Abstract
Genome-wide mRNA transcription profiles reveal widespread molecular sexual dimorphism or “sex-biased” gene expression, yet the relationship between molecular and phenotypic sexual dimorphism remains unclear. A major unresolved question is whether sex-biased genes typically perform male- and female-specific functions (whether these genes have sex-biased phenotypic or fitness consequences) or have similar functional importance for both sexes. To elucidate the relationship between sex-biased transcription and sex-biased fitness consequences, we analyzed a large data set of lethal, visible, and sterile mutations that have been mapped to the Drosophila melanogaster genome. The data permitted us to classify genes according to their sex-specific mutational effects and to infer the relationship between sex-biased transcription level and sex-specific fitness consequences. We find that mutations in female-biased genes are (on average) more deleterious to females than to males and that mutations in male-biased genes tend to be more deleterious to males than to females. Nevertheless, mutations in most sex-biased genes have similar phenotypic consequences for both sexes, which suggests that sex-biased transcription is not necessarily associated with functional genetic differentiation between males and females. These results have interesting implications for the evolution of sexual dimorphism and sex-specific adaptation.
Keywords: sexual dimorphism, sexual selection, sexual antagonism, deleterious mutation, fitness variation
Sex-specific selection can favor evolutionary divergence between males and females (Darwin 1871; Andersson 1994). However, because each sex develops from the same underlying genome (apart from gene-poor Y or W chromosomes), phenotypic divergence requires the differential utilization of shared genes (Ellegren and Parsch 2007; Arnold et al. 2009). Multiple, proximal genetic mechanisms can potentially underlie sexually dimorphic phenotypes. Males and females may use the same basic set of genes, with each sex exhibiting quantitative differences in the level of gene expression or utilizing alternative splice forms of a coding sequence. Alternatively, sex-specific selection may lead to the evolution of genes with male-specific and female-specific functions.
Although both mechanisms promote the evolution of sex-biased genes—genes that are differentially expressed at the molecular level—they make different predictions about the sex-specific functional consequences of mutations within these genes. If sex-biased genes have sex-specific functions, mutations in these genes should have sex-specific phenotypic and fitness effects. If sex-biased genes merely differ quantitatively in gene expression (including exon-specific, tissue-specific, or whole-body levels of gene expression analysis) and perform similar functions in both sexes, then mutations in these genes should similarly affect each sex. Genome-wide expression assays suggest that sex-biased gene expression is widespread among animal species (Ellegren and Parsch 2007), yet little is known about sex-specific mutational effects within genes that show male-biased and female-biased transcription.
A large number of spontaneous and induced mutations with well-characterized, fitness-related phenotypes have been mapped to the genome of Drosophila melanogaster (Tweedie et al. 2009). To characterize alleles with sex-limited and nonsex-limited effects, we exploit the observation that visible and lethal mutations generally have similar effects on both sexes, whereas sterility mutations typically exhibit pronounced sex-specific behavior (Lindsley and Lifschytz 1972; Ashburner et al. 2005). The availability of these mutation data, along with transcriptional profiles for associated genes, provides an opportunity to test whether sex-specific mutant phenotypes are indeed differentially associated with sexually dimorphic mRNA transcription levels. Given the nature of the data, we can ask two specific questions:
Do mutations with sex-specific phenotypes tend to occur in genes with sexually dimorphic expression, and if so, are male- and female-biased genes primarily associated with male- and female-limited mutational effects, respectively?
Do mutations involving nonsex-limited phenotypes tend to occur in genes that are similarly expressed by males and females?
Here, we address these questions and discuss their implications for sex-specific adaptation and the genetic basis of sexually dimorphic phenotypes.
Materials and Methods
Data
To identify genes with sex-limited and nonsex-limited functions, we searched FlyBase (Tweedie et al. 2009) for mutations within the following phenotypic categories: visible, lethal, semilethal, sterile, male sterile, and female sterile (searches used the TermLink section; http://flybase.org/static_pages/termlink/termlink.html). We trimmed the data set to include alleles associated with specific genes (although many alleles have been mapped to specific chromosomes and/or cytological bands, the mapping resolution for these cases was generally insufficient to be included within the final data set).
