Summary
Leavenworthia crassa is a rosette flowering species that differs from inflorescence flowering species, such as Arabidopsis thaliana, in having elongated pedicels and shortened interfloral internodes on the main axis. Based on previous experiments, we hypothesized that changes to the L. crassa TFL1 ortholog, LcrTFL1, were important in the evolution of rosette flowering.
We isolated LcrTFL1 and introduced a genomic construct into tfl1 mutant A. thaliana plants. We also generated and analyzed EGFP-LcrTFL1 reporter-fusion lines, and LcrTFL1/LcrLFY doubly transgenic lines.
The transgene rescued the mutant defects, but manifested gain-of-function phenotypes. However, LcrTFL1 lines differed from 35S:TFL1 lines in several regards. Defects in floral meristem identity establishment were observed, as was the production of flowers with extra petals. We also noted features that resemble rosette flowering: LcrTFL1 lines produced significantly shorter interfloral internodes and significantly longer pedicels than either wild-type or 35S:TFL1 plants.
Our data show that there are substantive differences in the regulation and/or function of TFL1 orthologs between A. thaliana and L. crassa. These may reflect changes that occurred during the evolution of rosette flowering in Leavenworthia, but, if so, our results show that additional, as-yet-unidentified genes were involved in this instance of architectural evolution.
Keywords: inflorescence evolution, LEAFY, meristem identity genes, molecular coevolution, plant architecture, TERMINAL FLOWER 1
Introduction
Plants exhibit a range of inflorescence architectures, which play an important role in adaptation to varied environments (Tucker & Grimes, 1999). Theoretical models of plant architecture have suggested that much of the current diversity of inflorescence form can be explained by modulation of very few developmental parameters (Prusinkiewicz et al., 2007). Specifically, if the identity of shoot meristems is controlled by two antagonistic gene products, developmental regulation of the levels of these gene products in parent (i.e. terminal) and daughter (i.e. axillary) meristems can explain almost all the inflorescence forms seen in angiosperms. The challenge is to connect these theoretical models with empirical data on the developmental genetic basis of inflorescence evolution.
Previous developmental work in model species has suggested that the two most important genes involved in the regulation of meristem identity are homologs of FLORICAULA/LEAFY (LFY), which promotes floral identity, and CENTRORADIALIS/TERMINAL FLOWER 1 (TFL1), which promotes vegetative identity (Bradley et al., 1996, 1997; Conti & Bradley, 2007). Genetic data show that early LFY expression during bolting activates TFL1 expression, whereas later in development the two genes act antagonistically such that floral meristems express LFY but not TFL1, and inflorescence meristems express TFL1 but not LFY (Ratcliffe et al., 1999; Benlloch et al., 2007). The most plausible mechanism by which TFL1 represses LFY is via competition with a close relative of TFL1, FLOWERING LOCUS T (FT). FT is part of a transcriptional complex that activates APETALA1 (AP1), which is an important activator of LFY expression (Ahn et al., 2006). Conversely, repression of TFL1 in floral meristems also appears to be mediated by AP1: the AP1 protein is thought to bind the 3′ noncoding region of the TFL1 locus and to directly repress its expression (Kaufmann et al., 2010). These data and models predict that inflorescence evolution will commonly involve changes at the LFY or TFL1 loci and/or genes such as AP1 that modulate their expression and/or genetic interactions (Coen & Nugent, 1994; Bradley et al., 1996, 1997).
Arabidopsis thaliana and most Brassicaceae produce all their flowers on elongated shoot systems, called inflorescences. Each flower has a short pedicel, and elevation of the flowers above the ground is primarily the result of the elongation of shoot internodes. We will call this architecture ‘inflorescence flowering’ (Fig. 1a). An alternative plant architecture, ‘rosette flowering’, also occurs within Brassicaceae. In rosette flowering species, flowers are borne singly on elongated pedicels that emerge from the rosette. This architecture is illustrated by Leavenworthia crassa, which initially produces a rosette containing solitary flowers borne on elongated pedicels (Fig. 1b). These flowers are produced in the axils of cryptic bracts (Bosch et al., 2008) on the flanks of an indeterminate shoot axis (lacking a terminal flower). However, L. crassa is best viewed as a partially rosette-flowering species because, if plants are grown in good conditions, they will later produce elongated inflorescences from the axils of basal rosette leaves. Nonetheless, because early development conforms to the rosette flowering architecture, and because Leavenworthia represents a clade that transitioned from being ancestrally inflorescence flowering to (partially) rosette flowering, L. crassa provides a valid model for studying the evolution of rosette flowering. It has been argued that rosette flowering in Brassicaceae is an adaptation to growing in low-competition environments with short and unpredictable growing seasons (Bosch et al., 2008).
Fig. 1.
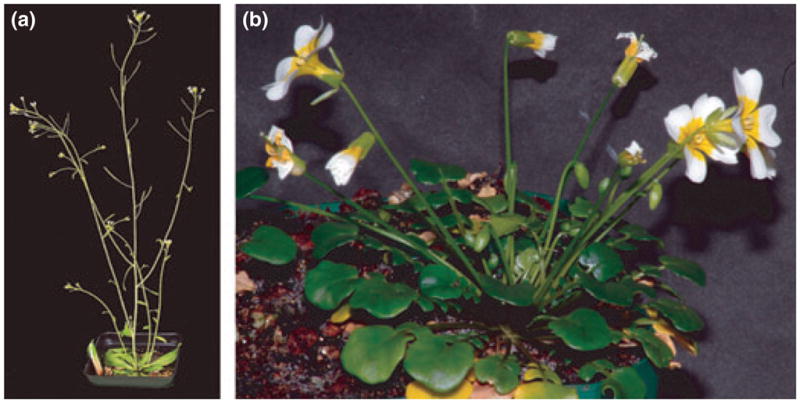
Comparison of the inflorescence-flowering species Arabidopsis thaliana (a) with the partially rosette-flowering species Leavenworthia crassa (b).
Previous work on the genetic changes underlying the evolution of rosette flowering has focused on independent transitions from inflorescence to rosette flowering in the genera Ionopsidium, Idahoa and Leavenworthia. Based on comparative expression (Shu et al., 2000; Sliwinski et al., 2007; Bosch et al., 2008), molecular evolutionary analysis (Baum et al., 2005), and interspecies transformation experiments (Yoon & Baum, 2004; Sliwinski et al., 2006, 2007), the regulation and/or activity of LFY homologs has been shown to have changed in each of the three lineages. However, in no case could the evolution of rosette flowering be attributed only to changes to LFY homologs, showing that other genetic players are involved.
Experiments with the L. crassa LFY ortholog (LcrLFY) suggested a possible role for this gene in internode compression (Sliwinski et al., 2006; Bosch et al., 2008). A. thaliana lfy mutants rescued with a LcrLFY genomic construct produced terminal flowers. This phenotype was attributed to the 5′ cis-regulatory region (‘promoter’) of the LcrLFY locus, which drives expression in the inflorescence meristem, apparently escaping TFL1-mediated repression (Yoon & Baum, 2004). At the same time, the coding region of LcrLFY (whether driven by the LcrLFY or LFY promoter) caused elevated early expression of TFL1, suggesting that the LcrLFY protein is more effective than LFY at activating TFL1 during early development (Sliwinski et al., 2006). Taken together, these data suggested the hypothesis that reciprocal up-regulation of LFY and TFL1 homologs in the Leavenworthia lineage contributed to the evolution of rosette flowering (Sliwinski et al., 2006).
In the work reported here, we tested this hypothesis by isolating the L. crassa TFL1 ortholog (LcrTFL1) and analyzing its function in an A. thaliana background. Our data are consistent with the prevailing models of LFY-TFL1 antagonism, and suggest that changes at the TFL1 locus played a role in internode compression and pedicel elongation. However, TFL1 and LFY are still not sufficient to explain all aspects of rosette flowering in L. crassa.
