Abstract
Mycobacterium tuberculosis (Mtb) has a highly complex cell wall, which is required for both bacterial survival and infection. Cell wall biosynthesis is dependent on decaprenyl diphosphate as a glyco-carrier, which is hence an essential metabolite in this pathogen. Previous biochemical studies indicated (E)-geranyl diphosphate (GPP) is required for the synthesis of decaprenyl diphosphate. Here we demonstrate that Rv0989c encodes the “missing” GPP synthase, representing the first such enzyme to be characterized from bacteria, and which presumably is involved in decaprenyl diphosphate biosynthesis in Mtb. Our investigation also has revealed previously unrecognized substrate plasticity of the farnesyl diphosphate synthases from Mtb, resolving previous discrepancies between biochemical and genetic studies of cell wall biosynthesis.
Keywords: Polyisoprenyl phosphates, Terpene Biosynthesis, Tuberculosis, Cell Wall biosynthesis, Genetic fitness
1. Introduction
Mycobacterium tuberculosis (Mtb) is the main intracellular pathogen responsible for the human disease Tuberculosis, which is particularly lethal in immune-deficient individuals. Evolution of drug resistant strains has renewed this pathogen’s threat on the general population, as these new strains are extremely pathogenic and untreatable by current antibiotic regimens [1]. Furthermore, decades of research have failed to uncover the pathogenic mechanism of Mtb, complicating drug design efforts. This elusive mechanism likely involves a multifactorial attack, employing attack by a variety of virulence factors over the course of the infection [2]. One set of these virulence factors involve the complex mycobacterial cell wall and lipids, which have been linked to pathogenesis and survival of the bacteria during infection [3].
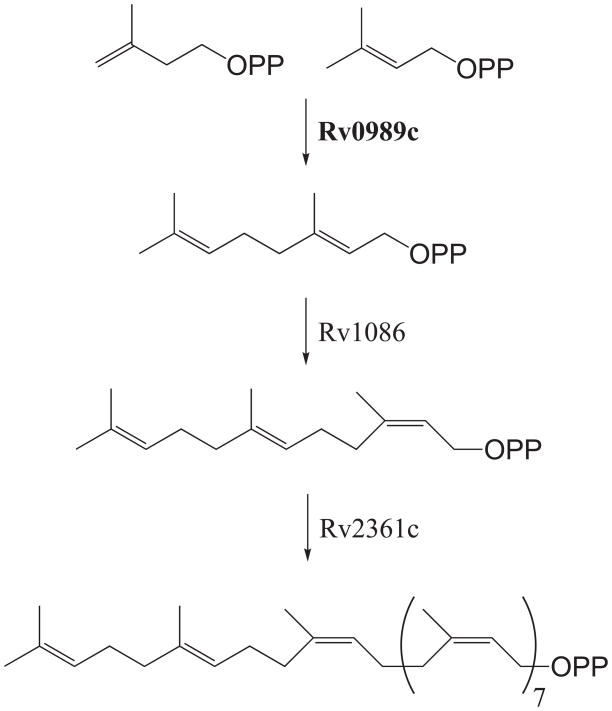
The mycobacterial cell wall is unique amongst bacteria. The cell wall consists of the peptidoglycan, arabinogalactan, and mycolipid layers, as well as a capsule-like layer separate from the core wall components [4] . Important in proper assembly of this complex structure is the glyco-carrier polyprenyl phosphate (Pol-P) [5]. Such Pol-P molecules are synthesized via the addition of isoprene units in the form of isopentenyl diphosphate (IPP) to a variety of allylic isoprenyl diphosphate acceptors, which are found in varying lengths, as well as various double bond isomers. Specifically, Mtb utilizes a 10 isoprene-unit decaprenyl diphosphate to assemble its complex cell wall [6], which is formed by the addition of seven IPP to (Z,E)-farnesyl diphosphate (FPP) by Rv2361c [7]. To synthesize (Z,E)-FPP, one unit of IPP is added to (E)-geranyl diphosphate (GPP) by Rv1086 [7]. Previous work has indicated that this process is dependent on GPP as the allylic acceptor of IPP, yet no GPP synthase has been characterized from Mtb [8] (Figure 1). In addition, while Rv2361c is essential for bacterial survival [9], neither Rv1086 nor the remaining uncharacterized prenyltransferases are similarly essential, complicating identification of the prenyltransferases involved in decaprenyl diphosphate biosynthesis.
