Abstract
Human plasminogen activator inhibitor type 1 (PAI-1) is a serine protease inhibitor with a metastable active conformation. The lifespan of the active form of PAI-1 is modulated via interaction with the plasma protein, vitronectin, and various metal ions. These metal ions fall into two categories: Type I metals, including calcium, magnesium, and manganese, stabilize PAI-1 in the absence of vitronectin, whereas Type II metals, including cobalt, copper, and nickel, destabilize PAI-1 in the absence of vitronectin, but stabilize PAI-1 in its presence. To provide a mechanistic basis for understanding the unusual modulation of PAI-1 structure and activity, the binding characteristics and conformational effects of these two types of metals were further evaluated. Steady-state binding measurements using surface plasmon resonance indicated that both active and latent PAI-1 exhibit a dissociation constant in the low micromolar range for binding to immobilized nickel. Stopped-flow measurements of approach-to-equilibrium changes in intrinsic protein fluorescence indicated that the Type I and Type II metals bind in different modes that induce distinct conformational effects on PAI-1. Changes in the observed rate constants with varying concentrations of metal allowed accurate determination of binding affinities for cobalt, nickel, and copper, yielding dissociation constants of ∼40, 30, and 0.09 μM, respectively. Competition experiments that tested effects on PAI-1 stability were consistent with these measurements of affinity and indicate that copper binds tightly to PAI-1.
Keywords: PAI-1, serpins, metals, vitronectin
Introduction
Recent surveys of the protein data bank (PDB) show that ∼40% of proteins contain metal cofactors that serve a structural and/or functional capacity.1 They assist in protein folding and stability or provide sites for substrate binding and catalysis. Metals give proteins functional flexibility because of their potential to bind to an assortment of ligands and perform quite diverse chemistries.2 Under physiological conditions, the most common metals bound to proteins are Na+, K+, Mg+2, Ca+2, Zn+2, Mn+2, Ni+2, Cu+1/+2, Fe+2/+3, and Co+2/+3.2 These metals were selected early in protein evolution because of their relatively high abundance in nature.3
The heavier metals are lower in abundance and are highly regulated. Most are bound to a series of carrier proteins that deliver the metals to specific organs/cell types. From there, they are passed along to intracellular transporters via additional carrier proteins and finally to the target. The metals are bound during protein synthesis or once a particular function is needed.4–6 In this way, the likelihood of protein/metal binding in the physiological environment is determined both by the metal binding affinity of the protein and local metal bioavailability because of the action of carriers and transporters.
Human plasminogen activator inhibitor-1 (PAI-1) is a serine protease inhibitor (serpin) that spontaneously undergoes a large conformational change, switching the protein from an active to latent form.7 At 37°C and neutral pH, the half-life (t1/2) of the active form is 1–2 h.8–17 Binding of PAI-1 to the plasma protein, vitronectin, extends its functional t1/2 by ∼50%.8,9,11,13,17 This is a high affinity interaction (Kd of ∼0.1–1 nM) such that vitronectin is often called a PAI-1 cofactor.18–23 These two proteins are located primarily in plasma and the extracellular space where they inhibit fibrinolysis or extracellular proteolysis, control cellular adhesion and migration, and assist in the inflammatory response.24–34 In our previous report35, we have shown that calcium, magnesium, and manganese (referred to as Type I metals) have a modest stabilizing effect on the t1/2 of PAI-1, whereas cobalt, copper, and nickel (referred to as Type II metals) effect a vitronectin- dependent modulation of the t1/2 of PAI-1. The Type II metals induce a substantial destabilization of PAI-1, reducing t1/2 to a few minutes. However, with vitronectin present in addition to the metals, the half-life of PAI-1 increases up to ∼40-fold.
In this accompanying study, we establish binding affinities for the Type II metals and demonstrate the differential conformational effects induced on PAI-1 by the Type I versus II metals. This was accomplished initially via steady-state binding experiments using surface plasmon resonance (SPR) and then was expanded to provide insight into the binding mechanism using transient-state kinetic measurements. Consistent with our previous metal categorization,35 the Type I and II metals promote different changes in PAI-1 intrinsic protein fluorescence upon binding, suggesting that each group of metals binds via unique interactions that induce differing effects on PAI-1 structure. These data also indicate that cobalt and nickel bind to PAI-1 with a dissociation constant in the low micromolar range, whereas copper binds with ∼200-fold stronger affinity. Thus, the binding of copper to PAI-1 occurs at concentrations that are well within the range of copper bioavailability. The ability of copper to modulate PAI-1 structure and activity represents a previously unrecognized control on the ability of this serpin to regulate plasminogen activation in the circulation and tissues.
Results
Both active and latent PAI-1 bind to immobilized nickel
The concentrations of metals used in our previous PAI-1 stability measurements were intentionally set to high values35 to ensure that the effects were measured under saturating conditions. Thus, it is now important to evaluate binding directly to determine whether metal affinities are in a physiological range. Because the Type II metals destabilize PAI-1 so that the active form converts to the latent form within minutes, typical steady-state titrations to measure metal binding are not practical. Therefore, to probe the PAI-1/metal binding affinity, we immobilized nickel onto a nitrilotriacetic acid (NTA) chip for SPR runs and then passed varying concentrations of active PAI-1 over the surface.
A plot of change in total response units at equilibrium versus the total concentration of active PAI-1 was used to quantify binding to the nickel-NTA chip.36 These data were best fit to a model that included both a hyperbolic function for high-affinity binding behavior, with an additional linear function due to a lower affinity nonspecific metal binding. This treatment gave the active PAI-1/nickel dissociation constant of 6.5 ± 0.2 μM. Replicate experiments with latent PAI-1 were fit directly to a single-site binding isotherm [Fig. 1(A)], yielding the latent PAI-1/nickel dissociation constant of 22 ± 3 μM. Active PAI-1 binds approximately threefold tighter than latent PAI-1, suggesting that the shortened t1/2 for the active form of PAI-1 with nickel present cannot be due to preferential binding of metal to the latent form. Instead, the perturbed half-life is more likely due to a specific, metal-induced conformational change in the active form of PAI-1 that favors conversion to the latent form.
Figure. 1.
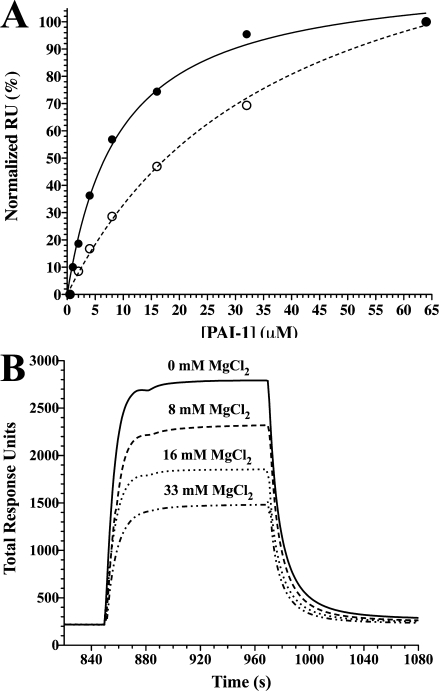
Evaluation of PAI-1 binding to nickel using surface plasmon resonance. Nickel was bound to an NTA-coupled chip, and PAI-1 binding was measured at equilibrium using SPR by injecting varying concentrations and flowing the solution over the chip (see Materials and Methods). In Panel A, the change in response units upon binding of active (•) or latent (○) PAI-1 to immobilized nickel was corrected for nonspecific binding, normalized, and fit to the equation for a hyperbolic binding function [Eq. (2)] with a single binding site. The dissociation constants for binding are calculated to be 6.5 ± 0.2 μM and 22 ± 3 μM for native and latent PAI-1, respectively. In Panel B, varying concentrations of magnesium chloride were mixed with PAI-1 (2 μM) and then passed over immobilized nickel on the NTA-chip.