Materials and Methods
Plant materials
Unless otherwise stated, A. thaliana seeds were originally obtained from the Arabidopsis Biological Recourse Center (The Ohio State University, Columbus, OH, USA). Transgenic LcrLFY A. thaliana were the lines described in Yoon & Baum (2004) and Sliwinski et al. (2006). 35S:TFL1 transgenic A. thaliana were kindly provided by Prof. Enrico Coen (John Innes Centre, Norwich, UK) and EGFP-TFL1 was kindly provided by Prof. Koji Goto (Research Institute for Biological Sciences, Okayama, Japan). The latter construct contains green flourescent protein (GFP) inserted at the start of exon 1 of a TFL1 genomic sequence, complete with non-coding sequences 1920 bp 5′ of the start codon and 5300 bp 3′ of the stop codon (K. Goto et al., pers. comm.). All Arabidopsis lines were grown as described previously (Sliwinski et al., 2006).
Leavenworthia crassa Rollins seeds were germinated and grown on prewetted and sterilized filter paper, then kept in a growth chamber at 15:9°C day:night under 16 h photoperiods until germination. Seedlings with two true leaves were transferred to soil and cultivated at 23°C under 16 h photoperiods.
Isolation of LcrTFL1 and sequence analysis
By using degenerate primers (TFL1-DGF, GNAGWGTK RTHGGAGA; TFL1-DGR, AAHDYDYKWGTGYTG AA), based on highly conserved regions of TFL1 genes, we isolated TFL1 homologs from L. crassa genomic DNA. Amplified products were cloned and sequenced to confirm their identity. We then conducted several rounds of genome walking (Clontech, Mountain View, CA, USA) in order to obtain several kilobases of flanking, non-coding sequence both 5′ and 3′ of the coding region. We identified potential cis-regulatory elements in the 5′- and 3′-intergenic regions of LFY and LcrLFY using the plant cis-acting element database, PLACE (http://www.dna.affrc.go.jp/PLACE/). We determined the dN:dS ratio using SNAP (http://www.hiv.lanl.gov/content/sequence/SNAP/SNAP.html).
To assess orthology, we built an alignment of TFL1 homologs, focusing on A. thaliana, papaya (Carica papaya (Cpa)), and poplar (Populus trichocarpa and P. balsamifera (Ptr and Pba)), which are all rosids. Homologs from Physcomitrella patens (PHYPADRAFT numbers 60404, 120777, 127945, and 134340 (PpaTFL1-like 1–4)) were used as outgroups. We downloaded coding sequences and built both DNA and protein alignments starting with an initial alignment in Clustal W (Thompson et al., 1994) using default settings, as implemented in MEGA (Kumar et al., 1994, 2008), followed by a manual adjustment in MacClade 4 (Maddison & Maddison, 2002). Nucleotide and protein alignments are provided (Supporting Information, Figs S1, S2). After removing truncated or partial sequences, we conducted phylogenetic analyses of both the DNA and protein matrices using MrBayes 3.1 (Huelsenbeck & Ronquist, 2001; Ronquist & Huelsenbeck, 2003) with the CIPRES 2.0 portal (Miller et al., 2009). We initiated two MCMC runs of two million generations, each comprising four linked chains (heat = 0.2) with sampling every thousand generations. The nucleotide data were analyzed using the Generalized Time Reversible model with rate heterogeneity modelled with a discrete approximation to a gamma distribution (GTR + γ). The protein data were analyzed using the Whelan and Goldman (WAG) model. The first 250 trees from the posterior sample of each run were discarded as burn-in, and the remaining trees were used to generate a majority rule consensus tree with clade posterior probabilities. Outgroups were pruned from the phylogeny to save space. The sequence names used for convenience are guided by preliminary phylogenetic results and do not reflect functional data: CpaTFL1-like, evm.TU.contig_45053.1; CpaFT-like, evm.TU.contig_32595.1; CpaBFT-like, evm.TU.supercontig_107.26; CpaE12A11-like, EST 1565745_5_G15_058 PY06; PtrTFL1-like, NC_008475.1; PbaTFL1-like, NW_1492762.1; PtrBFT-like, NC_008481.1; PtrFT-like-1, NC001491506.1; PtrFT-like-2, NW_1491690.1; PtrFT-like- 3, NC_008476.1; PtrMFT-like, NC_008480.1; TFL1, NM_120465; ATC (A. thaliana CENTRORADIALIS-like), NM_128315; FT (FLOWERING-LOCUS T ), NM_105222; TSF (TWIN SISTER OF FT ), NM_118156; MFT (MOTHER OF FT AND TFL1), NM_101672; BFT (BROTHER OF FT AND TFL1), NM_125597.
Cloning, transformation and phenotype scoring
Based on the genomic sequence, we designed primers to amplify the LcrTFL1 coding region and the entire 5′- and partial 3′-intergenic spacers. Four independent clones (two in each orientation) were generated using the binary vector pPZP211, which includes the NPTII selectable marker conferring kanamycin resistance. We also generated reporter constructs in which EGFP was translationally fused to LcrTFL1. The EGFP coding sequence was introduced between the 5′-untranslated region (UTR) and the start codon of the LcrTFL1 genomic sequence. The fusion construct was subcloned into the pCAMBIA3300 vector, which includes the Bar selectable marker conferring Basta resistance, and was introduced into tfl1-2 plants. T1 seedlings were identified by spraying with a 1:1000 dilution of Finale herbicide (AgrEvo Company, Wilmington, DE, USA).
Arabidopsis thaliana tfl1-2 Landsberg erecta (Ler) homozygous mutant plants were transformed with the four genomic clones using the floral dip method (Clough & Bent, 1998). Primary transformant (T1) plants were selected by seed germination on 0.5× Murashige and Skoog (MS) plates supplemented with 50 mg l−1 kanamycin. Each of the four constructs yielded c. 50 seedlings exhibiting kanamycin resistance. A subset of these sampled from the four constructs were transferred to soil at 23°C under long-day conditions (16:8 h day:night). The segregation ratio of kanamycin-sensitive (KanS) to kanamycin-resistant (KanR) offspring from T1 plants was used to estimate the number of transgene loci.
We identified five independent T1 lines in a wild-type background, each inferred to have a single transgene locus. T2 plants from each of these lines were grown on soil in a glasshouse, with a single plant in each 3.25 inch pot. At maturity (50–60 d after planting), plants were scored for the number of paraclades emerging from the elongated portion of the main axis, rosette diameter, height of the main axis, and length of the longest secondary inflorescence shoot. After scoring T2 plants, we harvested seed and grew a sample on kanamycin plates and used the ratio of Kanr:Kans as a proxy for genotype of the scored T2 plant (all resistant, homozygous for the transgene; all sensitive, wild-type; quarter sensitive, hemizygous).
Three single locus transgenic lines and three EGFP fusion lines, all in a tfl1 mutant background, were selected and selfed to yield homozygous lines for further phenotypic characterization in the T4 generation. For phenotype scoring experiments, seedlings were germinated on 0.5× MS medium after 2 d of cold treatment. Plants were transferred to soil and grown at 23°C and c. 120 μmol m−2 s−1 under 16:8 h day:night photoperiod. Statistical analyses were conducted in Excel (Microsoft Corp., Redmond, WA, USA) and SigmaStat (Systat Software Inc., San Jose, CA, USA). A sample of 25 homozygous, EGFP-LcrTFL1 plants in the T4 generation were examined using a confocal laser scanning microscope (Zeiss LSM 510 Meta) at a wavelength of 488 nm.
