPs) that increase susceptibility to T2D (2, 3). However, the actual molecular and physiological explanations for these well-replicated associations are uncertain.
Except for a few, these novel T2D-associated SNPs are noncoding, residing in either intronic or intergenic regions, are not in linkage disequilibrium (LD) with known nonsynonymous SNPs and are unlikely to alter translation or protein function. Instead, most associated SNPs identified in GWAS are expected to alter transcription or splicing (4–6). Nonetheless, the specific transcripts implicated by GWAS findings are often at considerable distance from the disease-associated SNPs, and functional data that may implicate any particular gene or transcript in the disease association is lacking. Many of these SNPs are also associated with insulin secretion traits and have been thought to act primarily to alter pancreatic β-cell function (7). However, several transcripts near T2D-associated SNPs are widely expressed, including in diabetes-relevant tissues (8). Thus, a much broader impact on T2D susceptibility is plausible.
Given these considerations, we hypothesized that novel T2D-associated SNPs act as cis regulatory elements or cis-expression quantitative trait loci (cis-eQTL) and modulate the transcription of nearby genes and expression of these transcripts in human adipose and muscle tissue. Further, these SNPs may correlate with metabolic traits, including insulin sensitivity. We tested the association of these SNPs with the expression of their nearest transcripts in transformed lymphocyte (TLs) from 82 Caucasian subjects. Association of these SNPs was also tested with the level of adjacent transcript in sc adipose and muscle from 168 individuals who spanned a broad range of insulin sensitivity and body mass index (BMI). We further tested the correlations between the expression of these GWAS-implicated transcripts in sc adipose and muscle with frequently sampled intravenous glucose tolerance test (FSIGT) derived measures of insulin sensitivity (SI), insulin secretion (AIRG, acute insulin response to glucose), and BMI. Finally, using allelic expression imbalance analysis we searched for the presence of other cis regulatory SNPs in those GWAS-implicated transcripts by comparing allele-specific expression in TLs from Caucasian and African American individuals.
Materials and Methods
Transformed lymphocyte cell line culture
We used total RNA extracted from Epstein–Barr virus-transformed lymphocytes (TLs) for evaluating the role of GWAS-associated SNPs in regulating transcript level expression of nearby genes. TLs used in our study were derived from blood samples of 82 Caucasian subjects (male/female = 35/48; Nondiabetic/T2D = 31/52) from Utah and Arkansas. Cells were grown under normoglycemic (5.6 mm glucose) standard culture conditions in RPMI-1640 culture media (Omega Scientific, Inc. Tarzana, CA) supplemented with 10% fetal bovine serum (Omega Scientific) (9). Further searches for cis-regulatory SNPs in T2D GWAS implicated genes were performed by allelic expression imbalance analysis in similarly cultured TLs from 188 Caucasian subjects (including 120 TLs from Hapmap CEU subjects) and 95 African American subjects from Arkansas.
Experimental subjects
We conducted our gene expression study in adipose and muscle tissue from 168 individuals who were recruited by advertisement for good general health, between 19–60 y of age, BMI between 19 and 45 kg/m2, and either self-determined European-American or African-American ancestry from Arkansas. A summary of our study cohort is provided in Supplemental Table 1 (published on The Endocrine Society's Journals Online web site at http://jcem.endojournals.org). Detailed methods for subject recruitment, physical examination, physiological experiments, and the method for obtaining biopsy samples were described previously (10). All participants had a screening visit, at which time measurements of height, weight, waist, and hip were taken, body fat determination was done by dual x-ray absorptiometry (DXA) scan, fasting blood samples for lipid measurements were taken, and a standard 75-g oral glucose tolerance test (OGTT) was done, with the measurement of glucose and insulin at baseline and 30-min intervals for 2 h. During the second visit, adipose and muscle biopsies were obtained from all subjects under fasting conditions, and 135 subjects with nondiabetic glucose tolerance tests completed FSIGT. Biopsies were obtained under local anesthesia (lidocaine) using a Bergstrom needle from abdominal sc fat and vastus lateralis muscle. Biopsy samples were rinsed immediately in sterile normal saline, quick frozen in liquid nitrogen, and stored at −80 C for further use. After the biopsy an insulin modified (0.04 U/kg) FSIGT was performed as described previously (11).
All study participants provided written, informed consent under protocols originally approved by the University of Arkansas for Medical Sciences (UAMS). Information on these subjects is de-identified and samples are now transferred from UAMS to Wake Forest University Heath Sciences (WFUHS). Our current study is approved by the WFUHS Institutional Review Board.
Laboratory measurements
Insulin was measured by the UAMS Clinical Research Center core laboratory using an immuno-chemiluminometric assay (Invitron limited, UK; formerly Molecular Lights Technology). Plasma glucose was measured by using a glucose oxidase method at LabCorp, Inc. (Burlington, NC).
SNP selection and genotyping
We selected novel T2D-associated SNPs from three initial GWAS studies published by the Finland-United States Investigation of NIDDM Genetics (FUSION) study, Wellcome Trust Case Control Consortium (WTCCC, UK), and Diabetes Genetics Initiative (DGI) on Finnish, British, and Swedish Caucasian populations (12–14). We further selected SNPs from a meta-analysis and large scale replication study published by the Diabetes Genetics Replication and Meta-analysis (DIAGRAM) Consortium (15). In all, we evaluated 23 SNPs in our study. Details of all selected SNPs and genotyping assays are described in Supplemental Methods and Supplemental Table 2.
Gene expression
Total RNA from TLs, adipose, and muscle were isolated using standard methods (see Supplemental Methods). Total RNA (1 μg) was reverse transcribed using Qiagen QuantiTect RT kit (QIAGEN Inc, Valencia, CA) using manufacturer's protocol, which includes a DNAse digestion step to remove genomic DNA contamination. Transcripts near T2D-associated SNPs reported by GWAS findings were measured by quantitative real time PCR using SYBR green chemistry (Applied Biosystems, Inc., Foster City, CA) and normalized to 18S ribosomal RNA as described previously (10, 16, 17). The standard curves were generated for absolute quantification using pooled cDNA from the samples assayed. Transcript-specific oligonucleotide primers for real-time PCR were designed to capture most known splice variants (see Supplemental Table 3 for details). Based on our previous study we evaluated TCF7L2 transcript isoforms containing exon 13a (18). GWAS-implicated transcripts were not equally expressed in all tissues tested. We initially selected expressed transcripts based on genome-wide expression data generated in an Agilent 44K expression array platform using eight TLs and 62 adipose and muscle RNA samples from our laboratory. Expression in each tissue was further validated by real time PCR assay. We successfully evaluated and analyzed 13, 19, and 13 GWAS-implicated transcripts in TLs, sc adipose, and skeletal muscle, respectively.