Individual genes can potentially have multiple alleles within the data set (though the majority of Drosophila genes were not associated with any alleles). We therefore classified each gene according to its range of mutant allele phenotypes, which fall between “entirely female-specific” to “entirely male-specific.” The genes were classified as follows:
Genes with female-limited fitness effects are those that contain female-sterile alleles and no other allele type;
Genes with female-biased fitness effects contain female-sterile alleles and any combination of visible, lethal, and semilethal alleles;
Genes with male-limited fitness effects contain male-sterile alleles and no other allele type;
Genes with male-biased fitness effects contain male-sterile alleles and any combination of visible, lethal, and semilethal alleles;
Genes without sex-biased fitness effects contain both male-sterile and female-sterile alleles, visible alleles, lethal alleles, and/or semilethal alleles.
Genes associated with sterility, but with neither sex specified (the underlying allelic data did not provide information about sex), were considered ambiguous and excluded from the analysis. The sample of sterile alleles that were included in the analysis is potentially heterogeneous because some studies examine fertility in only one sex rather than both. Nevertheless, the proportion of sex-limited and nonsex-limited steriles in our data set is consistent with independent experimental results that explicitly test male and female fertility (alleles associated with sex-specific sterility are roughly three times as common as alleles associated with sterility in both sexes; see Lindsley and Lifschytz 1972; Ashburner et al. 2005). This suggests that most genes and alleles classified as sex-limited are in fact associated with sex-limited sterility.
Molecular expression profiles were obtained from the Sex Bias Database (SEBIDA version 2.0: http://141.61.102.16:8080/sebida/index.php; Gnad and Parsch 2006). We downloaded male versus female expression ratios (M/F) from 15 different microarray studies (data were originally reported in: Parisi et al. 2003, 2004; Ranz et al. 2003; Gibson et al. 2004; Stolc et al. 2004; McIntyre et al. 2006; Goldman and Arbeitman 2007; Ayroles et al. 2009) and M/F ratios from a meta-analysis of several studies (details of the meta-analysis are described at SEBIDA). M/F ratios can potentially range from zero to infinity, with male-biased transcription for M/F > 1 and female-biased transcription for M/F < 1. To impose symmetry on sex-biased expression levels, we rescaled the data using an index of sex-biased expression: x = M/(M + F). This variable ranges between zero and one, with female-biased transcription for x < 0.5 and male-biased transcription for x > 0.5.
The final data set included 2,433 genes with M/F expression information from at least 1 of the 15 studies. Within the final data set, there were 1,955 genes with similar mutational effects on both sexes, 298 genes with female-biased fitness effects, 43 female-limited genes, 87 genes with male-biased fitness effects, and 50 male-limited genes (an additional 53 genes had ambiguous sex-specific sterility phenotypes). Supplementary table S1 (Supplementary Material online) provides a breakdown of the data set into phenotypic subcategories, including the mean and median number of alleles per gene, per phenotypic category.
Statistical Analysis
Two-tailed Mann–Whitney U tests (implemented in R; R Development Core Team 2005) were used to assess whether the distribution of sex-biased transcription levels differs between phenotypically defined gene categories. To examine whether different categories of sex-biased transcription have different compositions of phenotypes, we subdivided the data set into five expression categories, each with equal range: (1) 0 < x < 0.2; (2) 0.2 < x < 0.4; (3) 0.4 < x < 0.6; (4) 0.6 < x < 0.8; and (5) 0.8 < x < 1.0. Two-tailed Fisher’s exact tests were used to examine whether female-biased transcription categories (1, 2) were enriched for genes with female-specific phenotypes and whether male-biased transcription categories (4, 5) were enriched for genes with male-specific phenotypes.
The results presented below use meta-analysis expression profiles (from SEBIDA; see above) to transcriptionally categorize genes. The meta-analysis data set represents a composite of several independent microarray studies, which minimizes the likelihood of sex-biased transcription misclassification for each gene (compared with classifications based on single studies). The meta-analysis also includes data for a high proportion of the 2,433 genes (compared with single studies), which maximizes statistical power. Nevertheless, each analysis was also performed using transcription classifications from individual microarray studies. The results are consistent across studies, though the statistical power is often lower, due to decreased gene representation. Results for each platform are presented within the supplementary figs. S1 and S2 (Supplementary Material online).