Quantitative reverse transcription polymerase chain reaction (RT-PCR)
Total RNA was extracted with PureYield™ RNA Midiprep System (Promega, Madison, WI, USA). RNase-free DNase (Fermenta, Glen Burnie, MD, USA) was used to eliminate genomic DNA contamination. First-strand cDNA synthesis was performed using ImProm-II™ Reverse Transcription System (Promega) with the oligo(dT)15 primer. Real-time PCR was performed with the Brilliant SYBR Green QPCR Master Mix (Stratagene, Santa Clara, CA, USA) in a Stratagene MX3000P qPCR system. All PCR reactions were performed with the default program (94°C for 3 min, followed by 40 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 1 min). The PCR reactions were followed by a dissociation curve analysis to verify there was no primer self-amplification. Primer sequences (all 5′ to 3′) were as follows: LcrTFL1, ATATGAGAAGTAGAGTGG GAGATC and GTGTTCTTTTAGAAAGGGGTCAC TA; TFL1, ATAGACCCAGATGTTCCA and ATGTA TCCCTATGCTTGG. Relative concentrations were normalized to QPCR reactions using actin control primers (Act1 5′-GTATTGTGTTGGACTCTGGTGATGGTGT-3′ and Act2 5′-GATGGATCCTCCAATCCGACACTGTA-3′).
Scanning electron microscopy
Living specimens were fixed in FAA (5% formalin, 5% acetic acid, and 47.5% ethanol in distilled water) and subsequently stored in 70% ethanol. Tissue samples were then dehydrated in an ethanol series, critical-point dried, sputter-coated with gold, and studied at 20 kV in an Environmental Scanning Electron Microscope (FEI Quanta 450, Hillsboro, OR, USA).
Results
Isolation of LcrTFL1
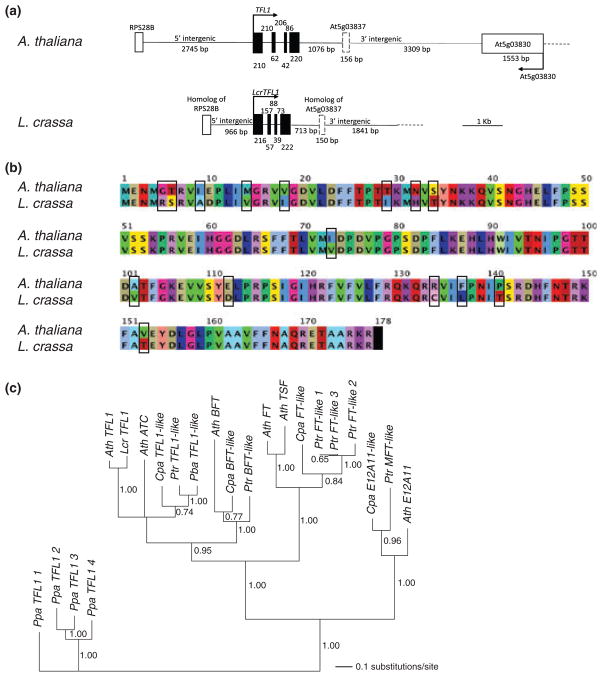
We searched the L. crassa genome for TFL1 (At5G03840) homologs using PCR with degenerate primers. A 727 bp fragment with high similarity to A. thaliana TFL1 was amplified and designated as LcrTFL1 (L. crassa Terminal Flower 1). We used genome walking (Clontech) to isolate the full-length genomic sequence of the LcrTFL1 locus (Genbank accession GU136396). The LcrTFL1 locus has a similar structure to the TFL1 locus (Fig. 2a). In the 5′ direction the closest gene to LcrTFL1 is homologous to At5G03850, which is also the gene that is directly upstream of TFL1. In the 3′ direction, the closest gene model to TFL1 is At5G03837, encoding a short hypothetical protein of 51 amino acids. Sequences 3′ of LcrTFL1 also showed similarities to At5G03837. The inferred amino acid sequence of the L. crassa At5G03837 homolog is highly divergent from At5G03837, except for one 20 amino acid domain that shows > 75 amino acid identity. However, even in this domain the dN:dS ratio is c. 1.0, pointing to a lack of purifying selection. Combined with the absence of evidence of any transcript, we conclude that neither At5G03837 nor the corresponding region in L. crassa encodes functional genes. The next gene 3′ of TFL1 is At5G03830, which is located just over 5 kb from the TFL1 stop codon. We sequenced a little > 2.7 kb 3′ of TFL1 but did not find any other open reading frames (ORFs).
Fig. 2. Leavenworthia crassa TFL1.
(LcrTFL1) is orthologous to the Arabidopsis thaliana gene TFL1. (a) Structure of the LcrTFL1 and TFL1 loci. (b) Alignment of the inferred protein sequences of LcrTFL1 and TFL1. The amino acid differences are boxed. (c) A phylogenetic tree estimate from the coding sequences (CDS) of various members of the TFL1 gene family. This tree is a majority-rule consensus tree based on the post-burn-in tree from two Bayesian Markov chain Monte Carlo runs of 2 000 000 generations each, applying the generalized time-reversible (GTR) + Γ model of molecular evolution. Values on branches are clade posterior probabilities. Branch lengths (substitutions/site) are the mean branch lengths across trees in the posterior that have the branch. Bayesian analysis of the amino acid sequence from the same genes (using the Whelan and Goldman (WAG) model of protein evolution) yielded a similar tree except that ATC was resolved as sister to Cpa TFL1-like with a 0.99 posterior probability.
Based on cDNA sequencing, it was determined that the intron–exon structure of LcrTFL1 is identical to TFL1, although LcrTFL1 introns are consistently shorter because of a number of small deletions (Fig. 2a). The cDNAs encode proteins of the same size (177 residues) that can be aligned without indels and with only 15 amino acid differences (Fig. 2b). The dN:dS ratio for the comparison of LcrTFL1 and TFL1 was 0.137. This value is much less than 1.0, showing that purifying selection has been acting on TFL1 and that positive selection has been either absent or restricted to a small subset of sites. For comparison, the dN:dS ratio for LcrLFY vs LFY was 0.091, implying that LFY homologs have been subject to stronger purifying selection than TFL1 homologs (or possibly that directional selection has acted on a greater subset of sites within TFL1 proteins).
To further confirm the orthology of LcrTFL1 and TFL1, we generated an exon alignment for these two genes and other members of the TFL1 gene family from a few other rosids. This alignment included additional homologs from A. thaliana (FT, TSF, ATC, MFT and BFT). Phylogenetic analysis of aligned DNA (Fig. S1) or amino acid sequences (Fig. S2) yielded a well-supported phylogenetic tree consistent with LcrTFL1 being orthologous to TFL1 (Fig. 2c).
In order to explore the potential for regulatory divergence between TFL1 and LcrTFL1, we compared their flanking noncoding sequences. The 5′-intergenic region of LcrTFL1 is only 966 bp, whereas that of TFL1 is 2745 bp. As a result, many potential 5′ cis-regulatory elements are absent from LcrTFL1. Interestingly, a potential LFY binding site located 379 bp upstream of the TFL1 start codon is missing in LcrTFL1. The 3′-intergenic regions of LcrTFL1 and TFL1 are highly divergent in sequence, making alignment impossible. We looked specifically for the two CArG boxes that have been implicated (in A. thaliana) as repressor sites that are bound by AP1 (Kaufmann et al., 2010). The more distal repressor site, situated 1756 bp downstream of the stop codon in A. thaliana, was identified in LcrTFL1 based on it having an identical sequence and sitting in the middle of a c. 120 bp region of high similarity between LcrTFL1 and TFL1 (Fig. S3). In LcrTFL1 this distal repressor is situated 1032 bp downstream of the stop codon, showing that there has been a net deletion of 724 bp in the 3′-region of LcrTFL1 relative to TFL1. This deletion may explain why the proximal AP1 repressor site, situated 1011 bp downstream of the stop codon in TFL1, is not present in LcrTFL1.
LcrTFL1 replaces the function of Arabidopsis TFL1 but causes novel phenotypes
The LcrTFL1 genomic locus, including the entire 5′-intergenic region and 2704 bp of 3′ noncoding DNA, was introduced into A. thaliana tfl1-2 mutant plants. We also generated constructs in which an EGFP domain was translationally fused to the 5′-end of the LcrTFL1 coding region. All 28 LcrTFL1 primary transformant (T1) plants carrying the genomic fragment showed full rescue of the mutant phenotype, with no terminal flowers observed. The 12 EGFP-LcrTFL1 lines examined showed either complete rescue (no terminal flowers) or partial rescue (much delayed production of terminal flowers). The observation that both the genomic and fusion constructs prevent or greatly delay terminal flower formation shows that the LcrTFL1 protein is functional in A. thaliana, as might be expected given the two proteins’ high amino acid similarity.