Allelic expression imbalance
We attempted to find additional cis-regulatory SNPs in T2D GWAS implicated genes by allelic expression imbalance (AEI) analysis. Unequal expression of alleles was sought as evidence for cis-regulatory variants by comparing peak heights in individuals heterozygous for SNPs in coding or untranslated (UTR) regions of these genes. One expressed SNP (eSNP) with minor allele frequency (MAF) ≥0.05 per gene was selected for initial analyses. We further selected additional SNPs in genes showing strong evidence of AEI. A total of 14 eSNPs in 11 genes were tested for AEI in TLs of 188 Caucasian (including 120 HapMap Caucasians) and 95 African-American subjects. Allele-specific quantitation of both cDNA and genomic DNA samples was determined using the same assay for pyrosequencing on a PSQ96 pyrosequencer (Qiagene) using our published protocol and peak height quantified using PyroMarkID v1 software (9, 19). Ratios observed between two alleles of a SNP are assay specific and may deviate from the expected ratio of 1 (50:50) due to uneven amplification of alleles which is not related to uneven transcription. We thus used ratios observed in gDNA and compared with ratios observed in cDNA of the same heterozygotes for determination of AEI after correcting for unequal amplification. Details of the PSQ AEI assays used in this study are provided in Supplemental Table 4. A large amount of cDNA is required (20 ng/SNP) to investigate AEI; therefore we were able to study AEI only in TLs.
Statistical methods
Insulin and glucose data obtained during FSIGT were used to determine insulin sensitivity (SI) using the MINMOD Millennium program (20), and acute insulin response to glucose (AIRG), disposition index (DI), and glucose-mediated glucose disposal (SG) were taken from the program output as described elsewhere (10). Glucose and insulin areas under the curve (AUCglucose and AUCinsulin) were calculated from OGTT data using the trapezoidal rule (11). Gene expression levels were normalized to 18S RNA, and the ratio was logarithmically transformed and used in all calculations. All skewed variables were ln-transformed to normality before analysis. Genotypic effect on gene expression was analyzed by ANOVA. Association between genotype and gene expression in adipose and muscle was further tested using mixed-effects, general linear regression models, and included age and BMI as covariates and genotype, gender, ethnicity, and T2D diagnosis as fixed factors (11). Partial correlation measures of glucose homeostasis (SI, AIRG) and obesity (BMI) traits with gene expression were calculated after controlling for age, gender, and ethnicity (10). For evaluation of allelic expression imbalance we compared the expression of one allele in cDNA with the quantity of the same allele in the genomic DNA of heterozygotes by using the nonparametric Wilcoxon sign rank test. We considered P < 0.05 to be significant without correcting for multiple analyses, based on strong prior hypotheses and a high correlation between tested traits. All statistical analyses were done in SPSS v.13 for windows (SPSS Inc., Chicago, IL).
Results
Association of T2D-associated SNPs with expression of nearby transcripts in transformed lymphocytes
In TLs from the 82 Caucasian subjects, the genotype of SNP rs9472138 was significantly (P = 0.037) associated with the expression of vascular endothelial growth factor A (VEGFA) transcript (Table 1). This association became statistically nonsignificant (Padj= 0.061) after adjustment for gender and T2D diagnosis. Homozygotes for the risk allele (TT) showed significantly lower expression compared with non-risk allele homozygotes (Supplemental Figure 1). Genotypes of 15 other SNPs were not associated with the expression of 12 other GWAS-implicated adjacent transcripts in TLs (Table 1).
Table 1.
Association analysis of SNPs from T2D GWAS findings with gene expression in transformed lymphocytes of Caucasian subjects
| Gene name | GWAS-SNP | Major/minor allele (D/d) | Genotype count (DD/Dd/dd) | Pa | P(adj)b |
|---|---|---|---|---|---|
| BCL11A | rs10490072 | T/C | 47/29/7 | 0.371 | 0.417 |
| CAMK1D | rs1277970 | A/G | 49/28/5 | 0.868 | 0.799 |
| CDC123 | rs11257622 | T/C | 56/20/7 | 0.592 | 0.546 |
| CDKAL1 | rs7754840 | G/C | 36/40/7 | 0.696 | 0.915 |
| CDKAL1 | rs10946398 | T/G | 36/40/7 | 0.696 | 0.915 |
| CDKN2A | rs10811661 | T/C | 57/24/2 | 0.456 | 0.528 |
| CDKN2A | rs564398 | G/A | 22/42/18 | 0.72 | 0.827 |
| DNAJC11 | rs12137794 | C/T | 68/15/0 | 0.424 | 0.228 |
| FTO | rs8050136 | C/A | 21/43/16 | 0.446 | 0.235 |
| IDE | rs1111875 | G/A | 33/43/7 | 0.798 | 0.958 |
| IDE | rs5015480 | C/T | 33/43/7 | 0.798 | 0.958 |
| PKN2 | rs6698181 | C/T | 32/35/15 | 0.304 | 0.322 |
| SMARCAD1 | rs728989 | C/G | 57/24/2 | 0.478 | 0.450 |
| THADA | rs7578597 | T/C | 70/12/1 | 0.442 | 0.465 |
| VEGFA | rs9472138 | C/T | 33/39/11 | 0.037 | 0.061 |
| WFS1 | rs10010131 | G/A | 30/39/12 | 0.481 | 0.381 |
Total number of individuals vary due to difference in success of genotyping each SNP and respective gene expression analysis.
P, statistical significance of genotypic association from analysis of variance analysis (ANOVA).
P(adj), statistical significance of genotypic association from mixed effects, general linear regression models and included genotype, gender, and T2D diagnosis as fixed factors.
Novel T2D-associated SNPs as cis eQTL in adipose and muscle
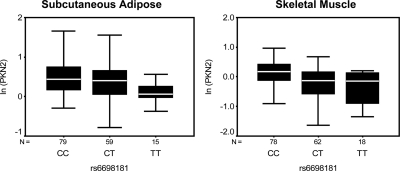
We examined the genotypic association of 20 novel T2D GWAS SNPs with the expression of 19 adjacent transcripts in human sc adipose tissue. We also sought to determine whether 14 T2D-associated SNPs acted as cis-eQTL of 13 transcripts in muscle. We found a significant association of protein kinase N2 (PKN2) expression with the genotype of SNP rs6698181 (P = 0.038) in adipose (Table 2). This association remained significant after adjustment for age, gender, ethnicity, T2D diagnosis, and BMI (P = 0.018) in the entire set of 153 adipose samples, and in the Caucasian subset (n = 103) after adjustment for age, gender, T2D diagnosis, and BMI (P = 0.002). SNP rs6698181 was also detected as a strong cis-eQTL (P = 0.00004) for PKN2 in muscle, and this association remained significant after adjustment for age, gender, ethnicity, T2D diagnosis, and BMI (P = 0.00027) in the entire set of 158 muscle samples (Table 3). In both adipose and muscle, T2D risk allele homozygotes (TT) showed lower expression of PKN2 transcript compared with nonrisk allele (CC) homozygotes (Figure 1). Significant associations were found between rs17036101 and synapsin II (SYN2) expression (P = 0.001) and rs7961581 with Tetraspanin 8 (TSPAN8) (P = 0.047) in adipose of Caucasian individuals after adjustment for age, gender, T2D diagnosis, and BMI. Similarly, expression of JAZF zinc finger 1 (JAZF1) transcript in muscle was significantly associated with the genotype of rs864745 (P = 0.011). Expression of several other transcripts including ADAMTS9, CAMK1D, CDC123, CDKAL1, DNAJC11, FTO, HHEX, IDE, NOTCH2, PPARG, SMARCAD1, TCF7L2 isoform with exon 13a, THADA, VEGFA, WFS1 in adipose, and/or muscle showed no significant association with the genotypes of adjacent T2D GWAS SNPs.