Results and Discussion
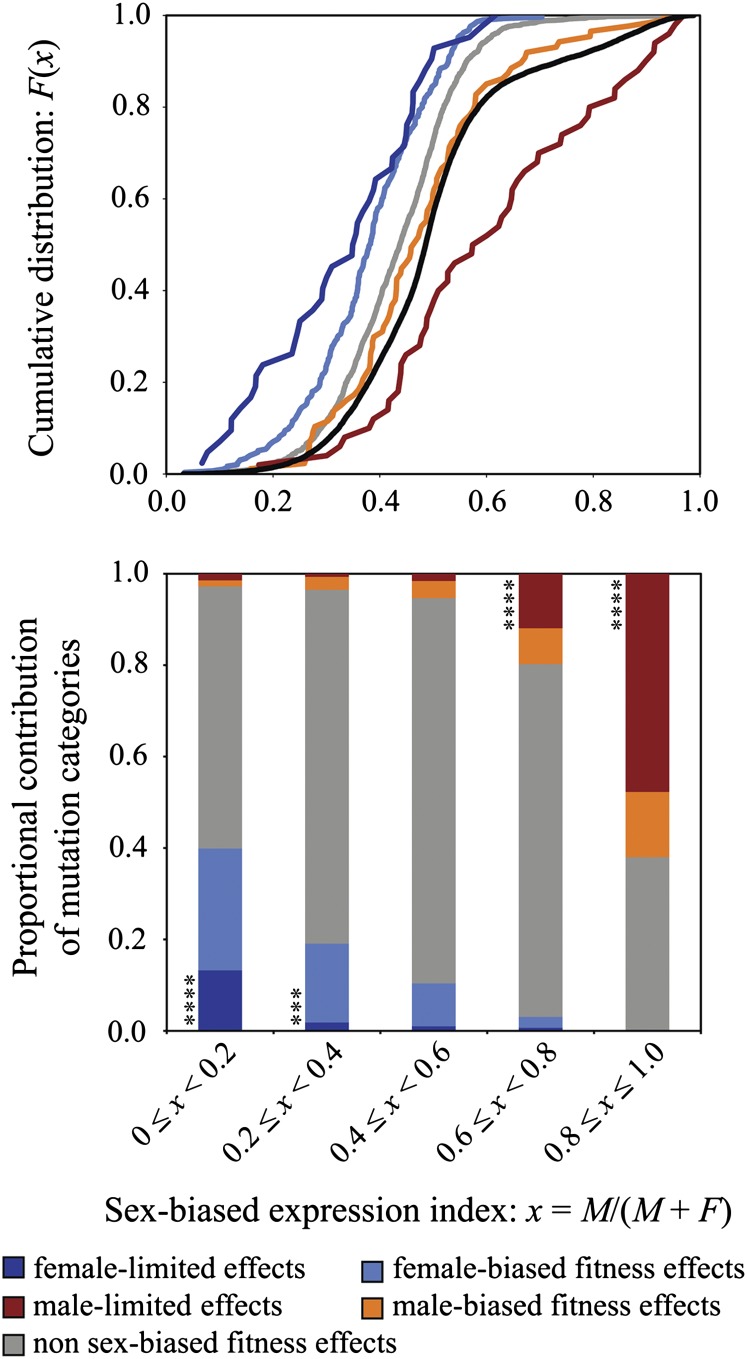
Genes with sexually dimorphic phenotypic effects tend to have relatively sex-biased mRNA expression levels (fig. 1; supplementary figs. S1 and S2, Supplementary Material online). Compared with genes with similar phenotypic effects in both sexes (those with visible, lethal, semilethal, and/or alleles for sterility in both sexes), genes with female-limited and female-biased fitness effects have higher mRNA expression in females (Mann–Whitney U tests: “female-limited effects” P = 1.51 × 10−5; “female-biased fitness effects” P = 3.30 × 10−13). Genes associated with male-limited and male-biased fitness effects have higher mRNA expression in males (“male-limited effects” P = 0.0105; “male-biased fitness effects” P = 1.03 × 10−10). These results largely extend to comparisons between individual phenotypic categories and the distribution of sex-biased expression throughout the entire Drosophila genome. Genes with greater phenotypic effects in females show greater female-biased mRNA expression (female-limited effects: P = 4.38 × 10−10; female-biased fitness effects: P < 2.2 × 10−16). Genes with male-limited phenotypic effects are more male biased in transcription (P = 6.78 × 10−6), though genes with male-biased fitness effects do not differ from the genome-wide distribution (P = 0.228). Unexpectedly, genes that similarly affect both sexes (e.g., lethal, visible, male, and female sterile) are relatively female biased (transcriptionally) compared with the genomic distribution (fig. 1, top panel; P < 2.2 × 10−16). Each of these results is highly consistent across individual microarray studies (supplementary figs. S1 and S2, Supplementary Material online).