Compared with wild-type A. thaliana, LcrTFL1 and EGFP-LcrTFL1 lines also displayed some new traits. These included an increased number of secondary inflorescences, conversion of flowers into paraclades, short internodes in the primary inflorescence, shorter paraclades, and increased rosette leaf number (Fig. 3). These phenotypes were also observed in 35S:TFL1 transgenic plants, suggesting that they most likely reflect ectopic or expanded expression.
Fig. 3.
Phenotypes of Arabidopsis thaliana plants transformed with Leavenworthia crassa TFL1 (LcrTFL1). Close-up photographs show various recurrent features of LcrTFL1 lines, including extra petals (a–g), production of elongated pedicels (b, c, g), production of occasional flower-shoot intermediates (c–d), and cauline leaves lacking associated paraclades (arrow in h). Comparison of LcrTFL1 and control lines (i) shows that the tfl1-2 mutation has been fully rescued and that LcrTFL1 lines have many similarities to 35S:TFL1 lines.
Although LcrTFL1 transformant plants resembled 35S:TFL1 lines in many regards, some phenotypes were observed consistently in LcrTFL1 lines but not in either 35S:TFL1 or wild-type A. thaliana: extra petals were formed in many flowers, specifically the earliest (lowermost) flowers produced on primary and secondary inflorescence axes (Fig. 3a–f); some flowers had elongated pedicels (Fig. 3b, c,g); some structures were formed that have features intermediate between flowers and paraclades (Fig. 3g); some cauline leaves lacked axillary paraclades (Fig. 3h); secondary inflorescence branches showed delayed elongation resulting in the transient formation of aerial rosettes (Fig. 3i). While the formation of aerial rosettes is not uncommon in A. thaliana, and seems to result from delayed elongation of paraclade shoots (Grbić & Bleecker, 2000), the other novel phenotypes suggest that the effects of LcrTFL1 might not result just from elevated expression levels, but may also reflect modifications to the expression pattern and/or changes in protein function.
To see if LcrTFL1 has a trans-dominant effect, we introduced the genomic construct into a wild-type A. thaliana background. To facilitate comparison with 35S:TFL1, which is in the Columbia ecotype (Ratcliffe et al., 1999), we used wild-type A. thaliana Col-0 as the recipient genome. The > 50 independent LcrTFL1/Col-0 transgenic lines were examined and all showed a phenotype that closely resembled LcrTFL1/tfl1-2 plants in the T1 generation. To quantify the phenotypic effects, we grew 30–50 seedlings from each of five T2 families without selection. A two-way ANOVA was used to assess whether the inferred genotype classes (wild-type, hemizygous, and homozygous for the transgene) differ significantly. A significant genotype effect was detected for all phenotypes, showing that the transgene does have a trans-dominant effect (Table S1). Post-hoc tests using Fisher’s least significant difference showed that there are significant differences between wild-type plants and plants with at least one transgene, but they did not detect a significant difference between homozygous and hemizygous T2 plants for any phenotype. This result suggests that once one copy of LcrTFL1 is present in the genome, the addition of a second copy has little additional impact on the phenotype. While quantitative variation was detected among T2 families, these differences were much smaller than the differences between transgenic plants and wild-type controls. This suggests that the transgenic phenotypes are relatively robust to between-line variation in expression level.
Plants from three homozygous LcrTFL1 transgenic single-locus lines in a Ler tfl1-2 mutant background, one LcrTFL1 transgenic single-locus line in a Col background, and three EGFP-LcrTFL1 transgenic lines in a Ler tfl1-2 mutant background were grown in controlled conditions alongside 35S:TFL1, EGFP-TFL1, tfl1-2 and wild-type Col and Ler lines. Table 1 summarizes some of the quantitative measurements that were made on these lines. LcrTFL1 plants resembled 35S:TFL1 lines for many phenotypes, including an increased number of secondary inflorescence shoots, an increased number of cauline leaves, and late flowering. For these phenotypes 35S:TFL1 plants showed the strongest deviation from wild-type plants, followed by LcrTFL1 lines, with EGFP-TFL1 lines being closer to, but still significantly different from, wild-type lines.
Table 1.
Effect of TFL1 and LFY transgenes on selected traits Arabidopsis thaliana
| Ecotype | Genotype | Petalsa (n = 9) | Pedicelb (cm) (n = 10) | Internodec (cm) (n = 5) | Inflorescence height (cm) (n = 16) |
|---|---|---|---|---|---|
| Ler | Wild-type | 4.04 ± 0.07 | 1.31 ± 0.04 | 1.40 ± 0.24 | 37.12 ± 0.35 |
| lfy-6 | 3.98 ± 0.05 | 1.01 ± 0.06** | 0.97 ± 0.21 | 31.96 ± 0.53** | |
| tfl1-2 | 4.18 ± 0.28 | 0.84 ± 0.10** | 1.13 ± 0.43 | 10.74 ± 0.62** | |
| LcrLFY L3-15.7 | 6.18 ± 0.57** | 0.70 ± 0.04** | 1.50 ± 0.42 | 25.26 ± 1.96** | |
| LcrTFL1 L1 | 4.98 ± 0.17** | 1.47 ± 0.08** | 0.50 ± 0.13** | 45.78 ± 0.46** | |
| LcrTFL1 L2 | 5.02 ± 0.23** | 1.55 ± 0.08** | 0.42 ± 0.13** | 46.59 ± 0.56** | |
| LcrTFL1 L16 | 4.84 ± 0.20** | 1.54 ± 0.12** | 0.67 ± 0.07** | 40.61 ± 1.07* | |
| eGFP-LcrTFL1 L2.5 | 4.80 ± 0.17** | 1.46 ± 0.06** | 0.61 ± 0.14** | 43.47 ± 1.21** | |
| eGFP-LcrTFL1 L41.2 | 4.96 ± 0.17** | 1.46 ± 0.06** | 0.47 ± 0.15** | 44.16 ± 1.17** | |
| eGFP-LcrTFL1 L24.1 | 4.84 ± 0.14** | 1.50 ± 0.06** | 0.47 ± 0.15** | 42.27 ± 1.31* | |
| Col-0 | Wild-type | 3.98 ± 0.05 | 1.57 ± 0.06 | 2.11 ± 0.39 | 57.08 ± 0.48 |
| 35S:TFL1 | 4.02 ± 0.05 | 1.38 ± 0.09** | 2.11 ± 0.17 | 74.19 ± 0.78** | |
| GFP-TFL1 | 4.0 ± 0.07 | 1.55 ± 0.04 | 2.25 ± 0.37 | 48.86 ± 0.63** | |
| LcrTFL1 | 5.1 ± 0.21** | 1.70 ± 0.09** | 1.02 ± 0.14** | 51.34 ± 1.03** | |
| Ecotype | Genotype | Secondary inflorescencesd (n = 16) | Cauline leavese (n = 16) | Flowering time (d) (n = 16) | Flowering nodef (n = 16) |
| Ler | Wild-type | 2.38 ± 0.13 | 2.31 ± 0.13 | 19.88 ± 0.22 | 12.50 ± 0.35 |
| lfy-6 | 2.88 ± 0.16 | 2.88 ± 0.16* | 20.06 ± 0.21 | 11.13 ± 0.26 | |
| tfl1-2 | 0.56 ± 0.18** | 1.94 ± 0.19 | 13.13 ± 0.29** | 8.38 ± 0.26** | |
| LcrLFY L3-15.7 | 1.81 ± 0.25 | 1.75 ± 0.27 | 21.00 ± 0.34 | 10.75 ± 0.36* | |
| LcrTFL1 L1 | 19.63 ± 0.43** | 10.50 ± 0.21** | 32.44 ± 0.50** | 19.06 ± 0.57** | |
| LcrTFL1 L2 | 27.75 ± 1.06** | 12.88 ± 0.41** | 31.94 ± 0.50** | 19.13 ± 0.36** | |
| LcrTFL1 L16 | 19.19 ± 1.74** | 11.06 ± 0.54** | 30.25 ± 0.60** | 18.25 ± 0.76** | |
| eGFP-LcrTFL1 L2.5 | 10.69 ± 0.51** | 6.50 ± 0.35** | 29.63 ± 0.42** | 15.88 ± 0.56** | |
| eGFP-LcrTFL1 L41.2 | 11.19 ± 0.46** | 7.19 ± 0.26** | 28.81 ± 0.39** | 17.50 ± 0.32** | |
| eGFP-LcrTFL1 L24.1 | 11.19 ± 0.42** | 6.38 ± 0.30** | 28.38 ± 0.46** | 16.63 ± 0.49** | |
| Col-0 | Wild-type | 2.94 ± 0.14 | 2.94 ± 0.14 | 22.75 ± 0.22 | 15.88 ± 0.46 |
| 35S:TFL1 | 52.75 ± 1.13** | 12.25 ± 0.47** | 46.69 ± 0.40** | 36.81 ± 0.40** | |
| GFP-TFL1 | 2.44 ± 0.18 | 2.31 ± 0.12* | 20.63 ± 0.24** | 14.25 ± 0.30* | |
| LcrTFL1 | 44.50 ± 1.56** | 11.79 ± 0.57** | 32.36 ± 0.35** | 19.81 ± 0.35 | |
Each value represents the mean ± 1 SEM. Asterisks indicate a significant difference between the transgenic line and the corresponding wild-type as judged using either two-way ANOVA (petal number, pedicel length, internode length) or Student’s t-test.