Figure 1.
Decaprenyl diphosphate biosynthesis in Mycobacterium tuberculosis. The roles of Rv1086 and Rv2361c have been demonstrated, yet biochemical analysis indicated they are dependent on a previously uncharacterized GPP synthase, which is demonstrated here to be encoded by Rv0989.
While studying the biosynthesis of the immune modulating diterpenoid, isotuberculosinol, we uncovered four previously uncharacterized putative trans-prenyltransferases (Rv0989c, Rv2173, Rv3383c, Rv0562) [10]. Of these prenyltransferases, only Rv0989c failed to synthesize the (E,E,E)-geranylgeranyl diphosphate (GGPP) required for isotuberculosinol or other diterpenoid biosynthesis. These initial assays were not set up to detect shorter chain isoprenoids (i.e. FPP or GPP), leading to a lack of observed activity with Rv0989c. After recognizing that a GPP synthase was expected, we altered our protocol to enable detection of ten and fifteen carbon isoprenoids, and found that Rv0989c acts as a GPP synthase, that seems to assist the production of FPP in Mtb. Furthermore, our analysis indicates that Rv1086 is able to produce small amounts of (Z,E)-FPP from dimethylallyl diphosphate (DMAPP), which may explain the non-essential nature of these prenyltransferases. Accordingly, we describe here the first bacterial GPP synthase, and further clarify the roles of the various isopentyl diphosphate synthases involved in production of the decaprenyl diphosphate required for mycobacterial cell wall biosynthesis, thus resolving the apparent contradictions between biochemical and genetic analysis of these enzymes.
2. Materials and methods
General
IPP, DMAPP, GPP, (E,E)-FPP, and GGPP were purchased from Isoprenoids, LC (Tampa, FL). Corresponding dephosphorylated alcohol standards for gas chromatography (GC) analysis were purchased from Sigma Aldrich (St. Louis, MO). Unless otherwise noted, all other chemicals were purchased from Fisher Scientific (St. Louis, MO) and all molecular biology reagents were purchased from Invitrogen (Carlsbad, CA). Sequence alignments were carried out using the AlignX program from the Vector NTI software package (Invitrogen), using standard parameters.
Cloning, expression, and purification
Rv0989c and Rv3398c were cloned from Mycobacterium tuberculosis CDC1551, while Rv1086 was obtained as a synthetic construct, codon-optimized for expression in Escherichia coli. These were cloned into pENTR/SD/D-TOPO prior to directional recombination into an expression vector containing a 6xHis N-terminal fusion (pDEST17, Gateway, Invitrogen). Proteins were expressed in E. coli strain C41 (Lucigen, Middleton, WI), by growing the recombinant bacteria in NZY liquid media at 37 °C to an OD600 of 0.6, transfer to 16 °C for 1 hr prior to induction with 0.5 mM IPTG, and subsequent 14–16 hr fermentation. Cells were harvested via centrifugation and resuspended in Lysis Buffer (10 mM Tris-Cl, 10% Glycerol, 1 mM Dithiothreitol) for sonication. The resulting lysates were clarified via centrifugation (10 min x 10,000g). The recombinant proteins were then purified over Ni-NTA Superflow resin (Novagen) following manufacturer protocols.