To assess whether Type I and Type II metals bind to the same or different sites on PAI-1, a direct test for competition for binding sites was taken using the SPR method. In this case, varying amounts of MgCl2 (Type I metal) were mixed with PAI-1 before injection over a nickel (Type II metal)-coated SPR chip. At high concentrations, MgCl2 is observed to inhibit PAI-1 binding to nickel [Fig. 1(B)], indicating that magnesium can bind to the Type II site but with very low affinity. This result suggests that a single metal site is able to accommodate binding of Type I and Type II metals, which have opposing effects depending on the metal bound.
Metal binding to PAI-1 can be monitored using measurements of changes in intrinsic fluorescence
Our previous experiments demonstrating differences in effects of metal binding on the stability of PAI-1 indicate that binding sites, affinities, and conformational effects may differ between Type I and Type II metals.35 The SPR binding screen worked well, but only with nickel binding because the NTA resin is designed for optimal binding to nickel, whereas other metals have lower affinity for the resin. Just as with the IMAC procedure,35 such results must be interpreted with caution and cannot be used to infer an intrinsic metal binding site. NTA is tetradentate and provides four coordination sites for Ni binding. Thus, Ni bound to NTA can coordinate with two additional functional groups (often histidine ligands) within a protein to fulfill its six coordination sites. Obviously, these conditions can be fulfilled without an intact coordination chemistry intrinsic to a protein structure. Furthermore, the SPR method gives no information about mechanisms of binding or effects on protein structure. Therefore, to measure binding directly, determine affinity, evaluate structural effects, and explore differences between Type I and II metal interactions with PAI-1, a spectroscopic method was used. We compared steady-state fluorescence emission spectra for active and latent PAI-1 in the absence of metals and observed a change in the intrinsic tryptophan fluorescence of the two forms of PAI-1, with latent PA-1 exhibiting a lower overall quantum yield (data now shown). This difference in tryptophan fluorescence between the two forms of PAI-1 allowed us to develop a stopped-flow method that relied on approach-to-equilibrium changes in intrinsic protein fluorescence upon rapid mixing of PAI-1 with metals. The kinetic method allowed for evaluation of the mechanism of metal binding and associated changes in conformation, which cannot be revealed from steady-state measurements.
Because typical PAI-1 preparations contain EDTA and its removal by dialysis is problematic,37,38 it was necessary to develop a purification protocol for PAI-1 without the addition of EDTA. As described in the Materials and Methods section, Chelex® resin was used to remove all metal during the modified purification procedure. This modification also ensured no EDTA was present to interfere with the stopped-flow binding experiments. The half-life (t1/2) of the active conformation of human PAI-1 has been determined on several occasions by different groups, with results that range from ∼1.5 h to over a day,8–17 likely attributable in part to varied buffer conditions for the experiments. In our companion article,35 we measured PAI-1 stability in Tris buffer at 37°C and pH 7.4, resulting in a t1/2 of 1–2 h. In our stopped-flow approach, we chose 20°C to extend the active lifespan of PAI-1 and readily observe multiple steps in the binding mechanism. We also compared measurements in MOPS and Tris buffers at pH 7.4.
The purified PAI-1 (free of chelators) exhibited measurable changes in intrinsic fluorescence in stopped-flow experiments in which Type I or II metals were mixed with PAI-1, providing a promising method to quantify binding and evaluate mechanistic details. In control experiments in which PAI-1 was mixed with buffer alone, the intrinsic protein fluorescence decreased slightly over time (data not shown), an effect that is likely due to photobleaching. To correct for this background loss of fluorescence, it was subtracted from all experiments with metals. The stopped-flow experiments were pursued with the Type II transition metals that had such dramatic effects on PAI-1 activity and stability. Under pseudo-first-order conditions, mixing of PAI-1 with CoCl2, NiCl2, or CuCl2 gave response curves that showed an initial drop in the intrinsic fluorescence of PAI-1, followed by an increase [Fig. 2(A–D)]. Because of known interactions between buffers and metal, we designed these experiments to be performed in buffers for which binding affinities have been explicitly determined and for which these interactions can be minimized.38–41 Stopped-flow measurements of changes in PAI-1 fluorescence in MOPS buffer are shown in Figure 2(A,B) for nickel and cobalt binding to PAI-1. Copper solubility in MOPS was problematic, so experiments with copper were pursued with Tris buffer, as shown in Figure 2(D). As a control for comparing results in the two buffers, we also measured approach-to-equilibrium binding of nickel to PAI-1 in Tris buffer, as shown in Figure 2(C). As can be seen by comparing Figure 2(A) with Figure 2(C), the changes that occur in the fluorescence of PAI-1 under these presteady-state conditions in the two buffers with nickel are comparable. For all three metals in either buffer, these changes in fluorescence were clearly multiphasic and fit well to three exponentials yielding the observed rate constants kobs,1, kobs,2, and kobs,final, respectively.
Figure. 2.
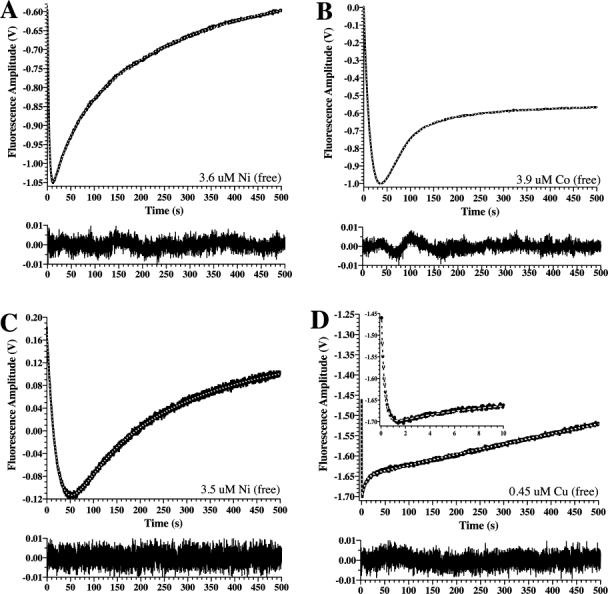
Approach-to-equilibrium changes in PAI-1 intrinsic protein fluorescence upon mixing with Type II metals. Panels A and B show representative results in MOPS buffer, whereas Panels C and D give results from Tris buffer (see Materials and Methods). PAI-1 was mixed 1:1 with NiCl2 (A), CoCl2 (B), NiCl2 (C), or CuCl2 (D). The free metal concentrations were calculated from the total metal concentration and the buffer/metal affinity constant using known metal:MOPS affinities (see Materials and Methods). Changes in intrinsic protein fluorescence emission at wavelengths above 320 nM were monitored continuously for 500 s. After subtraction of background (see Materials and Methods), data with all metals gave good fits to a triple exponential equation with acceptable residuals, shown in the bottom frame below each of the panels. The inset to Panel D shows an expanded view of the data at the earliest time points to more easily visualize the rapid decrease in fluorescence that occurs upon mixing with copper.
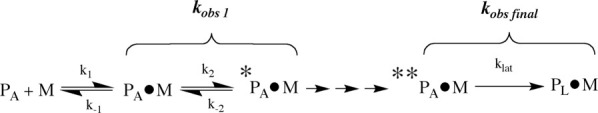
These stopped-flow experiments were performed over a broad range in metal concentrations to distinguish between binding events, which are concentration dependent, and conformational changes, which are not. When so evaluated using varied metal concentrations, it was determined that kobs,1 measured upon Type II metal binding to PAI-1 increased in a concentration-dependent manner (Fig. 3), whereas kobs,2 and kobs,final did not. The increase in kobs,1 approached a limiting rate (Fig. 3), a result consistent with a two-step binding process as illustrated in Scheme 1.42,43
Figure. 3.