Table 2.
Association of SNPs from T2D GWAS findings with gene expression in human adipose
| Gene name | SNP | Major/minor allele (D/d) | All |
Caucasian |
||||
|---|---|---|---|---|---|---|---|---|
| Genotype count (DD/Dd/dd) | Pa | P(adj)b | Genotype count (DD/Dd/dd) | Pa | P(adj)b | |||
| ADAMTS9 | rs4607103 | C/T | 85/58/10 | 0.096 | 0.142 | 63/36/7 | 0.817 | 0.791 |
| CAMK1D | rs1277970 | A/G | 109/38/4 | 0.834 | 0.791 | 72/30/3 | 0.705 | 0.502 |
| CDC123 | rs11257622 | T/C | 105/43/5 | 0.711 | 0.655 | 70/33/3 | 0.407 | 0.520 |
| CDKAL1 | rs7754840 | G/C | 59/63/30 | 0.498 | 0.336 | 55/39/11 | 0.425 | 0.784 |
| CDKAL1 | rs10946398 | T/G | 59/63/30 | 0.498 | 0.336 | 55/39/11 | 0.425 | 0.784 |
| DNAJC11 | rs12137794 | C/T | 128/24/0 | 0.546 | 0.601 | 84/22/0 | 0.386 | 0.218 |
| FTO | rs8050136 | C/A | 45/82/26 | 0.946 | 0.791 | 31/56/19 | 0.821 | 0.983 |
| HHEX | rs111875 | G/A | 68/55/30 | 0.818 | 0.963 | 38/43/25 | 0.836 | 0.478 |
| HHEX | rs5015480 | C/T | 58/61/34 | 0.914 | 0.656 | 37/44/25 | 0.937 | 0.641 |
| IDE | rs111875 | G/A | 68/55/30 | 0.63 | 0.463 | 38/43/25 | 0.267 | 0.053 |
| IDE | rs5015480 | C/T | 58/61/34 | 0.684 | 0.333 | 37/44/25 | 0.397 | 0.105 |
| JAZF1 | rs864745 | T/C | 50/74/29 | 0.447 | 0.670 | 21/60/25 | 0.754 | 0.481 |
| NOTCH2 | rs10923931 | G/T | 105/40/8 | 0.222 | 0.184 | 84/19/3 | 0.901 | 0.226 |
| PKN2 | rs6698181 | C/T | 79/59/15 | 0.038 | 0.018 | 44/48/14 | 0.009 | 0.002 |
| PPARG | rs1801282 | C/G | 120/32/1 | 0.094 | 0.233 | 76/29/1 | 0.136 | 0.376 |
| PPARG | rs17036101 | G/A | 138/15/0 | 0.791 | 0.722 | 95/11/0 | 0.169 | 0.054 |
| SMARCAD1 | rs728989 | C/G | 73/55/24 | 0.467 | 0.427 | 68/32/5 | 0.555 | 0.485 |
| SYN2 | rs17036101 | G/A | 138/15/0 | 0.111 | 0.119 | 95/11/0 | 0.001 | 0.001 |
| TCF7L2 (13a+) | rs7903146 | C/T | 70/69/13 | 0.087 | 0.168 | 52/45/8 | 0.358 | 0.751 |
| THADA | rs7578597 | T/C | 115/32/6 | 0.875 | 0.886 | 82/23/1 | 0.599 | 0.977 |
| TSPAN8 | rs7961581 | T/C | 72/64/12 | 0.226 | 0.200 | 51/42/10 | 0.177 | 0.047 |
| VEGFA | rs9472138 | C/T | 83/64/6 | 0.75 | 0.902 | 53/48/5 | 0.209 | 0.744 |
| WFS1 | rs10010131 | G/A | 52/84/11 | 0.101 | 0.115 | 39/56/8 | 0.136 | 0.179 |
Total number of individuals vary due to difference in success of genotyping each SNP and respective gene expression analysis.
P, statistical significance of genotypic association from analysis of variance analysis (ANOVA).
P(adj), statistical significance of genotypic association from mixed effects, general linear regression models and included age and BMI as covariate and genotype, gender, ethnicity, and T2D diagnosis as fixed factors.
Table 3.
Association of SNPs from T2D GWAS findings with gene expression in human muscle
| Gene name | SNP | Major/minor allele (D/d) | All |
Caucasian |
||||
|---|---|---|---|---|---|---|---|---|
| Genotype count (DD/Dd/dd) | Pa | P(adj)b | Genotype count (DD/Dd/dd) | Pa | P(adj)b | |||
| ADAMTS9 | rs4607103 | C/T | 88/58/12 | 0.664 | 0.274 | 66/38/9 | 0.518 | 0.360 |
| CAMK1D | rs1277970 | A/G | 108/44/4 | 0.762 | 0.724 | 73/36/3 | 0.712 | 0.727 |
| CDC123 | rs11257622 | T/C | 106/46/6 | 0.843 | 0.869 | 71/38/4 | 0.806 | 0.829 |
| CDKAL1 | rs7754840 | G/C | 61/68/29 | 0.534 | 0.835 | 57/45/11 | 0.232 | 0.295 |
| CDKAL1 | rs10946398 | T/G | 61/68/29 | 0.534 | 0.835 | 57/45/11 | 0.232 | 0.295 |
| DNAJC11 | rs12137794 | C/T | 133/25/0 | 0.601 | 0.603 | 90/23/0 | 0.67 | 0.590 |
| FTO | rs8050136 | C/A | 46/84/28 | 0.239 | 0.348 | 32/60/21 | 0.661 | 0.774 |
| JAZF1 | rs864745 | T/C | 53/75/30 | 0.020 | 0.011 | 25/61/27 | 0.098 | 0.086 |
| PKN2 | rs6698181 | C/T | 78/62/18 | 0.00004 | 0.00027 | 44/52/17 | 0.003 | 0.0021 |
| SMARCAD1 | rs728989 | C/G | 72/59/26 | 0.066 | 0.440 | 67/38/7 | 0.948 | 0.937 |
| THADA | rs7578597 | T/C | 116/35/7 | 0.297 | 0.249 | 84/27/2 | 0.091 | 0.058 |
| TSPAN8 | rs7961581 | T/C | 79/60/13 | 0.216 | 0.241 | 58/40/11 | 0.573 | 0.588 |
| VEGFA | rs9472138 | C/T | 85/64/9 | 0.388 | 0.435 | 56/49/8 | 0.282 | 0.392 |
| WFS1 | rs10010131 | G/A | 53/88/11 | 0.055 | 0.055 | 40/61/9 | 0.067 | 0.069 |
Total number of individuals vary due to difference in success of genotyping each SNP and respective gene expression analysis.