Prenyltransferase assay
The purified enzymes were characterized in Assay Buffer (50 mM sodium phosphate (pH 7.0), 10% glycerol, 5 mM MgCl2, 2 mM Dithiothreitol), with IPP and allylic isoprenyl diphosphate substrate (i.e. DMAPP, GPP, or FPP), in a final volume of 1 mL at 37 °C. Assays were stopped by the addition of 1 ml of Stopping Solution (9:1 Methanol: 3N Hydrochloric acid), which also dephosphorylates the substrates and products, and 1 mL of pentane was added immediately and mixed to allow for full partitioning of the isoprenoid alcohols to the organic solvent layer. This was removed and dried under a stream of N2 at room temperature prior to resuspension in 50 μL hexanes. Enzymatic products were identified on a gas chromatograph (GC) with mass spectrometry (MS) detection using a Varian 3900 GC with Saturn 2100T ion-trap MS (Varian Inc, Palo Alto, CA) with comparison to authentic standards, and were quantified via an Agilent 6890 GC with flame ionization detection (FID)(Agilent Technologies, Santa Clara, CA), using a previously published method [11].
Characterization of Rv0989c
Rv0989c was determined to produce GPP, and then assays with this enzyme optimized for protein concentration and incubation time to obtain initial linear reaction rates. It was necessary to use ≥ 0.5 μM enzyme to achieve linearity, with incubation times of 2 minutes then found to be optimal. Steady-state kinetic constants were determined by varying concentrations of either substrate, i.e. 10–300 μM IPP or 5– 200 μM DMAPP, with the other held constant at the highest relevant concentration, which initial analysis had determined was saturating. Concentrations of both the internal standard (cembrene) and product were determined by integration of peaks after separation and detection by GC-FID. The observed data was fit to the Michaelis-Menten equation (KaleidaGraph 4.0, Synergy Software; Reading, PA), resulting in R-values ≥ 0.97.
Coupled assays
Coupled assays were carried out by pre-incubating Rv0989c (5 μM) with DMAPP (100 μM) and IPP (100 μM) for 5 min. prior to the addition of Rv1086 (1 μM) or Rv3398c (0.35 μM), along with an additional 50 μM IPP, and the assay incubated for an additional 5 minutes before stopping. Control assays with Rv1086 or Rv3398c, in the absence of Rv0989c, using 100 μM of GPP or DMAPP as the allylic substrate, were run for 5 minutes. These all were analyzed by GC-MS, as described above, for production of the relevant isomers of FPP using separate methods, allowing for distinct separation of the resulting dephosphorylated farnesol (i.e. (E,E)-farnesol [12] or (Z,E)-farnesol [11]).
Characterization of Rv3398c and Rv1086
Rv3398c was kinetically characterized as described above for Rv0989c, using substrate concentrations ranging from 2.5–200 μM. Time course studies with Rv1086 were performed with 100 μM each of IPP and DMAPP. When Rv1086 was assayed alone, Triton X-100 was added at 0.3 % (v/v) to the Assay Buffer to mimic previously reported conditions [8].
3. Results
Characterization of Rv0989c as a GPP synthase
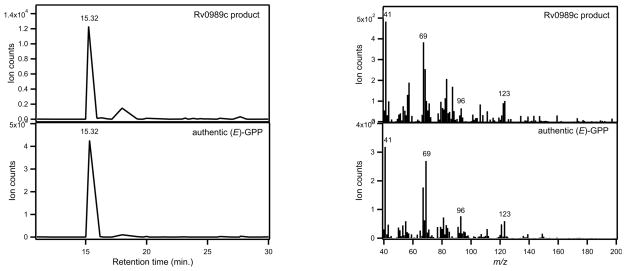
Rv0989c was initially screened for GGPP synthesis based on its homology to other GGPP synthases within the Mtb genome (Rv0562, Rv2173, Rv3383c) [10]. As Rv0989c failed to synthesize GGPP from any allylic precursor (DMAPP, GPP, or FPP), it was then assessed for activity as a GPP or FPP synthase, and found to produce GPP when incubated with DMAPP and IPP (Figure 2). Although production of (E,E)-FPP was occasionally observed in the presence of excess (>1 mM) substrate, this likely indicates non-physiologically relevant reuptake of product for further elongation under these conditions. Steady-state kinetic analysis of Rv0989c revealed unusual behavior. When varying DMAPP concentration in the presence of saturating amounts of IPP, kcat appeared to be 0.7 min−1, which is over 7.5-fold lower than that measured when varying IPP concentration in the presence of saturating amounts of DMAPP, where kcat appeared to be 5.1 min−1. Yet the pseudo-substrate binding constant KM suggests DMAPP is bound three-fold tighter than IPP (58 versus 163 μM, respectively). This may indicate non-catalytically relevant binding of two molecules of IPP at high concentrations, necessitating displacement by DMAPP that decreases the observed catalytic rate.