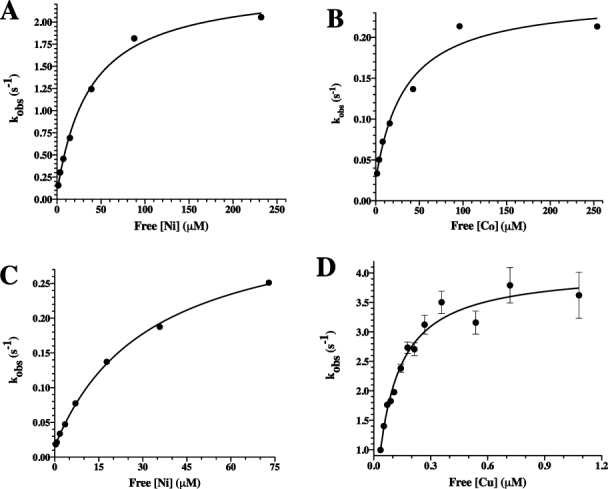
The rate constant, kobs,1, resulting from the approach-to-equilibrium binding studies of Type II metals to PAI-1, shows a hyperbolic dependence on metal concentration. PAI-1 was mixed 1:1 with increasing concentrations of nickel, cobalt, or copper. For nickel and cobalt experiments, data were collected for 500 s and were fit to a triple exponential equation after subtraction of background fluorescence changes, as described in the Materials and Methods section. For copper, the time window was reduced to 50 s, and the data were fit to a double exponential equation after subtraction of background (see Materials and Methods). Changes in the magnitude of kobs,1 versus free metal concentration were plotted for nickel (A) and cobalt (B) in MOPS buffer as well as nickel (C) and copper (D) in Tris buffer. kobs,1 increased in a hyperbolic fashion consistent with a two-step binding model in which there is an initial fast equilibrium binding step followed by a slower conformational change (Scheme 1). Nonlinear least-squares fits to Eq. (1) are shown.
Scheme. 1.

| (1) |
In this model, there is an initial rapid equilibrium established between active PAI-1 (PA) and metal (M), with forward and reverse rate constants (k1 and k−1) defining the binding affinity Kd, followed by a conformational change (*PA•M) described by the rate constants k2 and k−2 characterizing the forward and reverse conformational changes, respectively. The data (Fig. 3) fit well to the equation [Eq. (1)] describing this process, yielding both the dissociation constant for metal binding (Kd) and kinetic characterization of the conformational step (k2 and k−2) (Table I).
Table I.
Parameters from Fitting of kobs,1 versus Ligand Concentration Using Nonlinear Least Squares to Eq. (1) for Approach-to-Equilibrium Changes in PAI-1 Intrinsic Protein Fluorescence
| Metal | Buffer | Kd (μM) | k2 (s−1) | k−2 (s−1) |
|---|---|---|---|---|
| Cobalt | MOPS | 34 ± 13 | 0.22 ± 0.02 | 0.026 ± 0.013 |
| Nickel | MOPS | 38 ± 6 | 2.36 ± 0.09 | 0.074 ± 0.052 |
| Nickel | Tris | 32 ± 2 | 0.34 ± 0.01 | 0.015 ± 0.02 |
| Copper | Tris | 0.086 ± 0.027 | 4.4 ± 0.4 | −0.29 ± 0.51 |
Kd, k2, and k−2 represent the dissociation constant for the initial binding and forward and reverse rates for the subsequent conformational change that occurs in PAI-1 upon metal binding, as shown in Scheme 1. Errors shown come directly from the fit.
The similarity of the fluorescence changes within the Type II group indicates that these metals bind in a similar fashion and likely at the same site. These results also demonstrate that the affinity for copper is high, that is, ∼200-fold tighter than either for nickel or cobalt (Table I). Finally, they illustrate the importance of our approach to explicitly account for buffer:metal interactions and calculate the free metal concentration, validated by the similarity in the dissociation constants for nickel measured in the two buffers (Table I). As MOPS and Tris have quite different affinities for nickel, the two buffers require substantially different total metal concentrations to arrive at similar free metal concentrations (Table II); yet, both buffers gave a similar free metal concentration range over which kobs,1 approaches its limiting rate (Fig. 3), resulting in similar Kd values for PAI-1:nickel binding in the two cases.
Table II.
Calculated Dissociation Constants for Metal/Buffer Binding Interactions
| Metal | Buffer | β1a | Ka (M−1) | Kd (μM) |
|---|---|---|---|---|
| Magnesium | MOPS | 3.51 | 3.24 × 103 | 309 |
| Manganese | MOPS | 3.54 | 3.47 × 103 | 288 |
| Cobalt | MOPS | 3.41 | 2.57 × 103 | 389 |
| Nickel | MOPS | 3.45 | 2.82 × 103 | 355 |
| Nickel | Tris | 2.74 | 5.50 × 102 | 1820 |
| Copper | Tris | 4.05 | 1.12 × 104 | 89.1 |
The transient-state experiments demonstrate differences between Type I and II metal binding to PAI-1
Experiments with the Type I metals showed a much different effect compared to that observed with CoCl2, NiCl2, and CuCl2. Rapid mixing of MgCl2 or MnCl2 with PAI-1 showed an increase in fluorescence that fits well to a double exponential equation [Fig. 4(A,B)], but without the initial phase corresponding to the decrease in fluorescence observed with the Type II metals that directly reports binding of the metals to PAI-1. Consistently, using a similar time window, mixing with varying concentrations of either of these metals resulted in no significant changes in the observed rate constant for either exponential, indicating that these changes are conformational events that occur after binding. Thus, in contrast with what is observed with the Type II metals, the binding event for Type I metals is not detected by perturbations in any of four tryptophan residues (86, 139, 175, or 262), which are the main contributors to PAI-1 fluorescence using this method,44 so that the binding event is not reported directly. Instead, it appears that changes in the microenvironment of these residues are only observed subsequent to binding and are associated with conformational changes in PAI-1. These results, which show that Type I metal effects on PAI-1 upon binding are much different than observed with the Type II metals, are consistent with their differing effects on the rate of conversion of active PAI-1 to its latent form.
Figure. 4.
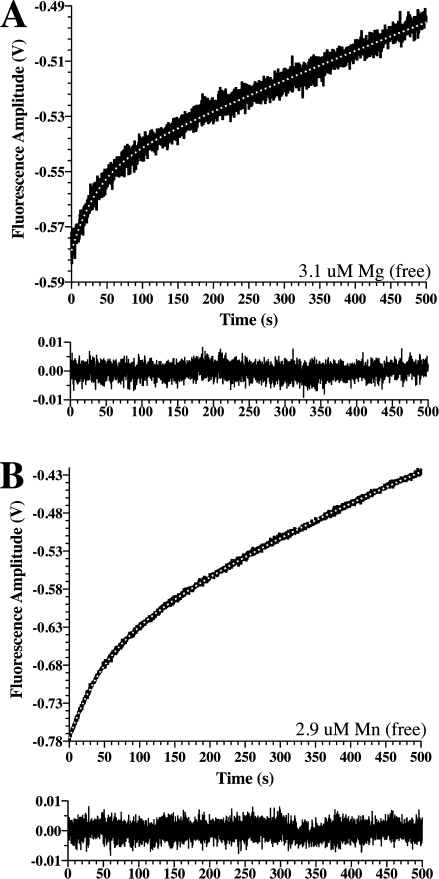
Approach-to-equilibrium changes in PAI-1 intrinsic protein fluorescence upon mixing with Type I metals. PAI-1 was mixed 1:1 with MgCl2 (A) or MnCl2 (B) in Tris buffer. Changes in intrinsic protein fluorescence emission at wavelengths above 320 nM were monitored continuously for 500 s. After subtraction of background (see Materials and Methods), the data were fit to a double exponential equation with acceptable residuals, represented in the bottom frame below each of the panels. The free metal concentration was calculated from the total metal concentration and the buffer/metal affinity constant (see Materials and Methods).
The timescale for klat corresponds to that for the final change in PAI-1 fluorescence measured in the stopped-flow experiments
As mentioned, because kobs,2 and kobs,3 do not vary significantly with Type II metal concentrations, these events appear to represent additional downstream conformational events that occur after metal binding. Although the conformational change associated with kobs,2 cannot be assigned at present to a known conformational event, we anticipated that the final exponential (i.e., kobs,3) was slow enough that it might correspond to the conformational transition of PAI-1 from the active to latent form. Thus, we determined whether this event occurs on a similar timescale as that for conversion of active PAI-1 to the latent conformation. To compare the rates directly, the t1/2 of PAI-1 was measured alone or in the presence of added metals in the MOPS buffer at 20°C (Table III). For experiments that used MOPS buffer, klat, for PAI-1 with added NiCl2 and CoCl2 matched very well with kobs,final determined from the mathematical fit of the approach-to-equilibrium binding data (Table III). It should be noted that accurate determination of kobs,final for CoCl2 required the approach-to-equilibrium experiments to be performed over a longer timescale, 2000 s, rather than the 500 s that was used in Figure 2. This was necessary because the initial timescale captured only a small segment of the final step, leaving it poorly defined [Fig. 2(B)]. At these longer timescales, it was determined that the final exponential actually corresponded to a decrease in fluorescence (data not shown).