FIG. 1.
Genes with sexually dimorphic mRNA transcription levels are associated with sex-biased fitness effects. The upper panel shows the cumulative distribution [Pr(X < x)] for sex-biased transcription among five phenotypic categories (color coded) and for the entire genome (the black curve). Each phenotypic category differs significantly from the genome-wide distribution (Mann–Whitney U; P < 10−5) except for genes with male-biased fitness effects (P = 0.228); each category significantly differed from the nonsex-biased phenotypic class of genes (partially male-limited: P = 0.0105; other categories: P < 0.00001). The lower panel shows the proportion of each phenotypic class within five sex-biased transcription categories. Two-tailed Fisher exact tests (***P < 0.001; ****P < 0.0001) were used to determine: 1) whether female-biased genes (0 < x < 0.2; 0.2 < x < 0.4) were enriched for female-biased or female-limited phenotypic effects and 2) whether male-biased genes were enriched for male-biased or male-limited phenotypic effects (0.6 < x < 0.8; 0.8 < x < 1.0).
Although these patterns validate the intuition that male-biased genes should be more important for male fitness and female-biased genes should be more important for female fitness, such associations are far from absolute. Genes with highly dimorphic transcription often have similar mutational effects on both sexes (fig. 1, within the upper and lower 20 percent tails for x: 60 percent of female-biased genes and 38 percent of male-biased genes had roughly equal fitness effects in both sexes; also see supplementary fig. S2B, Supplementary Material online). Genes with more moderate sex-biased transcription patterns are even less likely to have sex-limited fitness consequences. For example, among genes with 1.5-fold to 4-fold differential expression between the sexes (ranges 0.2 < x < 0.4 and 0.6 < x < 0.8), roughly 80 percent had similar mutational effects in each sex. Thus, although genes with male-biased and female-biased transcription are statistically associated with male-biased and female-biased fitness effects (respectively), the vast majority of sex-biased genes appear to be functionally important for both sexes (genome-wide, over 90 percent of genes fall within the range 0.2 < x < 0.8).
There are several evolutionary routes that may lead to sex-biased gene expression (Ellegren and Parsch 2007), yet only some are expected to generate associations between the relative transcription level of a gene and its fitness consequences in each sex. Sex-biased gene expression may be directly selected for when males and females have different gene expression optima. Because direct selection for expression dimorphism is not necessarily expected to alter the relative importance of the gene for either sex, sex-biased mutational effects may not accompany sex-biased transcription. Alternatively, the evolution of sex-biased expression may coincide with sex-specific selection for novel protein coding sequences or the differential incorporation of proteins into sex-specific genetic interaction networks (Arnold et al. 2009). This may involve sex-specific expression of previously noncoding sequence, gene duplication and sex-specific cooption of paralogs, or a change in a gene’s function (or subfunction) within one sex but not the other. Any of these processes can generate an association between sex-biased expression and sex-biased function and phenotypic consequence.