Significantly different from the wild-type control (P < 0.01);
significantly different from the wild-type control (P < 0.001).
The numbers of petals in the first five flowers of the primary inflorescence were counted.
The pedicel lengths of the first five flowers of the primary inflorescence were measured.
The lengths of the first three floral internodes in the primary inflorescence were measured.
Number of lateral shoots emerging from the main axis, whether from the rosette or inflorescence.
Number of leaves on the elongated portion of the main axis.
Nodes are defined as the number of leaves plus any lateral shoots that lack subtending leaves.
Despite the overall similarities between LcrTFL1 and 35S-TFL1 plants, there were some striking differences too. LcrTFL1 consistently caused the production of additional petals in the lower flowers of the main axis, and similarly these early flowers had significantly longer pedicels than wild-type plants. For both phenotypes, an ANOVA detected a systematic tendency for more basal flowers to have more petals and longer pedicels than more apical ones. By contrast, 35S:TFL1 plants produced a normal number of petals and had significantly shorter pedicels than wild-type plants. Additionally, the lowest three internodes on the elongated portion of the inflorescence were found to be significantly shorter than the wild-type in LcrTFL1 lines, but unchanged in 35S:TFL1 plants.
A more complex pattern was seen with regard to plant height: LcrTFL1 plants were, like 35S:TFL1 plants, significantly taller than wild-type plants in a tfl1 mutant Ler background, but they were significantly shorter than wild-type plants when in a wild-type Col background. This difference correlates with the fact that, in a wild-type Col background, the LcrTFL1 gene seems to reduce the number of cauline leaves, which is a proxy for the number of elongated internodes in the inflorescence.
LcrTFL1 shows expanded expression in A. thaliana transgenic lines
The broad similarities between LcrTFL1 and 35S:TFL1 A. thaliana plants led us to hypothesize that the LcrTFL1 gene would show an elevated expression level and/or an expanded expression domain relative to TFL1, which is restricted to the vicinity of the inflorescence meristem at both the mRNA and protein levels (Bradley et al., 1997; Conti & Bradley, 2007). Quantitative real-time RT-PCR (qPCR) showed that the quantity of transcript in the inflorescence of LcrTFL1 lines normalized to an actin control (1.4 ± 0.14) was significantly higher than the equivalent tissue in wild-type A. thaliana (1.0 ± 0.07).
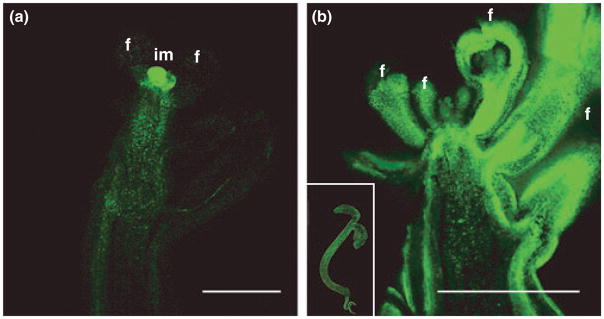
To assess the spatial pattern of expression of LcrTFL1 in an A. thaliana background, we looked at the distribution of GFP fluorescence in 12 independent EGFP-LcrTFL1 lines that showed complete or almost complete rescue of the tfl1-2 mutation. Confocal microscopic analysis detected GFP fluorescence not only in inflorescence meristems but also in the outer layers of the inflorescence axis, pedicels, and flowers (Fig. 4). This contrasts with fluorescence observed in Arabidopsis plants transformed with an EGFP-TFL1 construct (kindly provided by K. Goto), which show expression only in the inflorescence meristem (Fig. 4). Our construct, like the EGFP-TFL1 construct, includes the full 5′-intergenic spacer and introns. However, because the EGFP-TFL1 construct contains more sequences 3′ of the TFL1 gene (5.3 kb) than did our EGFP-LcrTFL1 construct (2.7 kb), we cannot rule out the possibility that we are missing a repressor element in this region of the L. crassa genome.
Fig. 4.
Localization of fluorescence in EGFP-TFL1 and EGFP-Leavenworthia crassa TFL1 (EGFP-LcrTFL1) lines. TFL1 expression was restricted to the inflorescence meristem (a). By contrast, LcrTFL1 was expressed in the inflorescence apex, floral organs, stem tissue (b) and throughout seedlings (b, inset). im, Inflorescence meristem; f, flower primordium. Bar, 500 μm.
Scanning electron microscope observations show that LcrTFL1 lines manifest a disrupted vegetative to reproductive transition
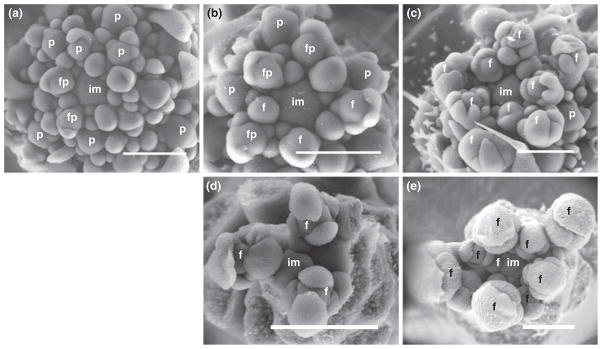
Scanning electron microscope (SEM) images of inflorescence meristems of LcrTFL1 lines showed that the establishment of floral identity was delayed (Fig. 5). In a plant that had not yet bolted, the shoot apical meristem was found to resemble an inflorescence meristem surrounded by numerous lateral primordia (Fig. 5a). In a pattern reminiscent of broccoli, the lateral primordia seem to have adopted neither a clearly floral nor an inflorescence identity (Carr & Irish, 1997). Later, when bolting is under way, many of the lateral primordia have clearly adopted a floral identity, as indicated by calyx development (Fig. 5b–c). However, many flowers at this stage were observed to have additional sepal lobes. The extra sepals seen in young floral primordia contrasts with mature LcrTFL1 flowers (Fig. 3a–b), which often have extra petals (Table 1) but were not observed to have extra sepals. While 35S:TFL1 inflorescence meristems are surrounded by more floral meristems at the time of bolting than are wild-type plants, these floral meristems proceed normally through the stages of flower development (Fig. 5d). These striking differences in early development between LcrTFL1 and 35S:TFL1 lines imply that the architectural and floral anomalies of LcrTFL1 lines cannot simply be explained by general ectopic expression of a TFL1 homolog but likely reflect more nuanced changes in expression and/or differences in protein function.