Table III.
Comparison of kobs,final from Approach-to-Equilibrium Changes in PAI-1 Intrinsic Protein Fluorescence with klat Measured from Kinetics Measuring the Half-Life of PAI-1 Using Similar Conditions for Buffer, pH, and Temperature
| Metal | Buffer | Total metal | Free metal | kobs final (s−1) | klat (s−1) |
|---|---|---|---|---|---|
| None | MOPS | NA | NA | NA | 8.42 (±0.20) × 10−5 |
| Cobalt | MOPS | 20 mM | 250 μM | 3.09 (±0.02) × 10−4 | 3.42 (±0.07) × 10−4 |
| Nickel | MOPS | 20 mM | 230 μM | 2.17 (±0.04) × 10−3 | 1.89 (±0.13) × 10−3 |
The values for kobs final come directly from the multiple exponential fitting, whereas that for klat is the mean and standard error for three replicate experiments.
Thus, because of the similarity with all of the Type II metals between experimental measurements of klat and the final observed rate constant, kobs final, in the approach-to-equilibrium measurements, we assign this step to the latency transition. Combining these results with the model in Scheme 1 gave an overall mechanism, Scheme 2, in which the last step is the conversion to latent PAI-1 (PL). This model shows intervening steps between kobs,1 and kobs final/klat, which at this point are unassigned and likely represent other conformational events.M
Scheme. 2.
Kinetic measurements of PAI-1 stability with mixtures of metals are consistent with a high-affinity copper interaction and suggest that Type I and II metals bind in the same location
Although our kinetic studies suggested that PAI-1 is promiscuous in its ability to interact with metals,35 there is likely a preference for the binding of one metal over another. Affinity measurements from approach-to-equilibrium studies suggest much tighter binding to copper than other metals. To confirm the high-affinity copper interaction and validate the determined binding constants, PAI-1 stability was evaluated using a mixture of NiCl2 and CuCl2 [Fig. 5(A)]. For each of these Type II transition metals, a characteristic t1/2 for conversion of PAI-1 to the latent form is observed,35 so it was reasoned that PAI-1 in the presence of a mixture of the two metals should give an intermediate t1/2 if the two metals bind to the same site on PAI-1. The experiment was designed such that the free metal concentration for each metal was more the twice the Kd; the free concentration of nickel was chosen in a 300-fold excess over the free concentration of copper to achieve conditions where competition for metals for the binding site(s) was feasible because of the ∼300-fold differential between the measured dissociation constants (Table IV). Thus, this experimental design was anticipated to give a ∼50% occupancy for each metal that was predicted to yield a t1/2 midway between that for each metal. The mixture of 0.135 mM CuCl2 and 2 mM NiCl2 (0.24 and 73 μM free metal, respectively) gave data that fit well to a single exponential [Fig. 5(A)] and yielded a t1/2 intermediate between that for the individual metals (Table IV), as predicted. This result confirms the affinity measurements determined with the stopped flow and indicates that these metals likely bind in the same site.
Figure. 5.
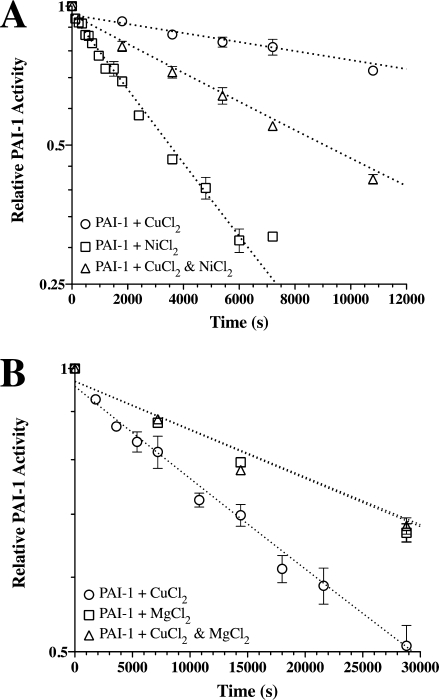
Kinetic measurements of PAI-1 half-life at 20°C in the presence of mixtures of metals. Panel A is a semilog plot showing a single rate associated with the loss of PAI-1 inhibitory activity in the presence of 73 μM free nickel and 0.24 μM free copper (Table IV) compared with the loss of PAI-1 inhibitory activity in the presence of the each metal alone at a similar metal concentration. Panel B shows a similar fit for the rate of loss of PAI-1 activity with a mixture of ∼400 μM free magnesium and 0.24 μM free copper (Table IV) compared with PAI-1 plus either of the metals fixed at the same metal concentration. All measurements were performed at 20°C, and the kinetic data represent the mean and standard error for three separate experiments.
Table IV.
Effects on the Rate of Conversion of Active PAI-1 to the Latent Form in the Presence of Mixtures of Type I and Type II Metals
| Additive | Total metal | Free metal | klat (s−1) | t1/2 (h) |
|---|---|---|---|---|
| MgCl2 | 500 μM | ∼400 μM | 1.23 (±0.01) × 10−5 | 15.7 (±0.1) |
| NiCl2 | 2000 μM | 73 μM | 20.5 (±0.8) × 10−5 | 0.94 (±0.03) |
| CuCl2 | 135 μM | 0.24 μM | 2.39 (±0.14) × 10−5 | 8.1 (±0.5) |
| CuCl2 and NiCl2 | Above | Above | 7.67 (±0.11) × 10−5 | 2.51 (±0.04) |
| CuCl2 and MgCl2 | Above | Above | 1.22 (±0.05) × 10−5 | 15.7 (±0.7) |
The values for klat and the associated t1/2 at 20°C were calculated from kinetic experiments that measure the rate of loss over time of PAI-1 antiprotease activity using tPA (Fig. 5).
In the SPR experiments, it appeared that magnesium perturbed nickel binding to PAI-1, but it was unclear whether magnesium (i.e., Type I) and nickel (i.e., Type II) bound to the same or separate sites. Thus, the kinetic approach to measure stability was performed to test for competition in binding between magnesium and the most tightly bound Type II metal, copper. Because the affinity of PAI-1 for magnesium was unknown, a 1600-fold excess of magnesium (Table IV) was added to readily detect competition for copper binding. Concentrations of 0.5 mM MgCl2 and 0.135 mM CuCl2 (∼400 and 0.24 μM free metal, respectively) were mixed with PAI-1, and the effect of the mixture of metals on the rate of conversion from the active to latent form was measured. The data again fit well to a single exponential with the resulting t1/2 virtually identical to that for PAI-1 plus MgCl2 [Table IV and Fig. 5(B)]. This result indicates that Type I and II metal vie for the same site with MgCl2 present in excess over CuCl2, as suggested by the SPR measurements.
Discussion
In this study, we have used equilibrium binding approaches, steady-state kinetics, and stopped-flow measurements to demonstrate that Type I and Type II metals invoke different structural changes in the protein upon binding. This observation is consistent with our work in the companion article in this series,35 which shows that Type I and Type II metals have dramatically different effects on the stability of PAI-1. The Type I metals used in this work, including magnesium and manganese, appear to have modest effects on the structure and function of PAI-1, with negligible effects on the stability of the protein measured by its t1/2. The effects of the Type II metals, including cobalt and nickel, are in stark contrast, leading to marked destabilization of PAI-1 and rapid conversion to the latent form.