P, statistical significance of genotypic association from analysis of variance analysis (ANOVA).
P(adj), statistical significance of genotypic association from mixed effects, general linear regression models and included age and BMI as covariate and genotype, gender, ethnicity, and T2D diagnosis as fixed factors.
Fig. 1.
Genotypic association of SNP rs6698181 with the expression of protein kinase N2 (PKN2) transcript in human adipose and muscle. The box plot shows transcript level expression of PKN2 for different genotypes for SNP rs6698181. PKN2 expression is shown after 18S normalization and ln transformation. N = no. of samples. The box represents the interquartile range, which contains 50% of the values. The whiskers are lines that extend the box to the highest and lowest values, excluding outliers. A line across the box indicates the median.
Correlation of metabolic traits with expression of T2D GWAS implicated transcripts
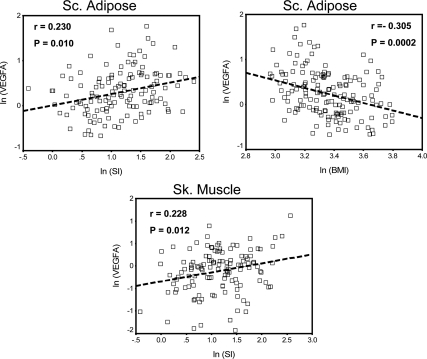
We examined the correlation of obesity traits (including BMI) and FSIGT-derived glucose homeostasis traits (including SI and AIRG) with the expression of T2D GWAS-implicated transcripts in the sc adipose and skeletal muscle tissues of Caucasian and African-American nondiabetic subjects spanning a range of BMI and SI to understand the putative metabolic role of these genes. Partial correlation analysis after controlling for age, gender, and ethnicity showed significant positive correlation of VEGFA transcript expression in both adipose (r = 0.230, P = 0.010) and muscle (r = 0.228, P = 0.012) with insulin sensitivity (SI). Adipose expression of VEGFA was also negatively correlated with BMI (r = −0.305, P = 0.0002) (Figure 2 and Supplemental Table 5). Expression of GWAS-implicated transcripts in our study showed no significant correlation with AIRG (Supplemental Table 5).
Fig. 2.
Expression of vascular endothelial growth factor A (VEGFA) in adipose and muscle is correlated with metabolic traits. VEGFA expression is shown after 18S normalization and ln transformation. Insulin sensitivity [SI (×10−4 · min−1 [μU/ml]−1)] and body mass index (kg/m2) data are shown after ln transformation. Sc. adipose, sc adipose; Sk. muscle, skeletal muscle; r, partial correlation coefficient; p, statistical significance of correlation.
Allelic expression imbalance in T2D GWAS-implicated genes
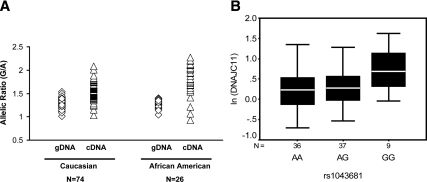
We observed that most of the T2D-associated novel SNPs from GWAS were not acting as cis-eQTLs for nearby transcripts. However, some of these transcripts showed significant correlations with metabolic traits. Using AEI analysis, we thus finally explored the presence of other cis-regulatory SNPs in those GWAS-implicated transcripts by comparing allele-specific expression in transformed lymphocyte cDNA of heterozygotes from 188 Caucasian and 95 African-American individuals. We initially examined AEI in 11 expressed SNPs in 11 GWAS-implicated genes. Additional SNPs were tested in genes showing significant AEI. Significant AEI was observed in 4/11 genes in both Caucasians and African Americans, while two genes showed AEI only in Caucasians (Table 4). SNP rs1043681 in 3′UTR of the DNAJC11 gene showed the strongest evidence of allelic expression imbalance (P = 3.5 × 10−12 in Caucasians and 2.6 × 10−5 in African Americans), with “G” allele showing higher expression in cDNA of heterozygotes than in gDNA(Figure 3A). Finally, SNPs showing AEI were examined for their association with total transcript expression (Supplemental Table 6). SNP rs1043681 showed significant AEI and was also significantly associated with total expression of DNAJC11 (P = 0.045). Homozygotes for the “G” allele of SNP rs1043681 showed higher expression compared with “A” allele homozygotes (Figure 3B).
Table 4.
Allelic expression imbalance in genes from T2D GWAS findings
| GWAS gene | SNP | Allele | Caucasian |
African American |
||
|---|---|---|---|---|---|---|
| Z | P | Z | P | |||
| BCL11A | rs7569946 | A | −1.1 | 0.271 | −0.865 | 0.387 |
| CAMK1D | rs1644394 | A | −4.754 | 1.99E-06 | −2.009 | 0.045 |
| CDC123 | rs1051055 | A | −4.176 | 3.00E-05 | −2.859 | 0.004 |
| CDKAL1 | rs1563727 | C | −1.338 | 0.181 | −0.254 | 0.8 |
| CDKN2A | rs3088440 | C | −0.135 | 0.893 | −0.241 | 0.809 |
| DNAJC11 | rs1043681a | G | −6.958 | 3.50E-12 | −4.204 | 2.60E-05 |
| DNAJC11 | rs11892 | C | −0.543 | 0.587 | −1.793 | 0.073 |
| PKN2 | rs786906 | A | −0.219 | 0.827 | −1.253 | 0.21 |
| SMARCAD1 | rs8336a | A | −6.42 | 1.40E-10 | −2.366 | 0.018 |
| SMARCAD1 | rs8026 | A | −4.697 | 2.64E-06 | −1.519 | 0.129 |
| THADA | rs11899823 | C | −2.697 | 0.007 | −1.027 | 0.304 |
| VEGFA | rs2010963a | C | −4.157 | 3.20E-05 | −1.48 | 0.139 |
| VEGFA | rs10434 | A | −1.451 | 0.147 | −1.882 | 0.06 |
| WFS1 | rs1046316 | T | −0.703 | 0.482 | −2.258 | 0.024 |
AEI was evaluated by comparing expression of one allele in cDNA with quantity of the same allele in genomic DNA of heterozygotes by using nonparametric Wilcoxon sign rank test. Z and P are Z-statistics and two-tailed asymptotic significance from Wilcoxon Signed Rank Test.