Figure 2. Confirmation of Rv0989c activity.
Comparison of dephosphorylated Rv0989c product to GPP-derived GOH. (A) GC-FID chromatograms. (B) GC-MS mass spectra.
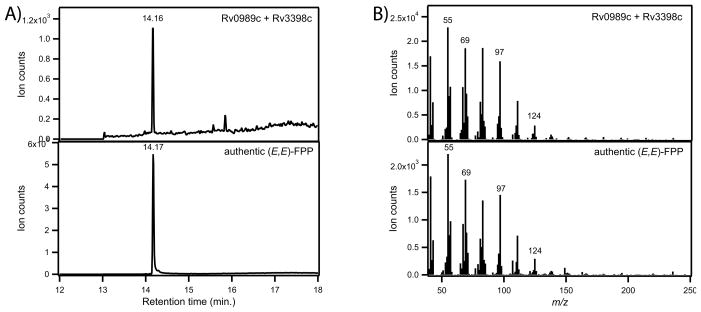
Rv0989c facilitates production of (Z,E)-FPP by Rv1086 and (E,E)-FPP by Rv3398c
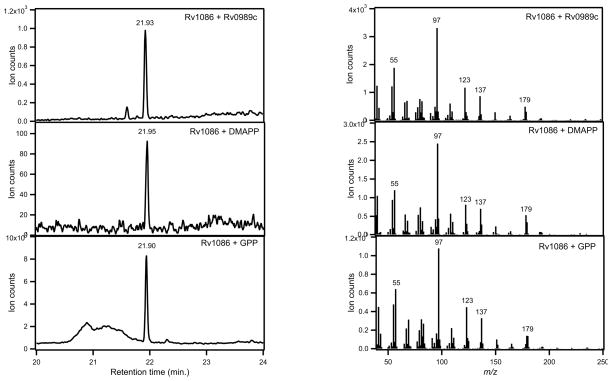
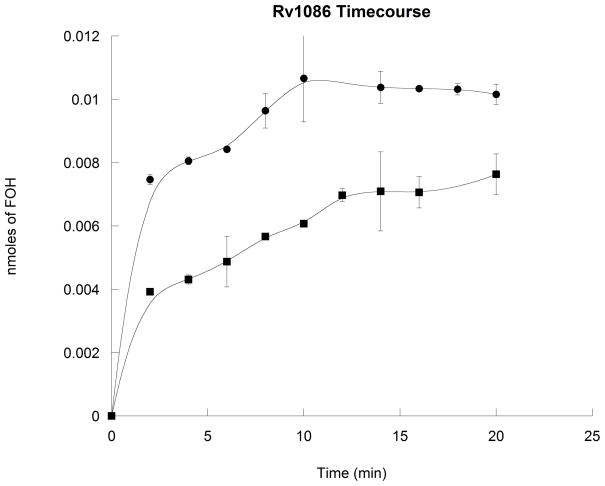
Decaprenyl diphosphate formation in Mtb proceeds through Rv1086, a (Z,E)-FPP synthase, which has been reported to use exclusively GPP as its allylic isoprenyl diphosphate substrate [8]. Similarly, Rv3398c, the Mtb (E,E)-FPP synthase, also has been reported to utilize GPP exclusively [13]. To demonstrate that Rv0989c synthesizes the required product for further elaboration into these important isoprenyl diphosphates, Rv0989c was assayed with each of these FPP synthases. Coupled assays with Rv0989c and Rv1086 led to production of significant amounts of (Z,E)-FPP (Figure 3), along with smaller amounts (<10%) of (E,E)-FPP (data not shown). Surprisingly, a small amount of (Z,E)-FPP also was produced by Rv1086 from IPP and DMAPP in the absence of Rv0989 (Figure 3). Nevertheless, time course measurements indicate the coupled reaction proceeds at a faster rate and nets higher yields of (Z,E)-FPP (Figure 4), supporting a role for GPP as the preferred substrate for this enzyme. Coupled assays with Rv0989c and Rv3398c produce (E,E)-FPP (Figure 5). However, again, when assayed only in the presence of DMAPP and IPP, Rv3398c is able to produce (E,E)-FPP at measureable rates. Steady state kinetic analysis of Rv3398c determined that kcat is 1.5 ± 0.1 min−1, KM for IPP is 2.6 ± 0.4 μM, while that for DMAPP is 20 ± 9 μM. Comparing to previous publications [13], this still suggests that GPP (KM = 10 μM) is the preferred allylic isoprenyl diphosphate substrate for Rv3398c.