Metals invoke a series of conformational changes in PAI-1
Clearly, there are differences in structural effects on PAI-1 induced by the two types of metals. These are most readily apparent from the stopped-flow approach-to-equilibrium measurement of changes in intrinsic tryptophan fluorescence upon metal binding to PAI-1. In the case of the Type II metals that have the most distinct effects on function of the serpin, there are three exponentials that characterize association of PAI-1. The fastest rate is associated with a decrease in fluorescence that approaches a limiting rate in a concentration-dependent fashion; thus, it corresponds to a metal binding event, followed by a conformational change. In contrast, interaction with Type I metals gives changes in PAI-1 structure that are associated with two events characterized by rates that do not vary with metal concentration. Likewise, the slower events observed in the transient kinetics with Type II metals are reminiscent of these changes that occur with Type I metals and are essentially concentration independent. These changes in fluorescence do not directly report the initial binding event, but instead indicate conformational effects that occur subsequent to binding. The slowest of these rates occurs on the same timescale as the conversion of PAI-1 from the active to latent form, so it is tentatively assigned to this transition.
The mechanism by which serpins inhibit proteases occurs via a series of carefully executed conformational changes. Although variability in reactive center loop orientation is the most obvious among the growing database of crystal structures on PAI-1,7,23,45–48 there are conformational changes that occur throughout the molecule. According to descriptions of movements that influence serpin function, which have resulted from a variety of investigations using structural and kinetic methods, such regions that have been dubbed as the “gate” region, the “hinge” region, the “shutter” region, and so forth.13,49–51 PAI-1 is particularly susceptible to adopting different conformational states as the active form is metastable.52 As the thermodynamically more stable form of PAI-1 is the latent conformation,52 the active conformation is adopted by folding in a local kinetic “trap,” with several other conformations that can be accessed with relatively little energetic cost en route to the global minimum that is achieved by reactive center loop insertion. RCL insertion can result either by forming the covalent serpin-protease acyl intermediate53,54 or the latent inhibitor.7 After either Type I or II metals associate with PAI-1, a series of conformational events occur that are detected by changes in the fluorescent tryptophan probes that are distributed throughout the molecule. Thus, metals perturb the metastable fold of PAI-1 to affect the half-life of the inhibitor. With reactive transition metals such as copper, oxidative damage to a protein may occur. Although this cannot be explicitly ruled out with PAI-1, the similar step-wise changes in tryptophan fluorescence from short to longer timescales with the three transition metals tested, which differ in oxidative capacity, are more consistent with changes in conformation rather than site-specific oxidation. Also, the enhancement of the stability of the active form of PAI-1 in the presence of these metals and vitronectin argues against oxidative damage.35
Copper binds to PAI-1 with the highest affinity
Steady-state SPR measurements indicate that active PAI-1 binds to nickel with a dissociation constant of ∼6 μM, whereas stopped-flow experiments give a slightly weaker affinity with a Kd in the 25–40 μM range (Table I). Similar experiments with cobalt result in an affinity at the high end of that range. Copper binds much tighter than either of these metals, with an affinity of between 60 and 115 nM (Table I). Kinetic experiments testing effects of mixtures of metal on PAI-1 stability are consistent with this conclusion, showing that copper dominates among metals in affecting PAI-1 function, so that a ∼300-fold higher concentration of free nickel is required to compete effectively for copper binding. Experiments with magnesium/copper mixtures also indicate that a large excess (∼1600-fold higher concentration) of free magnesium competes for binding at the same site. Thus, these data indicate a single metal-binding site on PAI-1 can bind both to Type I and Type II metals, either using different geometries or providing different side-chain or backbone atoms to accommodate different metals at the same site. These different binding modes invoke different conformational changes in PAI-1, as detected by the distinct changes in intrinsic fluorescence that are apparent from the stopped-flow measurements with the different classes of metals.
How do these dissociation constants compare with metal concentrations in vivo? The bioavailable pool of any metal includes the amount of free metal within a certain cellular/extracellular location, the amount bound to proteins within that location, the affinity of those proteins for the metal, and the ability of the bound metal to be transferred to another protein. Copper, likely the most studied of the Type II metals, provides a good example. Human plasma contains only about 0.1 pM free copper because almost all is bound to other proteins,55 whereas the intracellular copper pool is ∼8 μM.55,56 Within the brain, second only to the liver in copper abundance,57,58 the cerebrospinal fluid contains 0.5–2.5 μM of the metal, whereas the concentration within the synaptic cleft reaches up to 15 μM.56,59 In addition, dissociation constants for copper in the hypothalamus are 6–40 μM.59 These values in tissues are clearly in the range that would be significant given the low micromolar affinities for metals measured in this work.
What biological processes have potential roles for copper in regulating PAI-1 structure and activity?
PAI-1 and vitronectin are both present in the circulation and in the brain, and the dissociation constants measured in this study indicate that copper binding would occur at these physiological concentrations. For example, copper has been shown to have a role in vascular leakage, platelet–endothelium interactions, and vascular smooth muscle cell reactivity. Significantly, vitronectin and PAI-1 are colocalized in the ECM and fibrin matrices60,61 in a wide variety of these pathologies, where the proteins regulate cell–matrix interactions, cell migration, and tissue remodeling. For example, recent elegant studies using X-ray fluorescent microprobe analysis have revealed some key findings relative to copper and angiogenesis.62 This work has demonstrated that the distribution of copper is under exquisite spatial regulation.63 During angiogenesis, it is translocated from the perinuclear areas of the cell toward the tips of extending filopodia, where it is transported across the cell membrane into the extracellular space. In this work, the authors suggest that copper activates an extracellular target that is inactivated during copper chelation therapy. Also, our recently published work has shown a role for the proteins as regulators of the fibrinolytic cascade acting extracellularly to activate brain-derived neurotrophic factor in the suprachiasmatic nucleus, required for regulating circadian rhythms.64 The activity of the biological clock is mediated in part by the N-methyl-d-aspartate receptor, which has been shown to efflux copper upon activation.64 Additional work will be required to further evaluate metal effects in these various processes and determine how such effects link to PAI-1 and its binding partner, vitronectin.
Materials and Methods
Materials
The two-chain form of tPA (tc-tPA) was purchased from Molecular Innovations, Nova, MI. Spectrozyme tPA was purchased from American Diagnostics, Stamford, CT. Protease inhibitor cocktail P8849, Ultrapure MOPS, and NaCl were purchased from Sigma Aldrich, St. Louis, MO. Biotechnology-grade Chelex® resin was purchased from Bio-Rad Hercules, CA. All other reagents were of analytical grade and used without further purification.
PAI-1 expression, purification, and activity
Unless noted, PAI-1 was expressed and purified as described previously.35 For some experiments, it was necessary to modify standard procedures to purify PAI-1 free of any bound metals without using EDTA as a chelator. In these cases, PAI-1 was purified using the following modifications. Sigma protease inhibitor cocktail P8849 was used in place of P8465. The dialysis after the SP Sepharose column contained 5 g/L of Chelex® resin. Fractions eluted from the nickel Sepharose column were pooled, concentrated, and dialyzed against 50 mM MOPS and 300 mM NaCl (both ultrapure buffers), pH 6.5, containing 5 g/L Chelex® resin. The dialyzed protein was concentrated to 1–3 mL and then loaded onto the S-100 column. The column was washed with 50 mM MOPS and 300 mM NaCl (both ultrapure buffers, which were treated with Chelex® resin at 5 g/L for additional purity), pH 6.5. Fractions were pooled, concentrated, and dialyzed against the same buffer. Dialyzed protein was filtered through a 0.22-μm membrane and stored at −80°C. Active PAI-1 was converted to its latent conformation by dialysis into 50 mM Tris and 200 mM NaCl, pH 7.4, at room temperature, followed by incubation in the same buffer at 37°C for 3 days. The inhibitory activity of the PAI-1 preparation was tested by titration with tPA as described previously.35 PAI-1 stability in the presence of metals was monitored as described previously.35
SPR measurement of PAI-1 binding to immobilized metals
Experiments were performed at room temperature on a Biacore 3000 SPR instrument. Protein and metal were dissolved in 50 mM Tris and 200 mM NaCl, pH 7.4. Metal was loaded onto a NTA chip by injecting 7 μL of a 0.5 mM metal solution at a 10 μL/min. After a brief wash with 50 μM EDTA to ensure all unbound metal was flushed from the system, varying concentrations of PAI-1 were loaded onto the chip at 20 μL/min for 2 min, flowing over the cell with bound metal and a reference cell for comparison with no metal bound. The binding reached steady state during this time period, and the equilibrium dissociation constant (KD) was calculated according to the method outlined by Rich and Myszka.65 The sensorgram responses at equilibrium were plotted against PAI-1 concentration and fit by nonlinear least squares to the equation:
| (2) |
where R is the observed response, Rmax is the maximal response, C is the analyte concentration, and KD is the equilibrium dissociation constant. Dissociation was then monitored for 3 min. Following each PAI-1 injection, metal was stripped from the chip by injecting 10 μL of a 350 mM EDTA solution. For competition experiments, the concentration of PAI-1 that gave half-maximal binding to the nickel-loaded NTA chip was fixed and mixed with varying concentrations of magnesium. The PAI-1 plus magnesium solutions were then injected at a flow rate of 20 μL/mL for 2 min. Competition of magnesium for binding of PAI-1 to the solid-phase nickel bound to the chip was measured by decrease in steady-state binding in the experiment.