First cSNP tested.
Fig. 3.
rs1043681 is a cis-regulatory SNP for DNAJC11 expression in transformed lymphocyte. SNP rs1043681 in 3′UTR of the DNAJC11 gene allelic expression imbalance in both Caucasian and African-American TLs (A) and also regulates total expression of DNAJC11 (B). A, Allelic ratio of G and A alleles of rs1043681 in genomic DNA (gDNA) and cDNA from heterozygous individuals are shown as evidence of allelic expression imbalance. B, The box plot shows transcript level expression of DNAJC11 for different genotypes for SNP rs1043681. DNAJC11 expression is shown after 18S normalization and ln transformation. N = no. of samples. The box represents the interquartile range, which contains 50% of the values. The whiskers are lines that extend the box to the highest and lowest values, excluding outliers. A line across the box indicates the median.
Discussion
Genome-wide association studies in different Caucasian cohorts have identified several novel SNPs and replicated previously identified loci associated with T2D (12–15). Except for a few, the functional implication of most of these noncoding SNPs in T2D pathogenesis is largely unknown and unexplored. Most of the GWAS studies suggested that these SNPs have an effect on the regulation of genes in near proximity, but supportive data were lacking from those studies. The main objective of our study was to test the hypothesis that these SNPs modulate the transcription of nearby genes and may increase T2D susceptibility by acting as cis regulatory elements for those genes. We attempted to elucidate the function of genes in proximity to T2D-associated SNPs by examining the correlation between transcript level expression of these genes in adipose and muscle tissue with metabolic traits of well-characterized healthy human subjects. We also sought evidence for other cis-regulatory elements that might regulate expression of these genes. Our study suggests that cis-regulatory SNPs commonly (54%) regulate expression of these transcripts in TLs. However, in our study, T2D-associated GWAS SNPs rarely acted as a cis-regulator of nearby transcripts either in TLs or in diabetes-related tissues. Notable limitations of our study include: 1) limited statistical power of the cohort of 168 individuals for gene expression analyses may cause our results to be falsely positive or falsely negative; 2) biopsy samples were obtained only during a fasting state, and GWAS genes may play roles in other physiological conditions; 3) lack of access to tissues other than adipose and muscle from well-characterized and genotyped individuals limited our search to GWAS genes expressed only in available tissues; and 4) our study was limited only to identify the cis-regulatory role of GWAS associated SNPs.
Several studies have identified eQTLs in surrogate tissues including TLs from HapMap subjects and CEPH pedigrees (21–23). Using TLs from Caucasian subjects with or with out T2D we tested whether GWAS-confirmed T2D-associated SNPs act as a cis-eQTL to modulate the expression of nearby transcripts. Only one of 16 T2D-associated SNPs examined was associated with expression of a nearby transcript. A recently published genome-wide analysis of 1598 SNPs associated with different complex traits showed a general enrichment for eQTL among those traits associated with GWAS SNPs in HapMap TLs, but, consistent with our observations, a disease-specific analysis indicated no enrichment for eQTLs among T2D-associated SNPs (5). HapMap TLs in that study showed more enrichment for eQTLs among autoimmune disorder-associated SNPs and thus may indicate TLs cultured at basal condition as a poor proxy for human tissues relevant for metabolic disorders. Indeed, recent studies by Zhong et al. (24) using human liver and adipose samples indicated a 1.19-fold enrichment of T2D-associated GWAS SNPs among eQTLs from those tissues. We examined genotypic association of 20 novel T2D GWAS SNPs with the expression of 19 adjacent transcripts in human sc adipose, and 14 SNPs with 13 transcripts in muscle. Only one SNP was detected as an eQTL in both adipose and muscle (rs6698181 with expression of PKN2). T2D GWAS SNPs near SYN2 and TSPAN8 was detected as cis-eQTL in adipose, while a SNP near JAZF1 was detected as cis-eQTL in muscle. A study by Zhong et al. (25) detected T2D-associated SNPs, or a SNP in LD, with an associated SNP as a cis-eQTL for 4/18 GWAS genes, including NOTCH2, TSPAN8, and JAZF1 in liver and ADAMTS9 in omental adipose. Findings from this study partially overlap with our study. Differences may reflect sample ascertainment, sample size, and tissues chosen for analysis. It is important to consider that study the by Zhong et al. was done using liver samples from postmortem tissue and surgical resections from organ donors, or liver and adipose (sc and omental) samples from obese patients undergoing RXY gastric bypass surgery (24, 25). In contrast, we conducted our study on sc adipose and muscle of metabolically well-characterized, relatively healthy (mostly nondiabetic) individuals spanning a broad range of BMI and SI. Admittedly, our sample size was comparatively smaller.
Protein kinase N2 or PKC related kinase-2 (PKN2/ PRK2) is a serine threonine kinase and shows substrate specificity toward nucleic acid binding to basic proteins, and is induced by lipids including cardiolipin and unsaturated fatty acids (26, 27). Our study showed significant association of the SNP rs6698181 and the transcript level expression of PKN2 in sc adipose and muscle of Caucasian subjects as well as in the combined set. This SNP showed association with T2D in Caucasian populations but association was not replicated in African-American or Asian populations (28, 29). T2D risk allele (T) homozygotes showed lower expression of PKN2 transcript compared with nonrisk allele (C) homozygotes. The rs6698181 is located only 6.6Kb upstream of PKN2 and in silico analysis using MAPPER (http://mapper.chip.org/mapper/) indicates that this is the binding site of important human transcription factors, including PBX1 and PAX6, and that these sites are obliterated by the risk allele. Similarly SNP rs17036101, located 44.3 Kb downstream of SYN2 gene and 51.5 Kb upstream of PPARG gene, showed significant association with expression of SYN2 in adipose of our Caucasian subjects. The common risk allele (A) of this SNP showed significant association with T2D in Caucasian populations and showed lower expression in adipose of risk allele homozygotes compared with heterozygotes; this association was not observed in muscle. Thus, some of these T2D-associated SNPs possibly interact with specific transcription factors and may act as tissue specific cis-eQTL. Further functional studies will be required to understand the mechanism of transcriptional modulation and T2D pathogenesis mediated by these SNPs.