Figure 3. Production of (Z,E)-FPP-derived FOH by Rv0989c/Rv1086.
GC-MS analysis of product profiles from Rv1086 assayed in the presence of authentic GPP, GPP synthesized by Rv0989c, or DMAPP. (A) Total ion chromatograms. (B) mass spectra.
Figure 4. Time-dependent production of (Z,E)-FPP by Rv1086.
(Z,E)-FPP-derived FOH synthesis by Rv1086 assayed in the presence of 100 uM IPP and 100 uM DMAPP (squares) or 100 uM IPP, 100 uM DMAPP, and Rv0989c (circles). Smooth fit lines were applied.
Figure 5. Production of (E,E)-FPP-derived FOH by Rv0989c/Rv3398c.
GC-MS analysis of product profiles from Rv3398 assayed in the presence of authentic GPP and GPP synthesized by Rv0989c. (A) Total ion chromatograms. (B) mass spectra.
4. Discussion
The mycobacterial cell wall has been extensively studied as it is exceptionally reinforced and uniquely immunogenic [3]. Key to the assembly of this complex structure is the glyco-carrier decaprenyl diphosphate [6]. Though clearly required for cell growth, development, and viability [9], the biosynthetic pathways utilized in synthesis of decaprenyl diphosphate have frustrated scientists in recent decades. Genetic and biochemical analysis of the biosynthetic machinery has failed to completely characterize the pathway. While decaprenyl diphosphate production is required for cell wall biosynthesis [14], genetic analysis indicates that only the final step in the production of this important metabolite (that catalyzed by the gene product of Rv2361c) is absolutely required for bacterial viability [9]. While this decaprenyl diphosphate synthase will accept a range of allylic isoprenyl diphosphate substrates [7], it has been reported that the production of its preferred (Z,E)-FPP primer for elongation is completely dependent on the availability of GPP [8]. Previously, neither genetic nor biochemical analysis was able to identify the corresponding GPP synthase in Mtb, which our results now demonstrate is encoded by Rv0989c.
To the best of our knowledge, Rv0989c is the first characterized bacterial GPP synthase. While this does exhibit homology to other such trans-prenyltransferases, there are a number of surprising substitutions observed in Rv0989c. In particular, while readily aligned with other known GPP synthases (Figure 6), Rv0989c contains remarkable changes within two otherwise well-conserved aspartate-rich motifs (designated as the first and second, FARM and SARM, respectively). In Rv0989c, the second aspartate of the FARM and the first aspartate of the SARM are replaced by arginine, and the final aspartate of the SARM is replaced by an alanine, whereas the only other naturally found substitutions are asparagines. Indeed, site-directed mutations in the first and second aspartate of the SARM of other trans-prenyltransferases to alanine, specifically an (E,E)-FPP synthase from Saccharomyces cerevisiae, have demonstrated a drastic loss of activity [15]. The significance of the observed divergence in the FARM and SARM of Rv0989c is not readily apparent, but their expected deleterious effects may have been a contributing factor to the previous difficulty in identification of a GPP synthase in the Mtb genome.
Figure 6. Alignment of Rv0989c with confirmed GPP synthases.