Calculation of free metal concentrations from total metal levels using buffer/ metal affinity constants
PAI-1/metal measurements were made in either MOPS or Tris buffer based on metal solubility and availability of known metal/buffer binding constants. Using known first sphere buffer/metal affinity values, β1,39–41 the dissociation constant for metal in MOPS or Tris buffer was calculated using the equation: 10∧β1 = Ka = 1/Kd (Table II). The free metal concentration was then determined using the quadratic equation: Mf = Mt − [(Mt + Bt + Kd) − ((Mt + Bt + Kd)2 − 4•Bt•Mt)1/2]/2, where Mf is the free metal concentration, Mt is the total metal concentration, Bt is the total buffer concentration, and Kd is the buffer/metal dissociation constant.
Stopped-flow measurement of PAI-1 metal binding
Stock solutions of PAI-1 and metal chloride were made in ultrapure 50 mM MOPS or Tris and 150 mM NaCl, pH 7.4 (at 20°C). Buffer was treated with 5 g/L Chelex® resin and passed through a 0.22-μm filter before use. In an Applied Photophysics SX-20 stopped-flow spectrometer equilibrated to 20°C, the PAI-1 stock was rapidly mixed 1:1 with dilutions of the metal chloride stock. Samples were excited at 280 nM using a xenon lamp with one monochromator (slits set at 2 and 4 mm). Intrinsic protein fluorescence was monitored 90° to incident light using a 320 nM cutoff filter. Five to ten thousand data points were collected over the selected time window using oversampling. For long time-course experiments (500–2000 s), high concentrations of PAI-1 (100–200 nM) were used, and only three shots were averaged for each experiment. For 50-s experiments using 10 nM PAI-1, a 15-shot average was needed to achieve acceptable signal to noise. A baseline, recorded on samples of PAI-1 mixed with buffer only, was subtracted from all data on PAI-1 plus added metal chloride.
For magnesium, manganese, cobalt, and nickel in MOPS buffer, measurements were made over a 500-s timescale using 200 nM PAI-1, while varying the metal from ∼1 to 250 μM concentrations of free metal. For nickel in Tris buffer, measurements were performed over a similar timescale, but using 100 nM PAI-1 and varying the metal from ∼0.4 to 280 μM concentrations of free metal. Initial experiments with copper in Tris buffer were performed in a similar manner as those for nickel in the same buffer; however, because of the rapid drop in fluorescence observed for the first exponential, observed even at very low free copper concentrations, the timescale and protein/metal concentrations for these experiments were altered to maintain pseudo-first-order conditions and perform measurements with free copper concentrations that were below saturating conditions. The final conditions used to measure the copper effect on the observed rate constant for the first exponential were 10 nM PAI-1, with the free copper concentration varied from ∼0.04 to 1 μM. Free concentrations of metals in the buffer were calculated as described above.
PAI-1 stability assay
The rate of conversion of PAI-1 to the latent form was determined as previously described9 with the following alterations. A sample of 100 nM PAI-1 was equilibrated in 50 mM Tris-HCl buffer, pH 7.4, containing 150 mM NaCl at 20°C. These conditions were chosen to calculate rate constants for the conversion of PAI-1 to latent form in a comparable setting to that used for the stopped-flow measurements. Metal chloride samples were added to final concentrations given in Table IV, with calculated free concentrations shown also. Over time, aliquots of PAI-1 with or without metal were mixed with tc-tPA at a final concentration of 50 nM PAI-1 and 60 nM tPA. The relative amount of active PAI-1 was determined by its ability to inhibit functional tPA using 1 mM Spectrozyme tPA as a substrate and monitoring the release of p-nitroaniline by absorbance at 405 nM. Relative PAI-1 activity was normalized and fit to a single exponential to determine klat, the rate constant for conversion to the latent form. All data are the average of three separate experiments.
Glossary
Abbreviations:
- BSA
bovine serum albumin
- EDTA
ethylenediaminetetraacetic acid
- MOPS
3-(N-morpholino)-propanesulfonic acid
- NTA
nitrilotriacetic acid
- PAI-1
human plasminogen activator inhibitor type 1
- PDB
protein data bank
- serpin
serine protease inhibitor
- SP
methyl sulfopropyl
- SPR
surface plasmon resonance
- t1/2
half-life
- tc-tPA
two-chain tissue plasminogen activator
- Tris
tris(hydroxymethyl)aminomethane.
References
- 1.Dudev T, Lim C. Metal binding affinity and selectivity in metalloproteins: insights from computational studies. Annu Rev Biophys. 2008;37:97–116. doi: 10.1146/annurev.biophys.37.032807.125811. [DOI] [PubMed] [Google Scholar]
- 2.Lovell T, Himo F, Han WG, Noodleman L. Density functional methods applied to metalloenzymes. Coord Chem Rev. 2003;238:211–232. [Google Scholar]
- 3.Williams RJ. The natural selection of the chemical elements. Cell Mol Life Sci. 1997;53:816–829. doi: 10.1007/s000180050102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang AS, Enns CA. Iron homeostasis: recently identified proteins provide insight into novel control mechanisms. J Biol Chem. 2009;284:711–715. doi: 10.1074/jbc.R800017200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Turski ML, Thiele DJ. New roles for copper metabolism in cell proliferation, signaling, and disease. J Biol Chem. 2009;284:717–721. doi: 10.1074/jbc.R800055200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eide DJ. Homeostatic and adaptive responses to zinc deficiency in Saccharomyces cerevisiae. J Biol Chem. 2009;284:18565–18569. doi: 10.1074/jbc.R900014200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stout TJ, Graham H, Buckley DI, Matthews DJ. Structures of active and latent PAI-1: a possible stabilizing role for chloride ions. Biochemistry. 2000;39:8460–8469. doi: 10.1021/bi000290w. [DOI] [PubMed] [Google Scholar]
- 8.Mathiasen L, Dupont DM, Christensen A, Blouse GE, Jensen JK, Gils A, Declerck PJ, Wind T, Andreasen PA. A peptide accelerating the conversion of plasminogen activator inhibitor-1 to an inactive latent state. Mol Pharmacol. 2008;74:641–653. doi: 10.1124/mol.108.046417. [DOI] [PubMed] [Google Scholar]
- 9.Li SH, Gorlatova NV, Lawrence DA, Schwartz BS. Structural differences between active forms of plasminogen activator inhibitor type 1 revealed by conformationally sensitive ligands. J Biol Chem. 2008;283:18147–18157. doi: 10.1074/jbc.M709455200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wind T, Jensen JK, Dupont DM, Kulig P, Andreasen PA. Mutational analysis of plasminogen activator inhibitor-1. Eur J Biochem. 2003;270:1680–1688. doi: 10.1046/j.1432-1033.2003.03524.x. [DOI] [PubMed] [Google Scholar]