A recent bioinformatic analysis of publicly available gene expression profile data sets from four different tissues of human and animal models of T2D showed aberrant expression for 12/21 genes located in the vicinity of T2D GWAS SNPs (30). SNP rs9472138 at 57.5 Kb downstream of VEGFA was associated with T2D. In our study this SNP was detected as a cis-eQTL for VEGFA expression in TLs, but not in adipose or muscle. Interestingly, our study on metabolic traits of healthy human subjects indicates a significant correlation between VEGFA expression in adipose and muscle with insulin sensitivity (SI). In adipose, expression of VEGFA was also correlated with BMI. VEGF plays an important role in adipose tissue angiogenesis and modulates adipocyte differentiation (31). Consistent with our study, other recent studies also showed lower pO2, lower capillary density, and lower VEGF mRNA in adipose tissue of obese subjects compared with lean subjects (32, 33). Thus, despite the apparent discordance of gene expression and genetic association data, our study, along with other published studies, indicates an important role of the transcriptional modulation of VEGFA gene in T2D pathogenesis, and the SNP rs9472138 near VEGFA probably increases susceptibility to Type 2 Diabetes by a mechanism other than regulating transcription of VEGFA in adipose or muscle. Further studies will be required to illustrate this unidentified mechanism.
In our study only one of 16 T2D-associated GWAS SNPs appeared as cis-eQTL from quantitative analysis of total transcripts in TLs. Recent studies using TLs showed that variation in chromatin structure and transcription factor binding is allele-specific and heritable (34). Direct demonstration of cis activity of a variant requires measurement of allele specific expression. Genome-wide study in HapMap TLs showed sensitivity and specificity of AEI analysis to detect cis-effect on SNPs (35). Most T2D SNPs are located in intergenic or intronic regions and are not amenable to AEI analysis by pyrosequencing. To uncover the presence of other cis-regulatory SNPs we performed AEI analysis for SNPs located in coding or UTR regions of GWAS-indicated transcripts. We found evidence for cis-regulation in 6/11 GWAS genes tested in TLs by AEI analysis. Thus cis-regulation is common for these transcripts but only a small subset of GWAS SNPs may increase T2D susceptibility by modulating expression of adjacent transcripts as cis-eQTL. Most heritable gene expression traits are, however, complex and governed by a combination of cis- and trans-acting loci and nongenetic factors. Role of T2D-associated SNPs on transcripts at a distance on the same or another chromosome as a distal (trans) regulatory element cannot be disregarded from our study. Further genome wide analysis of expressed transcripts in a variety of human tissues will be required to explore these possibilities.
Supplementary Material
Acknowledgments
We thank the Clinical Research Center staff of University of Arkansas for Medical Sciences for their outstanding support of the physiologic studies and assistance with data management. We thank Prof. Siqun Zheng, Director, Genotyping Laboratory, Center for Human Genomics, Wake Forest University Health Sciences, for her extensive support in genotyping our samples using Sequenom MassARRAY system. We also thank Prof. Thomas DuBose, Chair, Department of Internal Medicine, Wake Forest University Health Sciences, for infrastructural and administrative support to complete this work. We thank Amanda Goode for a critical reading and editing of our manuscript.
This work was supported by grant DK039311 from the National Institutes of Health/National Institute of Diabetes and Digestive and Kidney Diseases. Clinical studies were supported by General Clinical Research Center Grant M01RR14288 from National Center for Research Resources, National Institutes of Health to the University of Arkansas for Medical Sciences.
Disclosure Summary: The authors have nothing to declare.
Footnotes
- AEI
- allelic expression imbalance
- AIRG
- acute insulin response to glucose
- AUC
- area under the curve
- BMI
- body mass index
- DI
- disposition index
- DXA
- dual x-ray absorptiometry
- eQTL
- expression quantitative trait loci
- eSNP
- expressed single nucleotide polymorphism
- FSIGT
- frequently sampled intravenous glucose tolerance test
- GWAS
- genome-wide association scan
- LD
- linkage disequilibrium
- MAF
- minor allele frequency
- OGTT
- oral glucose tolerance test
- PKN2
- protein kinase N2
- SG
- glucose-mediated glucose disposal
- SI
- insulin sensitivity
- SNP
- single nucleotide polymorphism
- T2D
- type 2 diabetes mellitus
- TL
- transformed lymphocyte
- UTR
- untranslated
- VEGFA
- vascular endothelial growth factor A.
References
- 1. Das SK, Elbein SC. 2006. The genetic basis of type 2 diabetes. Cellscience 2:100–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lyssenko V, Groop L. 2009. Genome-wide association study for type 2 diabetes: clinical applications. Curr Opin Lipidol 20:87–91 [DOI] [PubMed] [Google Scholar]
- 3. Stolerman ES, Florez JC. 2009. Genomics of type 2 diabetes mellitus: implications for the clinician. Nat Rev Endocrinol 5:429–436 [DOI] [PubMed] [Google Scholar]