Alignment of the Rv0989c gene product with that of the GPP synthases from Ips pini (IpGPS) [16], Aphis gossypii (AgGPS) [17], Arabidopsis thaliana (AtGPS) [18], and Citrus sinensus (CsGPS) [19]. The first- and second aspartate-rich motifs (FARM and SARM, respectively), are underlined, with the non-canonical substitution of arginine or alanine residues for otherwise conserved active site aspartate residues in Rv0989c indicated by * and # above the relevant position, respectively.
We have determined Rv0989c is able to provide the GPP precursor required for efficient production of both double bond isomers of FPP in Mtb, as demonstrated by coupled assays with the relevant FPP synthases (encoded by Rv1086 and Rv3398c). However, our enzymatic analysis also revealed that both of these are able to react with the shorter, universal allylic isoprenyl diphosphate substrate DMAPP. This clarifies the contradictory biochemical and genetic evidence regarding the decaprenyl diphosphate biosynthetic pathway in Mtb [8,9,13]. In particular, knockout analysis of these prenyltransferases in Mtb have demonstrated that they are not required for viability, indicating that cell wall biosynthesis was able to proceed in their absence, despite biochemical evidence indicating that decaprenyl diphosphate production depends on their activity/products. Nevertheless, these findings further highlight the need for development and study of functional, non-lethal knockouts in Mtb cell wall biosynthetic machinery, as many questions have yet to be answered regarding bacterial tolerance and flexibility for substitutions in this process. This important component of Mtb viability and pathogenesis needs to be completely understood before the puzzle that is the Mtb pathogenic mechanism will be solved.
Acknowledgments
We thank Dr. Niels Nieuwenhuizen of The New Zealand Institute for Plant and Food Research Limited for the kind gift of a synthetic version of Rv1086, and gratefully acknowledge support for this project from the NIH (GM076324 to R.J.P.).
List of abbreviations
- Mtb
Mycobacterium tuberculosis
- IPP
isopentenyl diphosphate
- DMAPP
dimethylallyl diphosphate
- GPP
geranyl diphosphate
- FPP
farnesyl diphosphate
- GGPP
geranylgeranyl diphosphate
- GC
gas chromatography
- Pol-P
polyprenyl phosphate
- FID
flame ionization detector
- MS
mass spectrometer
- FARM
first aspartate-rich motif
- SARM
second aspartate-rich motif
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Gandhi NR, Nunn P, Dheda K, Schaaf HS, Zignol M, van Soolingen D, Jensen P, Bayona J. Multidrug-resistant and extensively drug-resistant tuberculosis: a threat to global control of tuberculosis. Lancet. 375:1830–43. doi: 10.1016/S0140-6736(10)60410-2. [DOI] [PubMed] [Google Scholar]
- 2.Russell DG. Who puts the tubercle in tuberculosis? Nat Rev Microbiol. 2007;5:39–47. doi: 10.1038/nrmicro1538. [DOI] [PubMed] [Google Scholar]
- 3.Brennan PJ. Structure, function, and biogenesis of the cell wall of Mycobacterium tuberculosis. Tuberculosis (Edinb) 2003;83:91–7. doi: 10.1016/s1472-9792(02)00089-6. [DOI] [PubMed] [Google Scholar]
- 4.Varki A. Essentials of glycobiology. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, N.Y.: 2009. [PubMed] [Google Scholar]