- 11.Gils A, Pedersen KE, Skottrup P, Christensen A, Naessens D, Deinum J, Enghild JJ, Declerck PJ, Andreasen PA. Biochemical importance of glycosylation of plasminogen activator inhibitor-1. Thromb Haemost. 2003;90:206–217. doi: 10.1160/TH03-01-0034. [DOI] [PubMed] [Google Scholar]
- 12.Blouse GE, Perron MJ, Kvassman JO, Yunus S, Thompson JH, Betts RL, Lutter LC, Shore JD. Mutation of the highly conserved tryptophan in the serpin breach region alters the inhibitory mechanism of plasminogen activator inhibitor-1. Biochemistry. 2003;42:12260–12272. doi: 10.1021/bi034737n. [DOI] [PubMed] [Google Scholar]
- 13.Hansen M, Busse MN, Andreasen PA. Importance of the amino-acid composition of the shutter region of plasminogen activator inhibitor-1 for its transitions to latent and substrate forms. Eur J Biochem. 2001;268:6274–6283. doi: 10.1046/j.0014-2956.2001.02582.x. [DOI] [PubMed] [Google Scholar]
- 14.Vleugels N, Leys J, Knockaert I, Declerck PJ. Effect of stabilizing versus destabilizing interactions on plasminogen activator inhibitor-1. Thromb Haemost. 2000;84:871–875. [PubMed] [Google Scholar]
- 15.Mangs H, Sui GC, Wiman B. PAI-1 stability: the role of histidine residues. FEBS Lett. 2000;475:192–196. doi: 10.1016/s0014-5793(00)01656-2. [DOI] [PubMed] [Google Scholar]
- 16.Berkenpas MB, Lawrence DA, Ginsburg D. Molecular evolution of plasminogen activator inhibitor-1 functional stability. EMBO J. 1995;14:2969–2977. doi: 10.1002/j.1460-2075.1995.tb07299.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lindahl TL, Sigurdardottir O, Wiman B. Stability of plasminogen activator inhibitor 1 (PAI-1) Thromb Haemost. 1989;62:748–751. [PubMed] [Google Scholar]
- 18.Deng G, Curriden SA, Hu G, Czekay RP, Loskutoff DJ. Plasminogen activator inhibitor-1 regulates cell adhesion by binding to the somatomedin B domain of vitronectin. J Cell Physiol. 2001;189:23–33. doi: 10.1002/jcp.1133. [DOI] [PubMed] [Google Scholar]
- 19.Deng G, Curriden SA, Wang S, Rosenberg S, Loskutoff DJ. Is plasminogen activator inhibitor-1 the molecular switch that governs urokinase receptor-mediated cell adhesion and release? J Cell Biol. 1996;134:1563–1571. doi: 10.1083/jcb.134.6.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Deng G, Royle G, Wang S, Crain K, Loskutoff DJ. Structural and functional analysis of the plasminogen activator inhibitor-1 binding motif in the somatomedin B domain of vitronectin. J Biol Chem. 1996;271:12716–12723. doi: 10.1074/jbc.271.22.12716. [DOI] [PubMed] [Google Scholar]
- 21.Kamikubo Y, De Guzman R, Kroon G, Curriden S, Neels JG, Churchill MJ, Dawson P, Oldziej S, Jagielska A, Scheraga HA, Loskutoff DJ, Dyson HJ. Disulfide bonding arrangements in active forms of the somatomedin B domain of human vitronectin. Biochemistry. 2004;43:6519–6534. doi: 10.1021/bi049647c. [DOI] [PubMed] [Google Scholar]
- 22.Lawrence DA, Palaniappan S, Stefansson S, Olson ST, Francis-Chmura AM, Shore JD, Ginsburg D. Characterization of the binding of different conformational forms of plasminogen activator inhibitor-1 to vitronectin. Implications for the regulation of pericellular proteolysis. J Biol Chem. 1997;272:7676–7680. doi: 10.1074/jbc.272.12.7676. [DOI] [PubMed] [Google Scholar]
- 23.Zhou A, Huntington JA, Pannu NS, Carrell RW, Read RJ. How vitronectin binds PAI-1 to modulate fibrinolysis and cell migration. Nat Struct Biol. 2003;10:541–544. doi: 10.1038/nsb943. [DOI] [PubMed] [Google Scholar]
- 24.Czekay RP, Loskutoff DJ. Plasminogen activator inhibitors regulate cell adhesion through a uPAR- dependent mechanism. J Cell Physiol. 2009;220:655–663. doi: 10.1002/jcp.21806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lijnen HR. Pleiotropic functions of plasminogen activator inhibitor-1. J Thromb Haemost. 2005;3:35–45. doi: 10.1111/j.1538-7836.2004.00827.x. [DOI] [PubMed] [Google Scholar]
- 26.Webb DJ, Thomas KS, Gonias SL. Plasminogen activator inhibitor 1 functions as a urokinase response modifier at the level of cell signaling and thereby promotes MCF-7 cell growth. J Cell Biol. 2001;152:741–752. doi: 10.1083/jcb.152.4.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schvartz I, Seger D, Shaltiel S. Vitronectin. Int J Biochem Cell Biol. 1999;31:539–544. doi: 10.1016/s1357-2725(99)00005-9. [DOI] [PubMed] [Google Scholar]
- 28.Irigoyen JP, Munoz-Canoves P, Montero L, Koziczak M, Nagamine Y. The plasminogen activator system: biology and regulation. Cell Mol Life Sci. 1999;56:104–132. doi: 10.1007/PL00000615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Waltz DA, Natkin LR, Fujita RM, Wei Y, Chapman HA. Plasmin and plasminogen activator inhibitor type 1 promote cellular motility by regulating the interaction between the urokinase receptor and vitronectin. J Clin Invest. 1997;100:58–67. doi: 10.1172/JCI119521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tomasini BR, Mosher DF. Vitronectin. Prog Hemost Thromb. 1991;10:269–305. [PubMed] [Google Scholar]
- 31.Preissner KT, Wassmuth R, Muller-Berghaus G. Physicochemical characterization of human S-protein and its function in the blood coagulation system. Biochem J. 1985;231:349–355. doi: 10.1042/bj2310349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Park YJ, Liu G, Lorne EF, Zhao X, Wang J, Tsuruta Y, Zmijewski J, Abraham E. PAI-1 inhibits neutrophil efferocytosis. Proc Natl Acad Sci USA. 2008;105:11784–11789. doi: 10.1073/pnas.0801394105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dellas C, Loskutoff DJ. Historical analysis of PAI-1 from its discovery to its potential role in cell motility and disease. Thromb Haemost. 2005;93:631–640. doi: 10.1160/TH05-01-0033. [DOI] [PubMed] [Google Scholar]
- 34.Esmon CT. Crosstalk between inflammation and thrombosis. Maturitas. 2004;47:305–314. doi: 10.1016/j.maturitas.2003.10.015. [DOI] [PubMed] [Google Scholar]
- 35.Thompson LC, Goswami S, Ginsberg DS, Day DE, Verhamme IM, Peterson CB. Metals affect the structure and activity of human plasminogen activator inhibitor-1. I. Modulation of stability and protease activity. Prot Sci. 2011;20:353–365. doi: 10.1002/pro.568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Copeland RA. Enzymes: a practical introduction to structure, mechanism, and data analysis. 2nd ed. USA: Wiley-VCH; 2000. [Google Scholar]
- 37.O'Keeffe ET, Hill RL, Bell JE. Active site of bovine galactosyltransferase: kinetic and fluorescence studies. Biochemistry. 1980;19:4954–4962. doi: 10.1021/bi00563a003. [DOI] [PubMed] [Google Scholar]
- 38.Pereira-Mouries L, Almeida MJ, Ribeiro C, Peduzzi J, Barthelemy M, Milet C, Lopez E. Soluble silk-like organic matrix in the nacreous layer of the bivalve Pinctada maxima. Eur J Biochem. 2002;269:4994–5003. doi: 10.1046/j.1432-1033.2002.03203.x. [DOI] [PubMed] [Google Scholar]
- 39.Magyar JS, Godwin HA. Spectropotentiometric analysis of metal binding to structural zinc-binding sites: accounting quantitatively for pH and metal ion buffering effects. Anal Biochem. 2003;320:39–54. doi: 10.1016/s0003-2697(03)00281-1. [DOI] [PubMed] [Google Scholar]