- 4. Nica AC, Montgomery SB, Dimas AS, Stranger BE, Beazley C, Barroso I, Dermitzakis ET. 2010. Candidate causal regulatory effects by integration of expression QTLs with complex trait genetic associations. PLoS Genet 6:e1000895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Nicolae DL, Gamazon E, Zhang W, Duan S, Dolan ME, Cox NJ. 2010. Trait-associated SNPs are more likely to be eQTLs: annotation to enhance discovery from GWAS. PLoS Genet 6:e1000888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Voight BF, Scott LJ, Steinthorsdottir V, Morris AP, Dina C, Welch RP, Zeggini E, Huth C, Aulchenko YS, Thorleifsson G, McCulloch LJ, Ferreira T, Grallert H, Amin N, Wu G, Willer CJ, Raychaudhuri S, McCarroll SA, Langenberg C, Hofmann OM, Dupuis J, Qi L, Segre AV, van Hoek M, Navarro P, Ardlie K, Balkau B, Benediktsson R, Bennett AJ, Blagieva R, Boerwinkle E, Bonnycastle LL, Bengtsson BK, Bravenboer B, Bumpstead S, Burtt NP, Charpentier G, Chines PS, Cornelis M, Couper DJ, Crawford G, Doney AS, Elliott KS, Elliott AL, Erdos MR, Fox CS, Franklin CS, Ganser M, Gieger C, Grarup N, Green T, Griffin S, Groves CJ, Guiducci C, Hadjadj S, Hassanali N, Herder C, Isomaa B, Jackson AU, Johnson PR, Jorgensen T, Kao WH, Klopp N, Kong A, Kraft P, Kuusisto J, Lauritzen T, Li M, Lieverse A, Lindgren CM, Lyssenko V, Marre M, Meitinger T, Midthjell K, Morken MA, Narisu N, Nilsson P, Owen KR, Payne F, Perry JR, Petersen AK, Platou C, Proenca C, Prokopenko I, Rathmann W, Rayner NW, Robertson NR, Rocheleau G, Roden M, Sampson MJ, Saxena R, Shields BM, Shrader P, Sigurdsson G, Sparso T, Strassburger K, Stringham HM, Sun Q, Swift AJ, Thorand B, Tichet J, Tuomi T, van Dam RM, van Haeften TW, van Herpt T, Vliet-Ostaptchouk JV, Bragi WG, Weedon MN, Wijmenga C, Witteman J, Bergman RN, Cauchi S, Collins FS, Gloyn AL, Gyllensten U, Hansen T, Hide WA, Hitman GA, Hofman A, Hunter DJ, Hveem K, Laakso M, Mohlke KL, Morris AD, Palmer CN, Pramstaller PP, Rudan I, Sijbrands E, Stein LD, Tuomilehto J, Uitterlinden A, Walker M, Wareham NJ, Watanabe RM, Abecasis GR, Boehm BO, Campbell H, Daly MJ, Hattersley AT, Hu FB, Meigs JB, Pankow JS, Pedersen O, Wichmann HE, Barroso I, Florez JC, Frayling TM, Groop L, Sladek R, Thorsteinsdottir U, Wilson JF, Illig T, Froguel P, van Duijn CM, Stefansson K, Altshuler D, Boehnke M, McCarthy MI. 2010. Twelve type 2 diabetes susceptibility loci identified through large-scale association analysis. Nat Genet 42:579–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Florez JC. 2008. Newly identified loci highlight beta cell dysfunction as a key cause of type 2 diabetes: where are the insulin resistance genes? Diabetologia 51:1100–1110 [DOI] [PubMed] [Google Scholar]
- 8. Doria A, Patti ME, Kahn CR. 2008. The emerging genetic architecture of type 2 diabetes. Cell Metab 8:186–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Das SK, Sharma NK, Chu WS, Wang H, Elbein SC. 2008. Aryl hydrocarbon receptor nuclear translocator (ARNT) gene as a positional and functional candidate for type 2 diabetes and prediabetic intermediate traits: Mutation detection, case-control studies, and gene expression analysis. BMC Med Genet 9:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sharma NK, Das SK, Mondal AK, Hackney OG, Chu WS, Kern PA, Rasouli N, Spencer HJ, Yao-Borengasser A, Elbein SC. 2008. Endoplasmic reticulum stress markers are associated with obesity in nondiabetic subjects. J Clin Endocrinol Metab 93:4532–4541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Elbein SC, Chu WS, Das SK, Yao-Borengasser A, Hasstedt SJ, Wang H, Rasouli N, Kern PA. 2007. Transcription factor 7-like 2 polymorphisms and type 2 diabetes, glucose homeostasis traits and gene expression in US participants of European and African descent. Diabetologia 50:1621–1630 [DOI] [PubMed] [Google Scholar]
- 12. Scott LJ, Mohlke KL, Bonnycastle LL, Willer CJ, Li Y, Duren WL, Erdos MR, Stringham HM, Chines PS, Jackson AU, Prokunina-Olsson L, Ding CJ, Swift AJ, Narisu N, Hu T, Pruim R, Xiao R, Li XY, Conneely KN, Riebow NL, Sprau AG, Tong M, White PP, Hetrick KN, Barnhart MW, Bark CW, Goldstein JL, Watkins L, Xiang F, Saramies J, Buchanan TA, Watanabe RM, Valle TT, Kinnunen L, Abecasis GR, Pugh EW, Doheny KF, Bergman RN, Tuomilehto J, Collins FS, Boehnke M. 2007. A genome-wide association study of type 2 diabetes in Finns detects multiple susceptibility variants. Science 316:1341–1345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zeggini E, Weedon MN, Lindgren CM, Frayling TM, Elliott KS, Lango H, Timpson NJ, Perry JR, Rayner NW, Freathy RM, Barrett JC, Shields B, Morris AP, Ellard S, Groves CJ, Harries LW, Marchini JL, Owen KR, Knight B, Cardon LR, Walker M, Hitman GA, Morris AD, Doney AS, McCarthy MI, Hattersley AT. 2007. Replication of genome-wide association signals in UK samples reveals risk loci for type 2 diabetes. Science 316:1336–1341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Saxena R, Voight BF, Lyssenko V, Burtt NP, de Bakker PI, Chen H, Roix JJ, Kathiresan S, Hirschhorn JN, Daly MJ, Hughes TE, Groop L, Altshuler D, Almgren P, Florez JC, Meyer J, Ardlie K, Bengtsson BK, Isomaa B, Lettre G, Lindblad U, Lyon HN, Melander O, Newton-Cheh C, Nilsson P, Orho-Melander M, Rastam L, Speliotes EK, Taskinen MR, Tuomi T, Guiducci C, Berglund A, Carlson J, Gianniny L, Hackett R, Hall L, Holmkvist J, Laurila E, Sjogren M, Sterner M, Surti A, Svensson M, Svensson M, Tewhey R, Blumenstiel B, Parkin M, Defelice M, Barry R, Brodeur W, Camarata J, Chia N, Fava M, Gibbons J, Handsaker B, Healy C, Nguyen K, Gates C, Sougnez C, Gage D, Nizzari M, Gabriel SB, Chirn GW, Ma Q, Parikh H, Richardson D, Ricke D, Purcell S. 2007. Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science 316:1331–1336 [DOI] [PubMed] [Google Scholar]
- 15. Zeggini E, Scott LJ, Saxena R, Voight BF, Marchini JL, Hu T, de Bakker PI, Abecasis GR, Almgren P, Andersen G, Ardlie K, Bostrom KB, Bergman RN, Bonnycastle LL, Borch-Johnsen K, Burtt NP, Chen H, Chines PS, Daly MJ, Deodhar P, Ding CJ, Doney AS, Duren WL, Elliott KS, Erdos MR, Frayling TM, Freathy RM, Gianniny L, Grallert H, Grarup N, Groves CJ, Guiducci C, Hansen T, Herder C, Hitman GA, Hughes TE, Isomaa B, Jackson AU, Jorgensen T, Kong A, Kubalanza K, Kuruvilla FG, Kuusisto J, Langenberg C, Lango H, Lauritzen T, Li Y, Lindgren CM, Lyssenko V, Marvelle AF, Meisinger C, Midthjell K, Mohlke KL, Morken MA, Morris AD, Narisu N, Nilsson P, Owen KR, Palmer CN, Payne F, Perry JR, Pettersen E, Platou C, Prokopenko I, Qi L, Qin L, Rayner NW, Rees M, Roix JJ, Sandbaek A, Shields B, Sjogren M, Steinthorsdottir V, Stringham HM, Swift AJ, Thorleifsson G, Thorsteinsdottir U, Timpson NJ, Tuomi T, Tuomilehto J, Walker M, Watanabe RM, Weedon MN, Willer CJ, Illig T, Hveem K, Hu FB, Laakso M, Stefansson K, Pedersen O, Wareham NJ, Barroso I, Hattersley AT, Collins FS, Groop L, McCarthy MI, Boehnke M, Altshuler D. 2008. Meta-analysis of genome-wide association data and