Fig. 5.
Scanning electron micrographs of primary inflorescence apices show disruptions of the floral transition in Leavenworthia crassa TFL1 (LcrTFL1) lines compared with controls. In LcrTFL1 plants that have not yet bolted (a), dissection reveals that the primary inflorescence meristem is surrounded by numerous primordia, most of which resemble floral primordia in stage 2 (stages defined by Smyth et al., 1990). When the inflorescence shoots of LcrTFL1 lines are c. 3 cmin height (b), many lateral primordia around the meristem still resemble stage 2–3 floral primordia, whereas others have an aspect intermediate between a paraclade and a stage 4 flower. When LcrTFL1 lines reach 9 cmin height (c), most primordia resemble stage 4–6 floral primordia except that the sepal whorl is irregular and there are usually more than four sepal lobes. By contrast, when 3 cmbolts are observed in 35S:TFL1 (d) or wild-type lines (e), they are similar, each containing normal stage 5 and above floral primordia. im, Inflorescence meristem; f, flower primordium; p, paraclade; fp, shoot of uncertain, flower or paraclade identity. Bar, 200 μm.
LcrTFL1/LcrLFY double transgenic lines resemble LcrTFL1 lines, but show greater elongation of rosette-derived inflorescences
To explore the in vivo interaction between LcrTFL1 and LcrLFY, LcrTFL1/LcrLFY double transgenic lines were made in both the tfl1-2 and lfy Arabidopsis mutant backgrounds. There were made by transforming single-locus, homozgous LcrTFL1 tfl1-2 lines with LcrLFY and single-locus homozygous LcrLFY lfy-6 lines with LcrTFL1. In the lfy-6 mutant background, 19 of the 39 doubly transgenic T1 plants strong resembled lfy mutations (Fig. 6a,c), producing only flower-inflorescence intermediates (sometimes resembling broccoli), and lacking petals or stamens. Conversely, 22 of the 44 T1 lines in a tfl1 mutant background made terminal flowers and/or bracteate flowers, resembling tfl1 mutants (Fig. 6g). One hypothesis is that the balance of these two mutually antagonistic genes is finely balanced such that whichever gene starts at a lower background level of expression often becomes fully repressed, yielding plants that resemble the mutant.
Fig. 6.

Phenotypes of Leavenworthia crassa TFL1/L. crassa LFY ortholog (LcrTFL1/LcrLFY) double transgenic lines in a lfy (a–d) or tfl1 (e–h) mutant background. Phenotypes of LcrTFL1/LcrLFY/lfy transgenic lines are variable. Approximately half of the lines failed to form fertile flowers, instead forming compressed leafy axes (a) or broccoli-like inflorescences with apetalous flowers (c). In some cases, inflorescence stems lacked any paraclades (d). When flowers were formed, the earliest flowers tended to make extra petals (b). Phenotypes of LcrTFL1/LcrLFY/tfl1 transgenic lines exhibited primarily LcrTFL1-like phenotypes, including extra petals (e), conversion of flowers into shoots (f), and long pedicels (e– f). Some lines, however, produced terminal flowers (g).
The remainder of the double transgenic lines in both backgrounds resembled LcrTFL1 lines, having delayed flowering and increased numbers of paraclades. Those lines that produced more or less normal flowers also produced flowers with extra petals (Fig. 6b,e,g) and often had elongated pedicels (Fig. 6f), like LcrTFL1 single transgenic lines. While the double transgenic lines were qualitatively similar to LcrTFL1 single transgenic lines, they showed an enhanced tendency to suppress axillary shoots associated with cauline leaves on elongated portions of the primary inflorescence (Fig. 6d,h). This was especially true in a lfy mutant background, where axillary shoots were often completely suppressed (Fig. 6d). At the same time, rosette-derived shoots tended to show extensive elongation (again, especially in a lfy background).
Among the many double transgenic lines, we selected two representative lines from each background for quantitative analysis in the T3 generation, avoiding lines that resembled either lfy or tfl1mutants. The lines chosen were inferred to be homozygous at a single locus for both transgenes. The double lines were scored for the same phenotypic traits as the single transgenic lines (Table 2). For most phenotypes, the double transgenic lines were intermediate between LcrLFY single and LcrTFL1 single transgenic lines, although generally closer to LcrTFL1. This suggests that the ectopic activity of LcrTFL1 is sufficient to overcome the enhanced activity and expression reported for LcrLFY (Yoon & Baum, 2004; Sliwinski et al., 2006). However, the number of petals observed in the double lines was closer to LcrLFY single lines than to LcrTFL1 single lines.
Table 2.
Morphology of Leavenworthia crassa LFY ortholog/L. crassa TFL1 (LcrLFY LcrTFL1) doubly transgenic plants in either the tfl1-2 or lfy-6 mutant background
| Line | Genetic background | Petalsa (n = 9) | Pedicelb (cm) (n = 10) | Internodec (cm) (n = 5) | Inflorescence height (cm) (n = 16) |
|---|---|---|---|---|---|
| Ler | LFY TFL1 | 4.04 ± 0.07 | 1.31 ± 0.04 | 1.40 ± 0.24 | 37.12 ± 0.35 |
| L3L3 | lfy TFL1 | 5.98 ± 0.23** | 1.56 ± 0.12** | 0.27 ± 0.07** | nd |
| L3L4 | lfy TFL1 | 5.89 ± 0.16** | 1.26 ± 0.05 | 0.28 ± 0.04** | 29.62 ± 1.87* |
| G16 | LFY tfl1 | 5.73 ± 0.15** | 1.68 ± 0.08** | 0.63 ± 0.17** | 38.38 ± 1.24 |
| Y10 | LFY tfl1 | 5.29 ± 0.27** | 1.73 ± 0.07** | 0.64 ± 0.18** | 41.36 ± 0.78** |
| Line | Genetic background | Secondary inflorescencesd (n = 16) | Cauline leavese (n = 16) | Flowering time (d) (n = 16) | Flowering nodef (n = 16) |
| Ler | LFY TFL1 | 2.38 ± 0.13 | 2.31 ± 0.13 | 19.88 ± 0.22 | 12.50 ± 0.35 |
| L3L3 | lfy TFL1 | nd | nd | nd | nd |
| L3L4 | lfy TFL1 | 27.00 ± 1.87** | 15.31 ± 2.30** | 30.88 ± 0.34** | 20.13 ± 0.76** |
| G16 | LFY tfl1 | 9.94 ± 0.34** | 5.94 ± 0.24** | 29.56 ± 0.58** | 18.39 ± 0.26** |
| Y10 | LFY tfl1 | 12.00 ± 0.58** | 7.83 ± 0.57** | 30.06 ± 0.28** | 18.22 ± 0.55** |
Each value represents the mean ± 1 SEM. Asterisks indicate a significant difference between the transgenic line and the corresponding wild-type as judged using either two-way ANOVA (petal number, pedicel length, internode length) or Student’s t-test.
Significantly different from the wild-type control (P < 0.01);
significantly different from the wild-type control (P < 0.001).
The numbers of petals in the first five flowers of the primary inflorescence were counted.
The pedicel lengths of the first five flowers of the primary inflorescence were measured.
The lengths of the first three floral internodes in the primary inflorescence were measured.
Number of lateral shoots emerging from the main axis, whether from the rosette or inflorescence.
Number of leaves on the elongated portion of the main axis.
Nodes are defined as the number of leaves plus any lateral shoots that lack subtending leaves. nd, not determined.