The results indicate that male-biased and female-biased transcription does not necessarily equate with sex-specific importance or function. Phenotypically characterized and mapped mutations tend to similarly affect both sexes, despite their variable associations with sexually dimorphic gene transcription. Female-biased genes can even produce alleles that are more harmful to males than to females and vice versa, although such cases are rare. However, despite this broad pattern of shared importance, there is also a clear statistical association between transcriptional dimorphism and the probability that a gene has a sex-limited or sex-biased phenotypic effect. This implies that a fraction of male- and female-biased genes adopt sex-specific functions or become incorporated into sex-specific molecular pathways. This pattern is particularly striking when one considers the coarseness of the transcriptional data for each gene. The ratio of male to female gene expression is a composite signal from multiple tissues during the adult life-history stage and ignores the fine-scale spatial and temporal resolution that defines the process of Drosophila development (Meisel 2011). The association between sexually dimorphic gene expression and sex-biased phenotypic effects proves that such transcriptional assays represent a biologically and evolutionarily meaningful measurement.
These patterns may also have implications for debates over the population genetic consequences of sex-specific selection (e.g., Chippindale et al. 2001; Prasad et al. 2007; Morrow et al. 2008; Whitlock and Agrawal 2009; Agrawal 2011; Connallon et al. 2010; Mallet and Chippindale 2011). Theory predicts that stronger selection in males than females will enhance purifying selection and reduce the mutational load of females (Whitlock and Agrawal 2009), whereas stronger purifying selection in females and sexually antagonistic selection between the sexes increases the female mutational load (Day and Bonduriansky 2004; Bonduriansky and Chenoweth 2009). Considering patterns of selection across the genome, net fitness costs to females can potentially emerge when female-biased purifying selection or sexual antagonism operates across a relatively small fraction of the genome (Connallon et al. 2010). The results presented here suggest that, while a proportion of the genome is subject to much stronger selection in females than males, most genes with female-biased expression experience selection in males. This may either mitigate the female genetic load or expand the sequence space that can experience sexually antagonistic selection. Molecular evolutionary contrasts between genes in different phenotypic categories, including selection parameter estimates from resequencing data (using approaches described by Keightley and Eyre-Walker 2010), may shed additional light on the population genetic consequences of sex-specific selection.
One surprising observation is that alleles with similar effects on both sexes are associated with female-biased transcription, compared with the genomic distribution (fig. 1; fig. S1, Supplementary Material online). This phenotypic category is dominated by lethal alleles, which suggests that female-biased genes (at least those with moderately higher expression in females than males) have a higher proportion of essential functions than male-biased genes. These relatively severe mutational effects might partially explain why male-biased genes evolve more rapidly than female-biased genes (e.g., Zhang et al. 2004; Pröschel et al. 2006; Clark et al. 2007; Ellegren and Parsch 2007; Larracuente et al. 2008). Female-biased genes might have a smaller fraction of effectively neutral mutations or a higher degree of pleiotropy (Mank et al. 2008; Mank and Ellegren 2009; Meisel 2011), leading to decreased opportunities for neutral and adaptive evolution (e.g., Fisher 1958; Kimura 1983).
These conclusions should be considered tentative, as they may be sensitive to characteristics of the available data. For example, we analyzed mutations that produce relatively severe phenotypes. Lethal and sterile alleles reduce fitness to zero, whereas visible and semilethal alleles may have similarly strong effects on total fitness (e.g., due to strong mate discrimination against carriers of visible and/or subviable alleles; Grossfield 1975; Hollis et al. 2009). Because mutations with small fitness effects preclude direct laboratory measurement, a broader analysis of the sex-specific fitness consequences of sex-biased genes will require a different empirical approach—perhaps one that combines gene functional assays with molecular population genetics. Second, the data is Drosophila specific, with the clear possibility of taxon idiosyncrasies. Finally, phenotypic assays of mutations are not systematic, as they represent a collection of alleles characterized during the history of Drosophila genetics research. Future studies that systematically characterize genes and mutant phenotypes using targeted mutagenesis (e.g., P element insertions) or deletion mapping will further illuminate the connection between molecular and phenotypic sexual dimorphism.
Supplementary Material
Supplementary figures S1–S2 and table S1 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
We are grateful for discussion and comments from Rich Meisel, Akane Uesugi, and two anonymous reviewers. This work was supported by an National Institutes of Health grant (GM64590 to A.G.C. and A. Bernardo Carvalho).