large-scale replication identifies additional susceptibility loci for type 2 diabetes. Nat Genet 40:638–645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Das SK, Chu WS, Mondal AK, Sharma NK, Kern PA, Rasouli N, Elbein SC. 2008. Effect of pioglitazone treatment on endoplasmic reticulum stress response in human adipose and in palmitate-induced stress in human liver and adipose cell lines. Am J Physiol Endocrinol Metab 295:E393–E400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Das SK, Mondal AK, Elbein SC. 2010. Distinct gene expression profiles characterize cellular responses to palmitate and oleate. J Lipid Res 51:2121–2131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mondal AK, Das SK, Baldini G, Chu WS, Sharma NK, Hackney OG, Zhao J, Grant SF, Elbein SC. 2010. Genotype and tissue-specific effects on alternative splicing of the transcription factor 7-like 2 gene in humans. J Clin Endocrinol Metab 95:1450–1457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wang H, Elbein SC. 2007. Detection of allelic imbalance in gene expression using pyrosequencing. Methods Mol Biol 373:157–176 [DOI] [PubMed] [Google Scholar]
- 20. Boston RC, Stefanovski D, Moate PJ, Sumner AE, Watanabe RM, Bergman RN. 2003. MINMOD Millennium: a computer program to calculate glucose effectiveness and insulin sensitivity from the frequently sampled intravenous glucose tolerance test. Diabetes Technol Ther 5:1003–1015 [DOI] [PubMed] [Google Scholar]
- 21. Stranger BE, Nica AC, Forrest MS, Dimas A, Bird CP, Beazley C, Ingle CE, Dunning M, Flicek P, Koller D, Montgomery S, Tavare S, Deloukas P, Dermitzakis ET. 2007. Population genomics of human gene expression. Nat Genet 39:1217–1224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Cheung VG, Spielman RS. 2009. Genetics of human gene expression: mapping DNA variants that influence gene expression. Nat Rev Genet 10:595–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Morley M, Molony CM, Weber TM, Devlin JL, Ewens KG, Spielman RS, Cheung VG. 2004. Genetic analysis of genome-wide variation in human gene expression. Nature 430:743–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zhong H, Beaulaurier J, Lum PY, Molony C, Yang X, Macneil DJ, Weingarth DT, Zhang B, Greenawalt D, Dobrin R, Hao K, Woo S, Fabre-Suver C, Qian S, Tota MR, Keller MP, Kendziorski CM, Yandell BS, Castro V, Attie AD, Kaplan LM, Schadt EE. 2010. Liver and adipose expression associated SNPs are enriched for association to type 2 diabetes. PLoS Genet 6:e1000932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zhong H, Yang X, Kaplan LM, Molony C, Schadt EE. 2010. Integrating pathway analysis and genetics of gene expression for genome-wide association studies. Am J Hum Genet 86:581–591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Dettori R, Sonzogni S, Meyer L, Lopez-Garcia LA, Morrice NA, Zeuzem S, Engel M, Piiper A, Neimanis S, Frodin M, Biondi RM. 2009. Regulation of the interaction between protein kinase C-related protein kinase 2 (PRK2) and its upstream kinase, 3-phosphoinositide-dependent protein kinase 1 (PDK1). J Biol Chem 284:30318–30327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yu W, Liu J, Morrice NA, Wettenhall RE. 1997. Isolation and characterization of a structural homologue of human PRK2 from rat liver. Distinguishing substrate and lipid activator specificities. J Biol Chem 272:10030–10034 [DOI] [PubMed] [Google Scholar]
- 28. Tan JT, Ng DP, Nurbaya S, Ye S, Lim XL, Leong H, Seet LT, Siew WF, Kon W, Wong TY, Saw SM, Aung T, Chia KS, Lee J, Chew SK, Seielstad M, Tai ES. 2010. Polymorphisms identified through genome-wide association studies and their associations with type 2 diabetes in Chinese, Malays, and Asian-Indians in Singapore. J Clin Endocrinol Metab 95:390–397 [DOI] [PubMed] [Google Scholar]
- 29. Lewis JP, Palmer ND, Hicks PJ, Sale MM, Langefeld CD, Freedman BI, Divers J, Bowden DW. 2008. Association analysis in african americans of European-derived type 2 diabetes single nucleotide polymorphisms from whole-genome association studies. Diabetes 57:2220–2225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Parikh H, Lyssenko V, Groop LC. 2009. Prioritizing genes for follow-up from genome wide association studies using information on gene expression in tissues relevant for type 2 diabetes mellitus. BMC Med Genomics 2:72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ledoux S, Queguiner I, Msika S, Calderari S, Rufat P, Gasc JM, Corvol P, Larger E. 2008. Angiogenesis associated with visceral and subcutaneous adipose tissue in severe human obesity. Diabetes 57:3247–3257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Miranda M, Escote X, Ceperuelo-Mallafre V, Megia A, Caubet E, Naf S, Gomez JM, Gonzalez-Clemente JM, Vicente V, Vendrell J. 2010. Relation between human LPIN1, hypoxia and endoplasmic reticulum stress genes in subcutaneous and visceral adipose tissue. Int J Obes (Lond) 34:679–686 [DOI] [PubMed] [Google Scholar]
- 33. Pasarica M, Sereda OR, Redman LM, Albarado DC, Hymel DT, Roan LE, Rood JC, Burk DH, Smith SR. 2009. Reduced adipose tissue oxygenation in human obesity: evidence for rarefaction, macrophage chemotaxis, and inflammation without an angiogenic response. Diabetes 58:718–725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. McDaniell R, Lee BK, Song L, Liu Z, Boyle AP, Erdos MR, Scott LJ, Morken MA, Kucera KS, Battenhouse A, Keefe D, Collins FS, Willard HF, Lieb JD, Furey TS, Crawford GE, Iyer VR, Birney E. 2010. Heritable individual-specific and allele-specific chromatin signatures in humans. Science 328:235–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ge B, Pokholok DK, Kwan T, Grundberg E, Morcos L, Verlaan DJ, Le J, Koka V, Lam KC, Gagne V, Dias J, Hoberman R, Montpetit A, Joly MM, Harvey EJ, Sinnett D, Beaulieu P, Hamon R, Graziani A, Dewar K, Harmsen E, Majewski J, Goring HH, Naumova AK, Blanchette M, Gunderson KL, Pastinen T. 2009. Global patterns of cis variation in human cells revealed by high-density allelic expression analysis. Nat Genet 41:1216–1222 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.