For petal number, pedicel length, internode length, number of secondary inflorescences, and number of cauline leaves, the effects of LcrTFL1 were stronger in a lfy mutant background than in a tfl1 background. This was particularly marked for internode length, which was only c. 20% of wild-type in a lfy background and 48% of wild-type in a tfl1 background. These results suggest that the endogenous TFL1 gene makes a much larger contribution to these phenotypes than does the endogenous LFY gene. Double transgenic lines had shorter inflorescences than the wild-type when in a lfy background, but were similar or slightly taller than the wild-type when in a tfl1 background. This correlates with the data from single transgenic lines, where the presence of a functional endogenous copy of TFL1 (i.e. in a wild-type background) in addition to the LcrTFL1 transgene yielded shorter plants.
Discussion
We isolated LcrTFL1 from the rosette-flowering crucifer, L. crassa, and showed that it is orthologous to the A. thaliana gene TFL1, as evidenced by genomic colinearity, high similarity in protein sequence, and the genes’ sister-group relationship on the inferred phylogenetic tree. Introducing the LcrTFL1 genomic region into a tfl1 mutant, A. thaliana successfully rescued the terminal flower phenotype of tfl1 mutants. Taken together, these data suggest that the LcrTFL1 and TFL1 proteins share a conserved biochemical function.
Introduction of the LcrTFL1 genomic locus into A. thaliana mutants resulted in plants that resembled 35S:TFL1 transformants, consistent with the broad expression seen in EGFP-TFL1 reporter lines. There are two possible interpretations of this expanded expression. The first possibility is that the expanded expression is a consequence of the LcrTFL1 genomic fragment lacking an important repressor cis-regulatory element that is conserved between L. crassa and A. thaliana. In the 5′ direction, our LcrTFL1 construct is like the EGFP-TFL1 construct in that it includes the full 5′-intergenic spacer. This argues against there being a functionally and spatially conserved 5′ repressor element that could explain the additional expression of LcrTFL1. However, in the 3′ direction, the EGFP-TFL1 construct contains a larger portion of noncoding DNA than does the LcrTFL1 construct, so it is a formal possibility that the latter construct omits an important 3′ conserved repressor element. This could be tested with a chimeric construct that includes the full LcrTFL1 construct plus the additional portion of 3′ noncoding DNA that is present in the TFL1 construct but absent from the LcrTFL1 construct.
The second possible explanation for the resemblance between the LcrTFL1 and 35S:TFL1 plants is that there has been evolutionary divergence in the regulation of the expression of TFL1 genes; for example, a repressor element may have been lost or an enhancer element gained on the L. crassa lineage some time since its last common ancestor with A. thaliana. One candidate element is the proximal AP1 CArG repressor element, which is present in the 3′ noncoding region of TFL1 but is absent in LcrTFL1. Kaufmann et al. (2010) showed that the proximal AP1 repressor element, along with a more distal AP1 repressor element that is conserved between the two species, has functional significance in A. thaliana. A second candidate is a predicted LFY-binding site, which is found at −379 bp in the 5′ noncoding DNA of TFL1 but not in LcrTFL1. Future studies should examine whether the addition of the proximal AP1 CArG site and/or the 5′ potential LFY-binding site to LcrTFL1 is sufficient to cause expression that more closely resembles TFL1. This will be interesting because such a finding would render it plausible that the up-regulation of LcrTFL1 resulted from coevolution with interacting meristem identity genes (homologs of LFY and AP1), perhaps coincident with the evolution of rosette flowering.
While LcrTFL1 lines resembled 35S:TFL1 in many regards (e.g. late flowering, homeotic conversion of paraclades into flowers), there were also some striking differences. Two of these novel traits, the accumulation of arrested lateral primordia of intermediate flower/paraclade identity (Fig. 5) and the production of flowers with extra petals (Fig. 3), do not resemble the donor species, L. crassa. The temporary arrest of lateral primordia is best interpreted as resulting from defects in the establishment of floral identity. Of several possible explanations, one worth mentioning is that the LcrTFL1 protein is a better repressor of flower meristem identity genes than is TFL1. This would mirror the finding that LcrLFY is better than LFY at activating early expression of TFL1 (Sliwinski et al., 2006). While this hypothesis remains speculative, it would be worth using expression analysis to investigate whether LcrTFL1 is more effective than TFL1 at repressing floral meristem identity genes such as LFY and AP1. Ideally, this experiment should be done with plants transformed with a construct that contains the LcrTFL1 coding region surrounded by A. thaliana cis-regulatory sequences.
The production of extra petals in LcrTFL1 lines resembles neither the wild-type species nor 35S:TFL1 plants, all of which produce classic, four-merous, crucifer flowers. The extra petal phenotype was also seen in LcrLFY lines (Yoon & Baum, 2004). LcrTFL1/LcrLFY doubly transgenic lines did not show synergy, petal number being similar to LcrLFY single transgenic lines. What should we make of a phenotype that is induced by the introduction of either of two genes from the same donor species, given that those two genes are largely antagonistic in function? One possibility is that TFL1 and LFY genes work cooperatively via an additional, heretofore cryptic, pathway to regulate perianth development as a function of flower size. If this is true, introduction of either gene from the large-flowered L. crassa could cause proliferation of the perianth. This explanation does not seem compelling, however, given the lack of reports of TFL1 expression in the perianth. Another explanation worth considering is that the extra petals, which form in the lowermost flowers in inflorescence branches, are an indirect manifestation of defects in floral meristem identity establishment. Maybe these meristems transiently adopted a mixed flower-shoot identity resulting in some degree of proliferation of the outer parts of the primordia. Then, when floral identity was finally established, there may have been extra tissue that became assigned to new sepal lobe primordia (which later fuse to re-establish four organs) and new petal primordia. The latter hypothesis could be tested in part by conducting SEM investigations of LcrLFY lines to see if they too accumulate primordia of unclear identity during bolting. Additionally, it might be informative to drive expression of LcrLFY and LcrTFL1 by perianth-specific promoters.
In contrast to disrupted vegetative-reproductive transition and extra petals, two other novel traits observed in LcrTFL1 plants resemble the donor species: internode compression and pedicel elongation. Species in the Leavenworthia clade have compressed interfloral internodes on the main axis and elongated pedicels. We have previously hypothesized (Sliwinski et al., 2007; Bosch et al., 2008) that elevated expression of LFY genes contributed to internode compression along the main axis in rosette flowering species. The data presented here suggest that the Leavenworthia homologs of TFL1 may also play a role in internode compression along the main axis. LcrTFL1 plants had interfloral internodes that were significantly shorter than in wild-type plants. This effect is trans-dominant, being seen even in a wild-type background: LcrTFL1 Col plants have internodes that are about half as long as wild-type Col plants. Double transgenic lines also have shorter internodes than wild-type lines, especially in a lfy mutant background where internodes are c. 20% of wild-type. Because the internode compression effect is not observed in 35S:TFL1 lines, it cannot be attributed simply to ectopic TFL1 activity. The use of chimeric constructs that combine the coding region of TFL1 with the regulatory regions of LcrTFL1 (or vice versa) would help determine whether the internode compression phenotype is caused by amino acid or regulatory differences within the LcrTFL1 locus.
While we had no a priori reason to expect an effect of LcrTFL1 on pedicel elongation, we noticed that LcrTFL1 lines produced some flowers with unusually long pedicels. Quantitative analyses showed that the lower five flowers have significantly longer pedicels in singly or doubly transgenic LcrTFL1 lines than in wild-type lines, whereas 35S:TFL1 lines resemble wild-type lines. This effect was, again, trans-dominant, being seen in the presence of a functional endogenous TFL1 gene. This result suggests that the modulation of meristem identity genes not only can affect architecture in the narrow sense, but also can influence plant structure by affecting the relative elongation of different stem regions (pedicels vs internodes).