References
- Agrawal AF. Are males the more ‘sensitive’ sex? Heredity. 2011 doi: 10.1038/hdy.2010.156. doi: 10.1038/hdy.2010.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson MB. Sexual selection. Princeton: Princeton University Press; 1994. [Google Scholar]
- Arnold AP, van Nas A, Lusis AJ. Systems biology asks new questions about sex differences. Trends Endocrinol Metab. 2009;20:471–476. doi: 10.1016/j.tem.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M, Golic KG, Hawley RS. Drosophila: a laboratory handbook. 2nd ed. Cold Spring Harbor (NY): Cold Spring Harbor Laboratory Press; 2005. [Google Scholar]
- Ayroles JF, et al. Systems genetics of complex traits in Drosophila melanogaster. Nat Genet. 2009;41:299–307. doi: 10.1038/ng.332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonduriansky R, Chenoweth SF. Intralocus sexual conflict. Trends Ecol Evol. 2009;24:280–288. doi: 10.1016/j.tree.2008.12.005. [DOI] [PubMed] [Google Scholar]
- Chippindale AK, Gibson JR, Rice WR. Negative genetic correlation for adult fitness between sexes reveals ontogenetic conflict in Drosophila. Proc Natl Acad Sci U S A. 2001;98:1671–1675. doi: 10.1073/pnas.041378098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark AG, et al. Evolution of genes and genomes on the Drosophila phylogeny. Nature. 2007;450:203–218. doi: 10.1038/nature06341. [DOI] [PubMed] [Google Scholar]
- Connallon T, Cox RM, Calsbeek R. Fitness consequences of sex-specific selection. Evolution. 2010;64:1671–1682. doi: 10.1111/j.1558-5646.2009.00934.x. [DOI] [PubMed] [Google Scholar]
- Darwin C. The descent of man and selection in relation to sex. London: John Murray; 1871. [Google Scholar]
- Day T, Bonduriansky R. Intralocus sexual conflict can drive the evolution of genomic imprinting. Genetics. 2004;167:1537–1546. doi: 10.1534/genetics.103.026211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellegren H, Parsch J. The evolution of sex-biased genes and sex-biased gene expression. Nat Rev Genet. 2007;8:689–698. doi: 10.1038/nrg2167. [DOI] [PubMed] [Google Scholar]
- Fisher RA. The genetical theory of natural selection. 2nd ed. New York: Dover Publications Inc; 1958. [Google Scholar]
- Gibson G, et al. Extensive sex-specific nonadditivity of gene expression in Drosophila melanogaster. Genetics. 2004;167:1791–1799. doi: 10.1534/genetics.104.026583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gnad F, Parsch J. Sebida: a database for the functional and evolutionary analysis of genes with sex-biased expression. Bioinformatics. 2006;22:277–279. doi: 10.1093/bioinformatics/btl422. [DOI] [PubMed] [Google Scholar]
- Goldman TD, Arbeitman MN. Genomic and functional studies of Drosophila sex hierarchy regulated gene expression in adult head and nervous system tissues. PLoS Genetics. 2007;3:e216. doi: 10.1371/journal.pgen.0030216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossfield J. Behavioral mutants in Drosophila. In: King RC, editor. Handbook of genetics. Vol. 3. Invertebrates of genetic interest. New York: Plenum Press; 1975. pp. 679–702. [Google Scholar]
- Hollis B, Fierst JL, Houle D. Sexual selection accelerates the elimination of a deleterious mutant in Drosophila melanogaster. Evolution. 2009;63:324–333. doi: 10.1111/j.1558-5646.2008.00551.x. [DOI] [PubMed] [Google Scholar]
- Keightley PD, Eyre-Walker A. What can we learn about the distribution of fitness effects of new mutations from DNA sequence data? Philos Trans R Soc Lond B Biol Sci. 2010;365:1187–1193. doi: 10.1098/rstb.2009.0266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M. The neutral theory of molecular evolution. Cambridge: Cambridge University Press; 1983. [Google Scholar]