- 40.Anwar ZM, Azab HA. Ternary complexes in solution. Comparison of the coordination tendency of some biologically important zwitterionic buffers toward the binary complexes of some transition metal ions and some amino acids. J Chem Eng Data. 1999;44:1151–1157. [Google Scholar]
- 41.Scheller KH, Abel THJ, Polanyi PE, Wenk PK, Fischer BE, Sigel H. Metal ion-buffer interactions. II. Stability of binary and ternary complexes containing 2-[bis(2-hydroxyethyl)amino]-2(hydroxymethyl)-1,3-propanediol (Bis-Tris) and adenosine 5′-triphosphate (Atp) Eur J Biochem. 1980;107:455–466. [PubMed] [Google Scholar]
- 42.Morgenstern R, Svensson R, Bernat BA, Armstrong RN. Kinetic analysis of the slow ionization of glutathione by microsomal glutathione transferase MGST1. Biochemistry. 2001;40:3378–3384. doi: 10.1021/bi0023394. [DOI] [PubMed] [Google Scholar]
- 43.Kim YB, Kalinowski SS, Marcinkeviciene J. A pre-steady state analysis of ligand binding to human glucokinase: evidence for a preexisting equilibrium. Biochemistry. 2007;46:1423–1431. doi: 10.1021/bi0617308. [DOI] [PubMed] [Google Scholar]
- 44.Verheyden S, Sillen A, Gils A, Declerck PJ, Engelborghs Y. Tryptophan properties in fluorescence and functional stability of plasminogen activator inhibitor 1. Biophys J. 2003;85:501–510. doi: 10.1016/S0006-3495(03)74495-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stout TJ, Graham H, Buckley DI, Matthews DJ. Structures of active and latent PAI-1: a possible stabilizing role for chloride ions. Biochemistry. 2000;39:8460–8469. doi: 10.1021/bi000290w. [DOI] [PubMed] [Google Scholar]
- 46.Nar H, Bauer M, Stassen JM, Lang D, Gils A, Declerck PJ. Plasminogen activator inhibitor 1. Structure of the native serpin, comparison to its other conformers and implications for serpin inactivation. J Mol Biol. 2000;297:683–695. doi: 10.1006/jmbi.2000.3604. [DOI] [PubMed] [Google Scholar]
- 47.Jensen JK, Gettins PG. High-resolution structure of the stable plasminogen activator inhibitor type-1 variant 14-1B in its proteinase-cleaved form: a new tool for detailed interaction studies and modeling. Protein Sci. 2008;17:1844–1849. doi: 10.1110/ps.036707.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sharp AM, Stein PE, Pannu NS, Carrell RW, Berkenpas MB, Ginsburg D, Lawrence DA, Read RJ. The active conformation of plasminogen activator inhibitor 1, a target for drugs to control fibrinolysis and cell adhesion. Structure. 1999;7:111–118. doi: 10.1016/S0969-2126(99)80018-5. [DOI] [PubMed] [Google Scholar]
- 49.Dupont DM, Blouse GE, Hansen M, Mathiasen L, Kjelgaard S, Jensen JK, Christensen A, Gils A, Declerck PJ, Andreasen PA, Wind T. Evidence for a pre-latent form of the serpin plasminogen activator inhibitor-1 with a detached beta-strand 1C. J Biol Chem. 2006;281:36071–36081. doi: 10.1074/jbc.M606851200. [DOI] [PubMed] [Google Scholar]
- 50.Hagglof P, Bergstrom F, Wilczynska M, Johansson LB, Ny T. The reactive-center loop of active PAI-1 is folded close to the protein core and can be partially inserted. J Mol Biol. 2004;335:823–832. doi: 10.1016/j.jmb.2003.11.005. [DOI] [PubMed] [Google Scholar]
- 51.Aertgeerts K, De Bondt HL, De Ranter CJ, Declerck PJ. Mechanisms contributing to the conformational and functional flexibility of plasminogen activator inhibitor-1. Nat Struct Biol. 1995;2:891–897. doi: 10.1038/nsb1095-891. [DOI] [PubMed] [Google Scholar]
- 52.Baker D, Agard DA. Kinetics versus thermodynamics in protein folding. Biochemistry. 1994;33:7505–7509. doi: 10.1021/bi00190a002. [DOI] [PubMed] [Google Scholar]
- 53.Kvassman JO, Verhamme I, Shore JD. Inhibitory mechanism of serpins: loop insertion forces acylation of plasminogen activator by plasminogen activator inhibitor-1. Biochemistry. 1998;37:15491–15502. doi: 10.1021/bi9814787. [DOI] [PubMed] [Google Scholar]
- 54.Olson ST, Swanson R, Day D, Verhamme I, Kvassman J, Shore JD. Resolution of Michaelis complex, acylation, and conformational change steps in the reactions of the serpin, plasminogen activator inhibitor- 1, with tissue plasminogen activator and trypsin. Biochemistry. 2001;40:11742–11756. doi: 10.1021/bi0107290. [DOI] [PubMed] [Google Scholar]
- 55.Linder MC, Hazegh-Azam M. Copper biochemistry and molecular biology. Am J Clin Nutr. 1996;63:797S–811S. doi: 10.1093/ajcn/63.5.797. [DOI] [PubMed] [Google Scholar]
- 56.Kramer ML, Kratzin HD, Schmidt B, Romer A, Windl O, Liemann S, Hornemann S, Kretzschmar H. Prion protein binds copper within the physiological concentration range. J Biol Chem. 2001;276:16711–16719. doi: 10.1074/jbc.M006554200. [DOI] [PubMed] [Google Scholar]
- 57.Osterberg R. Physiology and pharmacology of copper. Pharmacol Ther. 1980;9:121–146. doi: 10.1016/0163-7258(80)90019-4. [DOI] [PubMed] [Google Scholar]
- 58.Nalbandyan RM. Copper in brain. Neurochem Res. 1983;8:1211–1232. doi: 10.1007/BF00963993. [DOI] [PubMed] [Google Scholar]
- 59.Hartter DE, Barnea A. Brain tissue accumulates 67copper by two ligand-dependent saturable processes. A high affinity, low capacity and a low affinity, high capacity process. J Biol Chem. 1988;263:799–805. [PubMed] [Google Scholar]
- 60.Podor TJ, Peterson CB, Lawrence DA, Stefansson S, Shaughnessy SG, Foulon DM, Butcher M, Weitz JI. Type 1 plasminogen activator inhibitor binds to fibrin via vitronectin. J Biol Chem. 2000;275:19788–19794. doi: 10.1074/jbc.M908079199. [DOI] [PubMed] [Google Scholar]
- 61.Podor TJ, Campbell S, Chindemi P, Foulon DM, Farrell DH, Walton PD, Weitz JI, Peterson CB. Incorporation of vitronectin into fibrin clots. Evidence for a binding interaction between vitronectin and gamma A/gamma' fibrinogen. J Biol Chem. 2002;277:7520–7528. doi: 10.1074/jbc.M109677200. [DOI] [PubMed] [Google Scholar]
- 62.Finney L, Vogt S, Fukai T, Glesne D. Copper and angiogenesis: unravelling a relationship key to cancer progression. Clin Exp Pharmacol Physiol. 2009;36:88–94. doi: 10.1111/j.1440-1681.2008.04969.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Finney L, Mandava S, Ursos L, Zhang W, Rodi D, Vogt S, Legnini D, Maser J, Ikpatt F, Olopade OI, Glesne D. X-ray fluorescence microscopy reveals large-scale relocalization and extracellular translocation of cellular copper during angiogenesis. Proc Natl Acad Sci USA. 2007;104:2247–2252. doi: 10.1073/pnas.0607238104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mou X, Peterson CB, Prosser RA. Tissue-type plasminogen activator-plasmin-BDNF modulate glutamate-induced phase-shifts of the mouse suprachiasmatic circadian clock in vitro. Eur J Neurosci. 2009;30:1451–1460. doi: 10.1111/j.1460-9568.2009.06946.x. [DOI] [PubMed] [Google Scholar]
- 65.Rich RL, Myszka DG. BIACORE J: a new platform for routine biomolecular interaction analysis. J Mol Recognit. 2001;14:223–228. doi: 10.1002/jmr.535. [DOI] [PubMed] [Google Scholar]