Our data are compatible with the claim that changes in LcrTFL1 and LcrLFY contributed to the evolution of several aspects of the derived architecture of Leavenworthia, most notably internode compression and pedicel elongation. However, even if this is so (which remains to be shown), our data also clearly show that LcrTFL1 and LcrLFY could not have been the sole target of developmental evolution. If they were, the double transgenic lines would closely approach the rosette-flowering form of L. crassa, which they certainly do not. The insufficiency of LcrTFL1 and LcrLFY to recapitulate rosette flowering in Leavenworthia shows that even if these two genes were important in the evolution of rosette flowering, other genes played additional essential roles. Further work on interacting genes, most obviously APETALA1 (Kaufmann et al., 2010), is needed to explain how an evolutionary change from inflorescence to rosette flowering was achieved without disrupting critical processes such as the formation of normal, fully functional flowers.
Supplementary Material
Fig. S1 Sequence alignment of LcrTFL1 and related genes from Arabidopsis thaliana, Populus trichocarpa, Carica papaya and Physcomitrella patens.
Fig. S2 Amino acid sequence alignment of LcrTFL1 homologs from Arabidopsis thaliana, Populous trichocarpa, Carica papaya and Physcomitrella patens.
Fig. S3 Alignment of a conserved domain in the 3′ noncoding region of TFL1 (AthTFL1) and LcrTFL1.
Table S1 Statistical analysis of the effect of LcrTFL1 in a wild-type background
Acknowledgments
The authors gratefully acknowledge the following for technical support and advice: Justin Bosch, Ivalú Cacho, Donna Fernandez, Ruth Litovsky, Abigail Mazie, Kevin Miller, Rebecka Pralle, R. Pulikesi, Sara Swanson and Amanda Teschke. Biological materials were kindly provided by Enrico Coen, Koji Goto, Angela Hay and Miltos Tsiantis. Assistance in the preparation of graphics was provided by Claudia Lipke and Kandis Elliot. This work was funded by the National Science Foundation (IOB-0641428).
References
- Ahn JH, Miller D, Winter VJ, Banfield MJ, Lee JH, Yoo SY, Henz SR, Brady RL, Weigel D. A divergent external loop confers antagonistic activity on floral regulators ft and tfl1. EMBO Journal. 2006;25:605–614. doi: 10.1038/sj.emboj.7600950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum DA, Yoon HS, Oldham RL. Molecular evolution of the transcription factor LEAFY in Brassicaceae. Molecular Phylogenetics and Evolution. 2005;37:1–14. doi: 10.1016/j.ympev.2005.07.002. [DOI] [PubMed] [Google Scholar]
- Benlloch R, Berbel A, Serrano-Mislata A, Madueño F. Floral initiation and inflorescence architecture: a comparative view. Annals of Botany. 2007;100:659–676. doi: 10.1093/aob/mcm146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch JA, Heo K, Sliwinski MK, Baum DA. An exploration of LEAFY expression in independent evolutionary origins of rosette flowering in Brassicaceae. American Journal of Botany. 2008;95:286–293. doi: 10.3732/ajb.95.3.286. [DOI] [PubMed] [Google Scholar]
- Bradley D, Carpenter R, Copsey L, Vincent C, Rothstein S, Coen E. Control of inflorescence architecture in Antirrhinum. Nature. 1996;379:791–797. doi: 10.1038/379791a0. [DOI] [PubMed] [Google Scholar]
- Bradley D, Ratcliffe O, Vincent C, Carpenter R, Coen E. Inflorescence commitment and architecture in Arabidopsis. Science. 1997;275:80–83. doi: 10.1126/science.275.5296.80. [DOI] [PubMed] [Google Scholar]
- Carr SM, Irish VF. Floral homeotic gene expression defines developmental arrest stages in Brassica oleracea L. vars. botrytis and italica. Planta. 1997;201:179–188. doi: 10.1007/BF01007702. [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant Journal. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Coen ES, Nugent JM. Evolution of flowers and inflorescences. Development. 1994;(Suppl):107–116. [Google Scholar]
- Conti L, Bradley D. TERMINAL FLOWER1 is a mobile signal controlling Arabidopsis architecture. Plant Cell. 2007;19:767–778. doi: 10.1105/tpc.106.049767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grbić V, Bleecker AB. Axillary meristem development in Arabidopsis thaliana. Plant Journal. 2000;21:215–223. doi: 10.1046/j.1365-313x.2000.00670.x. [DOI] [PubMed] [Google Scholar]
- Huelsenbeck JP, Ronquist F. MrBayes: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- Kaufmann K, Wellmer F, Muiño JM, Ferrier T, Wuest SE, Kumar V, Serrano-Mislata A, Madueño F, Krajewski P, Meyerowitz EM, et al. Orchestration of floral initiation by APETALA1. Science. 2010;328:85–89. doi: 10.1126/science.1185244. [DOI] [PubMed] [Google Scholar]
- Kumar S, Nei M, Dudley J, Tamura K. Mega: a biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief Bioinformation. 2008;9:299–306. doi: 10.1093/bib/bbn017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Tamura K, Nei M. Mega: molecular evolutionary genetics analysis software for microcomputers. Computer Applications in the Biosciences. 1994;10:189–191. doi: 10.1093/bioinformatics/10.2.189. [DOI] [PubMed] [Google Scholar]
- Maddison DR, Maddison WP. MacClade 4: analysis of phylogeny and character evolution. Sunderland, MA, USA: Sinauer; 2002. [DOI] [PubMed] [Google Scholar]
- Miller MA, Holder MT, Vos R, Midford PE, Liebowitz T, Chan L, Hoover P, Warnow T. [accessed on 4 August 2009];The CIPRES portal. CIPRES. 2009 [WWW document].URL http://www.phylo.org/sub_sections/portal. Archived by WebCite(r) at http://www.webcitation.org/5imQlJeQa.
- Prusinkiewicz P, Erasmus Y, Lane B, Harder LD, Coen E. Evolution and development of inflorescence architectures. Science. 2007;316:1452–1456. doi: 10.1126/science.1140429. [DOI] [PubMed] [Google Scholar]
- Ratcliffe OJ, Bradley DJ, Coen ES. Separation of shoot and floral identity in Arabidopsis. Development. 1999;126:1109–1120. doi: 10.1242/dev.126.6.1109. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Shu G, Amaral W, Hileman LC, Baum DA. LEAFY and the evolution of rosette flowering in violet cress (Jonopsidium acaule, Brassicaceae) American Journal of Botany. 2000;87:634–641. [PubMed] [Google Scholar]
- Sliwinski MK, Bosch JA, Yoon HS, Balthazar M, Baum DA. The role of two LEAFY paralogs from Idahoa scapigera (Brassicaceae) in the evolution of a derived plant architecture. Plant Journal. 2007;51:211–219. doi: 10.1111/j.1365-313X.2007.03148.x. [DOI] [PubMed] [Google Scholar]
- Sliwinski MK, White MA, Maizel A, Weigel D, Baum DA. Evolutionary divergence of LFY function in the mustards Arabidopsis thaliana and Leavenworthia crassa. Plant Molecular Biology. 2006;62:279–289. doi: 10.1007/s11103-006-9020-3. [DOI] [PubMed] [Google Scholar]
- Smyth DR, Bowman JL, Meyerowitz EM. Early flower development in Arabidopsis. Plant Cell. 1990;2:755–767. doi: 10.1105/tpc.2.8.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. Clustal W – improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Research. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker S, Grimes J. The inflorescence: introduction. The Botanical Review. 1999;65:303–316. [Google Scholar]
- Yoon HS, Baum DA. Transgenic study of parallelism in plant morphological evolution. Proceedings of the National Academy of Sciences, USA. 2004;101:6524–6529. doi: 10.1073/pnas.0401824101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Sequence alignment of LcrTFL1 and related genes from Arabidopsis thaliana, Populus trichocarpa, Carica papaya and Physcomitrella patens.
Fig. S2 Amino acid sequence alignment of LcrTFL1 homologs from Arabidopsis thaliana, Populous trichocarpa, Carica papaya and Physcomitrella patens.
Fig. S3 Alignment of a conserved domain in the 3′ noncoding region of TFL1 (AthTFL1) and LcrTFL1.
Table S1 Statistical analysis of the effect of LcrTFL1 in a wild-type background