- 5.Crick DC, Schulbach MC, Zink EE, Macchia M, Barontini S, Besra GS, Brennan PJ. Polyprenyl phosphate biosynthesis in Mycobacterium tuberculosis and Mycobacterium smegmatis. J Bacteriol. 2000;182:5771–8. doi: 10.1128/jb.182.20.5771-5778.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kaur D, Brennan PJ, Crick DC. Decaprenyl diphosphate synthesis in Mycobacterium tuberculosis. J Bacteriol. 2004;186:7564–70. doi: 10.1128/JB.186.22.7564-7570.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schulbach MC, Brennan PJ, Crick DC. Identification of a short (C15) chain Z-isoprenyl diphosphate synthase and a homologous long (C50) chain isoprenyl diphosphate synthase in Mycobacterium tuberculosis. J Biol Chem. 2000;275:22876–81. doi: 10.1074/jbc.M003194200. [DOI] [PubMed] [Google Scholar]
- 8.Schulbach MC, et al. Purification, enzymatic characterization, and inhibition of the Z-farnesyl diphosphate synthase from Mycobacterium tuberculosis. J Biol Chem. 2001;276:11624–30. doi: 10.1074/jbc.M007168200. [DOI] [PubMed] [Google Scholar]
- 9.Sassetti CM, Boyd DH, Rubin EJ. Genes required for mycobacterial growth defined by high density mutagenesis. Mol Microbiol. 2003;48:77–84. doi: 10.1046/j.1365-2958.2003.03425.x. [DOI] [PubMed] [Google Scholar]
- 10.Mann FM, Xu M, Davenport EK, Peters RJ. Genetic redundancy of terpene biosynthesis in pathogenic Mycobacterium tuberculosis. Tuberculosis (Edinb) 2011 (in preparation) [Google Scholar]
- 11.Schmidt A, Gershenzon J. Cloning and characterization of two different types of geranyl diphosphate synthases from Norway spruce (Picea abies) Phytochemistry. 2008;69:49–57. doi: 10.1016/j.phytochem.2007.06.022. [DOI] [PubMed] [Google Scholar]
- 12.Prisic S, Xu M, Wilderman PR, Peters RJ. Rice contains two disparate entcopalyl diphosphate synthases with distinct metabolic functions. Plant Physiol. 2004;136:4228–36. doi: 10.1104/pp.104.050567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dhiman RK, Schulbach MC, Mahapatra S, Baulard AR, Vissa V, Brennan PJ, Crick DC. Identification of a novel class of omega,E,E-farnesyl diphosphate synthase from Mycobacterium tuberculosis. J Lipid Res. 2004;45:1140–7. doi: 10.1194/jlr.M400047-JLR200. [DOI] [PubMed] [Google Scholar]
- 14.Mahapatra S, Yagi T, Belisle JT, Espinosa BJ, Hill PJ, McNeil MR, Brennan PJ, Crick DC. Mycobacterial lipid II is composed of a complex mixture of modified muramyl and peptide moieties linked to decaprenyl phosphate. J Bacteriol. 2005;187:2747–57. doi: 10.1128/JB.187.8.2747-2757.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Song L, Poulter CD. Yeast farnesyl-diphosphate synthase: site-directed mutagenesis of residues in highly conserved prenyltransferase domains I and II. Proc Natl Acad Sci U S A. 1994;91:3044–8. doi: 10.1073/pnas.91.8.3044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gilg AB, Bearfield JC, Tittiger C, Welch WH, Blomquist GJ. Isolation and functional expression of an animal geranyl diphosphate synthase and its role in bark beetle pheromone biosynthesis. Proc Natl Acad Sci U S A. 2005;102:9760–5. doi: 10.1073/pnas.0503277102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ma GY, Sun XF, Zhang YL, Li ZX, Shen ZR. Molecular cloning and characterization of a prenyltransferase from the cotton aphid, Aphis gossypii. Insect Biochem Mol Biol. 40:552–61. doi: 10.1016/j.ibmb.2010.05.003. [DOI] [PubMed] [Google Scholar]
- 18.Bouvier F, Suire C, d'Harlingue A, Backhaus RA, Camara B. Molecular cloning of geranyl diphosphate synthase and compartmentation of monoterpene synthesis in plant cells. Plant J. 2000;24:241–52. doi: 10.1046/j.1365-313x.2000.00875.x. [DOI] [PubMed] [Google Scholar]
- 19.Sharon-Asa L, et al. Citrus fruit flavor and aroma biosynthesis: isolation, functional characterization, and developmental regulation of Cstps1, a key gene in the production of the sesquiterpene aroma compound valencene. Plant J. 2003;36:664–74. doi: 10.1046/j.1365-313x.2003.01910.x. [DOI] [PubMed] [Google Scholar]