- Larracuente AM, et al. Evolution of protein coding genes in Drosophila. Trends Genet. 2008;24:114–123. doi: 10.1016/j.tig.2007.12.001. [DOI] [PubMed] [Google Scholar]
- Lindsley DL, Lifschytz E. The genetic control of spermiogenesis in Drosophila. In: Beatty RA, Gluecksohn-Waelsch S, editors. Edinburgh Symposium on the Genetics of the Spermatozoon. Copenhagen (Denmark): Bogtrykkeriet Forum; 1972. pp. 203–222. [Google Scholar]
- Mallet MA, Chippindale AK. Inbreeding reveals stronger net selection on Drosophila melanogaster males: implications for mutation load and the fitness of sexual females. Heredity. 2011 doi: 10.1038/hdy.2010.148. doi: 10.1038/hdy.2010.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mank JE, Ellegren H. Are sex-biased genes more dispensable? Biol Lett. 2009;5:409–412. doi: 10.1098/rsbl.2008.0732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mank JE, Hultin-Rosenberg L, Zwahlen M, Ellegren H. Pleiotropic constrain hampers the resolution of sexual antagonism in vertebrate gene expression. Am Nat. 2008;171:35–43. doi: 10.1086/523954. [DOI] [PubMed] [Google Scholar]
- McIntyre LM, et al. Sex-specific expression of alternative transcripts in Drosophila. Genome Biol. 2006;7:R79. doi: 10.1186/gb-2006-7-8-r79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meisel RP. Towards a more nuanced understanding of the relationship between sex-biased gene expression and rates of protein coding sequence evolution. Mol Biol Evol. 2011 doi: 10.1093/molbev/msr010. doi:10.1093/molbev/msr010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrow EH, Stewart AD, Rice WR. Assessing the extent of genome-wide intralocus sexual conflict via experimentally enforced gender limited selection. J Evol Biol. 2008;21:1046–1054. doi: 10.1111/j.1420-9101.2008.01542.x. [DOI] [PubMed] [Google Scholar]
- Parisi M, et al. Paucity of genes on the Drosophila X chromosome showing male-biased expression. Science. 2003;299:697–700. doi: 10.1126/science.1079190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parisi M, et al. A survey of ovary-, testis-, and soma-based gene expression in Drosophila melanogaster adults. Genome Biol. 2004;5:450. doi: 10.1186/gb-2004-5-6-r40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad NG, Bedhomme S, Day T, Chippindale AK. An evolutionary cost of separate genders revealed by male-limited evolution. Am Nat. 2007;162:29–37. doi: 10.1086/509941. [DOI] [PubMed] [Google Scholar]
- Pröschel M, Zhang Z, Parsch J. Widespread adaptive evolution of Drosophila genes with sex-biased expression. Genetics. 2006;174:893–900. doi: 10.1534/genetics.106.058008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team. R: a language and environment for statistical computing, reference index version 2.2.1. Vienna (Austria): R Foundation for Statistical Computing; 2005. [Google Scholar]
- Ranz JM, Castillo-Davis CI, Meiklejohn CD, Hartl DL. Sex-dependent gene expression and evolution of the Drosophila transcriptome. Science. 2003;300:1742–1745. doi: 10.1126/science.1085881. [DOI] [PubMed] [Google Scholar]
- Stolc V, et al. A gene expression map for the euchromatic genome of Drosophila melanogaster. Science. 2004;306:655–660. doi: 10.1126/science.1101312. [DOI] [PubMed] [Google Scholar]
- Tweedie S, et al. FlyBase: enhancing Drosophila Gene Ontology annotations. Nucleic Acids Res. 2009;37:D55–D59. doi: 10.1093/nar/gkn788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitlock MC, Agrawal AF. Purging the genome with sexual selection: reducing mutation load through selection in males. Evolution. 2009;63:569–582. doi: 10.1111/j.1558-5646.2008.00558.x. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Hambuch TM, Parsch J. Molecular evolution of sex-biased genes in Drosophila. Mol Biol Evol. 2004;21:2130–2139. doi: 10.1093/molbev/msh223. [DOI] [PubMed] [Google Scholar]